Abstract
We hypothesized that adequately engineered attenuated Salmonella enterica serovar Typhi strains can serve as multivalent mucosal live vector vaccines to immunize against unrelated human pathogens. Toward this ultimate goal, we have developed a novel genetic stabilization system for antigen-expressing plasmids, engineered to encode the single-stranded binding protein (SSB), an essential protein involved in DNA metabolism which was deleted from the live vector chromosome. We utilized full-length protective antigen (PA83) of anthrax toxin from Bacillus anthracis as a foreign antigen and expressed PA83 as a fusion with the ClyA export protein, which allows export of ClyA-PA83 to the surface of S. Typhi live vectors. A series of SSB-encoding multicopy expression plasmids were introduced into reengineered S. Typhi strains previously tested in clinical trials, i.e., CVD 908-htrA and its less attenuated parent CVD 908. Immunogenicity was examined using a mouse model of intranasal immunization with live vector, followed by parenteral boosting with purified PA83. PA-specific antibody responses markedly improved as the copy number of the SSB-encoding plasmids decreased, and this effect was dramatically enhanced when the foreign antigen was delivered by the less attenuated live vector CVD 908ssb. These results suggest that antibody responses to antigens delivered by S. Typhi live vectors are inversely related to the metabolic burden imposed by expression of the foreign antigen and that these responses can be improved when antigens are expressed from low-copy-number plasmids and exported out of the cytoplasm of less attenuated live vectors.
One of the hallmarks of molecular biology has been the engineering of multicopy plasmids for the expression of proteins from both prokaryotic and eukaryotic organisms. When expression plasmids encoding heterologous antigens are introduced into attenuated bacterial vaccine strains, the resulting multivalent live vectors can be employed to vaccinate against several unrelated human pathogens.
Historically, antibiotic resistance genes have been inserted into these expression plasmids for selection purposes after introduction into live vectors. One key disadvantage of selection with antibiotics is that the constant selective pressure required for maintaining potentially unstable plasmids within live vectors is absent in the host after immunization, increasing the probability of plasmid loss and reduced immune responses against foreign antigens.
Perhaps a more pressing concern with expression plasmids encoding resistance determinants relates to the containment of these plasmids within live vectors to prevent possible spread of antimicrobial resistance genes to other species, such as the commensal intestinal microbiota or transient pathogenic bacteria. Until recently, plasmids encoding resistance genes were considered to pose little or no risk for compromising clinical antimicrobial treatments because (i) such plasmids are engineered to be inefficiently mobilized from live vector donors to a recipient (27, 31), (ii) the live vectors themselves are also genetically attenuated and have a limited ability to persist in a host long enough to allow transfer of genetic information (23, 24, 26, 39), (iii) the genes encoding resistance to target antibiotics are rarely or never used in human medicine, and (iv) with no relevant antibiotic selective pressure, even rare plasmid transfers would not lead to de novo resistances becoming established within a new bacterial population (27, 31). However, accumulating evidence suggests that conditions within the human gastrointestinal tract are in fact conducive to the transfer of both self-transmissible and nonmobilizable plasmids. The human intestine is a reservoir of mobile genetic information that can be transferred between resident microbial flora and from commensal organisms to transient bacterial populations (34). Not surprisingly, plasmids play a prominent role in such genetic transfer of information and employ an astonishing variety of tools, including insertion sequences, transposons, and integrons, to capture genes and transfer them to other organisms (2, 3). Given sufficient population densities of both donor and recipient strains, transfer of self-transmissible resistance plasmids (encoding their own conjugation functions) has been demonstrated to occur within only a few days in the human gut, both in the presence (21, 27) and in the absence (40) of antibiotic selective pressure. Studies with experimental animal models suggest that gastrointestinal transfer frequencies can, in some cases, be higher than observed for in vitro conventional conjugations, particularly when there is antibiotic pressure (6, 10). It has also been demonstrated that transfer of engineered plasmids, in which all transfer and mobilization functions have presumably been removed, can occur in the presence of conjugative plasmids driving the transfer (27). Moreover, such conjugative plasmids do not have to be present in the same bacterium as the engineered plasmid for transfer to occur. Experiments in vitro have shown that nonmobilizable plasmids can be “captured” by other bacteria harboring a conjugative plasmid that initiates conjugation, in a process called retrotransfer (18, 36). Although this novel event has not yet been demonstrated to occur in the intestinal environment, it has been hypothesized that given such high densities of bacteria in an ever-changing microbiota, even rare events such as this are likely to occur and reoccur with the capacity to rapidly expand in the presence of appropriate selective pressure (2).
The growing appreciation of the genetic plasticity of plasmids and the possibility of unanticipated mobilization of genetic segments encoding drug resistance under unpredictable conditions have reinforced the need to move away from the use of antibiotic resistance genes in live vectors intended for oral immunization of humans. Here, we report the development and immunological evaluation of a nonantibiotic plasmid selection system which further improves the clinical acceptability of attenuated Salmonella enterica serovar Typhi live vector vaccines. This novel system takes advantage of the strict requirement of bacteria to synthesize single-stranded DNA-binding protein (SSB) to assure viability.
SSB is a noncatalytic 177-amino-acid protein with a molecular mass of 19 kDa, which binds to single-stranded DNA (ssDNA). The critical function of SSB is to prevent unstable ssDNA intermediates from adopting more energetically favored double-stranded configurations. SSB temporarily binds to ssDNA long enough to provide the required single-stranded substrate necessary for various catalytic proteins to efficiently carry out critical steps involved in DNA replication, recombination, and repair (4, 30). Here we have engineered plasmids encoding this essential SSB protein for introduction into attenuated S. Typhi strains no longer able to synthesize SSB, a strategy that theoretically allows for plasmid selection and maintenance both in vivo and in vitro. We report here that SSB plasmid maintenance combined with an antigen export system within an attenuated S. Typhi live vector strain surpasses the efficiency of conventional antibiotic resistance plasmids in inducing humoral immune responses in an intranasal mouse model of immunogenicity.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and molecular genetic techniques.
S. enterica serovar Typhi strain CVD 908-htrA is an auxotrophic derivative of wild-type strain Ty2, with deletions in aroC, aroD, and htrA, that proved well tolerated and immunogenic in phase 2 clinical trials (39). An earlier less attenuated version of this strain, CVD 908, carries chromosomal deletions only in aroC and aroD (28, 37). The Escherichia coli and Salmonella strains used in this study were grown in Luria-Bertani (LB) medium alone or supplemented with 2,3-dihydroxybenzoic acid (Sigma, St. Louis, MO) (14, 19). When grown on solid medium, plasmid-bearing derivatives of CVD 908-htrA were streaked from frozen (−70°C) master stocks onto 2XLB50 agar containing 2% (wt/vol) Bacto Tryptone, 1% (wt/vol) Bacto yeast extract, and 50 mM NaCl. Standard techniques were used for plasmid constructions. Taq DNA polymerase (Invitrogen, San Diego, CA) or Vent DNA polymerase (New England BioLabs, Beverly, MA) were used in PCRs. All plasmid constructions were recovered and maintained in E. coli DH5α (Invitrogen). Selection with carbenicillin and tetracycline was used, where appropriate, at concentrations of 50 μg/ml and 10 μg/ml, respectively. Live vector strains were electroporated with recombinant plasmids as previously described (15). Isolated transformants were swabbed onto supplemented 2XLB50 agar and incubated at 30°C for 20 h. Frozen master stocks were prepared by harvesting bacteria into SOC medium (Quality Biological, Gaithersburg, MD) without further supplementation and freezing at −70°C.
Construction of CVD 908-htrAssb and CVD 908ssb live vectors.
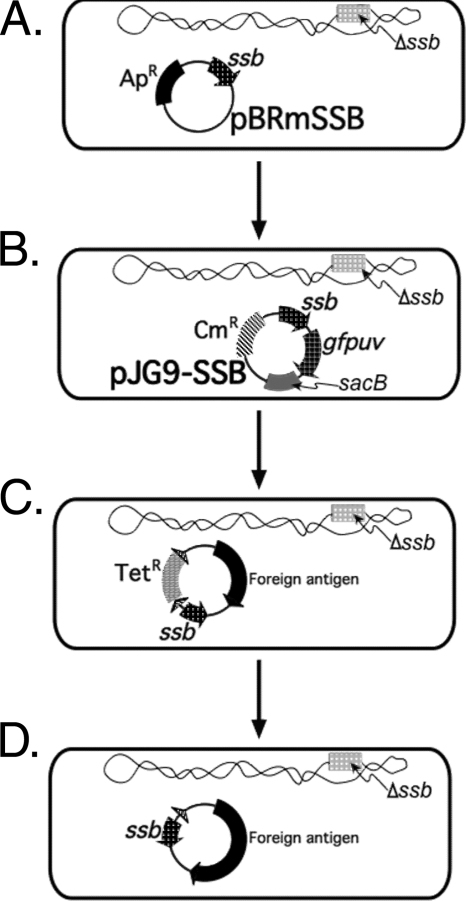
The SSB-based plasmid selection system is functionally similar to a “balanced lethal” system (11), which rests on the principle that if a gene encoding an essential protein is deleted from the host chromosome and placed instead on an expression plasmid, then plasmids encoding this essential protein will ensure cell viability after introduction into the mutated host bacterium. However, unlike most balanced lethal systems, SSB is not an enzyme and therefore produces no product that could be added to the growth medium for import into mutated bacteria. Since it was critical to ensure that SSB would still be present for proper metabolism during deletion of ssb from the live vector chromosome, it was therefore necessary to encode SSB on “maintenance plasmids” that would be present during the deletion of chromosomal ssb (Fig. 1). These maintenance plasmids could then be exchanged for ssb-stabilized expression plasmids expressing the foreign antigen of interest.
FIG. 1.
Schematic depiction of the implementation of an SSB-derived plasmid selection system. Details of this approach are thoroughly presented in Materials and Methods. Briefly, synthesis of SSB must be ensured throughout the construction of a live vector strain intended to deliver a foreign antigen from SSB-maintained plasmids. Therefore, a series of temporary “maintenance plasmids” were introduced to ensure the synthesis of SSB while critical steps in strain construction were accomplished, including the initial inactivation of chromosomal ssb (A), and establishing an intermediate live vector strain (B) into which any SSB-maintained expression plasmid encoding a foreign antigen could be easily introduced (C). Foreign-antigen-encoding plasmids temporarily encoded resistance to tetracycline, which was removed by using FLP recombinase (D) to yield the final live vector strain carrying only SSB-maintained plasmids and bearing no genes encoding resistance to antibiotics. Abbreviations: Apr, resistance to ampicillin, encoded by bla; Cmr, resistance to chloramphenicol, encoded by cat; Tetr, resistance to tetracycline, encoded by tetA; ssb, encodes SSB; gfpuv, encodes a prokaryotic codon-optimized GFPuv allele; sacB, encodes the counterselectable marker levansucrase from Bacillus subtilis. The bacterial chromosome is represented as an irregular wavy circle, with plasmids shown as uniform circles. Hatched triangles represent FRT recombination sites. The transcriptional direction of some plasmid-encoded genes is omitted for clarity.
λ Red-mediated site-directed mutagenesis (7) was utilized to delete ssb from the chromosomes of the S. Typhi vaccine strains. During the construction of these chromosomal ssb deletions, all strains were initially complemented for SSB using a pBRmSSB maintenance plasmid (Table 1). pBRmSSB was derived from pBR322 cleaved with SspI and PstI, treated with T4 polymerase, and religated to remove bla encoding β-lactamase and create pBRΔbla. A 734-bp fragment encoding ssb was synthesized by PCR using primers 1 and 2 (Table 2) with chromosomal template DNA from CVD 1208S, a derivative of attenuated Shigella flexneri 2a strain 2457T (25). The PCR product was recovered in pCR-BLUNT II-TOPO (Invitrogen), creating pCV546; the ssb cassette was then excised as a 750-bp EcoRI fragment and inserted into pBRΔbla digested with EcoRI, creating pBRmSSB. This maintenance plasmid was introduced into attenuated S. Typhi strains by electroporation (Fig. 1A).
TABLE 1.
Selected plasmids used in this study and relevant genotypes
| Plasmid | Size (kb) | Relevant genotype | Reference |
|---|---|---|---|
| pBRmSSB | 4.6 | oriE1 ssb tetA | This work |
| pCV546 | 4.3 | oriE1 ssb aph zeo | This work |
| pKD3 | 2.8 | oriR6Kγ FRT-cat-FRT bla | 7 |
| pKD46 | 6.1 | ori101 ParaB-gam-bet-exo repAtspar bla | 7 |
| pCP20 | 9.4 | ori101 cI857 PλpR-flp repAtspar bla cat | 7 |
| pJG9 | 7.0 | ori101 sacB repAtspar cat | 41 |
| pJG9-SSB | 8.8 | ori101 ssb gfpuv sacB repAtspar cat | This work |
| pGEN222 | 6.2 | ori15A gfpuv par bla hok-sok parA | 15 |
| pGEN222S | 7.0 | ori15A gfpuv par ssb bla hok-sok parA | This work |
| pGEN222SXbaI | 7.0 | ori15A gfpuv par ssb bla hok-sok parA | This work |
| pGEN222TS | 6.6 | ori15A gfpuv par ssb FRT-tetA-FRT parA | This work |
| pGEN222S2 | 5.3 | ori15A gfpuv par ssb FRT parA | This work |
| pGEN206 | 7.1 | ori101 gfpuv repA par bla hok-sok parA | 35 |
| pGEN206ΔSpeI | 7.1 | ori101 gfpuv repA par bla hok-sok parA | This work |
| pGEN206S | 7.9 | ori101 gfpuv repA par ssb bla hok-sok parA | This work |
| pGEN206SXbaI | 7.9 | ori101 gfpuv repA par ssb bla hok-sok parA | This work |
| pGEN206TS | 7.5 | ori101 gfpuv repA par ssb FRT-tetA-FRT parA | This work |
| pGEN206S2 | 6.2 | ori101 gfpuv repA par ssb FRT parA | This work |
| pSEC91 | 7.6 | ori15A clyA tetA par aph hok-sok parA | 13 |
| pSEC91SΔclyA | 4.6 | ori15A par ssb FRT parA | This work |
| pSEC91-83 | 8.5 | ori15A clyA::pa83 par aph hok-sok parA | 12 |
| pGEN222TS-83 | 9.0 | ori15A clyA::pa83 par ssb FRT-tetA-FRT parA | This work |
| pSEC91S-83 | 7.7 | ori15A clyA::pa83 par ssb FRT parA | This work |
| pGEN206TS-83 | 10.0 | ori101 clyA::pa83 repA par ssb FRT-tetA-FRT parA | This work |
| pSEC10S-83 | 8.7 | ori101 clyA::pa83 repA par ssb FRT parA | This work |
TABLE 2.
Primers used in the construction and testing of live vector strains and expression plasmids
| Primer | Sequencea | Template | Reference |
|---|---|---|---|
| 1 | 5′-CATATGATTGACCTGAATGAATATACAGTATTGGAA-3′ | ssb locus from S. flexneri 2a strain 2457T | 42 |
| 2 | 5′-GCTAGCTATTGTTTTAATGACAAATCAGAACGGAA-3′ | ssb locus from S. flexneri 2a strain 2457T | 42 |
| 3 | 5′-CTCGAGCTTGCCAGATTTTCCAGCGTTTTGGTGTGT-3′ | Ty2 | 8 |
| 4 | 5′-CATATGTTATTATTATTAGCTAGCTACTGTATATTCAAACAGGTTAAATTGTGT-3′ | Ty2 | 8 |
| 5 | 5′-CATATGCATTTTCGCTATAGTTCTCGTCTGCTGAAA-3′ | Ty2 | 8 |
| 6 | 5′-CTCGAGACTAGTTAGCTAATCATTGAAACTCTAAATCATTTT-3′ | Ty2 | 8 |
| 7 | 5′-CATATGAATATCCTCCTTAGTTCCTATTCC-3′ | pKD3 | 7 |
| 8 | 5′-GCTAGCGTGTAGGCTGGAGCTGCTTCGAAGTTCTA-3′ | pKD3 | 7 |
| 9 | 5′-TTCGGCGGATCGGAGAGATCGCAGACTTCG-3′ | Ty2 | 8 |
| 10 | 5′-AGACATCAATTATTGCACTAACTATATCTT-3′ | Ty2 | 8 |
| FRT-tetA-for | 5′-TCTAGAgaagttcctattctatatatagtataggaacttcGCTAGCTCATGTTTGAC AGCTTATCATCGATAAGCTTTAATGCGGTAGTTTATCAC-3′ | pSEC91 | 13 |
| FRT-tetA-rev | 5′-TCTAGAgaagttcctatactatatatagaataggaacttcGCTAGCCTATCAGGTCGAGGTG GCCCGGCTCCATGCACCGCGACGCAACGCGGGGAG-3′ | pSEC91 | 13 |
| gfpuv-for | 5′-CAGTGGAGAGGGTGAAGGTGATGC-3′ | pGEN222S2 | This work |
| gfpuv-rev | 5′-GTGTCGACAGGTAATGGTTGTCTG-3′ | pGEN222S2 | This work |
| ssb-for | 5′-GTCAGGACCCGGAAGTACGCTACATGCCAAATGGTGGCGCAG-3′ | pGEN222S2 | This work |
| ssb-rev | 5′-CGCCGCTGAACTGATTGCCACCCTGCGGCTGCTGAGGCTGAC-3′ | pGEN222S2 | This work |
| ori15A-for | 5′-GTCCTTCTTGAGTTTGTAACTGCTGCTG-3′ | pGEN222S2 | This work |
| ori15A-rev | 5′-CGAGGTAACTGGCTTGGAGGAGCGCAGTCAC-3′ | pGEN222S2 | This work |
Relevant restriction sites are underlined. The FRT recombination sites flanking the tetA gene are in lowercase.
Deletion of ssb from the chromosomes of both CVD 908(pBRmSSB) and CVD 908-htrA(pBRmSSB) was accomplished by the method of Datsenko and Wanner (7). Briefly, cassettes encoding upstream and downstream chromosomal sequences required for removal of ssb were constructed using primers 3 and 4 and primers 5 and 6, respectively (Table 2). These cassettes were used to exchange chromosomal ssb with a chloramphenicol resistance cassette, amplified from pKD3 using primers 7 and 8, and recombined into the chromosome using the λ Red recombination system encoded by pKD46. Final removal of the chloramphenicol resistance cassette was accomplished using FLP recombinase encoded by pCP20. We confirmed the integrity of the intended ssb mutations by DNA sequence analysis of the chromosomal locus from each strain using primers 9 and 10. Both CVD 908ssb(pBRmSSB) and CVD 908-htrAssb(pBRmSSB) were confirmed to carry the intended 709-bp chromosomal deletion of ssb, extending from 100 bp upstream of the uvrA start codon to 31 bp downstream of the ssb stop codon.
After chromosomal deletions were completed, pBRmSSB was replaced in all cases with a temperature-sensitive pJG9-SSB replicon derived from pJG9 (41) which carried ssb, in addition to a cat chloramphenicol resistance allele and a counterselectable sacB marker (Fig. 1B). To construct pJG9-SSB, the EcoRI site within the cat gene of pJG9 was changed from 5′-GAATTC-3′ to 5′-GAgTTC-3′ by site-specific mutagenesis using QuikChange (Stratagene, Cedar Creek, TX), creating pJG9dRI. A previously described 2,066-bp SpeI cassette consisting of a promoterless gfpuv allele and tetA (13) was then inserted into pCV546 cleaved with NheI to generate pCV546gfpuv-tetA. A 2,814-bp ssb-gfpuv-tetA cassette was then removed from pCV546gfpuv-tetA digested with EcoRI and inserted into pJG9dRI, partially cleaved with EcoRI, creating pJG9dRI-SGT. Final digestion of pJG9dRI-SGT with NheI and religation to remove tetA resulted in pJG9-SSB. This temperature-sensitive maintenance plasmid was then electroporated into both CVD 908ssb(pBRmSSB) and CVD 908-htrAssb(pBRmSSB), and chloramphenicol-resistant, tetracycline-sensitive colonies were recovered at 30°C (Fig. 1B). Loss of pBRmSSB was confirmed by PCR analysis using primers specific for tetA and oriE1 in separate reactions.
Construction of SSB-encoding expression plasmids.
To efficiently construct expression plasmids encoding both SSB and the desired foreign antigen, we engineered a system in which all SSB-encoding expression plasmids also encoded a removable tetA cassette conferring resistance to tetracycline (Fig. 1C). This would allow all expression plasmids, ultimately intended for use in S. Typhi live vectors, first to be constructed and routinely recovered in E. coli DH5α using tetracycline selection. To allow removal of tetA, we constructed a novel resistance cassette in which tetA was flanked by two direct repeats of FRT recombination sites, referred to hereafter as FRT-tetA-FRT. After engineering and selecting for plasmids in DH5α using tetracycline, plasmids could then be purified using routine procedures, electroporated directly into either CVD 908ssb(pJG9-SSB) or CVD 908-htrAssb(pJG9-SSB), and selected for using tetracycline at 42°C to remove pJG9-SSB. The final step to remove tetA could then be carried out exactly as for construction of the ssb chromosomal deletions, using the temperature-sensitive plasmid pCP20 encoding the FLP recombinase, and incubating again at 42°C to remove both the tetA allele (through FLP-mediated site-specific recombination between the FRT sites) and for curing of the temperature-sensitive FLP-encoding plasmid pCP20 (Fig. 1D).
(i) Expression plasmids encoding GFPuv.
An initial set of plasmids was constructed primarily to determine if ssb would ensure plasmid maintenance and stability. To monitor for plasmid stability, these first-generation plasmids encoded the green fluorescent protein GFPuv as a convenient marker to phenotypically assess plasmid retention. Both a medium-copy-number plasmid (∼15 copies per chromosomal equivalent) carrying the ori15A origin of replication from pGEN222 (15) and an isogenic low-copy-number plasmid (∼5 copies per chromosomal equivalent) carrying the ori101 origin from pGEN206 (35) were constructed.
To engineer the medium-copy-number plasmid, the ssb cassette used in the construction of the temporary maintenance plasmid pBRmSSB was first excised from pCV546 as a 798-bp XbaI-NheI cassette and inserted into a derivative of pGEN222, destroying the unique SpeI site and creating pGEN222S. Since ssb should effectively function as a postsegregational killing function in vivo, inclusion of hok-sok was no longer necessary, so the XhoI site 5′ proximal to hok-sok was changed by site-specific mutagenesis to an XbaI site, creating pGEN222SXbaI, which then allowed replacement of both hok-sok and bla with a FRT-tetA-FRT cassette by digestion with XbaI and NotI (Fig. 2). This FRT-tetA-FRT XbaI-NotI cassette was generated using FRT-tetA-forward and FRT-tetA-reverse primers (Table 2) with pSEC91 as the template and was recovered in pCR-BLUNT II-TOPO. This cassette was excised as a 1,397-bp XbaI-NotI fragment and ligated into XbaI-NotI-digested pGEN222SXbaI, replacing hok-sok and bla and creating pGEN222TS. pGEN222TS was then electroporated into CVD 908-htrAssb(pJG9-SSB), and tetA was removed by FLP recombinase to create the live vector CVD 908-htrAssb(pGEN222S2) expressing GFPuv from medium-copy-number plasmid pGEN222S2.
FIG. 2.
Genetic maps of selected SSB-maintained expression plasmids encoding foreign antigens used in this study. Key restriction sites used for construction are shown, and unique sites are in boldface. Hatched triangles represent FRT recombination sites. Abbreviations: PompC, modified osmotically controlled ompC promoter from E. coli; clyA::pa83, encodes the ClyA antigen export protein fused at its carboxyl terminus to the amino terminus of full-length PA83 from anthrax toxin; ori15A, origin of replication from p15A providing an expected copy number of ∼15 per chromosomal equivalent; ori101, origin of replication from pSC101 providing an expected copy number of ∼5 per chromosomal equivalent; repA, replication protein initiating replication at ori101; par, passive partitioning function from pSC101; T1, transcriptional terminator from the rrnB rRNA operon of E. coli; ssb, encodes SSB; parA, encodes the active partitioning system from pR1, composed of parM and parR.
To construct the low-copy-number isogenic plasmid, the replicon pGEN206 was first partially digested with SpeI and the site located 5′ proximal to repA within ori101 was removed by treatment with DNA polymerase I Klenow fragment and religation, creating pGEN206ΔSpeI. The above-mentioned 798-bp XbaI-NheI cassette encoding ssb was then inserted into the now unique SpeI site of a further derivative of pGEN206ΔSpeI to yield pGEN206S. As with the construction of pGEN222SXbaI, the XhoI site 5′ proximal to hok-sok was changed by site-specific mutagenesis to an XbaI site, creating pGEN206SXbaI (Fig. 2). FRT-tetA-FRT was then inserted as a 1,397-bp XbaI-NotI fragment to create pGEN206TS. pGEN206TS was electroporated into CVD 908-htrAssb(pJG9-SSB), and tetA was removed by FLP recombinase, creating the live vector CVD 908-htrAssb(pGEN206S2) expressing GFPuv from low-copy-number plasmid pGEN206S2.
(ii) Expression plasmids encoding full-length protective antigen (PA83) from anthrax toxin.
For construction of expression plasmids encoding both SSB and a foreign antigen relevant to vaccine development, we focused on expression of the 83-kDa cell-binding domain (PA83) of anthrax toxin from Bacillus anthracis. We elected to express PA83 as a fusion to the carboxyl terminus of the ClyA antigen export protein (13). ClyA-PA83 fusions encoded by a conventional medium-copy-number kanamycin resistance plasmid, pSEC91-83, were previously shown to be successfully exported to the surface of CVD 908-htrA live vectors and primed nonhuman primates to mount accelerated serum toxin neutralization activity responses within 1 week of boosting parenterally with a single dose of BioThrax or recombinant PA83 (12). As with the first-generation test constructs encoding GFPuv, two isogenic plasmids expressing ClyA-PA83 were engineered from medium- and low-copy-number replicons (Fig. 2). These PA83-expressing constructs were ultimately exchanged into both CVD 908ssb(pJG9-SSB) and CVD 908-htrAssb(pJG9-SSB) for immunogenicity studies with mice.
To construct pSEC91S-83, a 4,687-bp BglII-AvrII fragment encoding ClyA-PA83 was removed from pSEC91-83 and inserted into identically cleaved pGEN222TS, regenerating the parA active partitioning locus and creating pGEN222TS-83. After electroporation into live vectors, tetA was removed using FLP recombinase, creating live vectors with the final expression plasmid pSEC91S-83 (Fig. 2). For construction of low-copy-number pSEC10S-83, the identical 4,687-bp BglII-AvrII fragment encoding ClyA-PA83 was inserted into pGEN206TS digested with BglII and AvrII to yield pGEN206TS-83. tetA was then recombined out after electroporation of pGEN206TS-83 into live vectors, creating pSEC10S-83 (Fig. 2).
A final negative control plasmid, pSEC91SΔclyA, was also constructed. First, pSEC91 was digested with BamHI and NheI, and the termini were filled in with Vent DNA polymerase (New England Biolabs) and then religated together to remove clyA and create pSEC91ΔclyA. A 1,110-bp HpaI-AvrII fragment carrying the PompC promoter and the trpA terminator was then excised from pSEC91ΔclyA and used to replace the corresponding cassette of pGEN222SXbaI. Finally, the 1,397-bp FRT-tetA-FRT XbaI-NotI fragment was inserted, replacing the corresponding 1,790-bp XbaI-NotI cassette encoding hok-sok and bla, to yield pSEC91TSΔclyA. The tetracycline-sensitive pSEC91SΔclyA construct was recovered in CVD 908-htrAssb(pJG9-SSB) using FLP recombinase.
Analysis of in vitro plasmid stability.
To determine the persistence of SSB-maintained plasmids in attenuated S. Typhi live vectors grown in vitro, GFPuv-expressing bacteria were passaged for 5 days (120 h) in liquid culture. Frozen stocks were streaked onto 1XLA50 medium supplemented only with 2,3-dihydroxybenzoic acid (DHB) and incubated at 30°C for 24 h to obtain isolated colonies. Two or three fluorescing colonies were then inoculated into 20 ml of 1XLB50 liquid medium supplemented with DHB and incubated with shaking at 225 rpm overnight at 30°C (0-h starting cultures for serial passages). For comparison with strains carrying conventional expression plasmids, starter cultures of ampicillin-resistant live vectors containing either pGEN222 or pGEN206 were grown overnight under selection to ensure a uniform population of bacteria just prior to beginning passages. All overnight starter cultures were then diluted 1:1,000 into fresh supplemented nonselective liquid medium, incubated for 24 h at 37°C, and then serially passaged every 24 h for a total of 120 h. Viable counts on nonselective solid medium were determined at 0 h and every 24 h thereafter, and results were reported as the percentage of fluorescing colonies compared to the total CFU count. In a separate set of experiments, the generation times for these strains were determined under the same growth conditions of 1XLB50 liquid medium grown at 37°C and shaking at 225 rpm. Calculation of generation times based directly on viable counts was used to determine the number of generations that a given strain could be passaged without loss of fluorescence.
To investigate the hypothesis that nonfluorescing colonies carrying SSB-maintained plasmids still retained the plasmid after passage but suffered inactivating rearrangements in gfpuv, plasmids recovered from 18 nonfluorescing colonies arising after passage of CVD 908-htrAssb(pGEN222S2) were examined by PCR for genetic integrity using primer pairs specific for gfpuv (524-bp product), ssb (405-bp product), and ori15A (593-bp product). The sequences of the primers used are listed in Table 2.
Immunization and measurement of antibodies.
Groups of 10 to 12 female BALB/c mice (Charles River Laboratories, Wilmington, MA) 6 to 8 weeks old were inoculated intranasally (i.n.) by placing 5 μl of a vaccine suspension containing ∼2.5 × 109 CFU into the right and left nares at 0 and days 14 as previously described (13, 32). Inocula were analyzed for PA83 expression by Western immunoblot analysis as previously described (13). Mice were boosted intramuscularly on day 42 with 10 μg of recombinant PA83 (List Biological Laboratories, Campbell, CA) adsorbed to 0.5 mg of Alhydrogel and administered in a total volume of 50 μl. Serum samples were collected at 0, 13, 41, 48, 55, and 70 days and stored at −70°C until analyzed. PA83-specific serum immunoglobulin G (IgG) and toxin neutralization activity (TNA) titers were measured in individual animals as previously described (1, 12). All animal experiments and procedures were approved by the Institutional Animal Care and Use Committee of the University of Maryland Baltimore School of Medicine.
Statistical analysis.
IgG antibody responses for the last preboost bleed (day 41) were compared between groups of mice using the Student t statistic. Responses from the first postboost day (day 48) to day 70 were compared overall using Hotelling's T square statistic. In both cases, the comparisons were based on log10-transformed titers and the P value (two sided in the case of the t statistic) was determined from a randomization test on 100,000 random samples. Titers at one of the three specific days (day 48, 55, or 70) were comparing using the randomization test P value for the Student t statistic, with a Bonferroni adjustment (i.e., multiplying the P value by 3). Otherwise, no adjustment for multiple comparisons was made. TNA titers were compared similarly, using a randomization test with 100,000 samples on the T square statistic for untransformed values at day 55 and day 70. Correlation between IgG and TNA values was measured using the Spearman rank correlation coefficient. P < 0.05 was considered statistically significant. Statistical analysis was performed using NCSS 2007 (Number Cruncher Statistical Systems, Kaysville, UT).
RESULTS
Construction and in vitro stability testing of nonantibiotic expression plasmids.
In order to test the feasibility and efficiency of an SSB-based system for selection and maintenance of nonantibiotic expression plasmids, we first engineered a set of isogenic expression plasmids in which the green fluorescent protein (GFPuv) was expressed from a medium-copy-number (pGEN222S2, ∼15 copies per chromosomal equivalent) or a low-copy-number (pGEN206S2, ∼5 copies per chromosomal equivalent) expression plasmid (Table 1). Plasmids temporarily resistant to tetracycline were exchanged into S. Typhi vaccine strain CVD 908-htrAssb(pJG9-SSB), and the tetracycline resistance cassette was removed using FLP recombinase as described in Materials and Methods, resulting in GFPuv-expressing strains CVD 908-htrAssb(pGEN222S2) and CVD 908-htrAssb(pGEN206S2). As controls, we also included CVD 908-htrA(pGEN222) and CVD 908-htrA(pGEN206), in which isogenic medium- and low-copy-number conventional plasmids expressing resistance to ampicillin were used. The resulting four strains were tested in vitro for plasmid stability by passage at 37°C in liquid medium for 5 days in the absence of antibiotic selection. The percentage of fluorescent colonies compared to the total number of colonies is shown in Fig. 3. As previously reported (9, 15), conventional plasmid pGEN222 was extremely unstable and fluorescence was lost from the population within the first 24 h of passage. As the copy number is reduced from ∼15 copies to ∼5 copies per chromosomal equivalent, 100% of the bacteria carrying the lower-copy-number pGEN206 conventional plasmid were fluorescent for the first 24 h, but fluorescence was lost from ∼95% of the colonies recovered after 3 days. Interestingly, SSB-encoding medium-copy-number plasmid pGEN222S2 maintained fluorescence in at least 90% of the population of live vectors during the 5 days of passage, and the lower-copy-number pGEN206S2 plasmid maintained fluorescence in 100% of the live vectors through day 4, dropping to 92% by day 5.
FIG. 3.
In vitro stability of conventional (pGEN222 and pGEN206) and SSB-maintained (pGEN222S2 and pGEN206S2) plasmids expressing GFPuv in CVD908-htrA and CVD908-htrAssb live vectors, respectively. Live vector strains were passaged at 37°C from an overnight starter culture (grown without selection) every 24 h for 5 days (120 h) in antibiotic-free liquid medium. Plasmid stability is reported as the percentage of fluorescing colonies compared to the total CFU count plated on nonselective medium.
Since we are not aware of any homologues of ssb in the genome of S. Typhi that might theoretically allow the growth of plasmidless CVD 908-htrAssb strains, we hypothesized that nonfluorescent colonies recovered from the passage experiments still retained SSB-maintained expression plasmids but had suffered rearrangements that disrupted the open reading frame of gfpuv. To test this hypothesis, we examined 18 nonfluorescent colonies recovered from the passage of CVD 908-htrAssb(pGEN222S2) using PCR with forward and reverse primer pairs specific for gfpuv, ssb, and ori15A (Table 2). As expected, 100% of the PCRs targeting both ssb and the origin of replication (ori15A) for pGEN222S2 produced PCR products of the expected sizes of 593 and 405 bp, respectively, confirming the presence of the expression plasmid. However, for PCRs targeting the gfpuv allele, 16 of 18 reactions revealed amplicons either larger (∼850 bp) or smaller (∼50 bp) than the expected size (524 bp) or produced no product at all; the remaining 3 reactions produced the expected fragment.
Generation times were determined for the four strains passaged under the identical nonselective growth conditions used for the stability studies, and results are reported in Table 3. These data indicate that live vectors carrying medium-copy-number expression plasmids derived from pGEN222 grow slowly; surprisingly, shorter generation times were observed when these medium-copy-number plasmids encoded SSB. The apparent instability of the conventional antibiotic resistance plasmid pGEN222 was again observed in these experiments, with nonfluorescing colonies appearing after ∼6 h of growth in liquid medium without selection. Fluorescence for live vectors carrying SSB-maintained pGEN222S2 plasmids improved and was maintained for 41 generations. Reduction of copy number shortened generation times even further and improved apparent plasmid retention, with CVD 908-htrA(pGEN206) constructs maintaining fluorescence for 59 generations. However, the combination of a low copy number and expression of SSB provided the most dramatic evidence of plasmid retention, with no loss of fluorescence observed in live vectors carrying pGEN206S2 after passage for 241 generations in the absence of antibiotics.
TABLE 3.
Stability of SSB-selected plasmids within CVD 908-htrAssb live vectors passaged for 120 h without selection
| Strain | SSB selection | Expected copy no.a | Generation timeb (min) | No. of generations with 100% retention of fluorescence |
|---|---|---|---|---|
| CVD 908-htrA(pGEN222) | No | ∼15 | 48.7 | Unstablec |
| CVD 908-htrAssb(pGEN222S2) | Yes | ∼15 | 34.7 | 41 |
| CVD 908-htrA(pGEN206) | No | ∼5 | 24.5 | 59 |
| CVD 908-htrAssb(pGEN206S2) | Yes | ∼5 | 23.9 | 241 |
Per chromosomal equivalent.
Generation times were calculated for the total viable counts recovered.
For strains carrying plasmids pGEN222 and pGEN222S2, the lag phase lasted approximately 180 min. For pGEN222, ∼20% of the colonies recovered after 6 h were not fluorescent.
Construction of SSB-encoding plasmids expressing ClyA-PA83 protein fusions.
The results of the in vitro stability studies suggested that the use of SSB as a plasmid retention system can dramatically improve the stability of expression plasmids versus conventional plasmids encoding resistance to antibiotics. Therefore, we tested the hypothesis that the use of SSB-maintained plasmids could improve the immunogenicity of a clinically relevant foreign antigen. We constructed a set of isogenic medium- and low-copy-number expression plasmids in which PA83 was fused to the ClyA antigen export system, which was previously shown to express ClyA-PA83 fusions on the surface of S. Typhi live vectors (12). A secondary hypothesis was that any metabolic burden imposed by stabilized antigen-encoding plasmids on the live vectors could be ameliorated by using a less attenuated strain. An organism that is less attenuated could theoretically be more efficient at targeting antigens to immune cells, thereby inducing more robust immune responses. Therefore, medium-copy-number pSEC91S-83 and low-copy-number pSEC10S-83 expressing ClyA-PA83 fusions were exchanged into both CVD 908-htrAssb(pJG9-SSB) and CVD 908ssb(pJG9-SSB). As a control, we included CVD 908-htrA(pSEC91-83) carrying an isogenic medium-copy-number conventional plasmid expressing resistance to kanamycin, previously shown to be highly immunogenic in mice and nonhuman primates (12).
When examined in vitro by immunoblot analysis, the expression of the ClyA-PA83 fusion protein in CVD 908-htrAssb carrying SSB-stabilized medium-copy-number pSEC91S-83 appeared somewhat diminished compared to that of CVD 908-htrA carrying the isogenic conventional kanamycin resistance pSEC91-83 construct (Fig. 4, lane 3 versus lane 7). Interestingly, as the copy number was lowered, the expression levels of ClyA-PA83 in CVD 908-htrAssb(pSEC10S-83) increased (Fig. 4, lane 4 versus lanes 3 and 7). Expression of ClyA-PA83 in the less attenuated CVD 908ssb background was excellent, regardless of the copy number (Fig. 4, lanes 5 and 6).
FIG. 4.
Coomassie brilliant blue-stained (top) and Western immunoblot analysis (bottom) of whole bacterial lysates from live vectors expressing ClyA-PA83 fusions from SSB-maintained plasmids carried by CVD 908-htrAssb (lanes 2 to 4) and CVD 908ssb (lanes 5 and 6). Lysates from strains carrying medium-copy-number derivatives of pSEC91S are in lanes 2, 3, and 5; lysates from strains carrying low-copy-number derivatives of pSEC10S are in lanes 4 and 6. A lysate from the conventional strain CVD 908-htrA(pSEC91-83) expressing ClyA-PA83 from a medium-copy-number kanamycin resistance plasmid is in lane 7, and 125 ng of purified PA83 is in lane 1. Total protein from approximately 106 CFU was resolved in each lane. Detection of ClyA-PA83 fusions was carried out using a polyclonal goat anti-PA83 primary antibody. The values at the left are molecular masses in kilodaltons. The arrows on the right indicate the position of the ClyA-PA83 fusion protein, with an expected molecular mass of 117 kDa.
Serologic responses following mucosal priming with live vectors.
We first analyzed the serologic responses following two mucosal priming doses of live vectors expressing ClyA-PA83 from SSB-encoding plasmids (Table 4). These responses were recorded on day 41, 1 day before administration of the parenteral boost with PA83. Responses were compared with those of mice immunized i.n. with CVD 908-htrA(pSEC91-83), which expresses ClyA-PA83 from a conventional plasmid without ssb (group 5, comparator group). On day 41, the geometric mean titer (GMT) of IgG anti-PA83 in this group was 80 enzyme-linked immunosorbent assay units (EU)/ml, i.e., ∼3-fold over the baseline represented by groups 1 and 2 (GMT = 25 EU/ml). The same live vector with ssb deleted and carrying the SSB-encoding homologous plasmid pSEC91S-83 (group 3) showed no rise in serum anti-PA83 IgG (GMT = 25 EU/ml, day 41, Table 4). However, the deletion of ssb in less attenuated live vector strain CVD 908ssb carrying pSEC91S-83 (group 4) showed an anti-PA83 response (GMT = 491 EU/ml) that was ∼20-fold higher than the response elicited by CVD 908-htrAssb(pSEC91S-83) (P < 0.0001), demonstrating the relevance of the live vector background in the priming of immune responses. The GMT elicited by CVD 908ssb(pSEC91S-83) was sixfold higher than that for conventional CVD 908-htrA(pSEC91-83) (P = 0.014).
TABLE 4.
Anti-PA83 IgG responses in mice immunized with CVD 908-htrAssb and CVD 908ssb live vectors carrying SSB-encoding medium and low-copy-number expression plasmidsa
| Groupb | Strain | Plasmid | SSB selection | Expected copy no.c | IgG GMTd |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 days | 13 days | 41 days | 48 days | 55 days | 70 days | |||||
| 1 | CVD 908-htrAssb | pSEC91SΔclyA | Yes | ∼15 | 25 | 25 | 25 | 54 | 1,600 | 22,902 |
| 2 | CVD 908ssb | pSEC91SΔclyA | Yes | ∼15 | 25 | 25 | 25 | 46 | 4,546 | 45,020 |
| 3 | CVD 908-htrAssb | pSEC91S-83 | Yes | ∼15 | 25 | 25 | 25A | 1,355F | 59,234K | 228,828 |
| 4 | CVD 908ssb | pSEC91S-83 | Yes | ∼15 | 25 | 25 | 491B | 59,247G | 200,019L | 318,855O |
| 5 | CVD 908-htrA | pSEC91-83 | No | ∼15 | 25 | 25 | 80C | 13,806H | 119,779 | 401,214 |
| 6 | CVD 908-htrAssb | pSEC10S-83 | Yes | ∼5 | 25 | 35 | 2,322D | 156,330I | 267,944M | 362,382P |
| 7 | CVD 908ssb | pSEC10S-83 | Yes | ∼5 | 25 | 25 | 4,929E | 500,790J | 704,882N | 813,063Q |
Animals received primary immunizations on days 0 and 14 and booster immunizations on day 42 as described in Materials and Methods. Uppercase superscript letters following data are used to indicate groups compared to generate the following P values: B versus A, P < 0.0001; B versus C, P = 0.014; D versus A, P < 0.0001; D versus C, P < 0.0001; E versus B, P = 0.002; E versus C, P < 0.0001; H versus F, P = 0.0003; I versus F, P < 0.0001; M versus K, P = 0.0006; J versus G, P = 0.021; N versus L, P = 0.023; Q versus O, P = 0.017; G versus F, P = 0.001; L versus K, P = 0.033; J versus I, P = 0.053; N versus M, P = 0.030; Q versus P, P = 0.051.
n = 10 for groups 1, 2 and 5; n = 12 for groups 3, 4, 6, and 7.
Per chromosomal equivalent.
From twofold serial dilutions starting at 1:50; values are reported in enzyme-linked immunosorbent assay units per milliliter, and values of <50 were set to 25. Titers resulting from mucosal priming with live vector only (41 days) and 6 days after boosting with purified PA83 (48 days) are in boldface.
The copy number of the expression plasmid had an even greater influence on the serum antibody responses detected following mucosal immunization. CVD 908-htrAssb, which did not elicit anti-PA83 responses at day 41 when carrying the medium-copy-number SSB-encoding plasmid, was impressively immunogenic when carrying the ∼5-copy pSEC10S-83 plasmid compared with group 3 (group 6 versus group 3, P < 0.0001) and generated a GMT 29-fold higher than that of comparator group 5 (2,322 versus 80 EU/ml; P < 0.0001). Similarly, the GMT of mice immunized i.n. with CVD 908ssb(pSEC10S-83) (group 7) was 10-fold higher than that of mice immunized i.n. with CVD 908ssb(pSEC91S-83) (4,929 versus 491 EU/ml; P = 0.002). The impact of both the degree of attenuation and the reduced copy number of SSB-encoding plasmids on immune responses appeared to be synergistic, as the GMT of anti-PA83 in mice primed with CVD 908ssb(pSEC10S-83) was 62-fold higher than the GMT in the comparator group immunized with the conventional CVD 908-htrA(pSEC91-83) (4,929 versus 80 EU/ml; P < 0.0001). Thus, whether the live vector strain was CVD 908-htrAssb or CVD 908ssb, live vectors carrying the low-copy-number SSB-encoding plasmid elicited significantly higher antibody responses on day 41 following mucosal immunization compared with live vectors carrying the isogenic medium-copy-number plasmid.
Serologic responses following parenteral boosting with PA83.
Our proposed heterologous prime-boost vaccination strategy for protecting high-risk populations against anthrax is to prime immunologic memory in subjects mucosally vaccinated with S. Typhi live vector expressing PA83 so that they can mount a strong and rapid anamnestic response, even years later if needed, when boosted with a single dose of a PA83-based vaccine (12). Therefore, a primary readout in our experiments was the serum antitoxin response attained within 1 week after boosting. Within 6 days of the PA83 boost given on day 42, IgG titers rose ∼50- to 100-fold in all of the groups primed with live vectors expressing ClyA-PA83. No significant responses were observed in mice primed with empty vectors until about 2 weeks after the parenteral boost with PA83 (day 55), and these responses were still 13- to 440-fold lower than those of the ClyA-PA83-primed mice.
Following the parenteral boost with PA83, the IgG responses in mice primed with CVD 908-htrAssb(pSEC91S-83) were consistently lower than those of mice primed with conventional strain CVD 908-htrA(pSEC91-83) (group 3 versus group 5, P = 0.0005, Hotelling's T square), just as had been observed on day 41 following mucosal priming. On day 48, the IgG GMT in the group that received SSB-encoding pSEC91S-83 was 10-fold lower than that of mice that received conventional plasmid pSEC91-83 (1,355 versus 13,806, P = 0.0003), and these lower responses persisted until day 70 (P = 0.13).
When specifically comparing postboost serum responses as a function of the copy number of the expression plasmids, the low-copy-number SSB-maintained expression plasmids resulted in higher IgG GMTs following the parenteral boost compared with isogenic medium-copy-number plasmids. For CVD 908-htrAssb, the overall P value calculated for day 48 through day 70 was <0.0001 (Table 4, group 6 versus group 3). A low-copy-number plasmid carried by this strain was able to prime for higher antibody responses following the parenteral boost compared with the medium-copy-number plasmid, as observed on days 48 (P < 0.0001) and 55 (P = 0.0006). The differences were much smaller on day 70, when responses appeared to have reached a plateau (P = 0.39). For CVD 908ssb, the IgG responses in mice primed with the low-copy-number plasmid (group 7) were significantly higher after the boost compared with those of mice primed with the medium-copy-number plasmid (group 4) (overall, P = 0.049; day 48, P = 0.021; day 55, P = 0.023; day 70, P = 0.017).
When postboost serum responses were examined as a function of the attenuation of the live vector strain, responses improved when a given plasmid was carried by a less attenuated strain (CVD 908ssb), as opposed to a more attenuated derivative (CVD 908-htrAssb). PA83-specific IgG titers in mice immunized with CVD 908ssb(pSEC91S-83) were more than 40-fold higher on day 48 than those of mice immunized with CVD 908-htrAssb(pSEC91S-83) (group 4 versus group 3, P = 0.001). The GMTs were also higher on days 55 (P = 0.033) and 70, although at the latter time point, the difference between groups was not statistically significant (P = 0.74). When the copy number was reduced, mice primed with CVD 908ssb(pSEC10S-83) developed titers at all of the time points more than double the GMT of mice primed with CVD 908-htrAssb(pSEC10S-83), reaching their highest levels on day 70 with a GMT of 813,063 EU/ml (group 7 versus group 6: overall P = 0.085; day 48, P = 0.053; day 55, P = 0.030; day 70, P = 0.051).
Toxin-neutralizing activity of serum following immunization.
The functional capacity of these serum antibodies was examined by testing their ability to neutralize anthrax toxin in vitro (Table 5). No TNA activity was detected before boosting. However, significant TNA titers started to appear on day 55, reaching high levels on day 70, as responses progressively matured. The highest TNA GMTs for days 55 and 70 were seen in mice primed with CVD 908ssb(pSEC10S-83), although not significantly different overall from any group of mice receiving vaccine except those primed with CVD 908-htrAssb(pSEC91S-83) (group 7 versus group 3; P = 0.033). Despite the lack of apparent differences in toxin-neutralizing capacity among groups, the TNA titers paralleled the IgG responses, as shown by correlation analysis; the Spearman correlation coefficient between IgG and TNA titers was 0.62 at day 55 and 0.80 at day 70.
TABLE 5.
TNA responses in mice immunized with CVD 908-htrAssb and CVD 908ssb live vectors carrying SSB-encoding expression plasmidsa
| Group | Strain | Plasmid | TNA GMTb |
||||
|---|---|---|---|---|---|---|---|
| 0 days | 41 days | 48 days | 55 days | 70 days | |||
| 1 | CVD 908-htrAssb | pSEC91SΔclyA | 5c | 5 | 5 | 7 | 35 |
| 2 | CVD 908ssb | pSEC91SΔclyA | 5 | 5 | 5 | 9 | 42 |
| 3 | CVD 908-htrAssb | pSEC91S-83 | 5 | 5 | 6 | 40 | 198 |
| 4 | CVD 908ssb | pSEC91S-83 | 5 | 5 | 7 | 30 | 135 |
| 5 | CVD 908-htrA | pSEC91-83 | 5 | 5 | 7 | 58 | 268 |
| 6 | CVD 908-htrAssb | pSEC10S-83 | 5 | 5 | 8 | 57 | 230 |
| 7 | CVD 908ssb | pSEC10S-83 | 5 | 5 | 14 | 72 | 442 |
DISCUSSION
A variety of strategies have been used to engineer plasmid selection and maintenance systems that no longer rely on conventional techniques of selection using resistance to antibiotics. Direct selection has been reported with substances unrelated to antibiotics which are nevertheless lethal to bacteria in the absence of plasmid-encoded protective proteins. Examples include selection with small bactericidal peptides called microcins, where plasmids express a microcin-binding inhibitor (9), and selection using a biocide such as triclosan, which lethally inhibits an essential gene involved in fatty acid synthesis unless this essential target is overexpressed on a multicopy plasmid (17). A further variation of this theme of expressing essential genes on plasmids for selection and maintenance purposes is the conditional lethal approach, in which an essential gene is deleted from the live vector chromosome and placed onto multicopy plasmids. This powerful technique has many examples, including a widely used “balanced lethal” system in which chromosomally deleted aspartate β-semialdehyde dehydrogenase (Asd), an enzyme critical for the synthesis of the bacterial cell wall, is complemented by multicopy plasmids (11, 22, 26, 38). A variation of the balanced lethal approach involves a lactose repressor-titration control circuit in which repression of an essential chromosomal gene is prevented by plasmids which titrate off chromosomally bound repressor (5, 16).
In our view, an ideal plasmid selection system intended for use in live vector vaccines should fulfill a number of important requirements, including that (i) implementation be simple with few genetic manipulations of the vaccine strain, (ii) plasmidless bacteria can be grown on appropriately supplemented media, (iii) the selection system is invulnerable to mutations, (iv) selection also ensures maintenance of plasmids in vivo (i.e., after immunization), and (v) maintenance of multiple copies of an expression plasmid is favored. Arguably, the currently available selection system coming closest to meeting these requirements is the Asd balanced lethal system, in which chromosomal deletion of asd can be efficiently implemented by using newly available and highly efficient chromosomal mutagenesis techniques (7) and plasmidless mutants can be grown on media supplemented with the inexpensive metabolite diaminopimelic acid (DAP) prior to plasmid introduction. This system is also invulnerable to mutation, since inactivation of plasmid-encoded Asd causes lysis of bacteria in the absence of DAP. Since DAP is not available in mammalian tissues, plasmids are efficiently maintained in vivo after immunization. However, the Asd balanced lethal system relies on plasmid-based complementation of an enzyme with catalytic activity. Therefore, maintenance of only one copy of the plasmid is a concern unless additional modifications of the complementing gene are implemented to functionally reduce the synthesis of Asd and theoretically enhance the retention of more copies of the expression plasmid (20).
Here we described a novel attempt to sidestep the theoretical limitations of enzyme-based plasmid selection systems by developing a method that exploits the noncatalytic SSB protein, which is essential to DNA replication, recombination, and repair (4, 30). Plasmid selection using SSB was first reported more than 2 decades ago by Porter et al. (33) in experiments involving bioreactors. They observed that the frequency of plasmid loss was less than 1 × 10−7 in strains grown in continuous culture under nonselective conditions for >6 days; this frequency was independent of the copy number, as both lower-copy-number pACYC184 plasmids and very high-copy-number pUC19 plasmids were maintained efficiently. The plasmids, however, encoded a drug resistance marker in addition to the SSB protein.
Our work shows, for the first time, the efficient use of SSB for maintenance of plasmids expressing a relevant protective antigen in S. Typhi live vector-based vaccines. These candidate vaccine strains carry multicopy expression plasmids that are maintained in the absence of antibiotics and theoretically cannot be lost either in vitro or in vivo due to spontaneous mutation of the SSB gene.
We examined the stability in vitro of both medium-copy-number (ori15A replicons) and low-copy-number (ori101 replicons) plasmids encoding a green fluorescent reporter protein (GFPuv) and found that the percentage of fluorescing colonies increased as the copy number was reduced; interestingly, the generation times also improved (i.e., live vectors were able to grow more quickly). In addition, strains carrying SSB-encoding plasmids maintained their fluorescence capacity longer than strains carrying conventional plasmids encoding antibiotic resistance. If we accept the hypothesis that it is theoretically impossible for SSB-maintained plasmids to be lost from live vectors in which chromosomal ssb has been deleted, then loss of fluorescence during strain passage can only be explained by spontaneous inactivation of gfpuv, which was confirmed by PCR analysis of rearranged plasmids recovered from nonfluorescing colonies. To date, we have not been able to verify by PCR analysis the loss of SSB-maintained replicons from either strain CVD 908-htrAssb or CVD 908ssb. In fact, we believe that the stability of our expression plasmids may now enable us to detect previously rare plasmid rearrangements that were not observed from populations of bacteria carrying conventional plasmids.
Based on our in vitro analysis of antigen expression using immunoblot assays, we anticipated only slightly higher antibody responses when PA83 was delivered by live vectors carrying medium-copy-number conventional plasmids, as opposed to SSB-encoding medium-copy-number plasmids. Surprisingly, mice mucosally primed with the conventional CVD 908-htrA(pSEC91-83) strain and boosted with PA83 displayed PA-specific IgG titers 10-fold higher than those of mice primed with the new CVD 908-htrAssb(pSEC91S-83) strain. However, when the copy number was lowered from 15 to 5 per cell, the effect was reversed and the immune responses of the latter group increased 10-fold versus those of the conventional group, demonstrating that serologic responses can be improved if vaccine antigens are delivered by live vectors using lower-copy-number expression plasmids. This effect was further enhanced when using the less attenuated CVD 908ssb strain for antigen delivery. Indeed, given all of the strains and plasmid combinations tested in this study, the highest PA-specific IgG levels were achieved when low-copy-number pSEC10S-83 was carried by the less attenuated CVD 908ssb vaccine strain. It is important to note that CVD 908-htrAssb and CVD 908ssb are isogenic strains, differing only in the deletion of a single htrA chromosomal locus. The fact that pSEC10S-83 produced higher antibody responses in a less attenuated CVD 908ssb strain suggests that any metabolic burden still associated with pSEC10-83S may be ameliorated by the lower attenuation of CVD 908ssb.
In conclusion, we have constructed and tested a novel nonantibiotic plasmid stabilization system for live vectors based upon the synthesis of the essential SSB protein required for DNA metabolism. Since plasmid loss is lethal for transformed live vectors, the SSB system is expected to allow for stable maintenance of antigen expression in vivo following immunization. We demonstrated using SSB-stabilized plasmids that immune responses to a foreign antigen dramatically improve when the copy number is reduced and that this effect is further enhanced when the antigen is delivered by a less attenuated live vector. This newly developed expression platform is expected to improve the immunogenicity of otherwise problematic antigens that affect the fitness of live vectors in vivo. It may also prove an effective means to enhance immune responses to foreign antigens delivered by S. Typhi live vector vaccines in humans.
Acknowledgments
This research was supported by grant 5 RO1 AI29471, research contract NO1 AI45251, and Mid-Atlantic Regional Center for Excellence (MARCE) for Biodefense and Emerging Infectious Diseases Research grant U54 AI57168 (to M. M. Levine).
We thank Sharon Tennant for critical reading of the manuscript.
This work is dedicated to the memory of Ellen Galen and James F. Galen Jr., who always followed the research enthusiastically.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 2 November 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Baillie, L. W., A. L. Rodriguez, S. Moore, H. S. Atkins, C. Feng, J. P. Nataro, and M. F. Pasetti. 2008. Towards a human oral vaccine for anthrax: the utility of a Salmonella Typhi Ty21a-based prime-boost immunization strategy. Vaccine 26:6083-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, P. M. 2008. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153(Suppl. 1):S347-S357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 4.Chase, J. W., and K. R. Williams. 1986. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 55:103-136. [DOI] [PubMed] [Google Scholar]
- 5.Cranenburgh, R. M., K. S. Lewis, and J. A. Hanak. 2004. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 7:197-203. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, K. H., D. D. Mater, M. J. Flores, P. J. Johnsen, T. Midtvedt, G. Corthier, and A. Sundsfjord. 2007. Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptide selection. J. Antimicrob. Chemother. 59:478-486. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, C. M., J. Y. Wang, M. Chinchilla, M. M. Levine, W. C. Blackwelder, and J. E. Galen. 2008. Use of mchI encoding immunity to the antimicrobial peptide microcin H47 as a plasmid selection marker in attenuated bacterial live vectors. Infect. Immun. 76:4422-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feld, L., S. Schjorring, K. Hammer, T. R. Licht, M. Danielsen, K. Krogfelt, and A. Wilcks. 2008. Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J. Antimicrob. Chemother. 61:845-852. [DOI] [PubMed] [Google Scholar]
- 11.Galán, J. E., K. Nakayama, and R. Curtiss III. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94:29-35. [DOI] [PubMed] [Google Scholar]
- 12.Galen, J. E., M. Chinchilla, M. F. Pasetti, J. Y. Wang, L. Zhao, I. Arciniega-Martinez, D. J. Silverman, and M. M. Levine. 2009. Mucosal immunization with attenuated Salmonella enterica serovar Typhi expressing protective antigen from anthrax toxin (PA83) primes monkeys for accelerated serum antibody responses to parenteral PA83 vaccine. J. Infect. Dis. 199:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galen, J. E., L. Zhao, M. Chinchilla, J. Y. Wang, M. F. Pasetti, J. Green, and M. M. Levine. 2004. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun. 72:7096-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galen, J. E., O. G. Gomez-Duarte, G. Losonsky, J. L. Halpern, C. S. Lauderbaugh, S. Kaintuck, M. K. Reymann, and M. M. Levine. 1997. A murine model of intranasal immunization to assess the immunogenicity of attenuated Salmonella typhi live vector vaccines in stimulating serum antibody responses to expressed foreign antigens. Vaccine 15:700-708. [DOI] [PubMed] [Google Scholar]
- 15.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garmory, H. S., M. W. Leckenby, K. F. Griffin, S. J. Elvin, R. R. Taylor, M. G. Hartley, J. A. Hanak, E. D. Williamson, and R. M. Cranenburgh. 2005. Antibiotic-free plasmid stabilization by operator-repressor titration for vaccine delivery by using live Salmonella enterica serovar Typhimurium. Infect. Immun. 73:2005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh, S., and L. Good. 2008. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotechnol. 8:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haines, A. S., P. Akhtar, E. R. Stephens, K. Jones, C. M. Thomas, C. D. Perkins, J. R. Williams, M. J. Day, and J. C. Fry. 2006. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689-2701. [DOI] [PubMed] [Google Scholar]
- 19.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9:810-816. [DOI] [PubMed] [Google Scholar]
- 20.Kang, H. Y., and R. Curtiss III. 2003. Immune responses dependent on antigen location in recombinant attenuated Salmonella typhimurium vaccines following oral immunization. FEMS Immunol. Med. Microbiol. 37:99-104. [DOI] [PubMed] [Google Scholar]
- 21.Karami, N., A. Martner, V. I. Enne, S. Swerkersson, I. Adlerberth, and A. E. Wold. 2007. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J. Antimicrob. Chemother. 60:1142-1145. [DOI] [PubMed] [Google Scholar]
- 22.Karem, K. L., J. Bowen, N. Kuklin, and B. T. Rouse. 1997. Protective immunity against herpes simplex virus (HSV) type 1 following oral administration of recombinant Salmonella typhimurium vaccine strains expressing HSV antigens. J. Gen. Virol. 78:427-434. [DOI] [PubMed] [Google Scholar]
- 23.Khan, S., S. Chatfield, R. Stratford, J. Bedwell, M. Bentley, S. Sulsh, R. Giemza, S. Smith, E. Bongard, C. A. Cosgrove, J. Johnson, G. Dougan, G. E. Griffin, J. Makin, and D. J. Lewis. 2007. Ability of SPI2 mutant of S. typhi to effectively induce antibody responses to the mucosal antigen enterotoxigenic E. coli heat labile toxin B subunit after oral delivery to humans. Vaccine 25:4175-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotloff, K. L., M. F. Pasetti, E. M. Barry, J. P. Nataro, S. S. Wasserman, M. B. Sztein, W. D. Picking, and M. M. Levine. 2004. Deletion in the Shigella enterotoxin genes further attenuates Shigella flexneri 2a bearing guanine auxotrophy in a phase 1 trial of CVD 1204 and CVD 1208. J. Infect. Dis. 190:1745-1754. [DOI] [PubMed] [Google Scholar]
- 25.Kotloff, K. L., J. K. Simon, M. F. Pasetti, M. B. Sztein, S. L. Wooden, S. Livio, J. P. Nataro, W. C. Blackwelder, E. M. Barry, W. Picking, and M. M. Levine. 2007. Safety and immunogenicity of CVD 1208S, a live, oral ΔguaBA Δsen Δset Shigella flexneri 2a vaccine grown on animal-free media. Hum. Vaccin. 3:268-275. [DOI] [PubMed] [Google Scholar]
- 26.Kotton, C. N., A. J. Lankowski, N. Scott, D. Sisul, L. M. Chen, K. Raschke, G. Borders, M. Boaz, A. Spentzou, J. E. Galán, and E. L. Hohmann. 2006. Safety and immunogenicity of attenuated Salmonella enterica serovar Typhimurium delivering an HIV-1 Gag antigen via the Salmonella type III secretion system. Vaccine 24:6216-6224. [DOI] [PubMed] [Google Scholar]
- 27.Levine, M. M., J. B. Kaper, H. Lockman, R. E. Black, M. L. Clements, and S. Falkow. 1983. Recombinant DNA risk assessment studies in humans: efficacy of poorly mobilizable plasmids in biologic containment. J. Infect. Dis. 148:699-709. [DOI] [PubMed] [Google Scholar]
- 28.Levine, M. M., J. E. Galen, E. M. Barry, F. Noriega, C. O. Tacket, M. Sztein, S. Chatfield, G. Dougan, G. Losonsky, and K. Kotloff. 1997. Attenuated Salmonella typhi and Shigella as live oral vaccines and as live vectors. Behring Inst. Mitt. 98:120-123. [PubMed] [Google Scholar]
- 29.Li, H., S. D. Soroka, T. H. Taylor, Jr., K. L. Stamey, K. W. Stinson, A. E. Freeman, D. R. Abramson, R. Desai, L. X. Cronin, J. W. Oxford, J. Caba, C. Pleatman, S. Pathak, D. S. Schmidt, V. A. Semenova, S. K. Martin, P. P. Wilkins, and C. P. Quinn. 2008. Standardized, mathematical model-based and validated in vitro analysis of anthrax lethal toxin neutralization. J. Immunol. Methods 333:89-106. [DOI] [PubMed] [Google Scholar]
- 30.Lohman, T. M., and M. E. Ferrari. 1994. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63:527-570. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, B., S. Schluederberg, C. Tachibana, and S. B. Levy. 1981. Survival and transfer in the human gut of poorly mobilizable (pBR322) and of transferable plasmids from the same carrier E. coli. Gene 14:145-154. [DOI] [PubMed] [Google Scholar]
- 32.Pickett, T. E., M. F. Pasetti, J. E. Galen, M. B. Sztein, and M. M. Levine. 2000. In vivo characterization of the murine intranasal model for assessing the immunogenicity of attenuated Salmonella enterica serovar Typhi strains as live mucosal vaccines and as live vectors. Infect. Immun. 68:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter, R. D., S. Black, S. Pannuri, and A. Carlson. 1990. Use of the Escherichia coli ssb gene to prevent bioreactor takeover by plasmidless cells. Bio/Technology 8:47-51. [DOI] [PubMed] [Google Scholar]
- 34.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 35.Stokes, M. G., R. W. Titball, B. N. Neeson, J. E. Galen, N. J. Walker, A. J. Stagg, D. C. Jenner, J. E. Thwaite, J. P. Nataro, L. W. Baillie, and H. S. Atkins. 2007. Oral administration of a Salmonella enterica-based vaccine expressing Bacillus anthracis protective antigen confers protection against aerosolized B. anthracis. Infect. Immun. 75:1827-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szpirer, C., E. Top., M. Couturier, and M. Mergeay. 1999. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology 145:3321-3329. [DOI] [PubMed] [Google Scholar]
- 37.Tacket, C. O., D. M. Hone, G. A. Losonsky, L. Guers, R. Edelman, and M. M. Levine. 1992. Clinical acceptability and immunogenicity of CVD 908 Salmonella typhi vaccine strain. Vaccine 10:443-446. [DOI] [PubMed] [Google Scholar]
- 38.Tacket, C. O., S. M. Kelley, F. Schodel, G. Losonsky, J. P. Nataro, R. Edelman, M. M. Levine, and R. Curtiss III. 1997. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the Asd-balanced lethal vector system. Infect. Immun. 65:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tacket, C. O., M. Sztein, S. S. Wasserman, G. Losonsky, K. Kotloff, T. L. Wyant, J. P. Nataro, R. Edelman, J. G. Perry, P. Bedford, D. Brown, S. Chatfield, G. Dougan, and M. M. Levine. 2000. Phase 2 clinical trial of attenuated Salmonella enterica serovar Typhi oral live vector vaccine CVD 908-htrA in U.S. volunteers. Infect. Immun. 68:1196-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trobos, M., C. H. Lester, J. E. Olsen, N. Frimodt-Moller, and A. M. Hammerum. 2009. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J. Antimicrob. Chemother. 63:80-86. [DOI] [PubMed] [Google Scholar]
- 41.Wainwright, L. A., and J. B. Kaper. 1998. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol. Microbiol. 27:1247-1260. [DOI] [PubMed] [Google Scholar]
- 42.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]