Abstract
Previous work on Drosophila santomea suggested that its absence of abdominal pigmentation, compared to the other darkly-pigmented species, is based on mutations in the cis-regulatory region of tan inactivating the expression of that gene in the abdomen of D. santomea males and females. Our discovery that D. santomea males can produce viable hybrids when mated to D. melanogaster females enables us to use the armamentarium of genetic tools in the latter species to study the genetic basis of this interspecific difference in pigmentation. Hybridization tests using D. melanogaster deficiencies that include tan show no evidence that this locus is involved in the lighter pigmentation of D. santomea females; rather, the pigmentation difference appears to involve at least four other loci in the region. Earlier results implicating tan may have been based on a type of transgenic analysis that can give misleading results about the genes involved in an evolutionary change.
Keywords: genetics of adaptation, cis-regulation
INTRODUCTION
A recent debate in evolutionary genetics centers on whether adaptive changes in body form are caused by mutations that change the structure of proteins, including transcription factors (Hoekstra and Coyne, 2007) or by mutations that alter noncoding cis-regulatory elements (CREs; Carroll, 2005a; Carroll, 2005b; Gompel et al., 2005; Stern, 2000). Existing data show a preponderance of structural changes (about 80% of total changes among species), a figure that seems to have remained relatively constant over time (Stern and Orgogozo, 2008). But there is an ascertainment bias: cis-regulatory changes are much harder to detect than are coding changes in genes. Resolving this debate requires many additional studies of the genetics of interspecific differences in body form.
Recently, several studies have suggested that regulatory changes may be more important than structural changes as a source of adaptive morphological differences among species (Sucena et al., 2003; Colosimo et al., 2005; Clark et al., 2006; Shapiro et al., 2006, Wray 2007). Many of these studies have focused on pigmentation changes in humans, stickleback fish and Drosophila (Colosimo et al., 2005; Gompel et al., 2005; Wittkopp et al., 2003a; Wittkopp et al., 2002a; Wittkopp et al., 2002b, 2003b). One well-known example is the difference in abdominal pigmentation between the sister species D. santomea and D. yakuba. In D. yakuba, and seven of the other eight species in the well-studied Drosophila melanogaster species group, the posterior abdominal segments of both sexes are heavily pigmented. However, D. santomea, a species endemic to the island of São Tomé (an ancient volcanic island off the west coast of Africa) is unique in completely lacking dark abdominal pigment (Figure 1, Coyne et al., 2002; Lachaise et al., 2000; Llopart et al., 2002b).
FIGURE 1.

Abdominal pigmentation of D. melanogaster, D. santomea and D. yakuba. A. ♀ D. melanogaster, B. ♀ D. santomea, C. ♀ D. yakuba, D. ♂ D. melanogaster, E. ♂ D. santomea and F. ♂ D. yakuba.
Using QTL mapping, Carbone et al. (2005) located a genetic region on the X chromosome that has a large effect on the pigmentation difference in both sexes of these species. Recently, Jeong et al. (2008) studied the molecular genetics of this difference, and concluded that the light color of D. santomea was caused by the loss of expression of the tan locus (a gene that maps under the X-linked QTL peak) due to mutations fixed by selection in the cis-regulatory region. tan encodes a multifunctional enzyme that hydrolyzes both NBAD to dopamine, and carcinine (N-β-alanyl histamine) to histamine (True et al., 2005).
Jeong et al. (2008) adduce four lines of evidence to argue that tan is a major locus involved in the pigmentation difference. First, mutants of tan in D. melanogaster produce a lighter abdominal pigmentation—although not pigmentation as light as seen in D. santomea. Second, the tan gene is highly expressed in the heavily pigmented abdomens of male and female D. yakuba, but is not expressed in the lightly pigmented abdomens of male and female D. santomea. Third, sequencing of the tan locus and adjacent upstream promoter regions in both species show that they do not differ in protein sequence, but that D. santomea is polymorphic for three separate mutations in the cis-regulatory region of the gene, each of which could inactive it. Finally, the authors transferred the wild-type allele of the tan locus from (the darkly pigmented) D. melanogaster into D. santomea, and showed that this transfer darkened the abdomens of both males and females.
Here we report a failure, using direct genetic complementation tests, to confirm the results of Jeong et al. (2008). These results rests on our recent finding that D. melanogaster can produce viable female offspring when crossed with D. santomea. This allows us to use tan deletions in D. melanogaster to perform complementation tests of abdominal color in female hybrids with D. santomea. These tests are generally considered more reliable than transgenic analysis because they test the effect of the candidate gene in situ rather than when inserted into a random location in the genome. Moreover, showing that a wild-type allele involved in pigmentation produces darker color when transferred into another species does not unequivocally demonstrate that interspecific differences at that particular locus are responsible for observed color differences between the species. For example, transferring the gene for growth hormone from humans to chimps may produce larger chimps, but does not show that the difference in size between the two species maps to the growth-hormone locus.
Our direct genetic tests suggest that tan is not a major gene involved in the pigmentation difference between these species, at least in females. Additional deficiency mapping in the tan region, however, revealed the existence of five sub-regions that have a highly significant effect on pigmentation and do not encompass tan. Moreover, loss of expression of tan may have evolved after at the inactivation of other genes causing the species difference. At present, then, the tan gene cannot be considered a convincing example of the effect of cis-regulatory mutations on a major phenotypic difference.
EXPERIMENTAL PROCEDURES
We made a series of interspecific crosses involving several lines of D. melanogaster and seven different lines of D. santomea (See Supplementary Table 1 for the strains and results. Information about the lines can be found in Matute et al., 2008). Roughly 90% of the crosses between D. melanogaster females and D. santomea males produce offspring, all of which are sterile females (The reciprocal cross produces no offspring, and has never yielded matings or inseminated females). This unexpected and remarkable crossability between two distantly related species (10–15 million years old, Tamura et al. 2004) immediately makes it possible to use these species in genetic complementation tests.
Because tan mutations that reduce pigmentation are recessive in D. melanogaster, our strategy was to cross D. melanogaster females heterozygous for a tan mutation and a marked balancer chromosome (tan/Bal) to wild-type D. santomea males. If the light pigmentation of D. santomea females is caused by a mutation at tan, then two classes of female progeny should be produced, with those carrying the tan mutation being markedly lighter than those carrying the balancer with the tan + allele.
Drosophila stocks
Two stocks, w1118 P{XP}td07784 (a tan-like P-element insertion in the 5′ end of the first exon, henceforth called P[XP]), and Df(1)t20A, a null excision of tan, were provided by John True (SUNY at Stony Brook). We obtained other four tan mutants and a 7.5 kb Mi{ET1} insertion in the cis-regulatory element (CRE) of tan from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/). The tan strains (with their respective stock numbers) were t1 scp1 (110), t2 v1 f1 (131), t3 (132), and t5 v1 r12 (133, used by Jeong et al. in their study). The Mi{ET1} transposable element insertion (7.5 kb) in the CRE of tan (X-chromosome: position 9,120,883) was Mi{ET1}MB03163 (26107). All the mutants in the coding sequence showed the characteristic lightly pigmented phenotype of tan, while Mi{ET1}MB03163 was consistently darker than any other mutant. D. melanogaster ArkLa was made by combining the progeny of 6 isofemale lines collected in Arkansas and Louisiana (Llopart et al., 2002a).
As noted above, we crossed D. melanogaster females homozygous for the tan mutation to male carrying the Basc chromosome (In(1)scS1Lsc8R, sc8 scS1), a X-chromosome balancer lacking any mutations that affected pigmentation (Lindsley and Zimm, 1992). This crossed produced tan/Basc females that were then crossed to different types of males as described below.
The D. simulans Florida City (FC) isofemale line was collected by JAC in 1985 in Florida City, Florida. The D. mauritiana synthetic (SYN) stock was a wild-type line synthesized by combining six isofemale lines collected on Mauritius by O. Kitagawa in 1981.
We used two strains of D. yakuba: Taï 18 and yakSYN2005. Taï 18 was derived from the progeny of a single female collected in 1983 in the Taï rainforest on the border between Liberia and the Ivory Coast. yakSYN2005 line was made by combining progeny from six different isofemale lines collected in São Tomé in 2005. We also used two lines of D. santomea: STO.4 and sanSYN2005; STO.4 is an isofemale derived from a single female collected on São Tomé in 1998, while sanSYN 2005 derived from a combination of six isofemale lines collected in São Tomé in 2005 (see Matute et al., 2008 for further information).
We used quantitative deficiency complementation mapping to further resolve the locations of pigmentation QTL within the X-chromosome region previously identified by Carbone et al. Given that the QTL identification was done using a D. yakuba × D. santomea cross, and that there are many chromosome rearrangements differentiating D. yakuba and D. melanogaster (Ranz et al. 2008), we identified the homologous sub-regions of the target region (i.e., the chromosome region where tan is located) in D. melanogaster using previously published molecular cytologies (Ranz et al. 2008).
Stocks with deficiencies spanning the target cytological regions (8D-10A3, 14D-18B4 and 19B1-20F4) were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). We used minimal set of deficiencies covering the entire region. Deficiency breakpoints were provided by the donors of the strains.
Crossing scheme
Flies were raised in uncrowded cultures on standard cornmeal-yeast-agar medium, reared at 24°C under a 12-h light-dark cycle and maintained in large populations. To make interspecific crosses, ten virgin D. melanogaster tan/Basc females were crossed to ten males of either D. santomea, D. mauritania SYN or D. simulans FC (tan refers to all the tan mutants that we used). D. melanogaster tan/Basc (“mel tan/Basc”) females also were backcrossed to males from the original tan stock, or crossed to D. melanogaster ArkLa males. Using six different mutants of tan ensured that our results were not mutant-specific: two of these mutants (t1 and t5) have lesions resulting in amino acid replacements, one results from an insertion of a P-element in the 5′ end of the first exon, and the fourth was an excision of the whole chromosomal region that includes the tan promoter region (True et al., 2005). The other two mutants (t2 and t3) have unknown lesions but are presumed to be at the tan locus because of both their phenotype and their non-complementation with known tan mutants.
The crosses involving mel tan/Basc females produce two classes of progeny. For example if a mel tan/Basc female is crossed to a D. santomea male (san), the progeny will be tan/san or Basc/san. These two genotypes are distinguishable by the dominant marker Bar carried on the Basc chromosome, a marker that affects eye shape (Lindsley and Zimm, 1992). For all the crosses described below, we present the female genotype first. We followed the same experimental design to perform deficiency mapping and further study the region adjacent to tan as described by Carbone et al. (2005).
Scoring Pigmentation
D. melanogaster individuals and hybrid progeny were collected as virgins and kept in groups of twenty until four days old. To estimate the intensity of pigmentation on whole flies, we used a 0-to-4-point visual scale ranging from 0 (unpigmented areas) to 4 (dark and shiny black areas), with intermediate numbers representing intermediate degrees of pigmentation. We also measured the proportion of the area of each tergite that was pigmented (estimated to the nearest 0.1). To obtain the overall pigmentation score of each individual, we multiplied the percentage of the area of each of the three tergite covered by each shade of pigmentation by the intensity of that pigmentation, and these areas were summed (Carbone et al., 2005). All pigmentation scores are based on the sum of values for the A4, A5, and A6 tergites. The maximum pigment score for each tergite is therefore 4 (an intensity of 4, with 1.0 of the tergite covered with that pigment); the minimum is zero. Summing over the three tergites, then, the maximum possible score is 12 (most intense) and the minimum possible score is 0. The scoring was done blindly so that the scorer did not know either the species or genotype. In crosses that resulted in the segregation of two genotypes distinguishable by an eye-shape mutant, we removed the heads from the flies before scoring to conceal their genotype. For the crosses involving tan mutants and controls, we scored pigmentation in 100 individuals, and 50 flies per genotype in those crosses that produced two genotypes. For the deficiency mapping crosses, we scored 50 flies (25 per genotype).
Abdominal Cuticle Preparation
We prepared specimens for photography in a manner similar to that of Jeong et al. (2008). Four-day-old virgin female flies were killed with ether and immediately dissected or stored in individual vials at −80°C. Specimens were cut along the dorsal midline with a razor blade. Soft tissues were removed and the cuticles mounted in Canada balsam to dissolve any remaining soft tissue. The preparations were then flattened with a coverslip and incubated for 3 hr at 65°C (Kopp et al., 2000; Jeong et al., 2006, 2008).
Data analysis
We analyzed the total data (differences in pigmentation between genotypes of the same cross, including all the mutants) by fitting a random linear mixed model (Pinheiro and Bates, 2000) with genotype as a main effect (e.g., Basc/san vs. tan/san) and pigmentation as response. Differences between mutants were considered random effects when linear mixed models (LMM) were required for the analysis. We repeated the same procedure for each of the crosses (i.e., the different types of males crossed to tan/Basc females).
Additionally, to establish whether additional deficiencies spanning the X chromosome candidate region had a significant effect on the pigmentation levels of F1 hybrid females, we compared the pigmentation between flies carrying the deficiency and flies carrying Basc with a one-way ANOVA for each deficiency.
RESULTS
Pure species and tan mutants
We scored five species of the D. melanogaster group for pigmentation: four of them (D. melanogaster, D. simulans, D, mauritiana and D. yakuba) have black-pigmented abdomens while D. santomea is yellow, lacking black pigmentation. Supplementary Table 2 shows the mean pigmentation score for each species. While the pigmentation of the dark species ranges from 3.3 to 5.2, that of D. santomea is 0.22—less than 7% as intense as that of the least pigmented dark species (D. simulans).
We also scored each of the six tan mutants for pigmentation when homozygous. Their average pigmentation and the phenotypes of these mutants (as well as that of the Mi{ET1}MB03163 Mi{ET1} insertion allele) are given in Supplementary Table 3 and shown in Supplementary Figures 1 and 2. The six tan mutations were not significantly heterogeneous for pigmentation (F5,594 = 1.53, P = 0.179). All of these mutants are significantly darker than the pigmentation of wild-type D. santomea (F6,643 = 118.04, P < 10−5, HSD Tukey test).
Mi{ET1}MB03163, the stock having a large insertion of the Mi{ET1} transposable element in the CRE region of tan, showed a pigmentation score significantly higher than any of the tan mutants and lower than that of wild-type D. melanogaster (one-way ANOVA F2,297 = 925.71, p < 10−5; HSD Tukey test). This suggests that the disruption of the CRE may affect the pigmentation pattern of D. melanogaster far less than do lesions in the coding sequences of tan (Supplementary Figure 2). Note, however, that we surveyed only one wild-type strain of D. melanogaster.
Complementation tests within D. melanogaster
We used a large deletion containing the tan locus and its promoter (Df(1)t20A) to verify whether all the tan mutants were allelic. We crossed Df(1)t20A to four classical tan mutants (t1, t2, t3, and t5) as well as a P-insertion (P[XP]) allele, and scored 100 individuals from each cross, comparing their average pigmentation to a sample of 100 individuals of the pure strain. In all cases the large deletion failed to complement the tan mutants (Supplementary Figure 3, paired t-test against the parental line: t4 = −0.1183, p = 0.9116). All of the mutants, then, appear to reside in the region covered by tan deletion Df(l)t20A, confirming the known location of lesions in these strains.
Backcrosses of Basc/tan to D. melanogaster
When D. melanogaster females heterozygous for any of the tan mutants and the Basc balancer chromosome (mel tan/Basc) were backcrossed to tan males, we observed a bimodal distribution of pigmentation scores among female offspring (e.g., Figure 2) when crosses were scored blind. Figure 3 shows the difference in pigmentation between the two classes of females. To determine if the two modes represented the two genotypes segregating in this cross (Basc/tan and tan/tan), we analyzed the distribution of individual pigmentation by fitting a mixed linear model (Pinheiro and Bates, 2000) having genotype (whether or not the deficiency is present) as a main effect and specific mutants tested as a random effect. This analysis showed that the effect of genotype was highly significant (F1,593 = 2521.82, p < 0.0001; all the heavily pigmented flies had the Basc chromosome). These results show that tan/tan homozygotes can be clearly distinguished from Basc/tan heterozygotes in D. melanogaster (see Figure 2).
FIGURE 2.
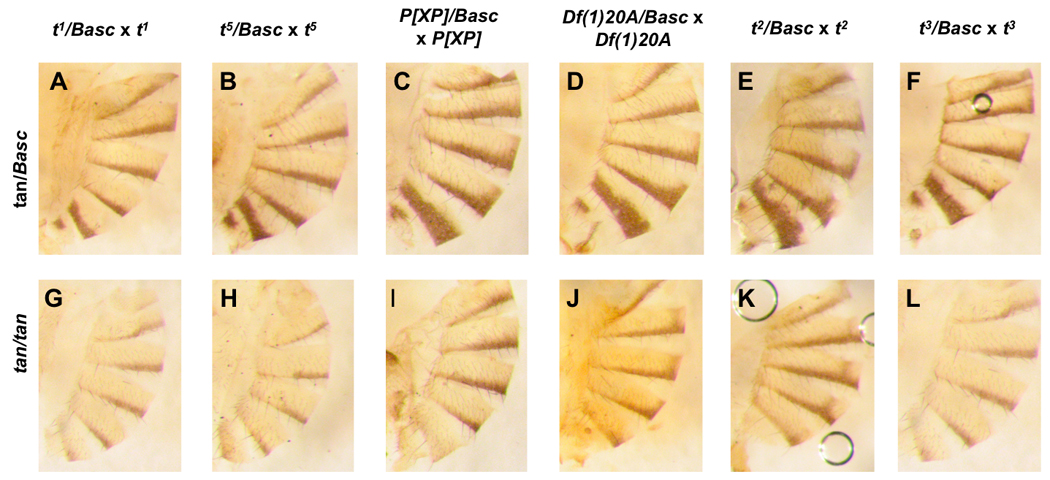
D. melanogaster females heterozygous for tan mutants and the Basc balancer chromosome (mel tan/Basc) backcrossed to tan males. In all cases Basc/tan heterozygotes (A–F) homozygotes (G–L) can be clearly distinguished from tan/tan homozygotes in D. melanogaster.
FIGURE 3.
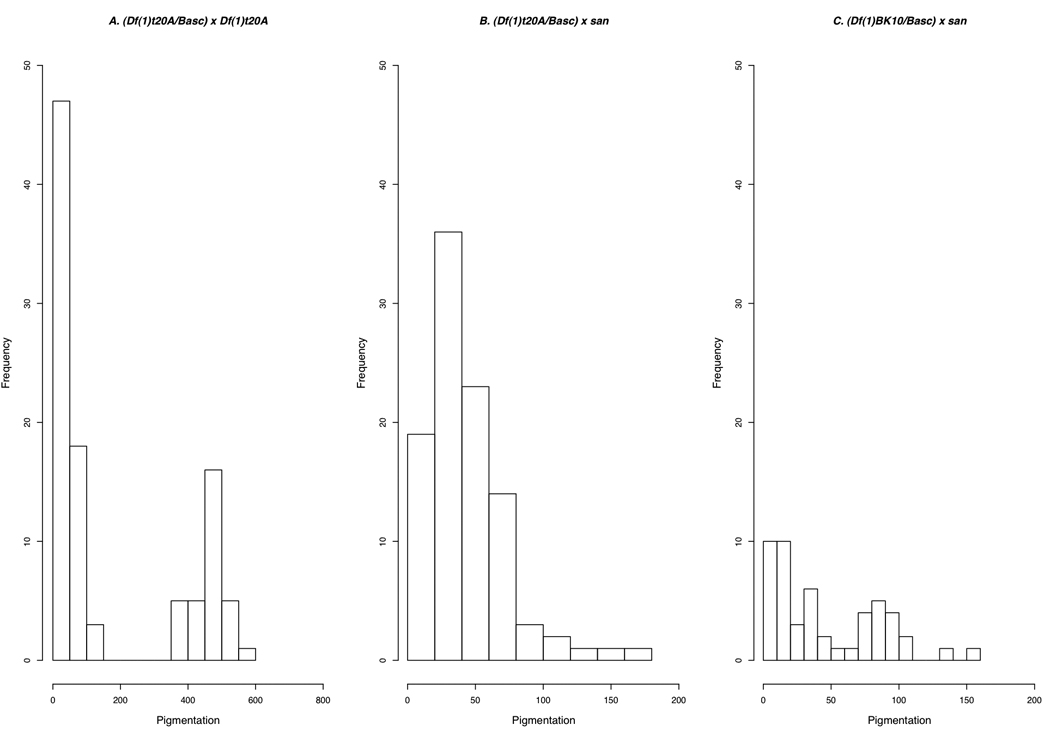
Phenotypic distribution of the progeny produced by the (Df(1)t20A/Basc) x Df(1)t20A (A) and (Df(1)t20A/Basc) × san (B) crosses.
We made five additional controls. To determine whether the tan stocks and the Basc stock differed in genetic backgrounds in a way that could affect pigmentation, we compared the pigmentation score of the tan/tan flies from the (mel tan/Basc) × tan cross to tan/tan flies from the (mel tan/tan) × tan cross. This comparison showed no significant difference between the genotypes (LMM, F1,6 = 0.089, p = 0.776). Second, we crossed D. melanogaster Basc/ArkLa) females to tan males to determine whether the balancer chromosome complemented the tan mutations as effectively as did the wild-type alleles. This cross too showed no significant difference in pigmentation between the tan/ArkLa and tan/Basc genotypes (LMM, F1,588= 1.3774, p = 0.241). These two results mean that Basc can restore the wild-type phenotype as effectively as can the tan + allele on wild-type chromosomes. Third, to determine whether the Basc chromosome could restore the wild-type pigmentation in females that carried the Mi{ET1}MB03163 allele, we crossed tan/Basc females to Mi{ET1}MB03163 males. The Mi{ET1} transposable element insertion in the promoter region of the D. melanogaster tan locus represents the closest match to the postulated CRE mutant allele in D. santomea discussed by Jeong et al. (2008). We found that for all the crosses we assayed, Basc rescues the D. melanogaster dark pigmentation when the Mi{ET1}MB03163 allele is present: Mi{ET1}MB03163/ tan females were consistently lighter than were Mi{ET1}MB03163/Basc females (LMM, F1,395= 442.445 P < 0.0001). These results show that Basc can complement mutations in both the coding sequences and the regulatory elements of tan.
Fourth, to check whether the Basc chromosome itself affected pigmentation score (i.e., whether it made its carriers darker), we crossed (Basc/mel) females to mel ArkLa males and compared the pigmentation of the two types of progeny (Basc/mel and mel/mel). We found no significant differences between the genotypes (LMM, F1,593= 0.001, p = 0.9747, Table 1). These results mean that the balancer has no unexpected effect on pigmentation compared to other chromosomes carrying the tan + allele. Finally, to make sure that the tan + allele of other species in the D. melanogaster subgroup could complement tan mutants of D. melanogaster, we crossed D. melanogaster tan/Basc) females to D. mauritiana and D. simulans males. These crosses showed that tan has no effect on the pigmentation of D. mauritiana and D. simulans (D. mauritiana: F1,593 = 0.080, p = 0.777; D. simulans: F1,593 = 4.776, p = 0.029, Table 1). Figure 4 shows representative females from these crosses.
TABLE 1.
Mean pigmentation (Standard Deviation) of the female progeny from the cross (tan/Basc) to different males on the D.melanogaster group of species. N = 50 for each genotype.
| scp1 t1 | t2v1f1 | t3 | t5v1r1 | P{XP}td07784 | Df(1)t20A | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | (+) | B | (+) | B | (+) | B | (+) | B | (+) | B | (+) | |
| tan/Basc × san | 0.470 (0.398) |
0.462 (0.259) |
0.408 (0.314) |
0.372 (0.234) |
0.412 (0.356) |
0.382 (0.256) |
0.503 (0.403) |
0.423 (0.303) |
0.522 (0.257) |
0.493 (0.397) |
0.544 (0.352) |
0.422 (0.304) |
| tan/Basc × mau | 5.134 (1.332) |
4.327 (1.433) |
4.148 (1.527) |
3.722 (1.684) |
3.633 (1.736) |
4.182 (1.476) |
4.113 (1.689) |
3.803 (1.573) |
3.975 (1.298) |
4.488 (1.452) |
4.074 (1.397) |
4.413 (1.332) |
| tan/Basc × sim | 3.158 (1.476) |
2.917 (1.156) |
3.478 (1.408) |
3.320 (1.233) |
3.537 (1.433) |
3.315 (1.249) |
3.606 (1.302) |
3.419 (1.316) |
3.394 (1.612) |
2.850 (1.598) |
3.671 (1.37) |
3.149 (1.332) |
| tan/Basc × mel | 4.028 (0.877) |
3.980 (0.730) |
4.114 (0.754) |
4.191 (0.700) |
4.613 (0.933) |
4.413 (0.604) |
4.469 (0.715) |
4.225 (0.805) |
4.553 (0.668) |
4.384 (0.825) |
4.446 (1.072) |
4.520 (0.924) |
| tan/Basc × tan | 3.942 (1.222) |
0.937 (1.048) |
4.232 (0.845) |
0.737 (0.380) |
3.992 (1.078) |
0.950 (0.631) |
4.203 (0.566) |
0.600 (0.347) |
4.259 (0.681) |
0.556 (0.234) |
4.035 (1.564) |
0.560 (0.502) |
FIGURE 4.
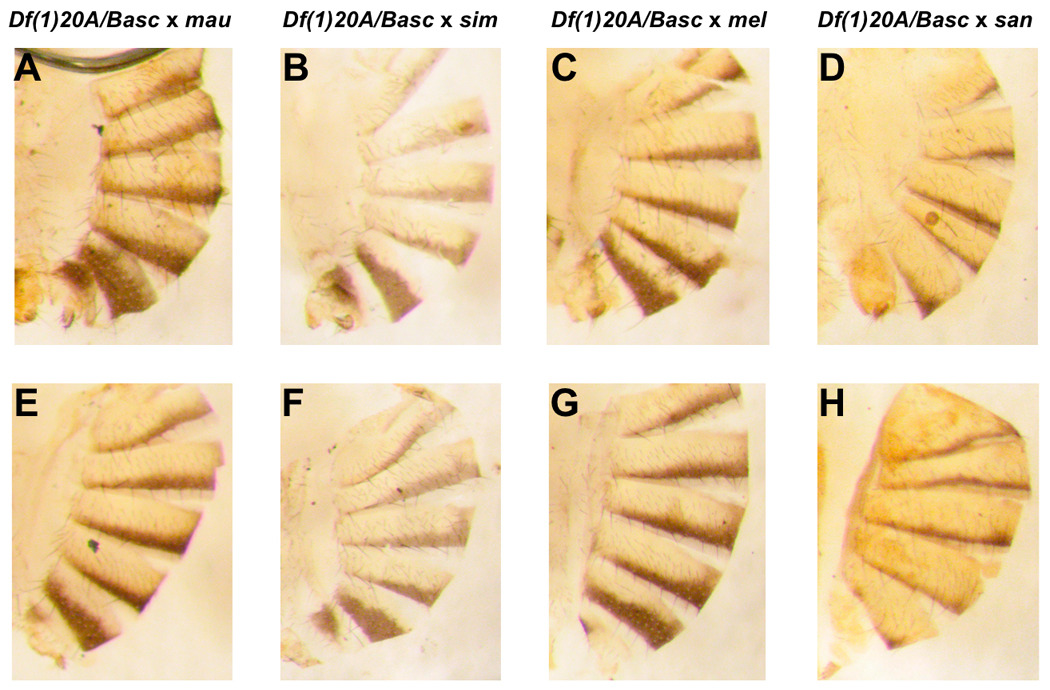
Interspecific complementation tests. D. melanogaster females heterozygous for tan mutants and the Basc balancer chromosome (mel tan/Basc) were crossed to males of four different species: D. mauritiana, D. simulans, D. melanogaster and D. santomea. Only the crosses involving Df(1)t20A/Basc females are shown but the results are the same with all the mutants studied. Top panels: F1 hybrid females carrying Basc. A.mau/Basc, B.sim/Basc, C.mel/Basc, D.san/Basc. Lower panels: F1 hybrid females carrying tan: E. mau/ Df(1)t20A, F. sim/ Df(1)t20A, G. mel/ Df(1)t20A, H. san/ Df(1)t20A.
Complementation crosses between D. melanogaster and D. santomea
The critical test of whether the lighter pigmentation of D. santomea is due to mutations at the tan locus involves crossing D. melanogaster tan/Basc females to D.santomea males. This cross produces two genotypes among hybrid females: Basc/san and tan/san. If the mutants in either the structural locus or the CRE of tan are largely responsible for the light abdominal pigmentation in D. santomea, then Basc will complement the D. santomea tan allele and restore the darkly pigmented phenotype in these F1 hybrids. We would then expect that this cross would show a bimodal distribution of pigmentation scores, similar to that seen among offspring of the (tan/Basc) × tan cross within D. melanogaster. Further, the two classes of interspecific hybrid females (Basc/san and tan/san) should differ significantly in pigmentation.
These crosses showed no statistically significant effect on the interspecific difference. In the combined data, the effect of genotype is not significant (F1,593 = 3.6742, p = 0.0557; Figure 3 and Figure 4), so that the two genotypes do not differ in their pigmentation (data given in Table 1). Clearly, tan does not have a large (or even statistically significant) effect on the pigmentation of D. santomea females.
Similarly, when we made interspecific crosses using Mi{ET1}MB03163, the stock with an insertion in the promoter region of tan, we found no effect of this insertion on the pigmentation of the two hybrid genotypes (one-way ANOVA: F1,98 = 4 × 10−4, p = 0.985). This insertion stock is probably the closest existing genetic parallel to the postulated CRE mutants in D. santomea that affect pigmentation.
All of these results strongly suggest that mutations in the tan locus, whether structural or regulatory, have little or no effect on the difference in pigmentation between females of D. santomea and females of the other members of the D. melanogaster subgroup.
Fine-scale mapping of the tan region
Because the tan region showed a large effect on pigmentation in the study of Carbone et al. (2005), we performed a more detailed analysis of this region with two aims: to find candidate alternative regions that might explain the difference in pigmentation between D. santomea and the other subgroup species, and to establish whether our methods had the power to detect regions (and genes) affecting pigmentation.
We studied 16 deficiencies spanning the QTL region described by Carbone et al. (Table 2): seven of these in the 14D-18B4 region; eight in the 8D-10A3 region, and one in the 19B1-20F4 region. We performed a one-way ANOVA to determine whether the differences between flies carrying Basc and flies carrying the deficiency were significant. Table 2 shows the results. First, our analyses of two additional deficiencies that encompassed the region which contain the tan locus (8D1) again reveal no effect of the region on pigmentation. This confirms our conclusion that tan is unlikely to be a major locus causing interspecific differences in pigmentation. Second, we found five deficiencies (two of them overlapping) that showed a significantly lighter pigmentation (Figure 5) than the balancer-carrying flies. There are, then, at least four strong candidate regions for genes involved in the pigmentation difference between D. santomea and its sister species.
TABLE 2.
Mean pigmentation (Standard Deviation) of the female progeny from the cross (df/Basc) for sixteen different deficiencies to D. santomea males. N = 25 for each genotype.
| Deficiency | Stock Number |
Cytological Location |
Mean pigmentation Basc/san |
Mean pigmentation df/san |
F1,49 | P value |
|---|---|---|---|---|---|---|
| Df(1)r-D17 | 991 | 14C1-15A6 | 0.556 (0.258) | 0.552 (0.303) | 0.0025 | 0.9601 |
| Df(1)BSC582 | 25416 | 15A1-15E2 | 0.512 (0.305) | 0.544 (0.258) | 0.1605 | 0.6905 |
| Df(1)B25 | 4741 | 15D3-16A6 | 0.496 (0.235) | 0.512 (0.285) | 0.0469 | 0.8295 |
| Df(1)BK10 | 4953 | 15F2-16C10 | 0.660 (0.388) | 0.288 (0.254) | 24.589 | 9.50 × 10−6 |
| Df(1)RR79 | 6217 | 16C-16F | 0.936 (0.455) | 0.648 (0.410) | 5.5173 | 0.02299 |
| Df(1)N19 | 970 | 17A1-18A2 | 0.732 (0.360) | 0.416 (0.300) | 11.373 | 0.00148 |
| Df(1)Exel6291 | 7754 | 18A2-18A3 | 0.556 (0.331) | 0.688 (0.410) | 1.5726 | 0.2159 |
| Df(1)ED6957 | 8033 | 8B5-8D9 | 0.720 (0.360) | 0.764 (0.142) | 1.947 | 0.1693 |
| Df(1)BSC538 | 25066 | 8C17-8E4 | 0.420 (0.200) | 0.456 (0.27 | 1.4157 | 0.2525 |
| Df(1)C52 | 952 | 8E4-9C4 | 0.644 (0.399) | 0.724 (0.411) | 0.4881 | 0.4881 |
| Df(1)v-L15 | 954 | 9B1-10A1 | 0.620 (0.342) | 0.304 (0.193) | 16.238 | 1.98 × 10 −4 |
| Df(1)BSC540 | 25068 | 9E8-10A3 | 0.624 (0.240) | 0.623 (0.267) | 8.28 × 10−31 | 1 |
| Df(1)BSC287 | 23672 | 10A10-10B11 | 0.504 (0.230) | 0.492 (0.287) | 0.0266 | 0.8711 |
| Df(1)BSC722 | 26574 | 10B3-10E1 | 0.524 (0.215) | 0.508 (0.269) | 0.054 | 0.8172 |
| Df(1)v[N124B] | 5707 | 9E3-10A8 | 0.524 (0.194) | 0.620 (0.333) | 1.5508 | 0.2191 |
| Df(1)DCB1-35b | 977 | 9E3-10A8 | 0.712 (0.286) | 0.488 (0.254) | 8.5703 | 0.00521 |
FIGURE 5.
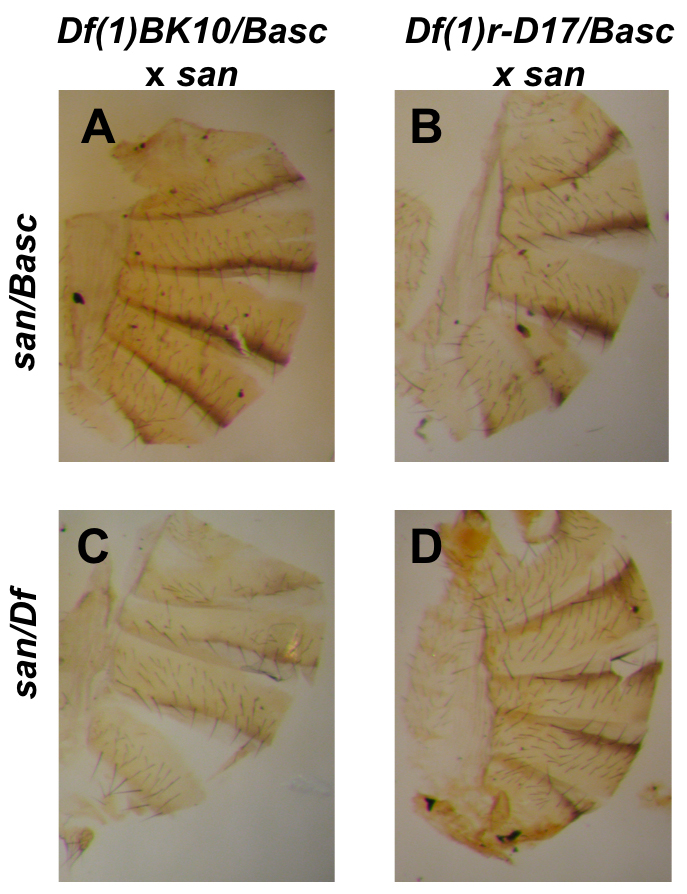
Deficiency analysis of QTL affecting pigmentation between D. melanogaster and D. santomea. Only two of the sixteen tested deficiencies are shown. D. melanogaster (Df(1)BK10) when crossed to a D. santomea male produced two kinds of progeny (Basc/san (A) and Df(1)BK10/san (C)) that showed a significant difference in pigmentation. The progeny of the cross mel (Df(1)r-D17) × san (Basc/san (B) and Df(1)r-D17/san (D)) showed no significant differences in their pigmentation levels. Top panels: F1 hybrid females carrying Basc. A. Basc/san, B. Basc/san Lower panels: F1 hybrid females carrying a deficiency C. Df(1)BK10/san, D. Df(1)r-D17/san
DISCUSSION
Our complementation tests using D. melanogaster deletions crossed to D. santomea show no significant effect of the tan locus on the pigmentation difference between these two species, and by inference on the difference between D. yakuba and D. santomea (The transgenic studies performed by Jeong et al. were between D. santomea and D. melanogaster). We thus cannot verify the conclusion of Jeong et al. (2008) that cis-regulatory mutations in the tan region play an important role in the pigmentation difference between the two species.
Before we discuss the discrepancy between the two studies, we must mention a few caveats. First, by necessity we were limited to studying genetic complementation in females, since tan is an X-linked locus. Most of Jeong et al.’s analysis, however, dealt with males, and it is possible that the genetic basis of the pigmentation difference between D. santomea and D. yakuba itself differs between males and females. However, we consider this unlikely. QTL analysis of both species (Carbone et al., 2005) shows that male and female “pigmentation” peaks are located in the same chromosome regions, implying that the same loci are involved in pigmentation differences of the two sexes. More important, Jeong et al.’s analyses show an effect of tan in both sexes, including the ability of transgenic tan+ alleles to restore some pigmentation in both male and female D. santomea (see, for instance, Figure 5D of Jeong et al.).
Second, Jeong et al. did not find that tan explained all of the pigmentation difference between D. melanogaster and D. santomea, only part of it—though a substantial part. A single transgenic copy of tan+ in D. santomea causes partial rescue of male pigmentation on the sixth (but not the fifth) abdominal segment of D. santomea (Jeong et al. figure 5B). In males, two copies of tan+ from D. melanogaster cause fuller but still brownish pigmentation of segments A6 and A5, and weaker pigmentation of A4 in males, while in pure D. yakuba these areas are larger, and are colored deep black instead of brown (Jeong et al. 2008, compare figure 5C vs. 1A). It is notable that although a single copy of the D. melanogaster tan+ transgene almost completely rescues dark pigmentation in the D. melanogaster tan mutant t5, it rescues D. santomea pigmentation far less well (Jeong et al. 2008, fig 4C vs. 5B for one copy, 5C for two). Rescue of pigmentation in D. santomea females by the tan+ transgene is also incomplete: as in males, the area of female pigmentation is reduced compared to that seen in D. yakuba, although distinct abdominal stripes are still present (Jeong et al. Figure 1E vs. 5D). Further, our failure to detect an effect of tan cannot be ascribed to our using only a single aberrant deletion or tan mutant, since the failure is seen using five structural mutants, one disruption of the CRE, and a deletion that encompasses the whole gene and its promoter.
Our results, then, seem to rule out a model in which recessive mutations at tan are a major cause of the yellow abdominal pigmentation in D. santomea. The low pigmentation scores in the F1 hybrids between D. melanogaster and D. santomea suggest the existence of at least one semi-dominant pigment-reducing locus somewhere in the D. santomea genome—a locus that must be different from tan.
Clearly, we have been unable to confirm the predicted importance of the tan locus to overall pigmentation change in D. santomea (Jeong et al. 2008). What is the explanation? The most likely is that the different outcomes reflect a difference in methodology: complementation mapping on our part versus transgenic analysis and expression differences/transgenic analysis in the study of Jeong et al. (2008). In other words, we suspect that tan is not a major genetic factor in the pigmentation difference, despite the results from previous QTL candidate gene analysis and transgenic studies (Jeong et al. 2008).
Complementation analysis using mutations or deletions has long been used to identify whether mutants are allelic or not, and whether a species difference is located in a particular region. For example, Sucena and Stern (2000) used a deletion in the ovo/shaven baby region to show that the recessive loss of hairs on the cuticle of D. sechellia (compared to the outgroup D. melanogaster) resided in that region. This is precisely the test that we have done with D. santomea. However, although complementation analysis is a well-accepted strategy for testing whether strains have mutations in the same or different genes, its power can be limited by the dominance of the tested alleles. Also, one occasionally finds complementation between different alleles of the same gene (intragenic complementation). We have tried to minimize the chance of such effects in the current experiments by using a variety of different tan alleles. Nevertheless, our tests also involve species that are not closely related, and we cannot exclude the possibility that the overall low level of pigmentation seen in F1 hybrids between D. santomea and D. melanogaster has masked a larger relative contribution of the tan locus to pigmentation changes that would be seen in intraspecific crosses with D.santomea, or in hybrids between D. santomea and other species in the melanogaster subgroup. We suspect that the tan+ allele from D. melanogaster adds pigmentation to D. santomea males and females not because the gene is involved in the species difference, but simply because transferring an allele involved in making pigment into a genetic background lacking that pigment may produce a darker phenotype, regardless of which mutations have produced the interspecific difference in pigmentation. The best to resolve this issue is, of course, through targeted gene replacement.
Jeong et al. (2008) also showed by in situ hybridization that tan is expressed in both adult abdomens and the posterior part of the pupa in D. yakuba, but not in D. santomea. While showing that there is a species-specific difference in tan expression, this does not prove that the observed pigmentation difference maps to that locus. In analysis of genetic pathways, it is not unusual to find that a transgene for one locus in a pathway will restore a phenotype affected by a different mutation in that pathway (e.g., Hyodo-Tagui et al.'s [1997] study of the pigmentation pathway in medakafish).
We have shown by complementation tests that tan seems to have little or no effect on the observed pigmentation difference between D. melanogaster and D. santomea (and, by inference, between D. yakuba and D. santomea), and that the large effect in pigmentation ascribed to the X chromosome is caused by at least four distinct chromosomal regions near—but not including—tan. Since males are hemizygous for tan, complementation tests are not useful for testing pigmentation differences in that sex. Here, targeted gene replacement of tan and its regulatory elements must be used.
Although tan maps under the QTL peak known to include a gene or genes with a large influence on species-specific pigmentation, tan itself is not the site of the lesion. Rather, tan appears to be closely linked to several genes with major effect—perhaps genes in the same pathway as tan (i.e., the core dopamine pathway). Beyond the five candidate regions near tan, we are using finer deletion mapping and fine-structure molecular QTL analysis to identify the genes responsible.
One other observation of Jeong et al. (2008) supports the idea that while tan may be involved in the biochemical pathway producing the pigmentation difference between D. yakuba and D. santomea, it does not carry mutations producing that difference. This is their finding that there is not one but three distinct cis-regulatory haplotypes that inactivate tan expression in natural populations of D. santomea. (One haplotype contains two nucleotide substitutions from the D. yakuba allele, while the other two have different deletions in or near these substituted regions). This polymorphism for three distinct haplotypes, each of which the tan gene, contradicts the idea that the derived loss of color in D. santomea resulted from a rapid selective sweep of a single adaptive mutation that reduced pigmentation. Indeed, Jeong et al. (2008) posit that the inactivation of the cis-regulatory region is due to relaxed selection on pigmentation in the D. santomea lineage followed by neutral evolution of the CRE. We agree, but suspect that the loss of tan expression resulted from changes at another locus that caused loss of pigmentation in this lineage. This loss would allow the tan+ allele, now lacking an effect on pigmentation, to accumulate mutations that inactivated its expression. At present, then, the genetic basis of this striking interspecific difference in pigmentation remains unclear.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jean Gladstone for technical assistance. This work was funded by NIH grant R01GM058260 to JAC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Carbone MA, Llopart A, deAngelis M, Coyne JA, Mackay TF. Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea. Genetics. 2005;171:211–225. doi: 10.1534/genetics.105.044412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Endless forms most beautiful : the new science of evo devo and the making of the animal kingdom. 1st edn. New York: W.W. Norton & Co; 2005a. [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS biology. 2005b;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villareal G, Dickson M, Grimwood J, Schmutz J, Myers R, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Kim SY, Chang AS, Lachaise D, Elwyn S. Sexual isolation between two sibling species with overlapping ranges: Drosophila santomea and Drosophila yakuba. Evolution; international journal of organic evolution. 2002;56:2424–2434. doi: 10.1111/j.0014-3820.2002.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution; international journal of organic evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hyodo-Taguchi Y, Winkler C, Kurihara Y, Schartl A, Schartl M. Phenotypic rescue of the albino mutation in the medakafish (Oryzias latipes) by a mouse tyrosinase transgene. Mechanisms of development. 1997;68:27–35. doi: 10.1016/s0925-4773(97)00128-7. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Lachaise D, Harry M, Solignac M, Lemeunier F, Benassi V, Cariou ML. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from São Tomé. Proceedings. 2000;267:1487–1495. doi: 10.1098/rspb.2000.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- Llopart A, Elwyn S, Coyne JA. Pigmentation and mate choice in Drosophila. Nature. 2002a;419:360. doi: 10.1038/419360a. discussion 360. [DOI] [PubMed] [Google Scholar]
- Llopart A, Elwyn S, Lachaise D, Coyne JA. Genetics of a difference in pigmentation between Drosophila yakuba and Drosophila santomea. Evolution; international journal of organic evolution. 2002b;56:2262–2277. doi: 10.1111/j.0014-3820.2002.tb00150.x. [DOI] [PubMed] [Google Scholar]
- Matute DR, Novak CJ, Coyne JA. Temperature-Based Extrinsic Reproductive Isolation in Two Species of Drosophila. Evolution; international journal of organic evolution. 2008;63:595–612. doi: 10.1111/j.1558-5646.2008.00588.x. [DOI] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. New York: Springer; 2000. [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LDW, Roote J, Ashburner M, Bergman C. Principles of genome evolution in the D. melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Stern DL. Tinker where the tinkering's good. Trends Genet. 2008;24:317–319. doi: 10.1016/j.tig.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 2006;103:13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. Evolutionary developmental biology and the problem of variation. Evolution; international journal of organic evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Sucena E, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura KS, Subramanian D, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21(1):36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou SR, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: Genetic mechanisms and the potential consequences of pleiotropy. Seminars in cell & developmental biology. 2008 doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003a;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development(Cambridge England) 2002a;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Vaccaro K, Carroll SB. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol. 2002b;12:1547–1556. doi: 10.1016/s0960-9822(02)01113-2. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Williams BL, Selegue JE, Carroll SB. Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2003b;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


