Abstract
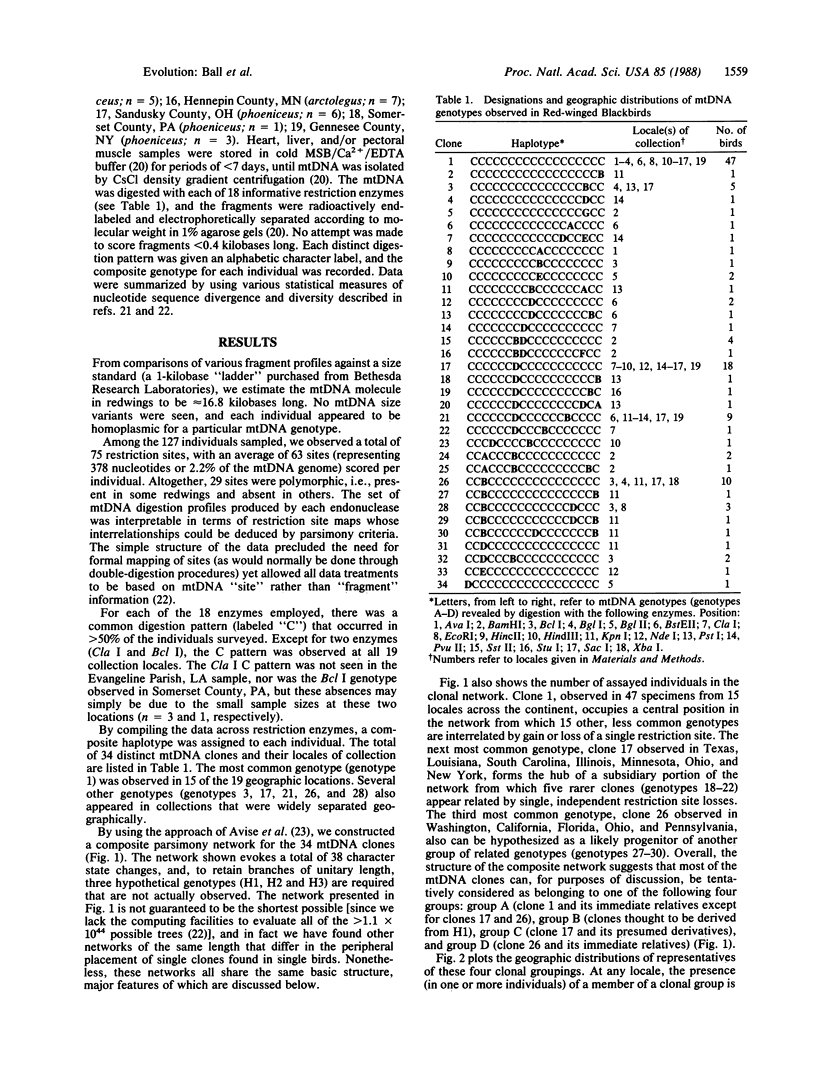
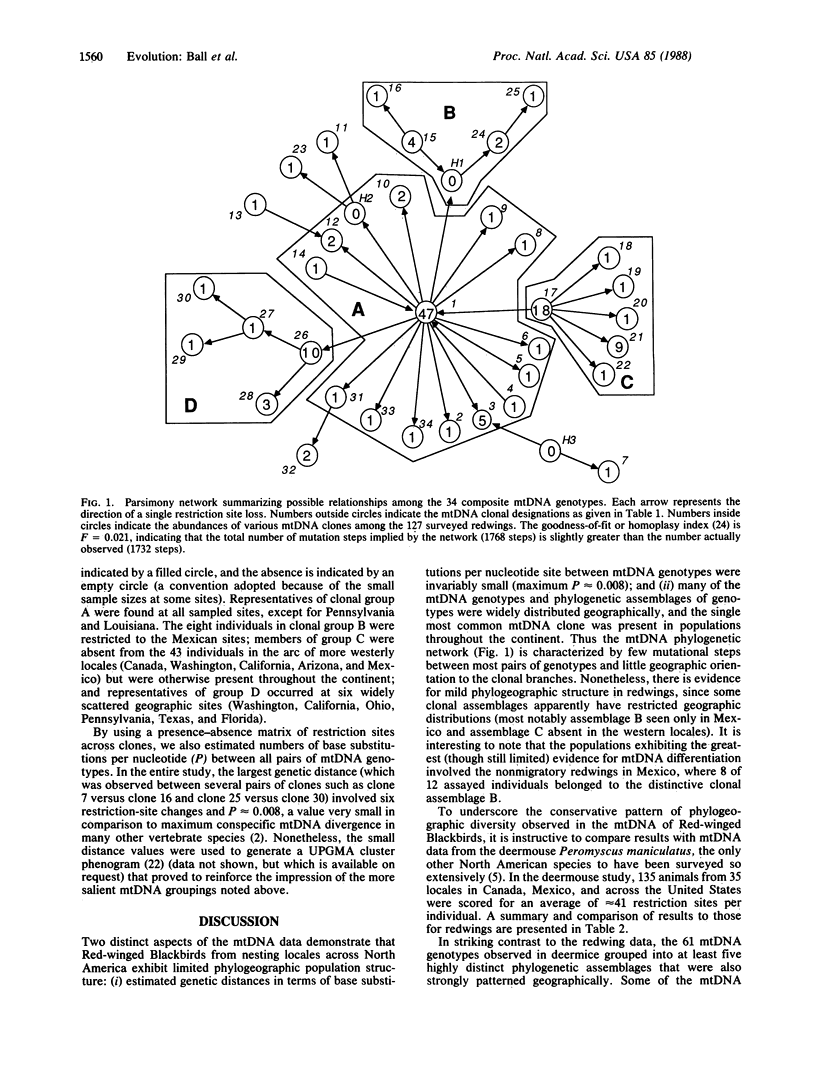
A continent-wide survey of restriction-site variation in mitochondrial DNA (mtDNA) of the Red-winged Blackbird (Agelaius phoeniceus) was conducted to assess the magnitude of phylogeographic population structure in an avian species. A total of 34 mtDNA genotypes was observed among the 127 specimens assayed by 18 restriction endonucleases. Nonetheless, population differentiation was minor, as indicated by (i) small genetic distances in terms of base substitutions per nucleotide site between mtDNA genotypes (maximum P ≈ 0.008) and by (ii) the widespread geographic distributions of particular mtDNA clones and phylogenetic arrays of clones. Extensive morphological differentiation among redwing populations apparently has occurred in the context of relatively little phylogenetic separation. A comparison between mtDNA data sets for Red-winged Blackbirds and deermice (Peromyscus maniculatus) also sampled from across North America shows that intraspecific population structures of these two species differ dramatically. The lower phylogeographic differentiation in redwings is probably due to historically higher levels of gene flow.
Keywords: intraspecific phylogeny, restriction sites, genetic distance, gene flow
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Giblin-Davidson C., Laerm J., Patton J. C., Lansman R. A. Mitochondrial DNA clones and matriarchal phylogeny within and among geographic populations of the pocket gopher, Geomys pinetis. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6694–6698. doi: 10.1073/pnas.76.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J. C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):325–342. doi: 10.1098/rstb.1986.0011. [DOI] [PubMed] [Google Scholar]
- Avise J. C., Neigel J. E., Arnold J. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol. 1984;20(2):99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- Bermingham E., Avise J. C. Molecular zoogeography of freshwater fishes in the southeastern United States. Genetics. 1986 Aug;113(4):939–965. doi: 10.1093/genetics/113.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Ferris S. D., Sage R. D., Prager E. M., Ritte U., Wilson A. C. Mitochondrial DNA evolution in mice. Genetics. 1983 Nov;105(3):681–721. doi: 10.1093/genetics/105.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm-Bychowski K. M., Wilson A. C. Rates of nuclear DNA evolution in pheasant-like birds: evidence from restriction maps. Proc Natl Acad Sci U S A. 1986 Feb;83(3):688–692. doi: 10.1073/pnas.83.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James F. C. Environmental component of morphological differentiation in birds. Science. 1983 Jul 8;221(4606):184–186. doi: 10.1126/science.221.4606.184. [DOI] [PubMed] [Google Scholar]
- Johnson M. J., Wallace D. C., Ferris S. D., Rattazzi M. C., Cavalli-Sforza L. L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19(3-4):255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- Kessler L. G., Avise J. C. A comparative description of mitochondrial DNA differentiation in selected avian and other vertebrate genera. Mol Biol Evol. 1985 Mar;2(2):109–125. doi: 10.1093/oxfordjournals.molbev.a040339. [DOI] [PubMed] [Google Scholar]
- Lansman R. A., Shade R. O., Shapira J. F., Avise J. C. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. III. Techniques and potential applications. J Mol Evol. 1981;17(4):214–226. doi: 10.1007/BF01732759. [DOI] [PubMed] [Google Scholar]
- Nei M., Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981 Jan;97(1):145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager E. M., Wilson A. C. Construction of phylogenetic trees for proteins and nucleic acids: empirical evaluation of alternative matrix methods. J Mol Evol. 1978 Jun 20;11(2):129–142. doi: 10.1007/BF01733889. [DOI] [PubMed] [Google Scholar]
- Shields G. F., Wilson A. C. Calibration of mitochondrial DNA evolution in geese. J Mol Evol. 1987;24(3):212–217. doi: 10.1007/BF02111234. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987 May 15;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]


