Abstract
Virus capsid assembly is a critical step in the viral life cycle. The underlying basis of capsid stability is key to understanding this process. Capsid subunits interact with weak individual contact energies to form a globally stable icosahedral lattice; this structure is ideal for encapsidating the viral genome and host partners and protecting its contents upon secretion, yet the unique properties of its assembly and intersubunit contacts allows for the capsid to dissociate upon entering a new host cell. The stability of the capsid can be analyzed by treating capsid assembly as an equilibrium polymerization reaction, modified from the traditional polymer model to account for the fact that a separate nucleus is formed for each individual capsid. From the concentrations of reactants and products in an equilibrated assembly reaction, it is possible to extract the thermodynamic parameters of assembly for a wide array of icosahedral viruses using well-characterized biochemical and biophysical methods. In this chapter we describe the basic analysis and provide examples of thermodynamic assembly data for several different icosahedral viruses. These data provide new insights into the assembly mechanisms of spherical virus capsids, as well as the biology of the viral life cycle.
Keywords: capsid, self-assembly, protein-protein interactions, thermodynamics, hepatitis B virus, cowpea chlorotic mottle virus
Introduction
A generic virus self-assembles within the cell, packaging the viral genome, viral proteins, and relevant host partners within a protein capsid for export from the cell. Spherical virus capsids are typically comprised of many copies of a few or even a single gene product. As such the virus uses relatively little of its genome to code for the capsid while the host generates the many copies of capsid subunit(s), thus exploiting the host resources to maximum effect with minimum cost to the virus. While this strategy leaves the virus dependent on the concerted assembly of hundreds of individual subunits that must recognize and specifically package the virus components with high fidelity, assembly usually proceeds to successfully form many complete and infectious virions.
Besides protecting the genome, virus capsids may also be involved with other steps in the virus lifecycle, interactions that are likely to be affected by capsid physical properties. Stability of protein-protein interactions is required for assembly. However, many viruses uncoat in response to exogenous signals to release their genome (e.g. poliovirus, Flockhouse virus). This requires sampling the environment, often accomplished by transiently exposing internal components of the virus. In many viruses (e.g. Hepatitis B virus) transiently exposed signals play a role in intracellular trafficking (Ganem and Prince, 2004). Thus, at odds with the need for stability, the transient exposure of internal residues implies that a capsid is a flexible, dynamic structure (Bothner et al., 1998; Hilmer et al., 2008; Lewis et al., 1998). The seemingly contradictory roles of “protection and release” imply a physical chemical explanation that should be evident in the thermodynamic stability of the capsid.
In this contribution we describe methods for determining a thermodynamic description of virus capsid assembly based on treating assembly as a polymerization reaction. Values determined experimentally are consistent with in silico assembly–master equations (Zlotnick et al., 1999), discreet event simulation (Zhang and Schwartz, 2006), and coarse grained molecular dynamics—that are outside the scope of this review (Hagan and Chandler, 2006; Nguyen et al., 2007; Rapaport, 2004). Within the scope of this review, we examine the thermodynamics of assembly of several viruses and emphasize the underlying physical chemistry.
The Structural Basis of Capsid Stability
Virus Capsid Geometry
Most small virus capsids take the shape of rods or spheres. Over fifty years ago it was first observed that tomato bushy stunt virus was icosahedral (Caspar, 1956), supporting the hypothesis that capsids are constructed of symmetrical repeating arrays of smaller subunits (Crick and Watson, 1956). It has since then been observed that almost all spherical viruses are based on icosahedral symmetry.
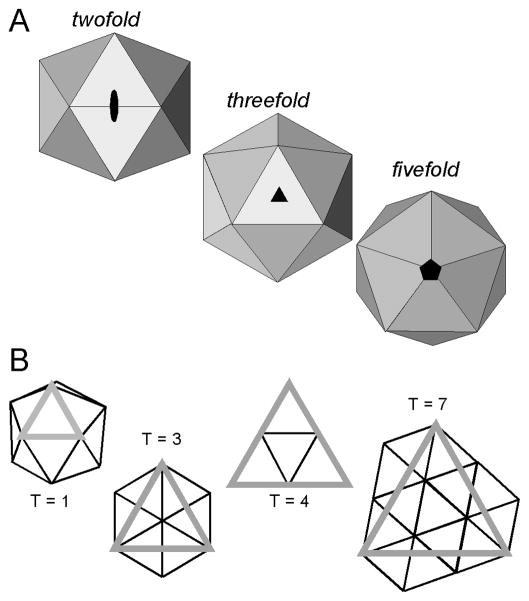
An icosahedron is a geometric solid comprised of 20 equilateral triangular facets (Figure 1A). An icosahedron has twelve vertices, each corresponding to a fivefold symmetry axis. Ten three-fold symmetry axes pass through the 20 triangular faces, and there are 15 two-fold axes that pass through the 30 edge-to-edge contacts between each facet. Viruses with no apparent common host or ancestry have independently evolved to form this structure.
Figure 1.
Icosahedral Symmetry in Virus Capsids (A) Icosahedra and Icosahedral symmetry, with views down symmetry axes (Reprinted from JMR 8(6):480 Copyright 2005 by Wiley Interscience). (B) Selected icosahedral facets, drawn to show quasi-equivalence in icosahedral assembly (Reprinted from PNAS, 101(44):15540, copyright 2004 by Proceedings of the National Academy of Sciences).
An icosahedron is a simple repeating lattice of a single subunit allowing a proportionally small surface area to enclose a large volume (Crick and Watson, 1956). For a virus, this means that a relatively small protein provides sufficient space to package the gene that encodes it as well as the rest of the genome. As a protein cannot have the intrinsic three-fold symmetry of an equilateral triangle, one facet of an icosahedral capsid must consist of at least three proteins. Thus, virus capsids are assembled from 60 subunits (3 proteins × 20 facets = 60 proteins) or a multiple thereof. The theory of quasi-equivalence (Caspar and Klug, 1962) provides a mechanism for building larger icosahedra: triangular facets can be extracted from a hexagonal lattice such that the vertices of each facet fall on lattice vertices, and each facet encompasses an integral number of repeating units. The resulting icosahedra display a mix of strict icosahedral fivefold axes and an array of quasi-sixfold axes (axes with local but not global sixfold symmetry). Quasi-equivalence is described in terms of a T-number (triangulation number), where T is the number of subunits in the repeating asymmetric unit, three of which form a triangular icosahedral facet (Figure 1B); T multiples of 60 asymmetric units make up the icosahedron. An icosahedral T number is restricted to an integer that fits the equation
| (1) |
Where h and k are integers, with h ≥ 1 and k ≤ 0; the restriction of the T number to fit this equation retains the quasi-equivalence of the virus structure
The subunits forming an icosahedral facet are termed quasi-equivalent, as the repeated protein is in similar but different environments (Caspar and Klug, 1962). The same contacts that form a hexagonal sheet can, through slight structural and environmental variation, form the fivefold contacts required to induce curvature in the otherwise flat hexagonal lattice (Caspar, 1980). With this freedom to arrange identical subunits into similar functional geometries, a relatively small protein can enclose a very large volume.
Some spherical viruses stretch the rules of quasi-equivalence. The polyoma- and papillomaviruses are assembled exclusively from pentamers, which cannot support the hexagonal repeat required for quasi-equivalence. And yet, upon the structure determination, it was shown that 12 pentamers occupied the predicted fivefold vertices and 60 pentamers occupied hexavalent positions predicted for a T = 7 quasi-equivalent lattice, with distinctly non-equivalent interactions of the identical chains within an icosahedral asymmetric unit (Baker and Caspar, 1984; Baker et al., 1989). Another variation was observed in the como- and picorna viruses; the subunits in the icosahedral asymmetric unit of these viruses are structurally similar but not identical, resulting in a pseudo-T=3, or P=3, lattice (Rossmann and Johnson, 1989).
Structures of virus capsid proteins
Evolutionary forces seem to have favored a few structural motifs in virus capsid proteins. Most small, non-enveloped viruses share common protein fold: an eight-strand antiparallel β-barrel. This “jelly roll” capsid protein is found across RNA picornaviruses, DNA parvoviruses, unrelated families of non-eveloped RNA plant viruses, DNA polyomaviruses and papillomaviruses, and some DNA bacteriophages (Rossmann and Johnson, 1989).
However, not all non-enveloped spherical viruses share this β-barrel subunit fold. The small RNA bacteriophage MS2 has a five-strand β-sheet flanked by two C-terminal alpha-helices (Golmohammadi et al., 1993); this fold is shared by other members of the Leviviridae family such as Qβ, R17, and PP7. Many large multi-component phages (and also the herpesviruses) share a common fold that was first discovered in bacteriophage HK97 (Jiang and Lin, 2003; Wikoff et al., 1998). In the Hepatitis B virus (HBV), an enveloped DNA virus, the capsid protein has a unique alpha-helical fold (Wynne et al., 1999); HBV has been one of the major systems used for investigating virus assembly thermodynamics.
Despite the advantages of stability and structure afforded by the icosahedral geometry, retroviruses, which also have a predominantly helical capsid protein, are notable for their irregular capsids. However, even these viruses display some measure of symmetry. The mature cores of the Human Immunodeficiency Virus (HIV) have the form of a fullerene cone (Ganser et al., 1999); however, the immature virions display a patchy hexagonal lattice suggestive of quasi-equivalence (Wright et al., 2007)
Interaction Specificity as a Means of Assembly Regulation
Despite the intersubunit contact degeneracy that allows a single subunit to form multiple types of interactions, a virus nonetheless must assemble with high fidelity in order to successfully form an infectious particle. In some cases changes to capsid geometry may be a means of assembly regulation (Maxwell et al., 2002). Alternatively, Flockhouse virus uses RNA-peptide as a switch for conformation change (Dong et al., 1998). In the case of cowpea chlorotic mottle virus, a conformational switch affects correct assembly, directing toward the correct T = 3 assembly products rather than toward aberrant T = 1 or pseudo T = 2 particles (Tang et al., 2006). Scaffold proteins in phages φX174 and P22 constrain conformational change and direct the coat into the correct icosahedral shape (Fane and Prevelige, 2003).
Correct capsid geometry and the interaction stability becomes not merely a matter of formation of a shell for packaging, but now also of assembly regulation, with subunits requiring a “switch” to activate them for assembly, package the correct material, and select the right conformation for the quasi-equivalent environment. Disruption of the native geometry thus becomes a serious liability to a virus, disrupting the biological timing of assembly, or resulting in aberrant assembly that fails to package the correct viral contents (Stray et al., 2005).
Subunit Interactions
By definition an icosahedron requires both hexameric and pentameric contacts in order to build a complete structure; contact domains must be capable of sustaining interactions in the necessarily different local geometries (Caspar, 1980). The assembly of virus capsids tends to be driven by the burial of hydrophobic surface area at the inter-subunit contact points (Bahadur et al., 2007); this predominance can be seen in VIPER database of virus structures (Reddy et al., 2001). The burial of the hydrophobic residues at the subunit interfaces is consistent with the observation that assembly (of at least those viruses that have been rigorously tested) is driven by an increase in system entropy, i.e. despite the ordering of the capsid subunits, the burial of the hydrophobic regions results in a more disordered solvent (Ceres and Zlotnick, 2002; Prevelige et al., 1994). On average a single capsid inter-subunit contact buries some 1750 Å2 of surface area; this is less than one would find in a typical homodimeric protein complex, but significantly more than a simple crystal contact (Bahadur et al., 2007). This relatively small contact area argues for the formation of inherently unstable assembly intermediates that are stabilized upon completion of a network of otherwise relatively weak interactions (Bahadur et al., 2007; Ceres and Zlotnick, 2002).
The Hepatitis B virus capsid is the system for which many of the thermodynamic models of assembly were first applied and will be used here as an example. The structure of this capsid has been solved to 3.3 Å (Wynne et al., 1999); HBV forms a T = 4 capsid based on hydrophobic contacts, with each asymmetric unit comprised of two copies of the homodimeric capsid protein (with a small population of 90-mer T = 3 capsids). Assembly is driven by increasing ionic strength, temperature, and/or capsid protein concentration. In the crystal structure, it was observed that the inter-subunit contacts were distinct and separate, facilitating an estimation of buried surface area and contact energy (Ceres and Zlotnick, 2002). On average a single HBV subunit-subunit contact buries ~1500 Å2.
Analysis of Capsid Stability
Macromolecular Polymerization and Classical Nucleation Theory
While the process of macromolecular polymerization has been studied extensively, the models for describing these infinite reactions fall short of describing virus capsid assembly. At the most basic level, models of assembly of macromolecular polymers are methods of describing a large population of discrete association reactions. Classical nucleation theory is built upon the theoretical platform of an open-ended, infinite polymer or lattice. Assembly begins with a nucleation event, and the polymer is then elongated through a series of faster and/or more stable additions of subunit, until an equilibrium between the polymer and the free subunits is reached (Frieden, 1985). As the ends of the polymer or lattice remain “open,” subunits can freely associate and dissociate from the polymer, reflecting a critical concentration of subunit that must be present in order to maintain the polymer as a unique phase (Zlotnick, 2005).
In a simplest case of classical polymerization, the initial rate of nucleus formation is determined by the rate of forward nucleation and the (assumed first order) rate of nucleus dissociation and can be expressed as follows:
| (2) |
Nuclei are the first species form in the reaction, and are subsequently depleted as they elongate into polymers.
Once a nucleus has been formed, elongation can proceed at the ends of the molecule by the step-wise addition of equivalent subunits.
| (3) |
When the polymerization reaction reaches steady-state, the net growth of the molecule is zero, thus the critical concentration of free subunit remaining in solution can be expressed as:
| (4) |
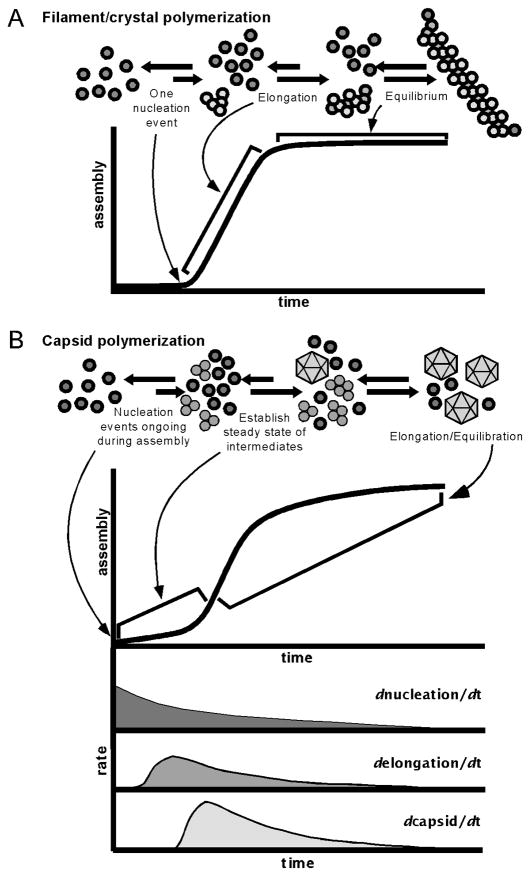
In this classical view of macromolecular polymerization, a polymer grows from a single nucleation event, and at equilibrium the final polymer’s “ends” remain in a state of flux but achieve no net growth. A reaction a single one nucleus gives rise to the sigmoidal kinetics (Figure 2A), where there is a lag phase while the nucleus forms, followed by rapid addition of subunits, and ending with a flat steady-state region wherein the critical subunit concentration has been reached.
Figure 2.
Modeling Capsid Assembly: (A) Classical nucleation model, wherein a single nucleus may give rise to a single infinite polymer. (B) Capsid kinetics, wherein each discrete capsid must arise from an individual nucleus. Top: Typical light-scattering trace of assembling capsid population. Bottom: Representation of reaction rate continuum within the population (A and B, top reprinted from JMR 8(6):483 Copyright 2005 by Wiley Interscience).
Superficially, capsid growth kinetics display a similar pattern (Figure 2B, top), and the classical polymerization model has been applied to capsid growth (Zandi et al., 2006). However, we suggest that a very different reaction is taking place requiring a different mathematical treatment. First, the very nature of assembly of a capsid population is distinct from the assembly of a single large polymer; capsids are discrete, individual units. Each individual capsid is the end point of a unique nucleus, rather than the entire population growing from a single point as with a classic polymer. Moreover, capsids do not exist in the same state of flux with open ends, but are closed, complete and distinct structures. In a classical infinite polymer, all “ends” are equivalent, whereas the gradual completion of the capsid requires a progressive change in the average number of ligands per subunit which engenders an apparent cooperativity of assembly. Lastly, nuclei and capsid intermediates continually form during the reaction, approximating a steady-state concentration of free subunits and intermediates through all but the initial stages of the reaction (Figure 2B). These distinct differences in capsid polymerization as compared to the classical infinite polymerization define a specific formalism for capsid assembly.
Thermodynamic Theory of Capsid Assembly
The capsid assembly reaction can be simply described as:
| (5) |
This reaction appears very similar to the assembly of an infinite polymer. The assembly of an individual capsid initially follows the classical nucleation scheme, with a single nucleation event followed by the addition of subunits until the icosahedron is completed. However, within a population of assembling capsids, these reactions are concurrent, but not synchronized and sum as shown in Figure 2B. With a classical polymer, the lag phase ends upon formation of the polymer nucleus. In capsid assembly, the apparently lag phase is characterized by the formation of many individual nuclei that then begin to elongate into progressively larger and more complete intermediates. The lag phase ends when an “assembly line” of intermediates is complete and capsids begin to accumulate.
The elongation phase of a classical polymer is characterized by the addition of subunits to the single nucleus. However, in capsid assembly, the rapid growth phase is characterized by continuing and concurrent nucleation and elongation with the subsequent formation of capsids. The reaction gradually slows as the free subunit population is consumed, eventually plateauing when the free subunit concentration reaches the pseudo-critical point.
The assembly of a population of capsids is thus characterized by concurrent reactions for nucleation, elongation, and capsid completion. The phases apparent in a light scattering experiment (Figure 2B) do not correspond to particular steps in the assembly reaction. All reaction steps have a certain sameness as the individual capsid assembly steps overlap through the reaction, with intermediates forming and then being consumed in the formation of progressively larger, more complete intermediates: all nuclei with one subunit added are used to form nuclei with two subunits added, and so forth to a complete capsid. Thus, the rate of accumulation of an intermediate of a given size is dependent on the rates of formation of smaller nuclei and the rates of dissociation of subunits of larger size, described as follows:
| (6) |
As these are reversible reactions and may follow multiple paths, “other terms” encompasses all of those other association and dissociation reactions that describe the reversible reactions leading to and from an intermediate of size nucleus + n subunits.
As the reaction proceeds, it approaches steady-state concentrations of intermediates. Ideally, assembly reactions gradually reach equilibrium with the final concentration of capsid dependent on stability, which for a capsid of N subunits can be described in terms of an overall association constant expressed as:
| (7) |
For practical reasons, Equation 7 is calculated in its logarithmic form.
Kinetic trapping is a critical barrier to reaction equilibration. This can result from conditions that (i) cause excessively high association energies, which in turn can deplete the subunit pool without successfully forming complete capsids (Endres and Zlotnick, 2002), (ii) where nucleation occurs so rapidly as compared to elongation that there are many metastable intermediates even in the presence of free subunits (Stray et al., 2004), or (iii) when off-path assembly leads to formation of metastable intermediates, intermediates that may even be icosahedral (Johnson et al., 2005; Tang et al., 2006).
Given equilibration, Equation 7 demonstrates a steep Nth power dependence on concentration; however, this does not limit the maximum concentration of free subunit, i.e., a critical concentration, but in practical terms it generates a pseudo-critical concentration. The value Kcapsid is a global capsid association constant, and thus is very large, in units of M−N, and impractical for calculations. From the value of Kcapsid, one can derive two terms that are much more convenient. The first is the apparent dissociation constant for a complete capsid, KD,apparent, which is approximately equal to the pseudo-critical concentration, and is defined as the point when the concentrations of subunit equal that of capsid, written as:
| (8) |
Alternatively, one can partition Kcapsid into the average association energy between two subunits; one now has in hand a small term that can be used to evaluate the stability of a single inter-subunit contact. If one assumes that all the contacts in the virus capsid are identical, then Kcapsid can be expressed in terms of a statistical factor accounting for reaction degeneracy and some power of the single contact equilibrium constant, Kcontact. The number of contacts forming a capsid, the exponential term for Kcontact, is cN/2, where N is the number of subunits, c is the number of contact surfaces per subunit, and the factor of one half is to account for the fact that a single subunit accounts for only half of each pairwise interaction. The statistical factor accounting for j-fold degeneracy with N subunits is of the general form jN−1/N. Thus, Kcontact can be expressed in terms of Kcapsid by the following equation:
| (9) |
The association energy per contact is:
| (10) |
Using an assembly-derived ΔGcontact, one can readily dissect the energy of association into its entropic and enthalpic components by van’t Hoff analysis, illuminating the driving forces behind capsid assembly.
Methods of Analysis
The thermodynamic parameters of capsid assembly can be easily determined from the concentration free subunits and assembled capsids in an equilibrated assembly reaction. However, it is necessary to verify that the reaction has in fact reached equilibrium. The ideal demonstration of equilibration would be comparison of assembly to disassembly to show that both reactions reach the same end point. Unfortunately, this generally does not work due to the hysteresis to dissociation, which is believed to derive from the difficulty of removing the first few subunits from the capsid (Singh and Zlotnick, 2003) as well as depletion of intermediates (Bruinsma et al., 2003). A time-course measurement of the amount reactants and products can give a strong indication that a reaction has reached steady state (e.g. Figure 4B). It is also necessary to ensure that the reaction is not kinetically trapped. Kinetic traps may be present when there is sufficiently high association energy such that very little free subunit remains in solution, when there are many intermediates but few capsids, or when there is off-path assembly. These last two possibilities can still be analyzed by treating the concentration of free subunit as a critical concentration (Bourne et al., 2008).
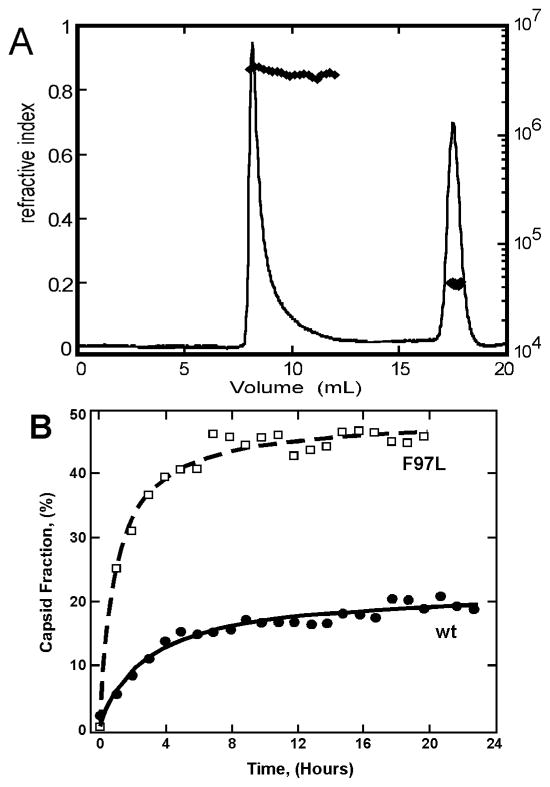
Figure 4.
Quantifying the thermodynamics of HBV capsid assembly. (A) SEC of a typical assembly reaction of 15uM HBV Cp, 0.3M NaCl. (C) Comparison of assembly kinetics of the wild-type capsid protein to the F97L mutation, which displays more rapid assembly into more stable capsids as compared to wild-type (Reprinted from J. Virology 78:9540 copyright 2004 by American Society for Microbiology).
Perhaps the most obvious and simple method for evaluating assembly reactions is size-exclusion chromatography (SEC) (Ackers, 1970). Provided that capsids do not dissociate during chromatography, an equilibrated assembly reaction can be separated into reactants and products (inset Figure 3A). Once the equilibrated reaction components have been separated, the integrated areas of the chromatogram peaks are proportional to the total protein concentration that is assembled or free in solution (Ceres and Zlotnick, 2002; Mukherjee et al., ; Zlotnick et al., 2000).
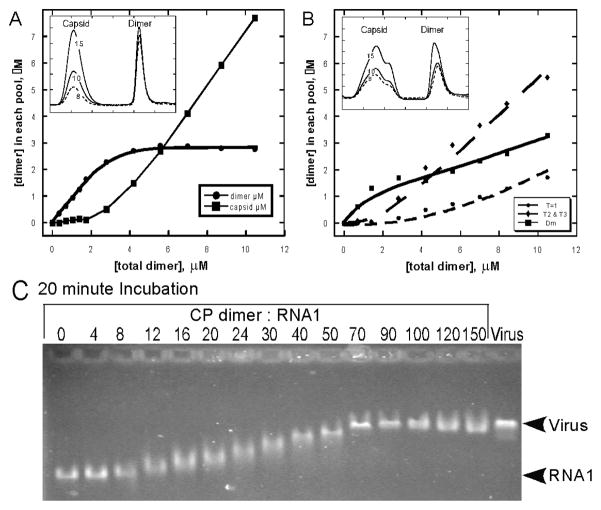
Figure 3.
The Mechanism of Assembly of CCMV. (A) Assembly induced by acidification illustrates the critical concentration of protein required for assembly, as quantified by SEC (inset) (Copyright © Elsevier, Nano Letters, 5(4):766, 2005). (B) Capsid assembly as observed by SEC (solid line) and light scattering (dashed line), showing two major peaks corresponding to 3.5 MDa T = 3 capsids and 43 kDa dimers, with the molecular weights estimated by Debye plots (diamonds); the right side of the capsid peak is polydisperse, trailing to a 200 kDa species corresponding to a pentamer of dimers (Reprinted from Virology, 277:453, copyright 2000 by Elsevier,). (C) Titration of CCMV RNA1 with CCMV CP showing weak cooperativity of assembly (Reprinted from JMB 335:456, copyright 2004 from Elsevier). (D) Assembly induced by acidification of the CCMV NΔ34 mutant capsid protein shows kinetic trapping and failure of the reaction to reach equilibrium (Reprinted from Nano Letters, 5(4):766, copyright 2005 by American Chemical Society).
SEC fractionation followed by quantitative gel densitometry was used to quantify the assembly products of multi-competent icosahedral virus-like particles of bacteriophage P22 (Parent et al., 2006b). After assembly, reactions were fractionated, and the concentration of free subunits and scaffold were quantified by densitometry of an SDS polyacrylamide gel. In the case of phages like P22, the role of the scaffold protein is an added concern for evaluation.
Differential scanning calorimetry (DSC), only rarely applied to viruses, was used very successfully with bacteriophage HK97. The energy required to dissociate a capsid can be directly measured. In the case of HK97, DSC was also used to monitor the stability of the intermediate forms of the capsid head found in the maturation process of HK97, allowing further quantification of energetics of capsid function that tend to be otherwise experimentally inaccessible (Conway et al., 2007). An advantage to this method is that the addition of heat can allow one to overcome the barriers of hysteresis to dissociation, allowing for measurement of both assembly and disassembly under similar conditions and without the use of denaturants. However, difficulties can arise when there is little separation of the transitions for disassembly and unfolding of the subunits themselves, as these two reactions can occur simultaneously (Galisteo and King, 1993; Wingfield et al., 1995). An added complication, particularly evident with phages, is that maturation may result in irreversible conformational changes that will skew the determination of capsid stability.
Another classic method of measuring protein stability, denaturant titration, is also fraught with these limitations when quantifying capsid stability. Titrations of HBV capsid with GuHCl or urea did show a two-phase transition, demonstrably the separate dissociation and unfolding reactions (Singh and Zlotnick, 2003). However, the calculated contact energies from the denaturant titrations returned contact energy values roughly twice those determined from assembly studies. This and the change of contact energy with subunit concentration indicated that denaturant titration did not effect a true equilibrium between capsids and subunits, but merely demonstrated the hysteresis of disassembly that prevented accurate measure of subunit contact energies (Singh and Zlotnick, 2003).
It should be noted that indirect signals have also been correlated with assembly and can be useful tools for examining these reactions. The intrinsic fluorescence and/or anisotropy of tryptophan and/or tyrosine may change during assembly correlating changes in the burial and/or mobility of these residues (Da Poian et al., 1993; Prevelige et al., 1994; Silva and Weber, 1988; Singh and Zlotnick, 2003). A recently described assay used proteins labeled with a self-quenching fluorophore, BoDIPY-FL, so that assembly correlated with the loss of fluorescence, thus generating a high throughput screen for assembly-directed antivirals (Zlotnick et al., 2007).
Applications of Thermodynamic Evaluation of Virus Capsid Stability
With the basic framework outlined, one can apply the described methods to a wide array of viruses. The results have shed light on certain aspects of virus capsid behavior that had previously been experimentally inaccessible.
Cowpea Chlorotic Mottle Virus
Cowpea chlorotic mottle virus (CCMV) is a non-enveloped RNA virus that forms T = 3 capsids comprised of 90 homodimeric subunits. In vitro the purified capsid proteins can be induced to form a variety of empty polymorphic structures by altering the chemical environment, specifically pH and ionic strength (Bancroft et al., 1967). This has made it a popular system for the study of icosahedral capsid assembly.
Early studies of CCMV assembly have been expanded and quantified. Purified dimer was induced to assemble by lowering pH with sodium acetate. Equilibrated reactions were then purified by SEC and capsid and dimer was quantified (Figure 3A, inset). Consistent with predictions, at very low concentrations capsids did not assemble. With increasing total protein concentrations, capsid was observed and free dimer reached a pseudo-critical concentration of approximately 6uM (Figure 3A). Lower concentration reactions near the pseudo-critical concentration tend to be slower to equilibrate and thus can result in less accurate measurement of Kcontact. From this SEC data, it was determined that the average pairwise association energy between subunits was very weak, only 3.1 kcal/mol at pH 5.25, −3.4 kcal/mol at pH 5.0, and −3.7 kcal/mol at pH 4.75. Nonetheless, in acidic solutions, capsids were globally stable.
These data can relate the free energy of the inter-subunit contacts to the biology of the virus. Under physiological conditions, the protein-protein interactions are very weak and interactions between protein and nucleic acid are relatively strong (Johnson et al., 2004; Mukherjee et al., 2006). Thus, interaction with viral RNA is expected to provide most of the support for assembly, and the cooperativity resulting from protein-protein interaction is expected to be weak. This was born out by an electrophoretic mobility shift assay where CCMV RNA was titrated with increasing amounts of capsid protein (Figure 3B) (Johnson et al., 2004). Increasing concentrations of capsid protein resulted in a gradual shift of the RNA band. Strong protein-protein interactions were predicted to lead to a highly cooperative assembly, resulting in capsids and bare RNA. However, intermediate concentrations of protein resulted in uniform intermediate migration of the RNA on the gel, and half-capsids were visible in micrographs.
Studies with CCMV also demonstrated the nature of kinetic traps (Johnson et al., 2005). While the complete capsid protein exclusively forms T = 3 particles, a mutant lacking the N-terminal 34 amino acid residues yields a mixed population of T = 1 (1.1 MDa), pseudo-T = 2 (2.0 MDa), and T = 3 capsids that can be observed by the uneven capsid peak (Figure 3C, inset). It was observed that the reaction never reached equilibrium, even after five days of incubation. There was clearly no critical concentration, seen by the absence of a plateau concentration of free subunit concentration (Figure 3C). It was proposed that the loss of this N-terminal domain results in a greater freedom of motion of the angles between the capsid subunits and a loss of control of intersubunit geometry. Interestingly, the pseudo-T = 2 particles are extremely heterogeneous, suggesting a broad and poorly-defined energy minimum (Tang et al., 2006).
Hepatitis B Virus
The Hepatitis B virus is the model system on which the bulk of our quantitative thermodynamic analyses have been performed. HBV is an enveloped DNA virus with an alpha-helical capsid protein that forms a T = 4 capsid (Ganem and Prince, 2004). In vitro a truncated form of the capsid protein consisting of only the assembly domain (residues 1–149) can be assembled into complete capsids of 120 homodimeric subunits. The rates and extents of reactions are dependent on ionic strength, capsid subunit concentration, and reaction temperature.
Amounts of reactants and products for an HBV assembly reaction were usually quantified by SEC (Figure 4A), though light scattering and fluorescence were also used semi-quantitatively (Bourne et al., 2008). A time course handily showed a reaction that was closely approaching equilibrium within 24 hours (Figure 4B). Results from assembly and van’t Hoff analyses shows assembly is entropy driven and heavily compensated, consistent with burial of hydrophobic surface (Table 1) (Ceres and Zlotnick, 2002).
Table 1.
Thermodynamics of HBV assembly: experimental and calculated values. (Data from Ceres and Zlotnick, 2002.)
| [NaCl], M |
|||||
|---|---|---|---|---|---|
| Energy Per contact | 0.15 | 0.3 | 0.5 | Calculated | |
| ΔG (kcal/mol) | −3.1 ± 0.1 | −3.7 ± 0.2 | −3.7 ± 0.2 | −4.0 ± 0.2 | −8.5 ± 0.8 |
| ΔH (kcal/mol) | +2.0 ± 1.0 | +4.3 ± 0.4 | +6.1 ± 0.8 | +6.2 ± 0.2 | +9.4 ± 2.6 |
| TΔS (kcal/mol) | +5.1 ± 1.1 | +8.0 ± 0.6 | +9.8 ± 1.0 | +10.1 ± 0.4 | +18 ± 3.7 |
| ΔS [cal/(mol·deg)−1] | +17 ± 3.7 | +27 ± 2.0 | +33 ± 3.3 | +34 ± 1.3 | +60 ± 12 |
| KD,apparent | 14μM | 1.9μM | 1.8μM | 0.77μM | 0.14pM |
These results demonstrated a surprising disagreement between experimental results and predictions based on structure. Structure-based estimation of association energy depends on buried surface area. The average value published on VIPER is −15 kcal/mol for each subunit of the capsid. A similar approach with different parameterization (Baker and Murphy, 1998) yielded values of about −9 kcal/mol. The calculated association energies suggest extraordinarily stable capsids. However, Table 1 shows that the experimentally determined average values for this contact energy is between −3 and −4 kcal/mol, values where both dimer and capsid concentrations are easily measured. Where does this the error in calculated association energy come from? It may be simply due to an incorrect parameterization in energy calculations; as described by Bahadur et al, virus contacts are not as tightly packed as a typical dimerization contact. Also, the experimental values may reflect the energetic cost of conformational change from a hypothetical solution structure to the observed capsid structure (Horton and Lewis, 1992). The intersubunit contact energies determined experimentally are so weak that, if taken on an individual basis, one would doubt their specificity. These results have led us to suggest that a virus is a dynamic structure, with the capsid stability maintained by its geometry and the sum of its icosahedral network of many weak individual contacts (Zlotnick, 2003).
The physiological basis of evolutionary selection in viruses can also have a thermodynamic basis. A common naturally occurring mutant of HBV, F97L, resulted in increased virion production and secretion of immature virus particles in vivo. Quantitative analysis showed that this conservative mutation led to greater enthalpy and compensating entropy per contact, resulting in a substantially more negative ΔGcontact at 37°C, than wildtype (Figure 4B). This result indicates a selective advantage for this mutation in chronic infections where viral protein production is attenuated (Ceres et al., 2004).
Thermodynamic analysis can contribute to our understanding of the mechanism of putative antiviral drugs that target the assembly pathway. The heteroaryldihydropryimidines (HAPs) are a class of compounds that decrease viral titer in cell culture (Deres et al., 2003). SEC showed that HAPs drove assembly, but resulted in the formation of aberrant products larger than normal capsids. Transmission electron microscopy showed that reaction products were large polymers dominated by hexagonal sheets of capsid protein. The association energy between subunits is increased in the presence of HAPs by as much as −2 kcal/mol (Bourne et al., 2008). Crystallographic studies of HBV capsids complexed with a HAP showed the small molecule filling a gap in the interdimer contact that led to a global alteration of quaternary structure—but not tertiary structure—that flattened quasi-sixfolds and caused fivefold vertices to protrude (Bourne et al., 2006). The thermodynamic effect of the HAPs is expected (and observed) to affect assembly kinetics as well as the extent of assembly. Combined with their assembly misdirecting effect, the HAPs may represent a new mechanism for antiviral therapeutics.
Human Papillomavirus
Papillomaviruses form icosahedral arrays of pentameric subunits in a T = 7 lattice, with pentamers occupying both the penta- and hexavalent environments. The C-terminal arms of the subunits interlock with adjacent pentamers and also participate in formation of disulfide crosslinks in the assembled capsid. Because it is not feasible to parse out the individual pairwise interactions, its was chosen to report the association energy per pentamer, which closely corresponds to the KD,apparent (Mukherjee et al., 2008). This average association energy per pentamer was between −8 and −10 kcal/mol—a very narrow range, but in agreement with the weakly associated capsid subunit model. Critical to this analysis, reversible assembly was induced under reducing conditions, indicating that formation of the disulfide crosslinks was not prerequisite to assembly.
Bacteriophages
Bacteriophage P22 is a tailed phage, but empty, icosahedral capsids can be assembled in vitro from coat and scaffold protein. These capsids are comprised of 420 coat protein molecules that assemble on a scaffold of between 100 and 300 scaffold protein molecules; these are a form of assembly chaperone and are unnecessary for the structure of the final assembled capsid (King et al., 1976). Prevelige et al. observed a pseudo-critical concentration for assembly, but analysis is complicated because of the energetic contributions from the scaffold protein, which binds non-stoichiometrically to the coat. The thermodynamic contributions of these scaffold proteins were examined through the use of SEC and gel densitometry, which were used as means to quantify reactants and products (Parent et al., 2006b). The observed exchange of free and bound coat protein in P22 capsids uniquely demonstrated that the assembly reaction is in equilibrium (Parent et al., 2006a). From these analyses it was determined that both the coat and scaffolding proteins contribute relatively weak association energies, but the network of all contributions result in a globally stable capsid, with each capsid protein subunit contributing −7.2 kcal/mol, and each scaffold protein contributing −6.1 kcal/mol. These values are per protein, rather than per contact, due to ambiguity in how to dissect protein-protein interactions. Like HPV, these per protein energies roughly correspond to the KD,apparent. At lower scaffold to capsid protein ratios, a small increase in the amount of scaffold present dramatically increased the number of capsids formed. However, at high ratios, scaffold concentration increases resulted in a markedly lower yield of capsids due to overnucleation (Parent et al., 2006b).
The thermodynamics of bacteriophage HK97 were analyzed not by their assembly but through their disassembly and denaturation by DSC, allowing for comparison between the two methods. This contrast in methods is particularly informative as the answers are surprisingly similar. In vitro assembled bacteriophage HK97 capsids are T = 7 icosahedra consisting of capsid subunits comprised of fused assembly and scaffold domains as well as a viral protease. After assembly of a functional prohead from a mix of pentamers and hexamers, the protease cleaves the scaffold domains, and the capsid undergoes a structural changes upon DNA packaging, expanding via multiple intermediates into a final mature head, a process which can be mimicked in vitro by acidification (Duda et al., 1995). Maturation is characterized first by the cleavage event, then by a series of reorganizations of the capsid subunits, and finally by the formation of covalent crosslinks (Ross et al., 2006).
DSC was used to examine the stability of each of the different prohead states and determine the thermodynamic contributions of the different assembly proteins. From the peak temperatures of each denaturation isotherm, it is apparent that each subsequent structure in the maturation process has a greater thermal stability. The contribution of the crosslinking step to the stability of the mature head was investigated using a K169Y mutant, which was unable to crosslink. This mutant had the same stability as the wild type for all steps prior to crosslinking, but significantly lower stability after that maturation step due to the loss of the crosslinks. The thermodynamic parameters of the virus structures were determined from the peak areas of the thermograms. The contact energy for Prohead I, the first assembly product, is around −2k cal/mol per pairwise interaction, in excellent agreement with the weak association model for virus assembly. Each subsequent structure in the maturation process had a lower free energy with a high barrier in between steps; thus, the lowered energies serve to lock the capsid in subsequent conformation (Ross et al., 2006).
Concluding Remarks
The thermodynamic stability of a virus capsid is the underlying means by which the capsid fulfills its roles in the virus; understanding the mechanisms of stability provide insight into mechanisms of capsid function in the virus life cycle, as well as providing a means for the design of new antiviral drugs. The WIN family of compounds bind poliovirus post-assembly to entropically stabilize it to, prevent genome release and infection (Tsang et al., 2000). More recent efforts focus on antivirals that block viral replication by affecting the stability of the capsid prior to and during assembly.
Challenges still remain in terms of completely and accurately describing virus assembly. Current methods tend towards treating all subunits as equal, but quasi-equivalence makes it clear that all subunits are not equal. Moreover, new analyses and techniques must be devised to analyze the energetics of assembly of the multi-component virus and phage capsids. Nonetheless, we have today a solid framework for the analysis of virus capsid stability.
References
- Ackers GK. Analytical gel chromatography of proteins. Adv Protein Chem. 1970;24:343–446. doi: 10.1016/s0065-3233(08)60245-4. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Rodier F, Janin J. A dissection of the protein-protein interfaces in icosahedral virus capsids. J Mol Biol. 2007;367:574–90. doi: 10.1016/j.jmb.2006.12.054. [DOI] [PubMed] [Google Scholar]
- Baker BM, Murphy KP. Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 1998;295:294–315. doi: 10.1016/s0076-6879(98)95045-5. [DOI] [PubMed] [Google Scholar]
- Baker TS, Caspar DL. Computer image modeling of pentamer packing in polyoma virus “hexamer” tubes. Ultramicroscopy. 1984;13:137–51. doi: 10.1016/0304-3991(84)90064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. The capsid of small papova viruses contains 72 pentameric capsomeres: direct evidence from cryo-electron-microscopy of simian virus 40. Biophys J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft JB, Hills GJ, Markham R. A study of the self-assembly process in a small spherical virus. Formation of organized structures from protein subunits in vitro. Virology. 1967;31:354–79. doi: 10.1016/0042-6822(67)90180-8. [DOI] [PubMed] [Google Scholar]
- Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- Bourne C, Finn MG, Zlotnick A. Global structural changes in hepatitis B capsids induced by the assembly effector HAP1. J Virol. 2006;80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. Small-Molecule Effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J Virol. 2008;82 doi: 10.1128/JVI.01360-08. published electronically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma RF, Gelbart WM, Reguera D, Rudnick J, Zandi R. Viral self-assembly as a thermodynamic process. Phys Rev Lett. 2003;90:248101. doi: 10.1103/PhysRevLett.90.248101. [DOI] [PubMed] [Google Scholar]
- Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DLD. Structure of tomato bushy stunt virus. Nature. 1956;177:476–477. doi: 10.1038/177475a0. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Ceres P, Stray SJ, Zlotnick A. Hepatitis B Virus Capsid Assembly is Enhanced by Naturally Occurring Mutation F97L. J Virol. 2004;78:9538–9543. doi: 10.1128/JVI.78.17.9538-9543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–31. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol. 2007;158:224–32. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC, Watson JD. The structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- Da Poian AT, Oliveira AC, Gaspar LP, Silva JL, Weber G. Reversible pressure dissociation of R17 bacteriophage. The physical individuality of virus particles. J Mol Biol. 1993;231:999–1008. doi: 10.1006/jmbi.1993.1347. [DOI] [PubMed] [Google Scholar]
- Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–6. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- Dong XF, Natarajan P, Tihova M, Johnson JE, Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J Virol. 1998;72:6024–33. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–35. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Endres D, Zlotnick A. Model-based Analysis of Assembly Kinetics for Virus Capsids or Other Spherical Polymers. Biophys J. 2002;83:1217–1230. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE., Jr . Mechanism of scaffolding-assisted viral assembly. In: Chiu W, Johnson JE, editors. Virus Structure. Vol. 64. Academic Press; San Diego: 2003. pp. 259–99. [DOI] [PubMed] [Google Scholar]
- Frieden C. Actin and tubulin polymerization: the use of kinetic methods to determine mechanism. Annu Rev Biophys Biophys Chem. 1985;14:189–210. doi: 10.1146/annurev.bb.14.060185.001201. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys J. 1993;65:227–35. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B Virus Infection — Natural History and Clinical Consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–3. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Golmohammadi R, Valegard K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J Mol Biol. 1993;234:620–39. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- Hagan MF, Chandler D. Dynamic Pathways for Viral Capsid Assembly. Biophys J. 2006 doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilmer JK, Zlotnick A, Bothner B. Conformational Equilibria and Rates of Localized Motion within Hepatitis B Virus Capsids. J Mol Biol. 2008;375:581–594. doi: 10.1016/j.jmb.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton N, Lewis M. Calculation of the free energy of association for protein complexes. Protein Sci. 1992;1:169–81. doi: 10.1002/pro.5560010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lin WB. Self-assembly of chiral molecular polygons. Journal of the American Chemical Society. 2003;125:8084–8085. doi: 10.1021/ja034402t. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, Zlotnick A. Regulating self-assembly of spherical oligomers. Nano Lett. 2005;5:765–70. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Willits D, Young MJ, Zlotnick A. Interaction with Capsid Protein Alters RNA Structure and the Pathway for In Vitro Assembly of Cowpea Chlorotic Mottle Virus. J Mol Biol. 2004;335:455–64. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc Lond B Biol Sci. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- Lewis JK, Bothner B, Smith TJ, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci U S A. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Pfeifer CM, Johnson JM, Liu J, Zlotnick A. Redirecting the coat protein of a spherical virus to assemble into tubular nanostructures. J Am Chem Soc. 2006;128:2538–9. doi: 10.1021/ja056656f. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Thorsteinsson MV, Johnston LB, DePhillips P, Zlotnick A. A Quantitative Description of In Vitro Assembly of Human Papillomavirus 16 Virus-Like Particles. J Mol Biol. 2008;381:229–237. doi: 10.1016/j.jmb.2008.05.079. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Reddy VS, Brooks CL., 3rd Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7:338–44. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Phage P22 Procapsids Equilibrate with Free Coat Protein Subunits. J Mol Biol. 2006a doi: 10.1016/j.jmb.2006.09.088. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Zlotnick A, Teschke CM. Quantitative Analysis of Multi-component Spherical Virus Assembly: Scaffolding Protein Contributes to the Global Stability of Phage P22 Procapsids. J Mol Biol. 2006b;359:1097–106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Jr, King J, Silva JL. Pressure denaturation of the bacteriophage P22 coat protein and its entropic stabilization in icosahedral shells. Biophys J. 1994;66:1631–1641. doi: 10.1016/S0006-3495(94)80955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DC. Self-assembly of polyhedral shells: A molecular dynamics study. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:051905. doi: 10.1103/PhysRevE.70.051905. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Natarajan P, Okerberg B, Li K, Damodaran KV, Morton RT, Brooks CL, 3rd, Johnson JE. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J Virol. 2001;75:11943–11947. doi: 10.1128/JVI.75.24.11943-11947.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, Steven AC, Duda RL. A Free Energy Cascade with Locks Drives Assembly and Maturation of Bacteriophage HK97 Capsid. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.08.048. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Silva JL, Weber G. Pressure-induced dissociation of brome mosaic virus. J Mol Biol. 1988;199:149–59. doi: 10.1016/0022-2836(88)90385-3. [DOI] [PubMed] [Google Scholar]
- Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem. 2003;278:18249–55. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A. 2005;102:8138–43. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray SJ, Ceres P, Zlotnick A. Zinc ions trigger conformational change and oligomerization of hepatitis B virus capsid protein. Biochemistry. 2004;43:9989–98. doi: 10.1021/bi049571k. [DOI] [PubMed] [Google Scholar]
- Tang J, Johnson JM, Dryden KA, Young MJ, Zlotnick A, Johnson JE. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J Struct Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Tsang SK, Danthi P, Chow M, Hogle JM. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J Mol Biol. 2000;296:335–40. doi: 10.1006/jmbi.1999.3483. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Duda RL, Hendrix RW, Johnson JE. Crystallization and preliminary X-ray analysis of the dsDNA bacteriophage HK97 mature empty capsid. Virology. 1998;243:113–8. doi: 10.1006/viro.1998.9040. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. Embo J. 2007;26:2218–26. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–80. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Zandi R, van der Schoot P, Reguera D, Kegel W, Reiss H. Classical nucleation theory of virus capsids. Biophys J. 2006;90:1939–48. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Schwartz R. Simulation study of the contribution of oligomer/oligomer binding to capsid assembly kinetics. Biophys J. 2006;90:57–64. doi: 10.1529/biophysj.105.072207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- Zlotnick A. Are weak protein-protein interactions the general rule in capsid assembly? Virology. 2003;315:269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- Zlotnick A. Viruses and the Physics of Soft Condensed Matter. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0406935101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A. Theoretical aspects of virus capsid assembly. J Mol Recognit. 2005;18:479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of Capsid Assembly for an Icosahedral Plant Virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Lee A, Bourne CR, Johnson JM, Domanico PL, Stray SJ. In vitro screening for molecules that affect virus capsid assembly (and other protein association reactions) Nature Protocols. 2007;2:490–498. doi: 10.1038/nprot.2007.60. [DOI] [PMC free article] [PubMed] [Google Scholar]