Introduction
The sequencing of the human genome, the identification of common SNPs and haplotype blocks, and advances in microarray technology has enabled the study of complex diseases at a level of detail not previously imaginable. These have aided in the design and analyses of association and linkage studies of many complex diseases including cardiovascular disease. Recent technological advances have enabled the undertaking of large-scale genome-wide association studies (GWAS) that can assay hundreds of thousands of polymorphic sites on hundreds to thousands of individuals in order to find genomic regions associated with disease. While results from these experiments enable the identification of smaller regions of association than previous studies, as with all linkage and association studies, there is the need for the further investigation of regions of interest for the causal genes or variants.
The purpose of this review is to present a detailed demonstration as to how publicly available resources can be utilized to easily guide more detailed research into genomic regions of interest identified in linkage and association study data. Large-scale projects, such as the Human Genome Sequencing project1, 2, have generated large volumes and varieties of annotated genomic data necessitating the development of Internet-based tools to organize and make practically available these public data. One important tool in human disease research is web-based graphical genome browsers that use the human genome sequence as the framework on which to organize genomic annotations providing various ways for researchers to view and extract important information. Currently, there are three human genome browsers that have been developed for public use: 1) the National Center for Biotechnology Information (NCBI) Map Viewer3; 2) the University of California Santa Cruz (UCSC) Genome Browser4; and 3) the European Bioinformatics Institute’s Ensembl system5. Although these genome browsers share common features and genomic information, each being built on top of the same reference genome sequence, they each have a different look and feel and provide unique capabilities6.
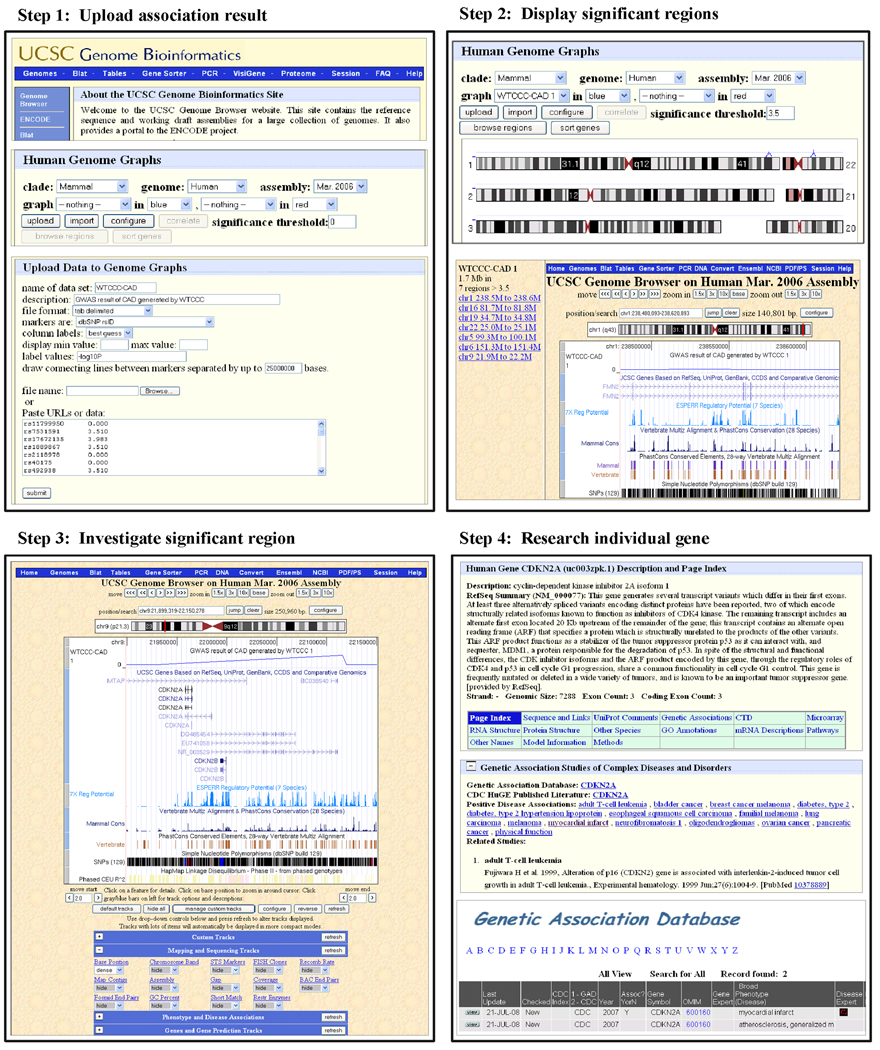
In particular, the UCSC Genome Browser has a tool called Genome Graphs that is especially suited for linkage and association study analyses. The following sections will demonstrate the capabilities of this tool focusing on a recently published GWAS result from the Wellcome Trust Case Control Consortium (WTCCC). As with all studies of this type, regions of disease association were identified encompassing large numbers of genes that are candidates for further studies. In order to prioritize future research, genes in each region need to be investigated for a possible role in a particular disease. The following step-by-step instructions demonstrate a straightforward and efficient method using the Genome Graphs tool within the UCSC Genome Browser that can help to prioritize a small number of meaningful candidates from a large-scale association study. Figure 1 provides an illustrated outline of this method with a more detailed description of each step below (Note: Each of the individual figures in Figure 1 is also available as larger figures in the Supplementary material).
Figure 1.
Flow chart of using UCSC genome browser to analyze GWAS results. Refer to text for details of each step.
Data
Numerous previous family-based linkage studies and case-control single marker association studies have indicated a strong genetic component to cardiovascular disease. Currently, about 40 quantitative trait loci (QTL) for human atherosclerotic disease have been identified by genetic linkage studies7. Large-scale association studies also identified several genes, such as LTA8, VAMP 8 and HNRPUL19, and CDKN2A and CDKN2B10, 11, 12.
Coronary artery disease (CAD) is one of the complex diseases studied by the WTCCC12. In their study, Affymetrix GeneChip 500K Mapping Arrays were used to identify seven regions of the genome showing evidence of association with CAD. The full dataset is only available with permission of the WTCCC, so we created a synthetic dataset (Supplemental file 1) for the NCBI build 36 human genome sequence based on the most significant SNP within each of the seven regions displayed in Tables 3 and 4 in their manuscript. Each of these SNPs is assigned its reported −log10 of P value to represent the statistical significance of the SNP and is calculated based on the allele distribution in cases and controls. To reflect the full extent of the identified region of association, we added SNPs at the edges of their identified regions to our dataset, each with a value of 3.51. Lastly, we added background SNPs (~2 Mb away from each side) with value 0. Readers are encouraged to download this synthetic dataset (Supplemental file 1) and to use it to actively perform each of the following steps in order to better understand the functionality of the Genome Graphs tool.
Step 1: Upload linkage/association results
First, proceed to the UCSC Genome Browser homepage (http://genome.ucsc.edu - Step 1, Figure 1, top image). Links to several tools available at this site can be seen in blue horizontal and vertical tool bars. For more information on the functionality of these other tools, we encourage exploring the FAQ and Help pages and reading a recent review describing features of this browser13.
Click on the Genome Graphs link on the left vertical pane. In the page that appears (Step 1, Figure 1, middle image), data from association or linkage studies can be input. Up to two datasets can be uploaded and displayed simultaneously. The bottom section of this web page describes briefly the page controls, and there is a link to the Genome Graphs User’s Guide that provides a more detailed set of instructions for this tool.
Click the upload button in the upper box to display the Upload Data to Genome Graphs page (Step 1, Figure 1, right image). On this page, information about the association data may be input such as a name and description. This tool will accept files of association or linkage data that are tab-delimited, comma delimited, or space delimited (see file format drop-down bar). Our test file is tab-delimited and simply consists of lines consisting of the name of a SNP and a corresponding value. The intent is that in general these values reflect some significance measure for that SNP, but there are no restrictions. This tool is aware of locations of several types of markers including SNPs denoted by rs values, and probes on several genome-wide genotyping microarrays from Affymetrix, Illumina, and Agilent (see markers are drop-down bar).
The association or linkage information to be displayed can be copied and pasted into the text box shown on this web page or can be uploaded as a file. The latter is recommend for very large datasets. Upload the test dataset and press the submit button. This will input the association results to be displayed in a graphical output page (Step 2, top figure – Note: This process may take a few minutes). By default, the range of the dataset to be plotted will be obtained from the minimum and maximum values in the data. Alternatively, this display range can be specified by setting display min value/max value on the Upload Data page, or can be adjusted later (see next step).
Step 2: Display significant regions
Once the data is uploaded, first a summary text page appears indicating how many (%) markers within the data file were successfully mapped to the genome. Click the OK button to proceed. Next, the main Genome Graphs page appears again where the uploaded data can be selected for display in a genome-wide manner. Using the graph drop-down menu, select the track name corresponding to the newly input data. This will cause these data to be displayed on this same web page as a line graph on top of ideograms of each chromosome (Figure 1, Step 2, top panel). Seven peaks corresponding to the regions of significance are displayed directly above the appropriate chromosomes for this specific dataset. The height of the peak indicates its statistical significance, in this case the −log10P value described above. Different features of this ideogram graph can be customized by clicking the configure button including the range of values to be shown. Images in Figure 1 and in the supplementary figure were generated using the default settings.
From this display, clicking on any point of interest on any chromosome will open the main Genome Browser tool (described in the next section) displaying a 1 Mb region around that chromosomal base pair position. Alternatively, regions of association can be displayed in the Genome Browser by first specifying a significance threshold (3.5 for this dataset), and then pressing the browse regions button. The Genome Browser tool is displayed with a pane on the left containing links to significant regions that are above the given significance threshold (Figure 1, Step 2, bottom panel). In this dataset, it includes a total of 1.7 Mb from seven regions sorted by their genomic positions. Each region can be displayed on the Genome Browser on the right pane by clicking on the corresponding link. The first region on the list is shown by default. Click on the link for the region chr9 21.9M to 22.2M, the region with the most significant association, to show this area of the genome in the browser.
Step 3: Investigate significant regions
The Genome Browser tool presents a graphical view of a wide variety of annotations, particularly those related to genes, for a specific span of genomic sequence in the form of horizontal annotation “tracks” (Figure 1, Step 3). The genome is as presented runs from left to right with the shorter p-arm on the left. Genes are represented by solid boxes (exons) connected by lines (introns) with arrows indicating the direction of transcription. Scrolling down this page shows numerous drop-down menus that control the multitude of tracks that can be displayed. Currently, there are more than 200 annotations, some developed using public data and/or research performed at UCSC, and others contributed from outside resources by third party researchers. These annotations are organized into categories, such as Genes and Gene Prediction, Regulation, Comparative Genomics, and Variation and Repeats13, 14. Annotation tracks most relevant to linkage and association studies include UCSC Genes, 7X Reg Potential (regulatory potential based on cross-species alignments), Conservation (can select what other species to view), Most Conserved, SNPs (129) (from dbSNP), and HapMap LD Phased (the association of alleles on chromosomes). Any specific track can be displayed by selecting any visibility option (dense, squish, pack, full) other than hide from the drop-down list under that track name and then pressing any of the refresh buttons to update the display. These options primarily control whether each element in the track is distinctly displayed or is summarized in a single line (Please see http://genome.ucsc.edu/goldenPath/help/hgTracksHelp.html#TRACK_CONT for more detailed descriptions of the different display options). If the Internet browser cookies are enabled, preferred track viewing options will be saved and automatically reset when returning to the Genome Browser in the future. In addition, navigation buttons are present along the top of this page that allow the zooming in and out of regions displaying more or fewer bases, and scrolling left and right along the chromosome. Clicking on the configure button to the right of the position text box allows more customizing of the appearance of the browser graphic in addition to allowing track display configuration. A useful configuration for those with larger monitors is to increase the width of the main image. By default, this is set to 620 pixels. For the browser images in Figure 1 and in the supplementary figures, the image width has been set to 1000 pixels. More detailed instructions concerning the functionality of this tool can be found in the Help section and also in recent reviews13, 14.
Individual genes in regions of interest can be easily investigated within the Genome Browser. The currently displayed region on chromosome 9 (Figure 1, Step 3) indicates that two well-annotated protein-coding genes, a pair of cyclin dependent kinase inhibitors, CDKN2A and CDKN2B, are fully contained in the corresponding sequence representing potential candidate genes related to CAD. In fact, the original WTCCC analysis of this region focuses on these two genes, and a replication study confirmed the significance of this region15. The multiple instances of each gene in the UCSC Genes track in the browser display correspond to alternatively spliced isoforms. Darker shades of blue indicate strong supporting evidence for the correct annotation of that isoform, while the lighter shades indicate less confidence. A general feature of the browser is that clicking on any element in any annotation track will display a more detailed description of that element. Genes in particular have a rich collection of information available including links to other databases and online resources as described in the following section.
In addition to these protein-coding genes, a non-coding gene, ANRIL (annotated as CDKN2BAS in the RefSeq Genes track, and DQ485454, EU741058, and NR_003529 in the UCSC Genes track), is contained in this region. The function of this gene is unknown at present, but should also be considered in further investigations. It is important to note that there are several gene annotation tracks in the browser, such as UCSC Genes and RefSeq Genes. In general, the UCSC Genes track is more comprehensive containing annotations of coding and non-coding genes and transcripts requiring a minimum level of biological support. RefSeq Genes, based on the Reference Sequence project at the NCBI16, has traditionally focused more on protein-coding genes, but the entries are well-supported and many are hand curated. It is advisable to review multiple gene annotations in follow-up research.
Not all linkage and association study results identify such small regions containing relatively few genes. In non-GWAS experiments, significant regions often span multiple megabases potentially containing hundreds of genes. In these cases, it may be beneficial to investigate linkage disequilibrium among top markers, recombination rates, and evidence of evolutionary selection pressure. Information about some of these can be found within other annotation tracks in the browser. In addition, several public tools are available that may be helpful such as SNAP (http://broad.mit.edu/mpg/snap/) that is specifically designed to query and display LD with GWAS results.
Step 4: Research individual genes
To investigate these genes of interest further, click on one of the genes (CDKN2A) within the graphic on the browser page to open the Human Gene Description and Page Index page (Figure 1, Step 4, top and middle). At the top of this page are a brief description of the gene and a summary of what is currently known about its biological function taken from the Reference Sequence (RefSeq) project16. In addition, to facilitate investigation into potential associations with the disease in question, several diverse types of detailed information are provided such as links to other genomic tools and databases, results from genetic association studies, microarray gene expression data, mRNA and protein structure models and information, Gene Ontology (GO) annotations, and biochemical and signaling pathways in which the gene participates. Each of these sections on this web page can be viewed either by scrolling down or using the Page Index table of links to directly jump to information of interest.
For CDKN2A and CDKN2B, the Genetic Association Studies of Complex Diseases and Disorders section (Genetic Associations link in the Page Index table) indicates that these have been previously linked to many types of disease including several cancers (Figure 1, Step 4, middle). Of particular interest, though, are that genetic association studies have linked both to myocardial infarction (click on “click on here to view complete list” link, see item 1210). To further understand the potential CAD association of CNKN2A and CNKN2B, clicking on the myocardial infarct link in the Positive Disease Associations list will open a page in the Genetic Association Database (GAD)17 (Figure 1, Step 4, bottom). The GAD contains several published independent studies describing both positive and negative associations between CAD and these two genes.
In addition to the GAD, several other publicly available resources contain valuable information related to disease association such as PubMed, Entrez Gene, OMIM, and GeneCards. OMIM (Online Medelian Inheritance in Man) is a comprehensive database of human genes and genetic phenotypes curated by researchers at Johns Hopkins University and the NCBI that specifically focuses on genetic disorders and the relationship between phentotype and genotype18 (http://www.ncbi.nlm.nih.gov/omim/). The intended audience of this resource is physicians and genetics researchers. GeneCards, developed at the Crown Human Genome Center and the Weizman Institute of Science, is likewise a human gene database that includes a wealth of information about all known and predicted genes including disease relationships, related drugs and compounds, and antibodies19 (http://www.genecards.org/index.shtml).
Information in these can be easily accessed through links provided within the UCSC Browser gene description web page (Figure 1, Step 4, top panel) under the Sequence and Links to Tools and Databases section. Other sections in this page that may also be of interest are Comparative Toxicogenomics Database (CTD) that reports what chemicals have been shown to interact with this gene, Microarray Gene Expression that displays in what tissues and cell types the gene is expressed, and Biochemical and Signaling Pathways that lists in what general cellular processes the gene is involved. Note, not all genes are necessarily as well-annotated as these and may not contain information in all of these sections.
In summary, by following the above described analysis pipeline within a genome browser, we quickly and easily find two meaningful candidate genes, CNKN2A and CNKN2B, in one of the seven regions with evidence of association generated from a GWAS. Obviously, the other regions could and should be further investigated in a similar manner, and further experimentation is necessary to confirm and better understand the potential role of any particular gene in the disease.
Discussion
The UCSC Genome browser is a powerful online tool that integrates a large and diverse set of genomic data efficiently and intuitively providing much needed support for biomedical research, especially in the age of large-scale data intensive experiments such as genome-wide association studies. Using a specific example, we have illustrated how to use this Genome Browser to obtain well-supported candidate genes from a GWAS concerned with coronary artery disease. Admittedly, not all investigations using this method will quickly yield such informative results and is largely dependent on the accuracy of the association or linkage data and the previous research and annotations of genes. Nonetheless, we feel that this provides a concrete way in which to integrate the results of GWAS with the wealth of publicly available genomic data for further discovery.
This tutorial has only briefly introduced some of the functionality of the UCSC Genome Browser. A recent review provides a more in-depth description of this browser13, 14, 20. We also do not describe other human genome browsers hosted at the NCBI and Ensemb13, 5, 21. These also provide rich sets of genomic annotations and functionality that greatly but not completely overlap those available at UCSC. General reviews comparing these three browsers are available6, 22. We encourage the further exploration of these websites to better understand these alternatives and their strengths. Researchers should decide for themselves the one that addresses their needs the best. We also caution that the quality of publicly available data displayed in the genome browsers and available in other public resources is highly variable. All these data should be viewed carefully and critically.
In the specific example we discussed here, we only included data in our synthetic results file from seven regions of the genome showing evidence of association with CAD. We surmise that researchers may want to upload more complete sets of raw or processed data generated from linkage and association studies to analyze within the Genome Graphs tool. Therefore, we need to point out that it takes a similar amount of time, about two minutes, to upload results that consist of 1 SNP or 500,000 SNPs. Therefore, analyses of large datasets is well within the capabilities of this tool.
We have recently learned of a new open access database of GWAS results23. This database consists of a collection of significant results (p < 10−3) from 118 GWAS articles for over 400 traits. The associated publication discusses the challenges of working with and standardizing annotations for GWAS results. Notably relevant to the current paper, a supplemental compressed file accompanying this paper contains individual GWAS results files spanning more than 400 traits formatted for UCSC Genome Graphs. These may be used for further experimentation with the Genome Graphs tool.
Supplementary Material
Acknowledgments
We would like to give special thanks to Elizabeth Hauser for her thoughtful comments and discussions of this manuscript.
Funding sources
This study was supported by the Duke Institute for Genome Sciences & Policy (IGSP), and NIH grants HL073389 (Hauser), MH059528 (Hauser) and HL73042 (Goldschmidt, Kraus).
Footnotes
Conflict of Interest Disclosures
TSF is a partner in the Genome Browser Authors partnership that licenses the UCSC Genome Browser software to for profit entities.
References
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:745–964. [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1145–1434. [Google Scholar]
- 3.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Gräf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kähäri A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furey TS. Comparison of human (and other) genome browsers. Human Genomics. 2006;2:266–270. doi: 10.1186/1479-7364-2-4-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Rollins J, Paigen B, Wang X. Genetic and genomic insights into the molecular basis of atherosclerosis. Cell Metab. 2007;6:164–179. doi: 10.1016/j.cmet.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 9.Shiffman D, Rowland CM, Louie JZ, Luke MM, Bare LA, Bolonick JI, Young BA, Catanese JJ, Stiggins CF, Pullinger CR, Topol EJ, Malloy MJ, Kane JP, Ellis SG, Devlin JJ. Gene variants of VAMP8 and HNRPUL1 are associated with early-onset myocardial infarction. Arterioscler Thromb Vasc Biol. 2006;26:1613–1618. doi: 10.1161/01.ATV.0000226543.77214.e4. [DOI] [PubMed] [Google Scholar]
- 10.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 11.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. UCSC genome browser tutorial. Genomics. 2008;92:75–84. doi: 10.1016/j.ygeno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC genome browser Database: update 2009. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn875. (Epub) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. WTCCC and the Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated nonredundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 18.Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information. Bethesda, MD: National Library of Medicine; 2009. Jan 5, [Google Scholar]
- 19.Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, Shmoish M, Peter Y, Glusman G, Feldmesser E, Adato A, Peter I, Khen M, Atarot T, Groner Y, Lancet D. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;1:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangan ME, Williams JM, Lathe SM, Karolchik D, Lathe WC., 3rd UCSC genome browser: deep support for molecular biomedical research. Biotechnol Annu Rev. 2008;14:63–108. doi: 10.1016/S1387-2656(08)00003-3. [DOI] [PubMed] [Google Scholar]
- 21.Spudich G, Fernández-Suárez XM, Birney E. Genome browsing with Ensembl: a practical overview. Brief Funct Genomic Proteomic. 2007;6:202–219. doi: 10.1093/bfgp/elm025. [DOI] [PubMed] [Google Scholar]
- 22.Baxevanis AD. Using genomic databases for sequence-based biological discovery. Mol Med. 2003;9:185–192. [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson AD, O’Donnell CJ. An open access database of genome-wide association results. BMC Med Gen. 2009 doi: 10.1186/1471-2350-10-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.