Abstract
In Salmonella enterica serovar Typhimurium, trxA encodes thioredoxin 1, a small, soluble protein with disulfide reductase activity, which catalyzes thiol disulfide redox reactions in a variety of substrate proteins. Thioredoxins are involved as antioxidants in defense against oxidative stresses, such as exposure to hydrogen peroxide and hydroxyl radicals. We have made a defined, complete deletion of trxA in the mouse-virulent S. Typhimurium strain SL1344 (SL1344 trxA), replacing the gene with a kanamycin resistance gene cassette. SL1344 trxA was attenuated for virulence in BALB/c mice by the oral and intravenous routes and when used in immunization experiments provided protection against challenge with the virulent parent strain. SL1344 trxA induced less inflammation in murine spleens and livers than SL3261, the aroA mutant, live attenuated vaccine strain. The reduced splenomegaly observed following infection with SL1344 trxA was partially attributed to a reduction in the number of both CD4+ and CD8+ T cells and B lymphocytes in the spleen and reduced infiltration by CD11b+ cells into the spleen compared with spleens from mice infected with SL3261. This less severe pathological response indicates that a trxA mutation might be used to reduce reactogenicity of live attenuated vaccine strains. We tested this by deleting trxA in SL3261. SL3261 trxA was also less inflammatory than SL3261 but was slightly less effective as a vaccine strain than either the SL3261 parent strain or SL1344 trxA.
Serovars of Salmonella enterica cause various diseases in diverse hosts. For example S. enterica serovar Typhi causes typhoid fever exclusively in humans, while domestic animals can be infected with many different Salmonella serovars, causing gastroenteritis, asymptomatic carriage, or an invasive disease, not dissimilar to typhoid. A number of serovars that infect animals can also cause food poisoning and gastroenteritis in humans. Prevention of salmonellosis in domestic animals would remove a major source of this organism from the ecosystem and would contribute to reducing disease in humans (22, 38).
Vaccination is a promising strategy for the control of Salmonella infections in humans and domestic animals. Current licensed typhoid vaccines include killed whole-cell vaccines, with a history going back to the 1890s (40), as well as the more recent Vi polysaccharide capsule subunit vaccine preparations (16, 20). A major area of research over several decades has been directed at the development of live attenuated typhoid vaccines, of which the only licensed example is Ty21a, developed in the 1970s by using chemical random mutagenesis (10). Some of the best studied and most widely used attenuating single-gene mutations are those that cause auxotrophy for aromatic amino acids. Mutants in these genes can be used for immunization and protection against subsequent challenge with the virulent parent strain (8, 9). We have used a proprietary technology, transposon-mediated differential hybridization to identify mutations in S. Typhimurium that are attenuated in a mouse model of typhoid infection (5, 6, 21). One such attenuated strain contained a trxA mutation.
Two genes encoding thioredoxins (trxA, encoding Trx1, and trxC, encoding Trx2) are present in the Escherichia coli and Salmonella genomes (17, 19, 25, 26). Trx1 is a small, soluble protein that catalyzes thiol disulfide redox reactions in a variety of substrate proteins. Compared with Trx1, Trx2 in E. coli is 5-fold less abundant and is 2.5-fold less efficient as an enzyme (26). In E. coli, thioredoxin is an electron donor for ribonucleotide reductase, methionine sulfoxide reductase, and 3′-phosphoadenylylsulphate reductase. Reduced Trx1 and Trx2 are regenerated by thioredoxin reductase (encoded by trxB), which is in turn maintained in a reduced state by NADPH (1). Three glutaredoxins have been found in E. coli, some of which can substitute for thioredoxin in some of its reactions (26), and a trxA trxC grxA (glutaredoxin 1) triple mutant of E. coli is not viable unless complemented by one of the three genes on a plasmid (36). Thioredoxins are involved as antioxidants in defense against oxidative stress, such as exposure to hydrogen peroxide and hydroxyl radicals. Kumar et al. (18) identified the Trx1-targeted proteome of E. coli, which comprises 80 proteins implicating the involvement of Trx1 in at least 26 distinct cellular processes, including the oxidative stress response.
Bjur et al. (2) showed that a trxA mutant of S. Typhimurium was less able to invade and replicate within MDCK or macrophage-like J774 cells than the parent strain, but interestingly this did not correlate with an increased sensitivity of the mutant to oxidative and nitrosative compounds in vitro. The trxA mutant was attenuated for virulence when injected intraperitoneally into BALB/c mice in competition experiments at a 1:1 ratio with the wild-type parent strain, unlike trxC or trxB mutants in similar experiments, showing that Trx1 contributes to the virulence of S. Typhimurium (2). Negrea et al. (29) have recently reported that thioredoxin 1 is required for the proper activity of the Salmonella pathogenicity island (SPI2) type III secretion system. However, these authors did not investigate the pathology caused by infection with a trxA mutant or whether the mutant offered protection against subsequent wild-type challenge.
Here, we present the characterization of SL1344 trxA in terms of its attenuation in BALB/c mice as a single infection and the ability of the mutant to confer protection against oral or intravenous challenge with the virulent parent and the comparison of these phenotypes with the aroA mutant vaccine strain SL3261. SL3261 is derived from SL1344 and is also a histidine auxotroph. We found that SL1344 trxA induced less inflammation in murine spleens and livers than SL3261. This less severe pathological response indicates that a trxA mutation might be used to reduce reactogenicity of live attenuated vaccine strains. We tested this by deleting trxA in SL3261. SL3261 trxA was also less inflammatory than SL3261 but was slightly less effective as a vaccine strain than either the SL3261 parent strain or SL1344 trxA.
MATERIALS AND METHODS
Generation and complementation of S. Typhimurium trxA.
An S. Typhimurium trxA deletion mutant was constructed by a modification of the one-step inactivation procedure (7, 28), replacing the entire trxA gene with a kanamycin resistance gene cassette from pUC4Kan (Amersham). PCR was used to amplify the antibiotic resistance cassette with 5′ and 3′ 60-bp homology arms complementary to the flanking regions of trxA in the S. Typhimurium chromosome (primer sequences are given in Table 1). The mutation was constructed in S. Typhimurium LB5010 (4) as described by Paterson et al. (31), and allelic replacement of trxA was confirmed by PCR and DNA sequencing of the product. The trxA mutation generated in S. Typhimurium LB5010 was transduced by bacteriophage P22 (34) into the mouse-virulent strain SL1344 and into SL3261, an aroA mutant of SL1344 that is attenuated for virulence in mice (13). Transductants were selected on LB agar with 25 μg/ml kanamycin and were screened for agglutination with anti-04 serotype antibodies (Remel Europe Ltd.); in addition, the presence of the mutation was verified by PCR and sequencing, as described above, and by Southern hybridization with a kanamycin resistance gene cassette probe. For complementation studies, trxA from S. Typhimurium SL1344 was amplified by PCR (primer sequences are given in Table 1) and cloned into EcoRV/BamHI-cut pBR322 (New England Biolabs), downstream of the plasmid's tetracycline resistance gene promoter in order for transcription of trxA to be driven from this promoter. The presence of the correct insert in the resultant plasmid (pBR322-trxA) was confirmed by DNA sequencing, and then pBR322-trxA was transformed into SL1344 trxA. Empty pBR322 was also transformed into SL1344 trxA as a vector-only control.
TABLE 1.
Primer sequences
| Primer function | Primer sequence (5′ to 3′) |
|---|---|
| Generation of the trxA mutation | TCAGCACCTCGTTGGTTAATGCTACACCAACACGCCAGGCTTATTCCTGTGGAGTTATATGCCGCCGTCCCGTCAAGTCAGCGa |
| TTCGTCGAATGACAGACGCCTGACCATACAGCGCCTTTGTCATTCGACGTATAAAAGGTAGGGAAAGCCACGTTGTGTCTCa | |
| Generation of the trxB mutation | TTTACGTCTGTAAATTCCCTACAATTCTGCTCATTGTCTGCCAACAACTATGGGGATCTCGCCGCCGTCCCGTCAAGTCAGCGa |
| ACGCAAAAATAAAGGCGACTTATAGTCGCCTTTTTTACTTTTGTTACTGATTTGTAAAAAGGGAAAGCCACGTTGTGTCTCa | |
| Generation of the trxC mutation | AACATTATAGAAACATCCCGCGCGTAGCGGGACGTCTTCCGACGTATTCAGAGGTTAGCTGCCGCCGTCCCGTCAAGTCAGCGa |
| CGGAGAAAAACGCTATTGTGGCAGAGGTGAAAACGGGGCACAAGATGCGCCCCGTGGCGTGGGAAAGCCACGTTGTGTCTCa | |
| Verification of trxA mutant (test primers) | ATCGTCGATCGCGGGATGAGAAAGT |
| CACACCGTCGCCAAAGATATCTTCG | |
| Verification of trxB mutant (test primers) | GCTATTACACATATTGTTAAC |
| GCTATTACACATATTGTTAAC | |
| Verification of trxC mutant (test primers) | AAACACATTGTTGTTTAGCACAG |
| AACTGAAGGACAGCGTTATTGG | |
| Cloning trxA for complementation | CTGCAGGATATCTGAGTGGAGTTATATATGAGCGATAAAATTATTb |
| AAGCTTGGATCCTTACGCCAGATTGGCGTCc |
Sequence underlined is homologous to the kanamycin resistance gene cassette.
Sequence underlined is an EcoRV restriction site.
Sequence underlined is a BamHI restriction site.
Virulence of mutants in a mouse model of infection and protection against virulent challenge.
Bacteria were inoculated into LB broth, supplemented with 25 μg/ml kanamycin as appropriate, and left to stand overnight at 37°C. Bacteria were harvested and resuspended in phosphate-buffered saline (PBS), pH 7.5, and diluted to 5 × 103 or 5 × 105 CFU ml−1 for intravenous (i.v.) infections or to 5 × 108 CFU ml−1 for oral infections, and the viable count of the inoculum was confirmed by plating serial dilutions on LB agar. BALB/c mice (Harlan), 6 to 8 weeks old, were inoculated with 0.2 ml of the appropriate dilution of the bacterial suspension via the tail vein for i.v. infections or by oral gavage under anesthesia for oral infections. Mice were euthanized by cervical dislocation at each time point postinfection. The spleens and livers of intravenously infected mice and spleens, livers, Peyer's patches, and mesenteric lymph nodes of orally infected mice were removed, placed in 10 ml of sterile distilled water, and homogenized using a Stomacher 80 Lab System (Seward). Viable bacterial counts were quantified by plating various dilutions of homogenized organs in LB agar. For some experiments, spleens and livers were weighed, half of each organ (known weight) was homogenized, dilutions were plated for viable counts, and half was used for histopathological and immunofluorescence studies (see below).
For protection experiments, 6- to 8-week old BALB/c mice (Harlan) were immunized i.v. with 1.0 × 105 CFU per mouse of SL3261, SL1344 trxA, or SL3261 trxA, grown as above, and later challenged with the virulent parent strain (SL1344), either i.v. (104 CFU per mouse) or orally (108 CFU per mouse). SL3261 is a well-studied vaccine strain and was used as a control (13). Mice were challenged at least 15 weeks after immunization, and 1 day before the challenge, two mice per group were euthanized, and spleen and liver homogenates were plated on LB agar to ensure that no immunizing bacteria remained. Nonimmunized, age-matched mice were challenged in the same way as the immunized mice. Organs from three or four mice per group at each time point postchallenge were plated for bacterial viable counts as above. All animal procedures were licensed under the Animals (Scientific Procedures) Act 1986 of the United Kingdom.
Analysis of spleen cell suspensions by flow cytometry.
Single-cell suspensions were obtained from pieces of spleen of known weight by enzymatic digestion with 0.5% collagenase in Hanks balanced salt solution at 37°C for 20 to 30 min with shaking, followed by passage through a 70-μm-pore-size nylon strainer (Becton Dickinson). The cell suspensions were washed in cold PBS, followed by centrifugation at 300 × g for 5 min and incubation of the cell pellet in red cell lysing buffer (Sigma) for 1 to 2 min on ice. Cells were washed and resuspended in cold PBS containing 0.1% bovine serum albumin (BSA) and 0.01% NaN3 (PBA) and passed through a 40-μm-pore-size nylon strainer. Splenocytes were counted by trypan blue exclusion, and the final concentration was adjusted to 5 × 106 cells/ml in PBA. Purified rat anti-mouse CD16/CD32, FcγIII/II receptor blocker, was added for 15 min to prevent nonspecific antibody binding. Fluorescein isothiocyanate (FITC)-conjugated anti-CD3 or anti-CD11c, phycoerythrin (PE)-Cy5-conjugated anti-CD4 or CD8α, and PE-conjugated anti-CD11b or anti-CD19 antibodies were used to stain splenocytes. All antibodies were purchased from BD Pharmingen. FITC-conjugated and PE- or PE-Cy5-conjugated antibodies were diluted 1:100 and 1:50, respectively, in PBS immediately prior to staining. Antibodies were added to splenocyte suspensions and left for 20 to 30 min on ice. After two further washes, cells were fixed with 1% paraformaldehyde for 10 min, washed again, and resuspended in PBA. Conjugated isotype standards were used as controls. Sixty thousand events were acquired on a FACSCalibur flow cytometer and analyzed with Cellquest software, version 3.3 (Becton Dickinson).
Histopathological analysis of the spleen and liver.
The left lateral or medial liver lobes and spleens of infected mice from the appropriate time points were harvested, weighed, and fixed in 10% phosphate-buffered formalin overnight before being embedded in paraffin. Sections (4 μm) were stained with hematoxylin and eosin (H&E) and examined by light microscopy. Inflammatory lesions in the liver formed by leukocyte infiltration were considered as foci of infection. Two sections 50 μm apart were examined, and the number of lesions per field was counted in 25 fields at ×200 magnification per section for each mouse. Twenty fields at ×10 magnification were examined to determine the lesion area. The lesion area was calculated using ImageJ software, version 1.37 (NIH public domain software). To determine the presence of specific cell types, liver tissues were fixed in 4% paraformaldehyde for 1 to 2 h, washed in PBS, and left in cold 20% sucrose in PBS at 4°C overnight (33, 35). Tissues were embedded in optimal cutting temperature (OCT) embedding medium (Raymond A. Lamb Laboratory Supplies) and frozen at −80°C. Sections (6 μm) were cut using a cryostat (Shandon) and were mounted onto X-tra Adhesive slides (SurgiPath) and left to air dry for 1 h. Sections were blocked in 10% normal goat serum (Dako) in PBS for 10 min before immunostaining. Rat anti-mouse CD45R/B220 (AbD Serotec), CD4, CD8α, and CD11b (BD Pharmingen) antibodies in 10% normal goat serum were incubated with sections at 4°C overnight. Primary antibodies were diluted as previously described by Whiteland et al. (39). The sections were washed over 1 h with three changes of PBS and then incubated with Alexa Fluor 568-conjugated goat anti-rat IgG (Molecular Probes, Invitrogen) for 45 min at room temperature. The sections were washed in PBS, excess fluid was blotted away, and they were mounted with Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories). Twenty-five random fields were analyzed from each of two sections at ×630 magnification using a fluorescence microscope (DM 6000B; Leica). The number of positively stained cells per field were counted. Isotype controls were used to confirm specific staining.
Anti-Salmonella antibodies assayed by ELISA.
Sonicated SL1344, the enzyme-linked immunosorbent assay (ELISA) capture antigen, was diluted in carbonate coating buffer (1.59 g/liter sodium carbonate, 2.93 g/liter sodium bicarbonate, pH 8.2) to 1 ×106 CFU/ml, based on the viable count of the original overnight culture. ELISA plates (Immunoplates; Nunc/Thermofisher Scientific) were coated overnight at 4°C with 100 μl of capture antigen solution. Plates were washed with washing buffer (PBS containing 0.05% [wt/vol] Tween 20), and wells were blocked with 300 μl/well of blocking buffer (PBS containing 0.05% [wt/vol] Tween 20 plus 1% bovine serum albumin) for 2 h at room temperature. Serial dilutions of heat-inactivated mouse serum were prepared in blocking buffer, and 100 μl was added to washed wells. Plates were incubated for 2 h at room temperature and washed; 100 μl/well biotinylated polyclonal goat anti-mouse immunoglobulins (Dako), diluted 1:1,000 in blocking buffer, was added to wells, and the plates were incubated for 1 h at room temperature. Following washing, 100 μl/well streptavidin (BD Bioscience), diluted 1:100 in blocking buffer, was added, and plates were incubated for 30 min at room temperature. Plates were then washed and developed with 100 μl of TMB (3,3′,5,5′-tetramethylbenzidine) substrate solution (BD Bioscience), and the reaction was stopped with the addition of 50 μl/well of 5N sulfuric acid. Absorbance was read at 450 nm.
Examination of LPS structure and activity.
Stationary-phase cultures (20 ml) were harvested and resuspended in 1 ml of PBS plus 250 μl of lipopolysaccharide (LPS) buffer 1 (0.1875 M Tris-HCl, pH 6.8, 6% [wt/vol] sodium dodecyl sulfate [SDS], 30% [wt/vol] glycerol), and 20 μl of this resuspended cell pellet was boiled for 5 min. Samples were allowed to cool, and 70 μl of LPS buffer 2 (0.0625 M Tris-HCl, pH 6.8, 0.1% [wt/vol] SDS, 10% [wt/vol] glycerol, 0.1% [wt/vol] bromophenol blue) was added along with proteinase K to a final concentration of 1.8 mg/ml. Samples were digested at 55°C overnight, separated by electrophoresis using Novex 16% Tricine gels (Invitrogen), and visualized by silver staining based on the method of Tsai and Frasch (37). Equal loading was based on the viable count of the original culture. Overnight cultures of SL1344 trxA or SL3261 in 10 ml of LB broth were washed in PBS, resuspended in 1 ml of PBS, and heated for 1 h 15 min at 70°C, and aliquots were stored at −80°C. Heat inactivation of bacteria was confirmed by spreading 200 μl of heated culture onto LB agar. A total of 2 × 105 immortalized bone marrow-derived macrophages from wild-type C57/BL6 mice or isogenic cells from tlr4−/− C57/BL6 mice (a gift of D. T. Golenbock and K. Hagpal) were treated with heat-killed bacterial cells at doses equal to multiplicities of infection (MOIs) of 0.01, 0.1, 1, 10, and 100, based on the viable count of the original culture prior to heat treatment. To determine inducible nitric oxide synthase (iNOS) activity, nitrite accumulation in the supernatant was measured by a Griess Reagent System (Promega) after 24 h. Cytotoxicity was measured by assaying lactate dehydrogenase release using a Cytotox 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the manufacturer's instructions.
Statistical analysis.
Data were analyzed using an unpaired Student's t test or one-way analysis of variance (ANOVA) as appropriate using GraphPad, version 4, with a P value of <0.05 considered as significant.
RESULTS
Generation of S. Typhimurium trxA mutants.
SL1344 trxA and SL3261 trxA mutants were constructed and then verified by PCR with primers 150 bp up- and downstream of the mutation (primer sequences are given in Table 1), followed by DNA sequencing of the wild-type and mutant PCR products and by Southern hybridization to confirm that there was only one insertion of the kanamycin resistance cassette into the bacterial chromosome (data not shown). SL1344 and SL1344 trxA colonies were identical on LB agar plates, and the two strains had identical growth kinetics in LB broth in both static and shaken cultures, as did SL3261 and SL3261 trxA (data not shown).
Attenuation, clearance, and complementation of SL1344 trxA in BALB/c mice.
Infection of BALB/c mice with 1.04 × 103 CFU of SL1344 by the i.v. route resulted in there being nearly 106 CFU in spleens and livers by 3 days postinfection (Fig. 1). In comparison, SL3261 (1.13 × 103 CFU per mouse) and SL1344 trxA (1.03 × 103 CFU per mouse) were attenuated for growth by days 1 and 3 postinfection. There was no statistically significant difference between the bacterial loads in the organs of mice infected with SL1344 trxA and those infected with SL3261 at either time point. trxB and trxC mutants constructed in the same way in SL1344 (primer sequences are given in Table 1) were not attenuated for virulence in this model of infection, in contrast to SL1344 trxA (data not shown). Clearance of SL1344 trxA following i.v. infection with 2 × 104 bacteria per mouse in comparison with SL3261 showed that SL3261 cleared from the spleen by day 49 and from the liver by day 35 postinfection but that clearance of SL1344 trxA, particularly from the spleen, took longer: bacteria were cleared from the livers of SL1344 trxA-infected mice by day 56 but were not cleared completely from the spleens by day 84 (12 weeks) postinfection (data not shown).
FIG. 1.
Attenuation of SL1344 by deletion of trxA. BALB/c mice were infected intravenously with 103 CFU of SL1344, SL3261, or SL1344 trxA. Bacterial counts in spleens and livers at days 1 and 3 postinfection are shown as the mean log10 CFU per organ with standard errors from one experiment that is representative of three experiments with similar results. *, P < 0.05; **, P < 0.001, versus infection with SL1344 by one-way ANOVA with Bonferroni's correction for multiple comparisons (n = 3).
A wild-type copy of trxA was cloned into pBR322 downstream of the tet promoter, and the resultant plasmid was transformed into SL1344 trxA. SL1344, SL1344 trxA, SL1344 trxA (pBR322-trxA), and SL1344 trxA (pBR322) were used to infect BALB/c mice with an i.v. dose of 103 CFU. Figure 2 shows the bacterial loads in the spleens and livers at days 1 and 3 postinfection. The presence of pBR322-trxA in SL1344 trxA restored its virulence to wild-type level. As expected, the vector-only control, SL1344 trxA (pBR322), was not restored to virulence and was as attenuated as the mutant strain.
FIG. 2.
Complementation of the SL1344 trxA mutation with a wild-type copy of trxA cloned into pBR322 (pBR322-trxA). BALB/c mice were infected intravenously with 103 CFU of SL1344, SL1344 trxA, SL1344 trxA (pBR322-trxA) (complemented strain), or SL1344 trxA containing pBR322 (vector control). Bacterial counts are shown as the mean log10 CFU per organ, with standard errors, in spleens (A) and livers (B). The experiment was repeated twice and gave similar results, which were then pooled. *, P < 0.05 versus SL1344 by one-way ANOVA with Bonferroni's correction for multiple comparisons, with n = 7 for all strains.
Oral infection with SL1344 trxA and complementation of SL1344 trxA with plasmid-borne trxA.
SL1344, SL1344 trxA, SL1344 trxA (pBR322-trxA), and SL1344 trxA (pBR322) were used to infect BALB/c mice with a dose of 108 CFU by oral gavage under anesthesia. The bacterial loads in spleens, livers, mesenteric lymph nodes, and Peyer's patches at a single time point (day 4) postinfection showed that SL1344 trxA was attenuated for virulence in all the organs in comparison with SL1344 and that complementation restored virulence (Fig. 3).
FIG. 3.
Complementation of the SL1344 trxA mutation after oral infection. BALB/c mice were infected orally with 108 CFU of SL1344, SL1344 trxA, SL1344 trxA (pBR322-trxA) (complemented strain), or SL1344 trxA containing pBR322 (vector control). Panels show the bacterial counts as the mean log10 CFU per organ, with standard errors, in spleens (A), livers (B), mesenteric lymph nodes (C), and Peyer's patches (PP; D) on day 4 postinfection. The experiment was repeated twice and gave similar results, which were then pooled. *, P < 0.05 versus SL1344 by one-way ANOVA with Bonferroni's correction for multiple comparisons with n = 7 to 10.
Reduced pathology in spleens and livers following infection with SL1344 trxA in comparison with SL3261.
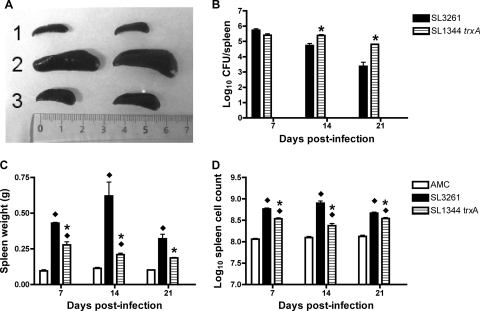
During experiments to characterize the virulence of SL1344 trxA, it was striking that the spleens of mice infected with SL1344 trxA were much smaller than those of mice infected with SL3261 although the bacterial counts in the organs were similar (Fig. 4). Spleen weight and spleen cellularity were compared between mice infected with SL1344 trxA or SL3261. Spleens were recovered from mice infected i.v. with 105 CFU of SL1344 trxA or SL3261 and age-matched uninfected controls, weighed, and plated for viable counts. At day 7 postinfection, there was no statistically significant difference between the mean bacterial loads per spleen of mice infected with SL1344 trxA or SL3261 (Fig. 4B), but there was a statistically significant difference between the mean weights of the spleens at this time point, with mice infected with SL3261 having heavier spleens than mice infected with SL1344 trxA (Fig. 4C). The difference in weights continued on days 14 and 21. However, at these time points there was also a statistically significant difference between the mean bacterial loads in spleens of mice infected with SL1344 trxA or SL3261. The mean bacterial count in spleens of mice infected with SL1344 trxA was higher than the count in those infected with SL3261 (Fig. 4B). Hence, at these time points, despite a higher bacterial load, the spleens of mice infected with SL1344 trxA were not as heavy, suggesting that they were infiltrated by fewer inflammatory cells. The total number of nucleated spleen cells per mouse showed that both groups of infected mice had significantly higher numbers of live spleen cells than age-matched controls (Fig. 4D). The total number of nucleated spleen cells was significantly increased in mice infected with SL3261 in comparison with those infected with SL1344 trxA at 7, 14, and 21 days postinfection. Examination of sections of spleen stained with H&E showed that increased cellularity in spleens was associated with a granulomatous inflammatory response within the marginal zone and that there were fewer inflammatory cells present in spleens of mice infected with SL1344 trxA than in the spleens of mice infected with SL3261 (data not shown). Granuloma-like lesions were reduced on days 7 and 14 postinfection in spleens infected with SL1344 trxA in comparison with those infected SL3261. From the planes of tissue examined, there was no structural damage in spleens of mice infected with SL3261 or with SL1344 trxA.
FIG. 4.
Reduced splenomegaly in mice infected with SL1344 trxA. (A) Gross pathology of spleens following intravenous infection with 105 CFU SL3261 or SL1344 trxA. Spleens were photographed 14 days postinfection. Row 1, age matched control (AMC) spleens, which weighed 0.099 g and 0.098g; row 2, SL3261-infected spleens (0.692 g and 0.831 g); row 3, SL1344 trxA-infected spleens (0.192 g and 0.260 g) (left and right, respectively). The bacterial counts (log10 CFU/spleen) from infected organs were (left and right, respectively) 4.45 and 4.60 for SL3261 and 4.99 and 5.00 for SL1344 trxA. (B) Bacterial loads as the mean log10 CFU per spleen (calculated from the bacterial counts and the known proportion of spleen used). (C) Spleen weights. (D) Mean log10 total live cell count per spleen (calculated from the cell counts and the known proportion of spleen used). Error bars indicate standard error (n = 3). The experiment was repeated twice and gave similar results. The SL1334 trxA value was significantly different (*) from that for SL3261 (P < 0.05); the SL1344 trxA or SL3261 values were significantly different (⧫) from values for the age-matched controls (AMC) (P < 0.05) in an unpaired t test.
Flow cytometry was used to analyze specific inflammatory cell populations in infected spleens (Fig. 5). There was a statistically significant increase in the numbers of CD3+, CD4+, CD8+, CD11b+, and CD11c+ cells that had infiltrated the spleen after SL3261 infection compared with age-matched controls on all days of the time course. However, at 7 days postinfection with SL1344 trxA, the numbers of CD3+ and CD3+ CD8+ stained cells were not significantly different from those in age-matched controls. Furthermore, the number of CD3+ CD4+ stained cells was significantly lower than the number of those cells in age-matched controls. At 14 days postinfection with SL1344 trxA, the numbers of CD3+, CD4+, CD8+, CD11b+, and CD11c+ stained cells were significantly higher than in age-matched controls, but, with the exception of CD4+ and CD8+ cells, the numbers were statistically significantly lower than in spleens infected with SL3261. By 21 days postinfection with SL1344 trxA, the numbers of CD4+ and CD8+ stained cells were not statistically significantly different from the numbers in age-matched controls, indicating a reduction in inflammation, while the number of these cells in spleens infected with SL3261 was still high. Anti-CD19+ antibodies were used to stain B-cell populations, which indicated that the number of B cells in the infected spleens was significantly higher than in age-matched controls over the time course but that the number of B cells in spleens infected with SL1344 trxA was significantly lower than in spleens infected with SL3261. This correlates with the level of anti-Salmonella antibodies in the serum at day 21 postinfection, where there was a significantly lower level of antibody in mice infected with SL1344 trxA than in mice infected with SL3261 although no difference between the antibody levels was seen at day 14 postinfection (data not shown).
FIG. 5.
Flow cytometric analysis of infected and uninfected spleen cell suspensions. Spleen cell suspensions from BALB/c mice infected intravenously with 105 CFU of SL3261 or SL1344 trxA or uninfected age-matched controls (n = 3) were stained with anti-CD3, anti-CD4, anti-CD8, anti-CD11b, anti-CD11c, and anti-CD19 antibodies. Total mononuclear cells were gated, and the percentage of stained cells in the gate was determined. The total number of each cell population in the spleen was calculated using the total number of cells in the spleen and the percentage of mononuclear cells stained with each stain. Data are presented as the mean log10 cell number with standard error. The experiment was repeated twice and gave similar results. The SL1334 trxA value was significantly different (*) from that for SL3261 (P < 0.05); the SL1344 trxA or SL3261 values were significantly different (⧫) from the value for the age-matched controls (P < 0.05) in an unpaired t test.
Livers were also recovered from infected mice. Half the organ was used for bacterial counts and half for H&E staining in order to examine the number and size of lesions produced after infection. The numbers of bacteria in the livers of mice infected with SL1344 trxA or SL3261 (Fig. 6A) were not statistically significantly different on days 14 and 21 postinfection, but on day 7, the bacterial load in livers infected with SL1344 trxA was significantly lower than the load in livers of mice infected with SL3261. The number of inflammatory lesions per field (Fig. 6B) and the area of the lesions (Fig. 6C) were both significantly higher in mice infected with SL3261 over the time course of the experiment than in mice infected with SL1344 trxA.
FIG. 6.
Reduced inflammation in the livers of mice infected with SL1344 trxA in comparison with SL3261. Bacterial loads (A), average number of liver lesions per field (B), and lesion area per field (C) in the liver following intravenous infection with 105 CFU of SL3261 or SL1344 trxA per mouse (n = 3). The viable counts of bacteria in the livers in panel A were comparable for each strain except on day 7, when the viable count of SL1344 trxA was significantly lower (*, P < 0.05) than for SL3261. Means with standard errors were compared in unpaired t tests (*, P < 0.05). The experiment was repeated twice and gave similar results.
The influx of T and B lymphocytes, neutrophils, and macrophages into the hepatic parenchyma and sinusoids during a 14-day infection was measured by counting the CD4+, CD8α+, CD11b+, and CD45R/B220+ cells per field (Fig. 7). There was statistically significantly more of each type of cell found in all infected livers in than in age-matched controls at both time points. On day 7 postinfection the numbers of CD4+ cells were similar in mice infected with SL3261 or SL1344 trxA, whereas at this time point infiltration of CD8α+ cells into SL1344 trxA-infected livers was significantly lower than in SL3261-infected livers. The infiltrating T-lymphocyte numbers decreased over the time course of infection with SL1344 trxA but remained elevated at day 14 in livers infected with SL3261. Significantly fewer CD11b+ cells infiltrated the livers of mice infected with SL1344 trxA on both days 7 and 14 than the livers of mice infected with SL3261. No statistically significant difference was observed in the CD45R/B220 cell populations in livers infected with either of the strains although there was a significant increase in B cells compared to age-matched uninfected control mice.
FIG. 7.
Inflammatory cell types infiltrating the liver following intravenous infection of BALB/c mice with 105 CFU of SL1344 trxA or SL3261. Fifty fields from two 6-μm liver sections from three mice per time point were stained with rat anti-mouse CD4, CD8α, CD11b, and CD45R/B220. Stained cells were counted, and the results are presented as mean numbers of cells per field with standard errors. The results shown are from one representative experiment from two independent experiments with similar results. The SL1334 trxA value was significantly different (*) from that for SL3261 (P < 0.05); the SL3261 and SL1344 trxA values were significantly different (⧫) from the value for the age-matched controls (P < 0.05) in an unpaired t test.
Structure and activity of LPS in SL1344 trxA.
It is possible that the reduced pathology and inflammation caused by infection with SL1344 trxA in comparison with SL3261 were due to alterations in the production or inflammatory activity of the LPS of SL1344 trxA. Analysis of silver-stained gels of cell lysates, prepared by digestion with proteinase K, revealed no gross difference in the LPS structures or production between SL1344 trxA and either SL1344 or SL3261 (data not shown). Furthermore, heat-killed SL1344 trxA or SL3261 induced similar levels of nitric oxide (NO) production from immortalized wild-type C57/BL6 bone marrow-derived macrophages (14) across a range of doses (MOI equivalents of approximately 0.01, 0.1, 1, 10, and 100) in assays to measure inducible NO synthase (iNOS) activity (12). NO production was abolished when isogenic cells derived from tlr4−/− mice were used, confirming that the main active component was LPS (data not shown). Likewise, comparison of SL1344 trxA with SL1344 revealed no significant difference in TLR-4-dependent NO production. There was no difference in toxicity between the bacterial strains when lactate dehydrogenase release from infected cells was measured. These data indicate that there are no overt differences in the functionality of the S. Typhimurium LPS molecule in response to trxA deletion.
Immunization with SL1344 trxA or SL3261 trxA and protection against challenge with the virulent parent strain.
The trxA mutation in SL1344 was transduced by bacteriophage P22 into SL3261, and the presence of the correct mutation was verified by PCR and DNA sequencing (data not shown). Similar to SL1344 trxA, SL3261 trxA was also less inflammatory than SL3261 following intravenous infection (data not shown). BALB/c mice were immunized i.v. with 1 × 105 CFU per mouse of SL1344 trxA, SL3261 trxA, or SL3261. After 20 weeks, prechallenge counts were performed on two mice per group to establish whether the infection had cleared. When no bacteria were present in the prechallenge counts, challenge with the virulent strain was undertaken. Immunized mice and age-matched, unimmunized controls were challenged with an i.v. dose of 1 × 104 CFU of SL1344 per mouse. Immunization with SL1344 trxA and SL3261 trxA resulted in protection of mice against this i.v. challenge (Fig. 8). Infections in unimmunized mice were allowed to proceed only to day 4 before mice were too ill to survive and were euthanized, and organs were recovered. In contrast, immunized mice were still well 14 days after challenge with fully virulent SL1344, showing that trxA mutants can be used as live attenuated vaccine strains when delivered intravenously to protect against a subsequent intravenous challenge. There was no statistically significant difference in the levels of anti-Salmonella antibody between all three immunizing strains at days 1 and 4 postchallenge, but there was a significantly higher level of antibody in the serum of mice immunized with SL1344 trxA at day 14 postchallenge (data not shown). The bacterial counts in the livers were statistically significantly lower on day 14 postchallenge in mice immunized with SL1344 trxA than in mice immunized with SL3261 trxA, showing that although SL3261 trxA is protective and the mice survived the challenge over the time course of the experiment, the double aroA trxA mutant provided somewhat lower protection. Hence, we investigated the potential only of SL1344 trxA to protect against oral challenge. Mice were immunized i.v. with 1.0 × 105 CFU of SL1344 trxA or SL3261 and challenged orally with 2 × 107 CFU of SL1344 per mouse 15 weeks later (Fig. 9). Infections in unimmunized mice were allowed to proceed only to day 7 before mice were too ill to survive and were euthanized. Immunized mice were well 14 days after oral challenge with SL1344, showing that SL1344 trxA can be used as a live attenuated vaccine strain to provide protection against an oral challenge with the virulent parent. In spleens and livers at day 14, there was no statistically significant difference between the mean bacterial counts in organs of mice immunized with SL1344 trxA or SL3261.
FIG. 8.
Intravenous immunization with SL1344 trxA, SL3261 trxA, or SL3261 and protection against subsequent intravenous challenge with virulent SL1344. Spleen (A) and liver (B) bacterial loads after immunization, followed 20 weeks later with an intravenous challenge with 104 CFU of SL1344. Bacterial counts are shown as the mean log10 and standard errors of one representative experiment from two with similar results. ⧫, all immunized groups were significantly different from the unimmunized group (P < 0.01); *, the SL3261-immunized group was significantly different from the groups immunized with the trxA mutants (P < 0.05), and the group immunized with SL3261 trxA was significantly different from the groups immunized with SL3261 or SL1344 trxA (P < 0.001); **, both the SL3261- and SL1344 trxA-immunized groups were not significantly different from each other (P > 0.05) but were significantly different from the group immunized with SL3261 trxA (P < 0.001). Data were analyzed using one-way ANOVA with Bonferroni's multiple comparison test with n = 4 or 5.
FIG. 9.
Intravenous immunization with SL1344 trxA or SL3261 and protection against subsequent oral challenge with virulent SL1344. Spleen and liver bacterial loads after immunization, followed 15 weeks later with an oral challenge with 2 × 107 CFU of SL1344. Bacterial counts are shown as the mean log10 and standard errors of one representative experiment from two with similar results. Data from days 4 and 7 were analyzed using one-way ANOVA with Bonferroni's multiple comparison test with n = 3, taking a P of <0.05 as significant. Viable counts in the spleens of the unimmunized group were significantly different (*) from those of the group immunized with SL3261 on day 4 postinfection. On day 4 in the liver, there was no significant difference between the counts from any of the groups. On day 7 in both organs, the unimmunized group was significantly different (⧫) from both immunized groups, which were not significantly different from each other. Data from day 14 were analyzed using an unpaired t test, which showed that there was no significant difference between the viable counts in the organs of mice immunized with SL1344 trxA or SL3261.
DISCUSSION
We have made a defined deletion of trxA in S. Typhimurium and investigated the virulence, pathology, and inflammation caused by this mutant and its potential as a live attenuated vaccine strain in a mouse typhoid model. SL1344 trxA was attenuated for virulence following intravenous or oral infection of BALB/c mice, resulting in a significant reduction in bacterial counts in mouse organs in comparison with infection with SL1344, the virulent parent strain. This is in agreement with the work of Bjur et al. (2), which showed that S. Typhimurium 14028 trxA was attenuated in competition with the parent strain in an intraperitoneal infection of BALB/c mice. We also showed that the attenuation phenotype of SL1344 trxA could be complemented in both intravenous and oral infections by the presence of a wild-type copy of trxA on plasmid pBR322. S. Typhimurium has two thioredoxin enzymes (Trx1 encoded by trxA and Trx2 encoded by trxC) and a thioredoxin reductase (encoded by trxB) (1). However, it is only mutation of trxA that results in attenuation of S. Typhimurium in this mouse infection model. S. Typhimurium trxB and trxC mutants are as virulent as their wild-type parent both in competitive intraperitoneal infection of BALB/c mice (2) and in our hands in the intravenous infection model used here (data not shown). In E. coli, Trx2 is both less abundant and less efficient as an enzyme than Trx1 (26), and the lack of trxA leads to increased transcription of trxB and trxC (32). But if this is also the case in S. Typhimurium, it does not make up for the effect of the loss of trxA on virulence.
A striking observation of the gross pathology of mice infected with SL1344 trxA was that the spleens were much smaller than those of mice infected with an equivalent bacterial load of SL3261. The weights of spleens from mice infected with SL3261 were significantly higher than those infected with SL1344 trxA, particularly 14 days postinfection, even though the bacterial load of SL3261 in the spleens was lower than that of SL1344 trxA. The total number of spleen cells in mice infected with SL3261 was also significantly higher 14 days postinfection than in mice infected with SL1344 trxA. SL1344 trxA took longer to clear from both spleens and livers than SL3261 over a time course of 84 days. This reduced rate of clearance could be because SL1344 trxA is less inflammatory than SL3261, leading to a reduction in recruitment and activation of mononuclear cells, resulting in inefficient clearance of bacteria from the organs.
To investigate the basis of the reduced splenomegaly and reduced pathological changes seen in the spleens and livers of mice infected with SL1344 trxA, we examined the inflammatory cell influx into these organs. Flow cytometric analysis of infected spleens was used to identify and quantify the different cell types present during infection. The number of spleen cells stained by T-cell marker antibodies against CD3, CD4, and CD8 was significantly higher in mice infected with SL3261 than in those infected with SL1344 trxA. We used a CD11b marker in conjunction with a CD11c marker to target the dendritic cell population. S. Typhimurium can reside in CD11b+ cells (24), and splenic dendritic cell populations are activated during S. Typhimurium infection (41). The CD11b+ and CD11b+/CD11c+ populations were significantly increased in spleens infected with SL3261 in comparison with those from animals infected with SL1344 trxA. Consistent with this finding was the presence of more granuloma-like inflammatory lesions in H&E-stained sections of spleens infected with SL3261, especially on days 7 and 14 postinfection, than in those infected with SL1344 trxA (data not shown).
Protective immunity to S. Typhimurium infection depends to some extent on B cells (23). In this study, we observed a significant increase in cells stained with anti-CD19, a B-cell marker, in spleens infected with SL1344 trxA or SL3261 in comparison with uninfected animals, but the recruitment of CD19+ B cells was significantly higher following infection with SL3261 than with SL1344 trxA.
Previous studies have reported the occurrence of splenomegaly and increased spleen cellularity after infection with S. Typhimurium in mice (15, 27, 30), and, consistent with our findings, that maximal splenomegaly coincides with the start of bacterial clearance (15). It has been proposed that splenomegaly and concomitantly increased numbers of T cells and macrophages are required for protection against subsequent challenge (30); an aroA mutant which induced splenomegaly was protective against virulent challenge, whereas a single purA mutant and a double aroA purA mutant induced much less splenomegaly and were not protective. Although SL1344 trxA infection resulted in reduced splenomegaly and less inflammation in spleens and livers than infection with SL3261, using SL1344 trxA as a live attenuated vaccine strain conferred protection against a lethal infection with wild-type bacteria, delivered orally or intravenously. The less severe pathological response to infection with the trxA mutant suggests that a trxA mutation could be used to reduce reactogenicity associated with live attenuated vaccine strains. However, SL3261 trxA is less effective as a vaccine strain than SL3261 or SL1344 trxA, showing that the combination of aroA and trxA mutations in a live attenuated vaccine strain may not be ideal, whereas a single trxA mutant protects as well as a single aroA mutant against intravenous and oral challenge with the wild-type strain.
In the liver, there were significantly more lesions per field, and the area of the lesions was significantly larger in mice infected with SL3261 than in mice infected with SL1344 trxA over a time course of 21 days. The influx of T (CD4+ or CD8α+) lymphocytes into the liver was consistent with the findings in the spleen, with increased numbers in mice infected with either SL1344 trxA or SL3261 compared to uninfected controls. The expansion of T-cell populations was particularly pronounced in response to infection with SL3261 14 days postinfection. The CD11b+ (neutrophil and macrophage) population in the livers was significantly increased during infections with SL3261 in comparison with infections with SL1344 trxA, whereas the number of B cells was similar in livers infected with either SL3261 or SL1344 trxA.
The much-reduced inflammation, both grossly and microscopically, in mice infected with SL1344 trxA indicates that deleting trxA alters the relationship of the bacteria with the host in terms of the generation of proinflammatory signals. Fewer lesions were formed in the liver by SL1344 trxA than by SL3261. We are continuing to study the distribution of bacteria in these lesions and the differences in dissemination within the organ between the different mutants in the light of our recent work on within-host dynamics and mathematical modeling of Salmonella infection in mice (3, 11, 35).
Acknowledgments
A BBSRC Applied Genomics Link Grant, a BBSRC Project Grant, and a National University of Singapore Scholarship to E.S.D.B. funded this work.
We acknowledge the work of Elizabeth Maxim during the early stages of this project. We are grateful to Madeline Fordham and Tessa Hoather for assistance with immunohistochemical staining, T. J. McKinley and Olivier Restif for help with the statistical analysis of the data, and D. T. Golenbock and K. Hagpal, University of Massachusetts Medical School, Worcester, MA, for supplying the immortalized BMDM from wild-type and tlr4−/− mice.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Aslund, F., and J. Beckwith. 1999. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J. Bacteriol. 181:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjur, E., S. Eriksson-Ygberg, F. Aslund, and M. Rhen. 2006. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 74:5140-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. P., S. J. Cornell, M. Sheppard, A. J. Grant, D. J. Maskell, B. T. Grenfell, and P. Mastroeni. 2006. Intracellular demography and the dynamics of Salmonella enterica infections. PLoS Biol. 4:e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullas, L. R., and J. I. Ryu. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charles, I. G., and D. J. Maskell. September 2001. Transposon mediated differential hybridization. International patent WO2001/007651.
- 6.Chaudhuri, R. R., S. E. Peters, S. J. Pleasance, H. Northen, C. Willers, G. K. Paterson, D. Cone, A. G. Allen, P. J. Owen, G. Shalom, D. Stekel, I. G. Charles, and D. J. Maskell. 2009. Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougan, G., S. Chatfield, D. Pickard, J. Bester, D. O'Callaghan, and D. Maskell. 1988. Construction and characterization of vaccine strains of Salmonella harboring mutations in two different aro genes. J. Infect. Dis. 158:1329-1335. [DOI] [PubMed] [Google Scholar]
- 9.Dougan, G., D. Maskell, D. Pickard, and C. Hormaeche. 1987. Isolation of stable aroA mutants of Salmonella typhi Ty2: properties and preliminary characterisation in mice. Mol. Gen. Genet. 207:402-405. [DOI] [PubMed] [Google Scholar]
- 10.Gilman, R. H., R. B. Hornick, W. E. Woodard, H. L. DuPont, M. J. Snyder, M. M. Levine, and J. P. Libonati. 1977. Evaluation of a UDP-glucose-4-epimeraseless mutant of Salmonella typhi as a live oral vaccine. J. Infect. Dis. 136:717-723. [DOI] [PubMed] [Google Scholar]
- 11.Grant, A. J., O. Restif, T. J. McKinley, M. Sheppard, D. J. Maskell, and P. Mastroeni. 2008. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, L. C., D. A. Wagner, J. Glogowski, P. L. Skipper, J. S. Wishnok, and S. R. Tannenbaum. 1982. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 126:131-138. [DOI] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 14.Hornung, V., F. Bauernfeind, A. Halle, E. O. Samstad, H. Kono, K. L. Rock, K. A. Fitzgerald, and E. Latz. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killar, L. M., and T. K. Eisenstein. 1985. Immunity to Salmonella typhimurium infection in C3H/HeJ and C3H/HeNCrlBR mice: studies with an aromatic-dependent live S. typhimurium strain as a vaccine. Infect. Immun. 47:605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kossaczka, Z., F. Y. Lin, V. A. Ho, N. T. Thuy, P. Van Bay, T. C. Thanh, H. B. Khiem, D. D. Trach, A. Karpas, S. Hunt, D. A. Bryla, R. Schneerson, J. B. Robbins, and S. C. Szu. 1999. Safety and immunogenicity of Vi conjugate vaccines for typhoid fever in adults, teenagers, and 2- to 4-year-old children in Vietnam. Infect. Immun. 67:5806-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotani, H., and K. Nakajima. 1992. Cloning and sequence of thioredoxin gene of Salmonella typhimurium LT2. Nucleic Acids Res. 20:1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, J. K., S. Tabor, and C. C. Richardson. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 101:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurent, T. C., E. C. Moore, and P. Reichard. 1964. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239:3436-3444. [PubMed] [Google Scholar]
- 20.Lin, F. Y., V. A. Ho, H. B. Khiem, D. D. Trach, P. V. Bay, T. C. Thanh, Z. Kossaczka, D. A. Bryla, J. Shiloach, J. B. Robbins, R. Schneerson, and S. C. Szu. 2001. The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 344:1263-1269. [DOI] [PubMed] [Google Scholar]
- 21.Maskell, D. J., I. G. Charles, A. Allen, and P. Owen. December 2003. Transposon. International patent WO2003/074700.
- 22.Mastroeni, P., J. A. Chabalgoity, S. J. Dunstan, D. J. Maskell, and G. Dougan. 2001. Salmonella: immune responses and vaccines. Vet. J. 161:132-164. [DOI] [PubMed] [Google Scholar]
- 23.Mastroeni, P., C. Simmons, R. Fowler, C. E. Hormaeche, and G. Dougan. 2000. Igh-6−/− (B-cell-deficient) mice fail to mount solid acquired resistance to oral challenge with virulent Salmonella enterica serovar Typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect. Immun. 68:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui, H., M. Eguchi, and Y. Kikuchi. 2000. Use of confocal microscopy to detect Salmonella typhimurium within host cells associated with Spv-mediated intracellular proliferation. Microb. Pathog. 29:53-59. [DOI] [PubMed] [Google Scholar]
- 25.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 26.Miranda-Vizuete, A., A. E. Damdimopoulos, J. Gustafsson, and G. Spyrou. 1997. Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J. Biol. Chem. 272:30841-30847. [DOI] [PubMed] [Google Scholar]
- 27.Mittrucker, H. W., A. Kohler, and S. H. Kaufmann. 2002. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect. Immun. 70:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo, E., S. E. Peters, C. Willers, D. J. Maskell, and I. G. Charles. 2006. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Microb. Pathog. 41:174-182. [DOI] [PubMed] [Google Scholar]
- 29.Negrea, A., E. Bjur, S. Puiac, S. E. Ygberg, F. Aslund, and M. Rhen. 18 September 2009. Thioredoxin 1 participates in the activity of Salmonella enterica serovar Typhimurium pathogenicity island 2 type III secretion system. J. Bacteriol. doi: 10.1128/JB.00532-09. [DOI] [PMC free article] [PubMed]
- 30.O'Callaghan, D., D. Maskell, J. E. Beesley, M. R. Lifely, I. Roberts, G. Boulnois, and G. Dougan. 1988. Characterisation and in vivo behaviour of a Salmonella typhimurium aroA strain expressing Escherichia coli K1 polysaccharide. FEMS Microbiol. Lett. 52:269-274. [Google Scholar]
- 31.Paterson, G. K., H. Northen, D. B. Cone, C. Willers, S. E. Peters, and D. J. Maskell. 2009. Deletion of tolA in Salmonella Typhimurium generates an attenuated strain with vaccine potential. Microbiology 155:220-228. [DOI] [PubMed] [Google Scholar]
- 32.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 33.Richter-Dahlfors, A., A. M. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 35.Sheppard, M., C. Webb, F. Heath, V. Mallows, R. Emilianus, D. Maskell, and P. Mastroeni. 2003. Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell Microbiol. 5:593-600. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, E. J., F. Aslund, and J. Beckwith. 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 17:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 38.Wallis, T. S. 2004. Host-specificity of Salmonella infections in animal species. In P. Mastroeni and D. Maskell (ed.), Salmonella infections: clinical, immunological and molecular aspects. Cambridge University Press, Cambridge, United Kingdom.
- 39.Whiteland, J. L., S. M. Nicholls, C. Shimeld, D. L. Easty, N. A. Williams, and T. J. Hill. 1995. Immunohistochemical detection of T-cell subsets and other leukocytes in paraffin-embedded rat and mouse tissues with monoclonal antibodies. J. Histochem. Cytochem. 43:313-320. [DOI] [PubMed] [Google Scholar]
- 40.Wright, A. E., and D. Semple. 1897. Remarks on vaccination against typhoid. Br. Med. J. 1:256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yrlid, U., M. Svensson, A. Hakansson, B. J. Chambers, H. G. Ljunggren, and M. J. Wick. 2001. In vivo activation of dendritic cells and T cells during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 69:5726-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]