Abstract
Recent emergence of highly pathogenic avian (H5N1) influenza viruses, their epizootic and panzootic nature, and their association with lethal human infections have raised significant global health concerns1,2. Several studies have underscored the importance of non-structural protein NS1 in the increased pathogenicity and virulence of these strains3,4. NS1, which consists of two domains, a dsRNA binding domain (RBD)5,6 and the effector domain (ED)7 separated through a linker, is an antagonist of antiviral type-I interferon (IFN) response in the host8,9. Here we report the x-ray structure of the full length NS1 from a H5N1 (A/Vietnam/1203/2004) strain that was associated with 60% of human deaths in an outbreak in Vietnam1,2. Compared to the individually determined structures of RBD and ED from non-H5N1 strains, the RBD within H5N1 NS1 exhibits modest structural changes, while the H5N1 ED shows significant alteration particularly in the dimeric interface. Although both domains in the full-length NS1 individually participate in dimeric interactions, an unexpected finding is that these interactions result in the formation of a chain of NS1 molecules instead of distinct dimeric units. Three such chains in the crystal interact with one another extensively to form a tubular organization of similar dimensions observed in the cryo-EM images of NS1 in the presence of dsRNA. The tubular oligomeric organization of NS1 in which residues implicated in dsRNA binding face a 20 Å wide central tunnel provides a plausible mechanism for how NS1 sequesters varying lengths of dsRNA, to counter cellular antiviral dsRNA response pathways, while simultaneously interacting with other cellular ligands during an infection.
NS1 of influenza virus, is a multifunctional protein associated with a number of regulatory functions during a viral infection10, including conferring resistance to antiviral interferon (IFN) induction3,4. Although, the mechanisms for how NS1 antagonizes IFN response remains to be fully understood, both RBD11,12 and ED13-15 have been implicated in this activity. Our goal was to determine the X-ray structure of the full length NS1 protein to provide a more complete structural framework for understanding the functional aspects of NS1, including the effects of various mutations in H5N1 strains. In contrast to RBD and ED, which were individually amenable for crystallographic analysis6,7, full length NS1 readily aggregates and precipitates at the concentrations needed for crystallographic studies. We determined that two mutations, R38A and K41A, completely abrogated aggregation and allowed full length NS1 to be sufficiently concentrated for crystallographic studies. The H5N1 NS1 structure was determined using molecular replacement techniques with previously reported structures of the RBD6 and ED7 as search models (see Methods and Table S1).
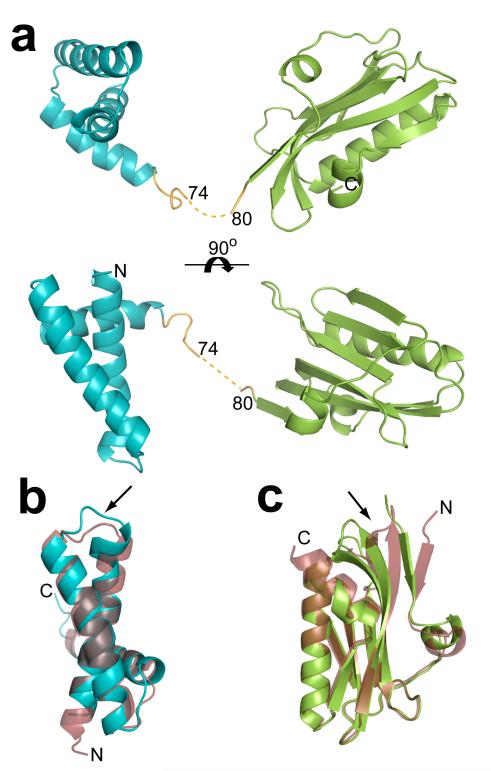
In the full length H5N1 NS1 monomer structure, the RBD and ED are formed by residues 1-72 and residues 84-220, respectively (Fig. 1). The density corresponding to the five residues (75-79) in the linker region is not well defined indicating the flexible nature of the linker. The electron density is relatively weak for the residues 67-74 and these residues exhibit higher than average temperature factors (~70 Å2). The proposed connectivity between RBD and ED (Fig. 1), although not entirely unequivocal (see Fig. S1 for alternative possibility), is based on reappearance of the density in the 2Fo-Fc and Fo-Fc maps after removing these residues. The H5N1 NS1 RBD consists of three large α-helices connected by short loops as seen in the A/Udorn/72 H3N2 NS1 RBD structure5,6. The amino acid sequences in the RBD of these two strains show differences at 9 positions (Fig. S2). One of the changes in the sequence, V22F, appears to have resulted in a minor but noticeable structural alteration. F22, located at the C-terminus of the first α-helix causes a slight movement of the loop that connects this α-helix and the second main α-helix (Fig 1b). Except for this minor alteration, the two structures are invariant, with a root mean square deviation (RMSD) of 1.2 Å, including at the locations where the two mutations, R38A and K41A, were introduced to make the protein behave well for crystallographic studies.
Figure 1. H5N1 NS1 structure.
a. Cartoon representation of the H5N1 NS1 structure as viewed looking across (top) and down onto (bottom) the main α-helix of the RBD (aquamarine) that is implicated in dsRNA binding. The ED is colored in green, and the linker region in orange along with the 5 residues (75-79) not well defined in the electron density map shown in dotted line. b. Structural alignment of H5N1 NS1 RBD (aquamarine) with H3N1 NS1 RBD6 (ruby, pdb id: 1AIL), and (c) alignment of the H5N1 NS1 ED (green) with H1N1 NS1 ED7 (ruby, pdb id: 2GX9). In both b and c the alignments are oriented to display the areas of greatest deviation, the V22F conformational change in a loop region observed in the H5N1 RBD in panel b, and the movement of the β-sheets in the H5N1 ED in panel c, are indicated by the black arrows.
The overall polypeptide fold in the H5N1 ED, with seven β-strands and three α-helices, is similar to the α-helix β-crescent fold7 observed in the previously reported structure of the A/PR8/34 H1N1 ED despite sequence differences in 15 positions and a C-terminal truncation in the H5N1 ED at residue 220 (Fig. S2). The RMSD of 1.6 Å between the two ED structures is slightly higher than that observed for the RBD indicating more pronounced structural changes in the ED domain than those observed in the RBD. These structural changes, although not correlated specifically with any amino acid change, include an en-mass shift in three β-strands (by about 8.0 Å at the point of greatest deviation) formed by residues 83-86, 121-132 and 135-146 (Fig 1c). These three β-strands in the H5N1 NS1 ED, compared to H1N1 ED7, are more twisted toward the center of the ED monomer, and shifted away from the ED dimeric interface.
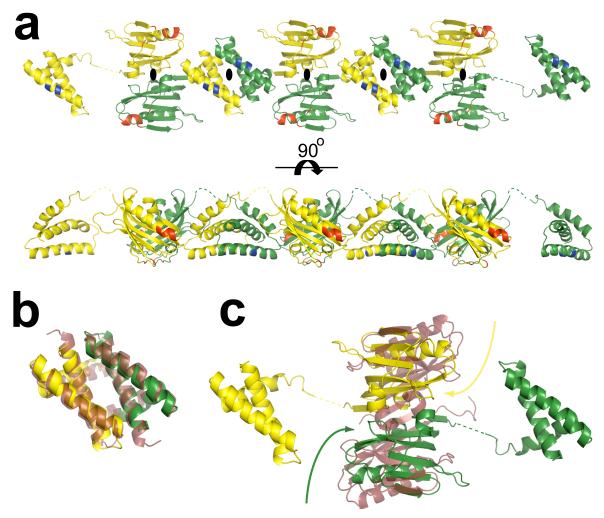
In the crystal structure of the full-length H5N1 NS1, both RBD and ED participate in dimeric interactions as expected from the individual crystal structures of these domains from non-H5N1 strains6,7. However, in the full-length crystal structure, surprisingly, the two domains of each NS1 molecule, instead of participating in a dimeric interaction with another NS1 molecule to form a single dimeric unit, they separately interact with their respective domains from the neighboring NS1 molecules, related by crystallographic 2-fold axes, resulting in the formation of a chain of NS1 molecules with alternating RBD and ED dimers (Fig. 2a, Fig. S3). The orientation of the subunits and their interactions in the H5N1 RBD dimer is remarkably identical to that observed in the previously reported structure of the H3N1 RBD6 despite differences in the crystallization conditions and crystal forms (Fig. 2b). Formation of such a structurally conserved RBD dimer is consistent with the observation that RBD dimerization is critical for dsRNA binding activity of the NS116.
Figure 2. RBD and ED dimer formation and NS1 chain.
a. The RBD and ED of each NS1 molecule separately interact with their respective domains from the neighboring NS1 molecules, related by crystallographic 2-fold axes (perpendicular to plane of the paper indicated the black ovals), resulting in the formation of a chain of NS1 with alternating RBD and ED dimers. The 2-fold related NS1 molecules are colored separately in yellow and green. The residues critical to dsRNA (residues 38, and 41)16 and CPSF (conserved residues F103, M106, and GLEWN183-187) binding14,15,19 are colored in blue and red respectively. b. Superposition of H5N1 RBD dimer with the H3N2 RBD6 dimer (in ruby, pdb id: 1AIL), each protomeric subunit in the H5N1 RBD dimer is colored differently in yellow and green. c. Structural alignment of the H5N1 dimer and H1N1 NS1 ED7 dimer (in ruby, pdb id: 2GX9) demonstrating the twisting motion (curved arrows) of the H5N1 ED monomers, with respect to H1N1 ED, toward their RBDs. Each monomer in the H5N1 NS1 dimer is colored as in panel a. The dimeric interface of the H5N1 NS1 ED, with a buried surface area of ~1600 Å2, consists of a series of electrostatic interactions: a salt bridge between K131 and E97, hydrogen bonding involving the side chains of T91 and R193, E196 and R200, E152 and the amide group of L95, as well as a back bone hydrogen bond between the E96 amide group and the E152 carbonyl group. In contrast, the dimeric interactions in the H1N1 NS1 ED consists of primarily strong hydrophobic interactions along the continuous anti-parallel β-sheet involving residues L90, V136, and L1417.
The ED dimer in our full-length H5N1 structure exhibits a large buried surface area of ~1600 Å2, which is similar to that observed in the previously published H1N1 ED dimer7. However, despite some resemblance, the H5N1 ED dimer exhibits substantial differences when compared to H1N1 NS1 ED dimer7. The superposition of these two ED dimers indicates that the protomeric subunits in the full-length ED dimer are significantly more twisted with respect to each other (Fig. 2c). As a result, the dimeric interface of the H5N1 NS1 ED consists of a series of electrostatic interactions in contrast to the dimeric interactions observed in the H1N1 NS1 ED, which consists of primarily hydrophobic interactions. The residues that participate in the dimeric interactions of the H5N1 NS1 ED dimer are conserved within H1N1 NS1 and vice versa. Thus, one distinct possibility is that the conformational difference in the H5N1 NS1 ED dimer is due to the presence of the RBD and the formation of the RBD dimer. It appears that without the RBD, ED can adopt alternative dimeric states as shown by the recent structural analysis of ED from an avian H12N5 strain17. In addition to the constraints imposed by the RBD dimerization, another important factor that influences the extent of the conformational changes, or the degree of twisting between the subunits in the ED dimer, may be the length of the linker between RBD and ED in the full-length NS1. Compared to either H1N1 or H3N2 NS1, H5N1 NS1 has a shorter linker due to a five residue deletion18. As a result of the shorter linker, greater stress is induced within the full-length H5N1 NS1 structure upon RBD dimerization. Thus, the N-termini of the ED subunits in the dimer are pulled toward their respective RBD causing more pronounced twisting between the ED subunits. Whereas within H1N1 or H3N2 NS1 the additional five residues contained in the linker may limit the stress induced and thus the degree of ED twisting upon RBD dimerization.
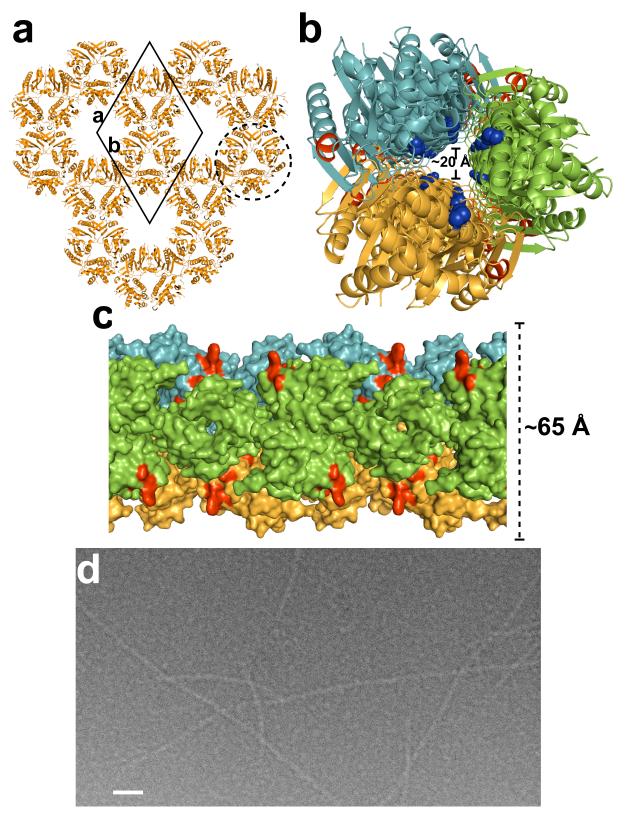
An unexpected and a novel observation from our crystallographic analysis of full-length H5N1 NS1, which has implications in the function of NS1, is the formation of long chains of NS1 molecules with alternating RBD and ED dimers. Despite the involvement of crystallographic 2-fold axes in the formation of these dimers, conservation of the RBD dimeric interactions, as mentioned above, and the large buried surface area in the ED dimer, which is typical of protein homodimers, strongly suggest that such a chain formation is more a consequence of the intrinsic ability of NS1 molecules to oligomerize than crystal packing. In the crystal, three such chains related by the crystallographic 31-screw axis parallel to the c axis, interact with one another to form a ~65 Å wide tubular organization with a 20 Å central tunnel (Fig 3a,b,). This tubular organization can be described as a helical structure with a pitch of 69.6 Å (unit cell dimension along c-axis) consisting of three NS1 dimers in one turn of the helix. The tubular NS1 organization is stabilized by inter-chain interactions involving ED dimers interlocking into saddle-like depressions between the RBD and ED dimers from other NS1 chains (Fig 3c). The RBD dimers within the tubular structure are positioned such that the conserved residues, R38 and K41, implicated in the dsRNA binding16 would project into the tunnel to interact with dsRNA (Fig. 3b, Fig. S4), whereas all the known ligand binding sites including a conserved Trp residue critical for binding cleavage and polyadenylation specificity factor (CPSF)14,15,19 are clearly surface exposed (Fig. 3c).
Figure 3. NS1 tubular structure.
a. A thin slice of the crystal packing of H5N1 NS1 molecules (orange) as viewed along the crystallographic c-axis. The crystallographic unit cell (solid black diamond) along with the a and b axes are shown for reference. One of the tubular structures formed by the interaction of three NS1 chains related by the crystallographic 31-screw axis (perpendicular to the plane of the paper) is shown surrounded by a broken circle. b. NS1 tubular structure (diameter of ~65 Å) as viewed down its long axis showing the ~20 Å wide tunnel. Each of the three chains is colored differently in green, orange and light blue. The residues 38 and 41 critical for dsRNA binding16 are shown as blue spheres, demonstrating their location within the central tunnel, the residues critical to CPSF binding14,15,19 are colored red. c. A surface representation of the tubular structure (diameter ~65 Å) viewed across its long axis. The CSPF binding sites colored in red are surface exposed. Each chain is colored as in panel b. d. A cryo-electron micrograph of native H1N1 NS1 in the presence of dsRNA. Long tubular structures of diameter ~70 Å can be clearly visualized. Scale bar, ~500 Å.
Presently it is not known how NS1 interacts with dsRNA. One mechanism that has been proposed for the IFN-antagonist activity of the NS1 RBD is the sequestration of dsRNA that would otherwise activate the IFN-inducing PKR12,20 and 2′ 5′-oligoadenylate synthetase/RNase L11 pathways. It has been shown that during a viral infection, dsRNA of at least 30bp is required to activate these pathways, with maximum activation occurring with a dsRNA greater than 80bp21. Thus, NS1 must be able to sequester varying lengths of dsRNA and completely prevent exposure of the dsRNA to any interactions with cellular dsRNA binding proteins involved in activating the type-I IFN response in the cell. Previous mutational analysis has implicated both K41 and R38 of NS1 as essential to dsRNA binding16. While mutation K41A lowers dsRNA affinity, mutation R38A completely abrogates the binding of dsRNA16. Elimination of ED is shown to significantly alter the dsRNA binding affinity of NS1 suggesting the involvement of this domain also in the dsRNA interaction22.
A plausible model that emerges from our crystallographic analysis for the dsRNA binding activity of full-length NS1 is that NS1 cooperatively oligomerizes in the presence of dsRNA to from a tubular structure completely encasing any length of dsRNA within the observed central tunnel. The ability of full-length NS1 to form chains through ED and RBD dimerization, instead of individual dimeric units, provides a robust mechanism not only to completely protect the dsRNA but also to adapt to any structural variations in dsRNA, including changes in the helical pitch, using the inherent flexibility in the linker between the ED and RBD domains. In this model, the orientation of the RBD with R38 projecting into the central tunnel, and the involvement of ED are consistent with previous mutational studies16,22 mentioned above.
Cryo-EM images of native full-length NS1 (A/PR8/34) in the presence of dsRNA further substantiate the tubular model for how NS1 sequesters dsRNA. These images show formation of smooth tubular structures of similar width to that observed in the crystal structure (Fig 3d). As mentioned earlier, the native full-length NS1 precipitates readily, but in presence of dsRNA it remains in solution forming such tubular structures. However, based on our crystallographic analysis, full-length NS1 with R38A and K41A mutations mimics formation of the tubular structure even in the absence of dsRNA indicating thereby that these charged residues in the native protein need to be neutralized by the bound dsRNA or mutated to uncharged residues for the native protein to remain in solution and form tubular structures. The formation of the tubular structure by the mutated full-length protein is concentration dependent; consistent with the suggestion that NS1 cooperatively binds to dsRNA depending on its length. At lower concentrations, as shown by size exclusion chromatography mutated H5N1 NS1 is a dimer (Fig. S5), and with increasing concentrations it forms higher order oligomers as indicated by dynamic light scattering (data not shown).
Our crystallographic and cryo-EM studies on full-length NS1 bring a novel perspective to the structure-function relationship of NS1. Formation of chains of NS1 and their association to form a tubular structure provides an attractive model for how NS1 could effectively compete with cellular dsRNA-binding proteins to sequester varying lengths of dsRNA. Such a tubular organization does not preclude interactions with other host proteins as all of the binding sites for the known cellular proteins remain exposed on the outer surface of the NS1 tubular structure. Our model also provides an explanation for the recent perplexing data indicating that despite overlapping binding sites, NS1 simultaneously interacts with both dsRNA and α-importin23. The cooperative interactions leading to NS1 tubular formation in the presence of dsRNA could allow for RBD, which is not involved in dimer interaction, at either end of the tubular structure to bind α-importin. Remarkably, a consistent feature in a majority of the recent H5N1 strains is the deletion of ~5 residues in the region of NS1 that links RBD and ED18. It is possible that this linker region depending upon its length, residue composition, and flexibility allows modulation of NS1 affinity for dsRNA by influencing the positioning and spacing of the RBD dimers in our proposed model for dsRNA sequestration. Considering also that the observed conformational changes in the ED dimeric interface in our crystal structure of the full-length NS1 could have resulted from a shortened linker, further studies could test the hypothesis that this shortening of the linker region in H5N1 NS1 contributes to increased virulence either by modulating its dsRNA binding affinity or its interaction with host proteins.
Methods Summary
The expression of H5N1 NS1 was done using a construct containing the mutations R38A and K41A, in BL-21 E.coli cells. The clarified lysate was then bound to Qiagen-NTA agarose beads using the batch method. Once eluted, the protein was filtered and put through an HQ column followed by size exclusion chromatography with a 50 mM NaCl, 10 mM Hepes pH 8.0 column buffer. Crystals of the H5N1 NS1 were grown via the hanging drop method using a 1:1 ratio of well solution to protein at 5-8 mg/ml. The well solution was 17% PEG 3350, 225-275 mM KSCN, 100 mM Hepes pH 6.8-7.2. Hexagonal crystals were seen after 2-3 weeks, reaching a maximum size of 0.1-0.2 mm after a month. Data was collected at the Argonne National Laboratory using SBC-19-ID beamline. Diffraction data were merged and scaled using D*TREK24 with an Rmerge of 10% (Table S1). Although crystals diffracted to ~2.2 Å, the diffraction intensities were weak between this resolution and 2.7 Å with I/σ < 2. The crystals belong to the primitive hexagonal space group (P 65 2 2), as confirmed by analyzing the unmerged data in P1 using POINTLESS25. The structure of H5N1 NS1 was determined by molecular replacement techniques with PHASER26 using the H1N1 ED7 and the H3N2 RBD6 subunits as search models and refined using REFMAC527 as implemented in the CCP428 package. The specimen for cryo-EM imaging was prepared by incubating dsRNA with a saturating amount of purified A/PR8/34 H1N1 NS1 for 45 minutes at room temperature followed by a size exclusion chromatography to separate NS1 bound to dsRNA from the unbound NS1.
Methods
Protein Expression and Purification
Expression of H5N1 NS1 was done using a construct containing the mutations R38A and K41A, placed in pET-46 Ek/LIC (Novagen) with an engineered thrombin site to remove the HIS tag. The construct was expressed in BL-21 E.coli cells at 25° C overnight at an ODA600 of 0.6-0.8 with .05 mM IPTG. Each liter of bacterial pellet was resuspended in 20 ml of lysis buffer, and lysed using a microfluidizer. The clarified lysate was then bound to 2-3 ml of Qiagen-NTA agarose beads using the batch method. The beads were then washed twice within a 50 ml conical tube with a 50mM NaH2PO4, 300 mM NaCl, 30mM imidazole wash buffer, and then washed 3 times with 50 mM NaCl, 10 mM Hepes pH 8.0 buffer. The beads were then suspended in 20 ml of 50 mM NaCl, 10 mM Hepes pH 8.0 buffer and cut off the beads with thrombin at room temperature overnight on a rocker. The protein was then filtered and put through HQ column to remove the thrombin, then size exclusion chromatography was performed with a 50 mM NaCl, 10 mM Hepes pH 8.0 column buffer which was the final crystallization buffer.
Crystallization
Crystals of the H5N1 NS1 were grown by the hanging drop method by using a 1:1 ratio of well solution to protein at 5-8 mg/ml. The well solution was 17% PEG 3350, 225-275 mM KSCN, 100 mM Hepes pH 6.8-7.2. Hexagonal crystals were seen after 2-3 weeks, reaching a maximum size of 0.1-0.2 mm after a month.
Data Collection and Processing
Crystals of H5N1 NS1 were cryoprotected by shock soaking for ~10 seconds in 35% sucrose, 17% PEG 3350, 225-275 mM KSCN, 100 mM Hepes pH 6.8-7.2. The crystals were then flash frozen in liquid nitrogen and shipped to the synchrotron facility. Data was collected at the Argonne National Laboratory using SBC-19-ID beamline using a 1.0° oscillation angle. Diffraction data were merged and scaled using D*TREK24 with an Rmerge of 10% (Table S1). Although crystals diffracted to ~2.2 Å, the diffraction intensities were weak between this resolution and 2.7 Å with I/σ < 2. The crystals belong to the primitive hexagonal space group (P 65 2 2), as confirmed by analyzing the unmerged data in P1 using POINTLESS25, with unit cell dimensions of a = 106.12 Å, b = 106.12 Å, c = 69.63 Å, α = 90°, β = 90°, γ = 120°.
Structure Determination and Refinement
The structure of H5N1 NS1 was determined by molecular replacement techniques with PHASER26 as implemented in the CCP428 package using the H1N1 ED7 and the H3N2 RBD6 subunits as search models. Some of the missing regions in the model were identified by computing composite-omit maps computed using CNS29. Following iterative cycles of model building using COOT30, the model was refined using Refmac527 (Table S1). A 5% data were set aside for Rfree31 calculations. The final structure has an Rfree of 29% and a final R-factor of 27% using all the data to 2.7 Å resolution. During the course of the refinement, and following the final refinement, stereochemistry of the structure was checked using PROCHECK32. The buried surface area for the dimer complex was calculated with CNS29. The structural alignments and calculations of RMSD were carried out using PyMOL (http://pymol.sourceforge.net), which was also used for making all the figures.
Cryo-EM imaging
Single stranded, complementary RNA, 12 bases in length (GAGAGAGAGAGA and CUCUCUCUCUCU) were ordered from Dharmacon Products. The strands of RNA were then deprotected, mixed together in equal amounts, and boiled for 1 minute. The reaction was then allowed to cool overnight to allow for annealing and formation of dsRNA. The chosen sequence of the complimentary strands with GA and CU repetitions allows for the formation of long dsRNA strands, which would be suitable for cryo-EM visualization following addition of NS1. The dsRNA mixture was then mixed with a saturating amount of purified A/PR8/34 H1N1 NS1. Expression of H1N1 NS1 (A/PR8/34) was done using pET-46 Ek/LIC (Novagen). The construct was expressed in BL-21 E.coli cells at 25° C overnight at an ODA600 of 0.6-0.8 with .05 mM IPTG. Each liter of bacterial pellet was resuspended in 20 ml of lysis buffer, and lysed using a microfluidizer. The clarified lysate was then bound to 2-3 ml of Qiagen-NTA agarose beads using the batch method. The beads were then washed twice within a 50 ml conical tube with a 50 mM NaH2PO4, 300 mM NaCl, 30 mM immidazole wash buffer (pH 8.0), and then eluted with a 50 mM NaH2PO4, 300 mM NaCl, 250 mM immidazole elution buffer (pH 8.0). The NS1-dsRNA binding reaction was done in the elution buffer, was incubated for 45 minutes, and then NS1 bound to dsRNA was separated from unbound NS1 via size exclusion chromatography before preparing the specimen for cryo-EM imaging. Specimen for cryo-EM was embedded in vitreous ice on holey carbon films using standard procedures33, and was imaged on a JEOL1200 EX transmission electron microscope operated at x 30000 magnification using 100-kV electrons and a beam dose of ~5 electons/Å2. Several attempts to duplicate this procedure using A/Vietnam/1203/2004 H5N1 NS1 were unsuccessful due the noticeably accelerated aggregation rate of the wild-type protein when compared to A/PR8/34 H1N1 NS1. The increased instability of the wild-type H5N1 NS1 is likely a result of the observed five amino acid deletion within the linker domain.
Supplementary Material
Acknowledgements
We thank P. Palese for providing us clones of A/Vietnam/1203/2004 H5N1 and A/PR8/34 (H1N1) NS1. This work was supported by grants from the NIH (AI36040) and the Robert Welch Foundation (to BVVP). ZB acknowledges support from an NIH virology training grant (AI07471). We thank Peter Palese, Andy Rice, Michael Schmid, and Berenice Carrillo for useful discussions and comments on the manuscript, Hua Chen for technical assistance with cryo-EM. We acknowledge the use of cryo-EM facilities at National Center for Macromolecular Imaging at Baylor college of Medicine supported by grants from the National Institutes of Health (NIH) RR002250 to W. Chiu, the Center for Advanced Microstructures & Devices (CAMD) Baton Rouge, LA and Dr. Henry Bellamy, and the SBC-CAT 19ID beam line at the Advanced Photon Source (supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38) and its staff for their help during data collection.
References
- 1.Abdel-Ghafar AN, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 2.Beigel JH, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 3.Seo SH, Hoffmann E, Webster RG. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 2004;103:107–113. doi: 10.1016/j.virusres.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 5.Chien CY, et al. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 7.Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, et al. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y, Wambach M, Katze MG, Krug RM. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 13.Hale BG, et al. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci U S A. 2006;103:14194–14199. doi: 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemeroff ME, et al. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 15.Twu KY, et al. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol. 2006;80:3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. Rna. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale BG, Barclay WS, Randall RE, Russell RJ. Structure of an avian influenza A virus NS1 protein effector domain. Virology. 2008 doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Li KS, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 19.Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 20.Diebold SS, et al. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li WX, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melen K, et al. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 25.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 33.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.