Abstract
Acquisition of complement regulator factor H (CFH) and factor H-like protein 1 (CFHL1) from human serum enables Borrelia spielmanii, one of the etiological agents of Lyme disease, to evade complement-mediated killing by the human host. Up to three distinct complement regulator-acquiring surface proteins (CRASPs) may be expressed by serum-resistant B. spielmanii, each exhibiting an affinity for CFH and/or CFHL1. Here, we describe the functional characterization of the 15-kDa CRASPs of B. spielmanii, members of the polymorphic Erp (OspE/F-related) protein family, that bind two distinct host complement regulators, CFH and factor H-related protein 1 (CFHR1), but not CFHL1. CFH bound to the B. spielmanii CRASPs maintained cofactor activity for factor I-mediated C3b inactivation. Three naturally occurring alleles of this protein bound CFH and CFHR1 while a fourth natural allele could not. Comparative sequence analysis of these protein alleles identified a single amino acid, histidine-79, as playing a significant role in CFH/CFHR1 binding, with substitution by an arginine completely abrogating ligand binding. The mutation of His-79 to Arg did not inhibit binding of plasminogen, another known ligand of this group of borrelial outer-surface proteins.
Lyme disease, caused by spirochetes of the Borrelia burgdorferi sensu lato complex, is the most prevalent vector-borne anthropozoonosis in Eurasia and North America (6). Throughout their complex enzootic life cycle, Lyme Borrelia spp. utilize an array of strategies to successfully colonize their tick vectors and rodent reservoirs, survive in diverse environments, and overcome host innate and adaptive immune responses. Regarding the human host, certain genospecies of the B. burgdorferi sensu lato complex, in particular B. burgdorferi sensu stricto (hereafter referred to as B. burgdorferi), Borrelia afzelii, and Borrelia spielmanii are resistant to the alternative pathway of complement activation in vitro (7, 21, 26, 32, 51). Protection against complement-mediated lysis is strongly associated with the ability of spirochetes to bind host fluid-phase complement-regulatory proteins such as factor H (CFH) and/or factor H-like protein 1 (CFHL1) (4, 20, 28).
CFH and CFHL1, which is the product of an alternatively spliced transcript of the cfh gene, attenuate alternative pathway activation by acting as cofactors for factor I-mediated inactivation of C3b to iC3b and by inhibiting the formation and enzymatic activity of the C3bBb convertase, thereby protecting self-surfaces from harmful complement attack (29, 30, 40, 54). Both structurally related glycoproteins are organized into individually folding protein domains termed short consensus repeats (SCRs), of which the four N-terminal SCRs of CFH and CFHL1 exhibit the main complement-regulatory activity (55). Upon complement activation, both proteins ensure careful regulation and modulation of the complement cascade. More recently, Heinen et al. (19) demonstrated that factor H-related protein 1 (CFHR1), a distinct member of the CFH protein family, displays complement-regulatory activity by acting upon C5 convertase and terminal complex formation.
As many as five different outer surface proteins that bind distinct members of the CFH protein family, collectively termed complement regulator-acquiring surface proteins (CRASPs), have been identified in individual serum-resistant B. burgdorferi and B. afzelii bacteria (17). Isolates of B. spielmanii, a recently identified human pathogenic genospecies, produce up to three CRASP proteins. B. spielmanii CRASP-1 (BsCRASP-1) was characterized on the molecular level to be the major receptor for CFH and CFHL1 (22). The 15-kDa BsCRASP-3 protein of B. spielmanii is of a size similar to CRASPs of B. burgdorferi and B. afzelii that are members of the polymorphic Erp protein family (20, 25, 53; also P. Kraiczy, unpublished data). Several Erp proteins have been demonstrated to also bind host plasmin(ogen) and are hypothesized to play roles in dissemination and invasion of host tissues (8). To further examine the host-interactive properties of B. spielmanii outer surface proteins, we have identified and functionally characterized BsCRASP-3 molecules, demonstrating them to be similar to, but unique from, CFH/CFHR1-binding CRASPs of B. burgdorferi and B. afzelii.
(This research was conducted by A. Seling and C. Siegel in partial fulfillment of the requirements for a Ph.D. from the Institute of Medical Microbiology and Infection Control, University Hospital of Frankfurt.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. spielmanii tick isolates PC-Eq17, TIsar2, and TIsar3; B. spielmanii human isolate A14S; B. burgdorferi skin isolate LW2; B. afzelii skin isolates FEM1-D15 and EB1; and B. garinii cerebrospinal fluid (CSF) isolate G1 were cultured until mid-exponential phase (5 × 107 cells per ml) at 33°C in modified Barbour-Stoenner-Kelly (BSKII) medium as described previously (27). Cultures of Escherichia coli strain JM109 and Rosetta pLysS were propagated routinely in 2× YT (yeast extract-tryptone) medium supplemented with ampicillin (100 μg/ml).
Proteins, antibodies, and DNA primers.
Nonimmune human serum (NHS) was used as a source of CFH for ligand affinity blotting. Purified CFH and the polyclonal goat anti-CFH antiserum were purchased from Calbiochem, Bad Soden, Germany. The cloning, expression, and purification of CFH constructs consisting of distinct SCR domains (CFH consisting of SCR domains 1 to 5 [CFH1-5], CFH1-6, CFH8-20, CFH15-20, CFH19-20, and CFH15-19), CFHL1, and CFHR1 have been described previously (16, 29, 30). Polyclonal rabbit antiserum to SCR1 to SCR4 (anti-SCR1-4) or the monoclonal antibody (MAb) B22 was used for detection of CFHL1 (29), and the MAb VIG8 was applied to specifically detect CFH (41). MAb L41 1C11 was used for the detection of borrelial FlaB (18), and a polyclonal anti-glutathione S-transferase (GST) antibody was from GE Healthcare, Freiburg, Germany. Unless otherwise stated, antibodies were used at the following final dilutions: 1/1,000 for L41 1C11 and anti-SCR1-4 and 1/2,000 for anti-GST. MAbs VIG8 and B22 were used undiluted. High-performance liquid chromatography (HPLC)-purified DNA primers (Table 1) used for site-directed mutagenesis were purchased from Sigma-Genosys, Steinheim, Germany.
TABLE 1.
Oligonucleotides used in the course of the study
| Primer function and name | Sequence (5′ to 3′)a |
|---|---|
| PCR | |
| BsCRASP-3 145(+) BamHI | GCTGTTTTTGCACTGTTTGGATCCTGTGGAAATTTTAC |
| BsCRASP-3 3nc(−) XhoI | ATTCATAATTATTCTCTTCTCGAGTTTGAATTTCTA |
| TISAR32 | TTCATAATTATTCTCTTCTCGAGTTTG |
| TISAR33 | CACCTGTGGAAATTTTACAAGTAGTTTA |
| pGEX(+) | GGGCTGGCAAGCCACGTTTGGTG |
| pGEX(−) | CCGGGAGCTGCATGTGTCAGAGG |
| Site-directed mutagenesis | |
| BsCRASP-3 E77G(+) | CGGGTTTAAATGCTACTGGGGGGCATTCAGCTACGTTATTTTC |
| BsCRASP-3 E77G(−) | GAAAATAACGTAGCTGAATGCCCCCCAGTAGCATTTAAACCCG |
| BsCRASP-3 H79R(+) | GTTTAAATGCTACTGAGGGGCGTTCAGCTACGTTATTTTCATTAG |
| BsCRASP-3 H79R(−) | CTAATGAAAATAACGTAGCTGAACGCCCCTCAGTAGCATTTAAAC |
| BsCRASP-3 E131K(+) | GAGATAATAACAAAGATGAAAACCATTGATGGTTCTGAAC |
| BsCRASP-3 E131K(−) | GTTCAGAACCATCAATGGTTTTCGTCTTTGTTATTATCTC |
| BsCRASP-3 Q115E(+) | GTTTATATTATGGATATAGGGAAGAGAGCACTGAAAAGGGTATCC |
| BsCRASP-3 Q115E(−) | GGATACCCTTTTCAGTGCTCTCTTGCCTATATCCATAATATAAAC |
Underlining indicates introduced substitution(s) of the respective residue(s).
Identification of B. spielmanii Erp-encoding genes.
A set of diverse oligonucleotides specific for erp and for circular DNA elements of approximately 32 kb (cp32), previously used to characterize the arrangements of erp genes of B. burgdorferi strains (48), were selected for amplification of the orthologous erp genes of B. spielmanii isolate PC-Eq17. Amplicons were purified using Centricon-100 filters (11) and cloned into pCR2.1 (Invitrogen, Carlsbad, CA) and subsequently sequenced. Orthologous genes of B. spielmanii strains TIsar2 and TIsar3 were identified using the oligonucleotide primers BsCRASP-3 145(+) BamHI and BsCRASP-3 3nc(−) XhoI based upon the sequence of the strain PC-Eq17 gene (Table 1). One gene was identified in PC-Eq17 (erp60, encoding the BsCRASP-3 of strain PC-Eq17 [Erp60]), two genes were identified in TIsar2 (erp61 and erp62, encoding Erp61 and Erp62, respectively), and one gene was identified in TIsar3 (erp63, encoding Erp63).
Construction of expression plasmids and purification of recombinant borrelial proteins.
The above-described erp genes of B. spielmanii isolates PC-Eq17, TIsar2, and TIsar3 were subcloned by PCR using primers BsCRASP-3 145(+) BamHI and BsCRASP-3 3nc(−) XhoI into the pGEX 6P-1 expression vector, resulting in plasmids pGEX N38PC-Eq17, pGEX N38TIsar2.1, pGEX N38TIsar2.2, and pGEX N38TIsar3 (Table 1). All of these recombinant proteins contained amino-terminal GST tags, with the Erp segment beginning with that protein's first amino acid following the cysteine lipidation site. The GST-Erp fusion proteins were expressed in E. coli JM109, and affinity purification over glutathione-Sepharose columns was performed as recommended by the manufacturer (GE Healthcare, Freiburg, Germany).
For analyses of plasminogen binding, a polyhistidine-tagged recombinant form of Erp63 was produced. The above-described clone was PCR amplified using oligonucleotide primers TISAR32 and TISAR33 (Table 1), and the resulting amplicon was cloned into pET200 (Invitrogen, Carlsbad, CA). Recombinant His-tagged Erp63 was expressed in E. coli Rosetta pLysS (Novagen, San Diego, CA) and purified using Magne-His Ni-particles (Promega, Madison, WI).
Site-directed mutagenesis.
To introduce single or double amino acid substitutions into erp genes, a site-directed mutagenesis approach was conducted as described previously (44). Briefly, PCRs were carried out for 18 cycles (95°C for 30 s, 55°C for 30 s, and 68°C for 13 min) using 100 ng of expression vector pGEX N38 PC-Eq17, 125 ng of each mutagenic primer, and 2.5 U of Pfu DNA polymerase (Stratagene) in a final volume of 50 μl. Before transformation of E. coli, the reaction mixtures were treated with 10 U of DpnI (New England Biolabs, Frankfurt, Germany) for 1 h at 37°C. All mutations introduced into the erp genes were verified by DNA sequencing of both DNA strands.
SDS-PAGE, ligand affinity blotting, and Western blot analysis.
Whole-cell lysates obtained from respective borrelial isolates or purified recombinant proteins (500 ng per lane) were subjected to 10% Tris-Tricine-SDS-PAGE under reducing conditions and transferred to nitrocellulose as previously described (27).
ELISAs.
Binding of CFH, CFHR1, and CFHL1 to borrelial proteins was analyzed by enzyme-linked immunosorbent assays (ELISAs) as described previously (44). Briefly, purified GST fusion proteins were immobilized, and unspecific binding sites were blocked with 0.2% gelatin in phosphate-buffered saline (PBS). CFH (Calbiochem, Darmstadt, Germany), CFHR1, or CFHL1 (5 μg/ml each) was added to the wells and left overnight at 4°C. Protein complexes were identified using a polyclonal anti-CFH antibody following a secondary peroxidase-conjugated anti-goat IgG antibody. The reaction was developed with 1,2-phenylenediamine dihydrochloride (Sigma-Aldrich).
Plasminogen-binding affinities were measured by ELISA as described previously (8). Briefly, Maxisorp 96-well plates (Nalge-Nunc, Rochester, NY) were coated overnight with 10 μg/ml recombinant Erp63, B. burgdorferi strain B31 ErpP protein, or gelatin in PBS. Unspecific binding sites were blocked with 0.1% (mass/vol) gelatin in PBS. Afterwards 100 μl/well of 50 μg/ml plasminogen (Sigma-Aldrich) was added and incubated for 2 h at 37°C. Wells were washed and then incubated with goat anti-human plasminogen (Novus Biologicals, Littleton, CO). After incubation with horseradish peroxidase (HRP)-conjugated protein A (GE Healthcare, Little Chalfont, United Kingdom) (1:5,000), the reaction was developed with tetramethylbenzidine substrate.
Cofactor assay.
Cofactor activity of CFH was analyzed on immobilized recombinant Erp proteins by measuring factor I-mediated conversion of C3b to iC3b as described by Herzberger et al. (21). In brief, fusion proteins (20 μg/ml) immobilized on a microtiter plate were incubated with purified CFH overnight at 4°C. After a washing step, human C3b (Calbiochem, Bad Soden, Germany) and human factor I (Calbiochem) were added to the wells, and the reaction mixtures were incubated for 15 min at 37°C. The samples gathered from the wells were then analyzed by SDS-PAGE and Western blotting.
Immunofluorescence assay.
For detection of deposited complement components on the bacterial surface, an immunofluorescence assay was performed as previously described (21). Briefly, spirochetes (6 × 106) were incubated for 30 min at 37°C with gentle agitation in 25% NHS or, as a control, in 25% heat-inactivated NHS. Ten microliters of cell suspension was spotted on glass slides, allowed to air dry overnight, and fixed in methanol. After a 1-h incubation at 37°C with polyclonal antibodies directed against the complement components C3 (Calbiochem) or C6 (Calbiochem) or with a MAb directed against C5b-9 (Quidel, San Diego, CA), slides were washed and subsequently incubated with an Alexa Fluor 488-conjugated anti-goat antibody or Alexa 488 Fluor-conjugated anti-mouse antibody (Molecular Probes). After a washing step, the slides were mounted with ProLong Gold antifade reagent (Molecular Probes) containing DAPI (4′,6′-diamidino-2-phenylindole). Slides were visualized at a magnification of ×1,000.
Nucleotide sequence analysis and accession numbers.
The deduced amino sequences of the B. spielmanii Erp orthologs were aligned using the DNAstar Lasergene 99 software package. The secondary structure prediction was obtained using GOR4 (12) available at http://www.expasy.org.
Statistical analysis.
ELISA data were analyzed by a two-tailed unpaired Student's t test. Calculated P values of ≤0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The erp gene sequences reported in this paper have been deposited in the EMBL database under accession numbers FN433925 (erp60 of PC-Eq17), FN433926 (erp61 of TIsar2), FN433927 (erp62 of TIsar2), and FN433928 (erp63 of TIsar3).
RESULTS
Serum susceptibility testing and complement activation of B. spielmanii.
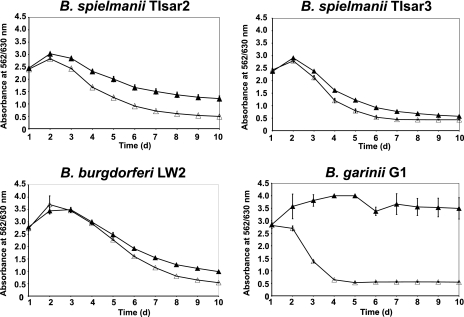
To assess susceptibility to complement-mediated lysis, B. spielmanii isolates TIsar2 and TIsar3 were incubated for up to 10 days in medium containing either 50% normal human serum (NHS) or 50% heat-inactivated NHS (hiNHS) (Fig. 1). Serum-resistant strains B. burgdorferi LW2 and serum-sensitive B. garinii G1 were also examined as positive and negative controls, respectively (27). Both of the B. spielmanii isolates resisted complement-mediated killing, as indicated by continuous growth over the whole cultivation time. Thus, TIsar2 and TIsar3 were classified as being serum resistant. As expected, B. burgdorferi LW2 also showed a serum-resistant phenotype, while growth of the serum-sensitive B. garinii isolate G1 was strongly inhibited by normal serum. None of the analyzed borrelial strains was affected by incubation in the presence of hiNHS.
FIG. 1.
Serum susceptibility among borrelial isolates. Growth inhibition assays of susceptibility to human serum of B. spielmanii TIsar2 and TIsar3, as well as control strains B. burgdorferi LW2 and B. garinii G1 (27). Spirochetes were incubated in either 50% NHS (filled triangles) or 50% heat-inactivated NHS (open triangles) over a cultivation period of 10 days at 33°C. Color changes were monitored by measurement of the absorbance at 562/630 nm. All experiments were performed three times in which each test was done five times with very similar results. For clarity only data from representative experiments are shown. Error bars represent ± standard deviations (SDs). d, day.
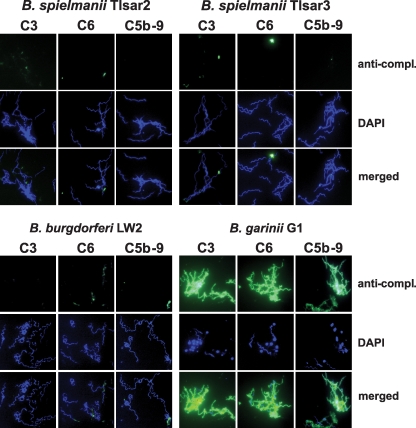
We also analyzed deposition of activated complement components C3, C6, and C5b-9 (terminal complement complex) on the surface of serum-resistant B. spielmanii TIsar2 and TIsar3, B. burgdorferi LW2, and serum-sensitive B. garinii G1. After incubation, deposition of complement components on the spirochetal surfaces was assayed by immunofluorescence microscopy. In addition, counterstaining with DAPI was performed to identify all spirochetes. No fluorescent staining was observed for the majority of the cells of serum-resistant isolates TIsar2, TIsar3, and LW2 (Fig. 2). The few strongly fluorescent spirochetes observed might represent dying cells that are found in every bacterial cell population and that, provoked by morphological change of the outer surface, can strongly activate complement. In contrast, large amounts of C3, C6, and C5b-9 were deposited on the majority of the cells of serum-sensitive B. garinii G1. Complement-positive B. garinii cells showed signs of morphological changes including larger bleb formation, as also observed by dark-field microscopy. These serum-treated cells were irregularly shaped, and their movements were strongly affected. Interestingly, most of the complement-coated B. garinii cells stained negative with DAPI, indicating that these spirochetes might represent cell ghosts from which the DNA had been lost. In contrast, blebs showed a strong fluorescence for deposited complement components as well as in the DAPI stain, thus suggesting that the borrelial DNA was highly concentrated within these structures. An increased formation of blebs is a strong indication of continuous cell destruction as they were observed only with serum-susceptible cells. In control studies, none of the spirochetes incubated with hiNHS exhibited fluorescent labeling (data not shown). These results are similar to those obtained previously for the B. spielmanii type strain, PC-Eq17 (21).
FIG. 2.
Deposition of complement components C3, C6, and C5b-9 on the surface of borreliae. Activated complement components deposited on the surface of serum-resistant B. spielmanii isolates TIsar2 and TIsar3, serum-resistant B. burgdorferi LW2, and serum-sensitive B. garinii isolate G1, detected by indirect immunofluorescence microscopy. Spirochetes were incubated with either 25% NHS or hiNHS for 30 min at 37°C with gentle agitation, and bound C3, C6, and C5b-9 were analyzed with specific antibodies against each component and appropriate Alexa Fluor 488-conjugated secondary antibodies. For visualization of the spirochetes in a given microscopic field, the DNA-binding dye DAPI was used. The spirochetes were observed at a magnification of ×100. The data were recorded via a DS-5Mc charge-coupled-device camera (Nikon) mounted on an Olympus CX40 fluorescence microscope. Panels shown are representative of at least 20 microscope fields.
Identification of complement regulator-acquiring surface proteins in B. spielmanii.
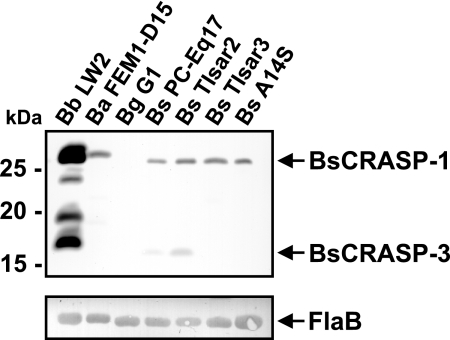
Ligand affinity blotting (27) was used to identify potential CFH-binding CRASPs produced by strains TIsar2 and TIsar3. Following incubation with purified CFH and a specific antibody, two CFH-binding proteins, designated BsCRASP-1 and BsCRASP-3, were identified in B. spielmanii isolates PC-Eq17 and TIsar2 while a single CFH-binding protein, BsCRASP-1, was found in B. spielmanii strains TIsar3 and A14S (Fig. 3). Lysates from serum-resistant B. burgdorferi LW2 showed up to five distinct CRASPs (B. burgdorferi CRASP-1 [BbCRASP-1] to BbCRASP-5). In concordance with our previous finding, the 27.5-kDa BbCRASP-1/CspA protein bound very strongly to CFH while BbCRASP-2/CspZ showed a somewhat weaker binding intensity (27). BbCRASP-4/ErpC and BbCRASP-5/ErpA cannot be distinguished as single proteins due to their similar molecular masses of 18.5 kDa and 17.7 kDa, respectively. In addition, a single CFH-binding B. afzelii CRASP, BaCRASP-1, was clearly visible in B. afzelii FEM1-D15. As expected, no CRASP proteins were detected in the serum-sensitive B. garinii isolate G1.
FIG. 3.
Identification of factor H and FHL-1 binding proteins expressed among borrelial isolates. Protein extracts (30 μg each) obtained from B. burgdorferi LW2, B. afzelii FEM1-D15, B. garinii G1, and B. spielmanii PC-Eq17, TIsar2, TIsar3, and A14S were separated by 10% Tris-Tricine-SDS-PAGE and transferred to nitrocellulose. The membrane was incubated with purified factor H (2.5 μg/ml), and binding of the proteins was detected with MAb VIG8 specific for SCR 20 of factor H. For detection of FlaB as a control, MAb L41 1C11 was applied. The identified CRASP proteins are indicated on the right, and the mobility of the marker proteins is indicated on the left.
Cloning and identification of the BsCRASP-3 proteins of diverse B. spielmanii isolates.
Based upon the proteins' sizes and previous studies of other Lyme disease-associated Borrelia spp., we hypothesized that the genes encoding BsCRASP-3 might be members of the polymorphic erp family. We therefore employed oligonucleotides that had previously been used to characterize other borrelial erp genes for PCR amplification of PC-Eq17 DNA. Sequence analysis of one amplicon revealed an open reading frame of 504 bp that showed strong similarity to the erpA, erpC, and erpP genes of B. burgdorferi B31 (data not shown). This PC-Eq17 gene, erp60, encodes a unique protein of 168 amino acid residues and a calculated molecular mass of 18.6 kDa. The predicted N terminus of this protein showed significant homology to the consensus signal peptidase II cleavage sequence (14).
Based upon the PC-Eq17 erp60 gene sequence, expression vectors were constructed from erp60 and from DNA of B. spielmanii isolates TIsar2 and TIsar3 to produce GST-tagged fusion proteins. One amplicon of 534 bp was obtained from isolate TIsar3, and two amplicons of 507 bp and 534 bp were obtained from isolate TIsar2. Lyme spirochetes naturally maintain as many as 10 distinct cp32 plasmids, each with an erp locus, so the finding of two similar erp genes within strain TIsar2 was not unexpected (48, 49, 50). All four of these B. spielmanii erp genes were predicted to produce proteins with an overall sequence identity of 92.6 to 100% with each other, with identities between 55.7 and 64.9% to ErpA, ErpC, and ErpP from B. burgdorferi type strain B31 and between 55.7 and 69.6% to other borrelial CFH-binding Erp proteins (Fig. 4 and data not shown). Each protein was therefore designated as being an Erp allele. Alignment of the predicted Erp60 protein of isolate PC-Eq17 sequences indicates that Erp61 and Erp62 each contain a six- and three-amino-acid insertion near their amino termini (Fig. 4 and data not shown).
FIG. 4.
Alignment of predicted amino acid sequences of the erpA-encoded homologs of B. spielmanii. Amino acid residues identical to those in the sequence of Erp60 from PC-Eq17 are indicated by a dot. Blank spaces in the N terminus of Erp homologs indicate a region for which sequence was not determined due to the cloning procedure used to generate fusion proteins. The lipid-modifiable cysteine residue at position 20 is indicated by an asterisk.
Erp proteins of B. spielmanii bind to distinct complement regulators.
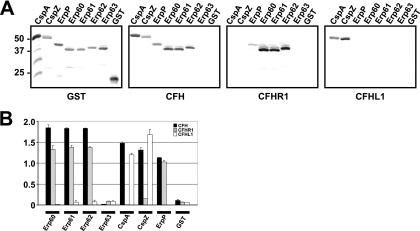
Binding of CFH, CFHR1, and CFHL1 to purified Erp proteins was assessed by ligand affinity blotting and by ELISA. The Erp60, Erp61, and Erp62 proteins of strains PC-Eq17 and TIsar2 bound CFH and CFHR1 but not CFHL1 (Fig. 5A). Unexpectedly, none of the complement regulators showed detectable binding to Erp63 of TIsar3. CspA, CspZ, and ErpP proteins of B. burgdorferi LW2 served as positive controls showing the reported binding specificities to CFH, CFHL1, or CFHR1 (17). Purified GST protein by itself did not bind to any of the complement regulators.
FIG. 5.
Analysis of the binding capabilities of CFH, CFHR1, and CFHL1 to recombinant Erp homologs of B. spielmanii. Binding capabilities of CFH, CFHR1, and CFHL1 to recombinant Erp proteins were analyzed by ligand affinity blotting (A). Purified Erp fusion proteins of isolates PC-Eq17, TIsar2, and TIsar3 (500 ng/lane) were subjected to 10% Tris-Tricine-SDS-PAGE and blotted to nitrocellulose membranes. GST fusion proteins were detected by using an anti-goat GST antibody. For detection of CFH and CFHR1 bound to CRASP proteins, membranes were incubated with NHS as a source for CFH or with purified CFHR1. Protein complexes were then visualized using MAb VIG8 or JHD8, respectively. Binding of CFHL1 was detected using MAb B22 specific for the N-terminal region of CFHL1. The CFH/CFHL1-binding CspA and CspZ proteins, the CFH/CFHR1-binding ErpP protein, and purified GST served as controls. Binding was also assayed by ELISA using recombinant Erp60, Erp61, Erp62, Erp63, CspZ, and GST (B). Proteins were immobilized on a microtiter plate and incubated with either CFH, CFHR1, or CFHL1. For detection of protein complexes a polyclonal anti-CFH antiserum was used. All experiments were performed at least three times in which each test was done twice or three times with very similar results, but for clarity data from a representative experiment are shown. Error bars represent standard deviations.
To extend these data, the binding capabilities of each recombinant Erp protein to CFH, CFHR1, and CFHL1 were also assessed using ELISAs (Fig. 5B). Consistent with the ligand affinity blot analyses, strong binding of Erp60, Erp61, and Erp62 to both CFH and CFHR1 was detected. Neither borrelial protein bound CFHL1. None of the complement regulators interacted with Erp63. CspA and CspZ used as controls showed strong binding to CFH and CFHL1 but did not bind to CFHR1. As did the Erp proteins of PC-Eq17 and TIsar2, ErpP bound to CFH and CFHR1 but not to CFHL1.
Identification of the amino acid residues responsible for the interaction of Erp homologs of B. spielmanii with complement regulators.
We next addressed the lack of interactions between Erp63 and the complement regulators. Sequence alignment revealed differences at four positions (E77, H79, Q115, and E131) among the CFH/CFHR1-binding Erp62 and the nonbinding Erp63 (Fig. 4 and data not shown). The TIsar3 erp63 gene was independently PCR amplified and sequenced three times, always with the same result, confirming that the reported sequence is accurate. Both Erp62 and Erp63 contain identical insertions near their amino termini, making it unlikely that those additional amino acids negatively impacted CFH and CFHR1 binding by Erp63.
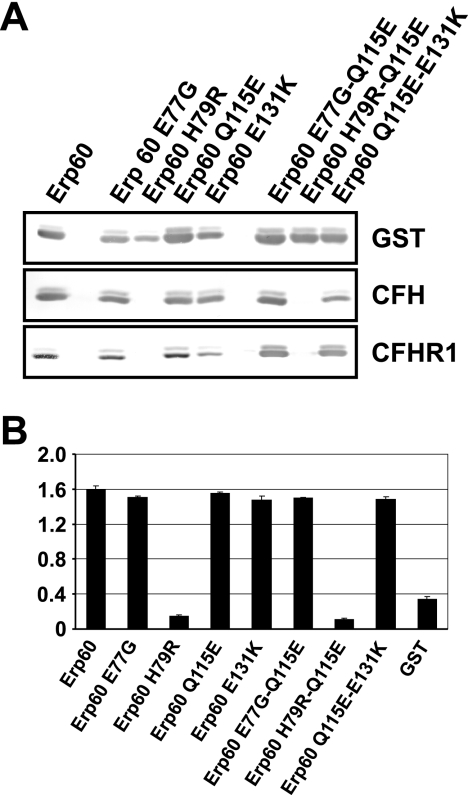
Next, an in vitro mutagenesis approach was conducted to identify the residue(s) responsible for binding of complement regulators by sequential substitution of residues 77, 79, 115, and 131 of the Erp60 protein. Wild-type and mutant recombinant proteins were assayed for CFH and CFHR1 binding using ligand affinity blotting (Fig. 6A). Replacing amino acids E77, Q115, and E131 with the corresponding amino acid of Erp63 did not affect Erp60 binding of either complement regulator. In addition, no differences in binding were observed among mutant proteins carrying two mutations (Erp60 E77G Q115E and Erp60 Q115E E131K). In contrast, binding to CFH was completely abrogated using mutants Erp60 H79R and the double mutant Erp60 H79R Q115E. These data were confirmed by analyzing CFH binding to mutant proteins with ELISAs (Fig. 6B). Taken together, these results demonstrate that the histidine at position 79 of Erp60 is crucial for CFH and CFHR1 binding.
FIG. 6.
Identification of amino acid residues within Erp homologs of B. spielmanii responsible for binding of CFH, CFHR1, and CFHL1. Ligand affinity blotting (A) and ELISAs (B) were performed to detect binding of CFH and/or CFHR1 to BsCRASP-3 mutants as described in the legend of Fig. 2.
Cofactor activity of CFH bound to Erp proteins of B. spielmanii.
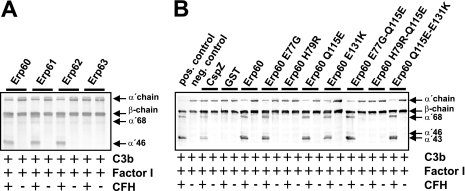
The complement-regulatory activity of CFH bound to recombinant Erp proteins of B. spielmanii was assayed by factor I-mediated conversion of C3b. Appearance of specific C3b-degradation products (α′-68-, α′-46-, and α′-43) indicated cofactor activity of CFH bound to Erp60, Erp61, and Erp62 (Fig. 7A). In contrast, cleavage products were not detected when CFH, factor I, and C3b were added to Erp63. Functional activity of CFH was also assessed with the mutated Erp60 proteins (Fig. 7B). CFH maintained cofactor activity when bound to all mutants, with the exception of those proteins that contained the H79R mutation. Taken together, these results indicate that Erp60 utilizes the cofactor activity of CFH to promote C3b degradation.
FIG. 7.
Cofactor assay of CFH bound to Erp homologs of B. spielmanii. Erp homologs (A) or Erp mutants (B) immobilized on microtiter plates were used to capture CFH. After sequential addition of C3b and factor I, bound CFH retained cofactor activity by enabling factor I-mediated cleavage of C3b to iC3b. Following incubation, the mixture was separated by SDS-PAGE under reducing conditions and transferred to nitrocellulose, and C3b and degradation products were analyzed using a C3 antiserum (Calbiochem, Darmstadt, Germany). CspZ was used as a positive control for factor I-mediated degradation of C3b. GST and proteins incubated without CFH served as negative controls. The mobility of the α′- and the β-chain of C3 and the cleavage products of the α′ chain the α′-68, α′-46 and α′-43 are indicated.
Mapping of the domains within CFH responsible for binding of Erp proteins of B. spielmanii.
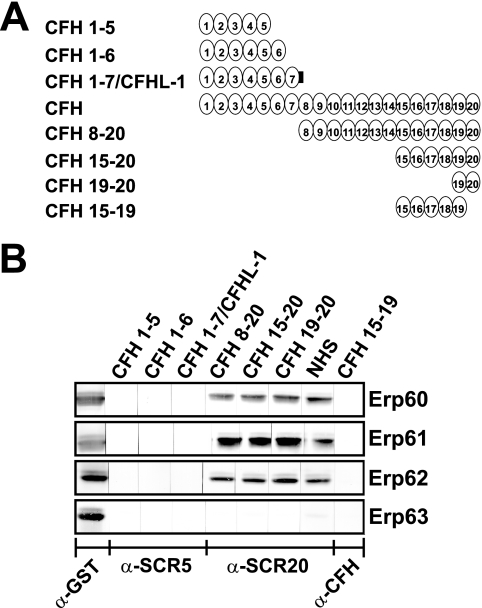
Previous studies demonstrated that CFH and CFHR1 bound to the CFH/CFHR1 binding CRASPs of B. burgdorferi via their C-terminally located SCR domains 19 to 20 (16, 17, 27). Due to the high similarity of Erp alleles of B. spielmanii to ErpA, ErpC, and ErpP of B. burgdorferi, we aimed to localize the binding domains of CFH to recombinant B. spielmanii Erp homologs. Erp60, Erp61, and Erp62 of isolates PC-Eq17 and TIsar2 strongly bound CFH as well as CFH constructs representing C-terminal SCR domains CFH8-20, CFH15-20, and CFH19-20 (Fig. 8). None of the B. spielmanii Erp proteins bound the C-terminal construct CFH15-19, nor did they interact with CFHL1 and the N-terminal CFH constructs CFH1-5 and CFH1-6. These finding indicate that the C-terminal SCR domain 20 is involved with binding of CFH to these B. spielmanii proteins.
FIG. 8.
Mapping of the domains of CFH responsible for binding to Erp homologs of B. spielmanii. Schematic representation of the CFH protein and CFH deletion constructs (A). Purified recombinant Erps were separated by 10% Tris-Tricine-SDS-PAGE and transferred to nitrocellulose. The membrane strips were incubated with either several constructs of CFH consisting of SCR domains 1 to 5 (CFH1-5), CFH1-6, CFH1-7 (which represents CFHL1), CFH8-20, CFH15-20, CFH19-20, or CFH15-19 or with NHS as a source of CFH (B). Bound proteins were visualized using MAb B22 specific for SCR5 of CFH and CFHL1, MAb VIG8 specific for SCR20 of CFH, or a polyclonal CFH antiserum for detection of bound SCR15-19. GST fusion proteins were detected by using an anti-goat GST antibody. α, anti.
Plasminogen binding by Erp63.
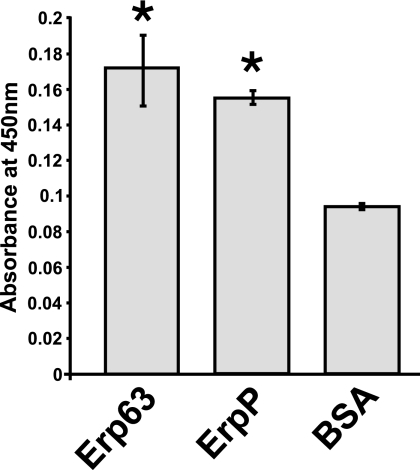
ErpA, ErpC, and ErpP, three proteins of B. burgdorferi strain B31 that share >55% identical amino acids with the above-analyzed Erp proteins of B. spielmanii, all bind human plasminogen (8). The conclusion that Erp63 cannot bind CFH or CFHR1 led us to examine whether or not that protein could bind plasminogen. An ELISA demonstrated that Erp63 has an affinity for plasminogen comparable to that of ErpP (Fig. 9).
FIG. 9.
Erp63 of TIsar3 binds human plasminogen. Results of ELISAs measuring adherence of plasminogen to recombinant Erp63 from B. spielmanii TIsar3, ErpP from B. burgdorferi B31, and bovine serum albumin (BSA). Error bars represent standard deviations. Asterisks indicate ELISA results significantly different from those obtained when BSA was used (P < 0.05).
DISCUSSION
A wide range of pathogenic bacteria utilize a multitude of strategies to overcome the complex network of the host immune defenses, in particular, recognition and elimination by complement, generally regarded as the first line of defense against intruding microorganisms (33). The Lyme disease spirochetes B. burgdorferi, B. afzelii, and B. spielmanii bind the complement regulators CFH, CFHL1, and CFHR1 to help protect against complement-mediated killing (4, 19, 20, 27, 43). In the present study, we report characterization of BsCRASP-3s as receptors for CFH and CFHR1 and their identification as members of the polymorphic, infection-associated Erp protein family. Following convention, the identified BsCRASP-3 molecules were renamed as Erp proteins. Important insight into the mechanism of complement regulator binding by such proteins was provided by the finding that substitution of a single amino acid residue completely blocked binding of both CFH and CFHR1. The Erp63 of isolate TIsar3, the naturally occurring allele that could not bind either CFH or CFHR1, possessed affinity for mammalian plasminogen, suggesting that conservation of that function may have selected for retention of the otherwise defective protein in nature. These results also indicate that binding of CFH/CFHR1 and plasminogen by these Erp family members occurs through distinct amino acid residues and are consistent with the observed simultaneous binding of both CFH and plasminogen (8).
Distinct members of the Erp family, e.g., ErpA, ErpC, and ErpP of B. burgdorferi B31 (8, 36, 45); OspE of B. burgdorferi N40 (20); OspE and p21 of B. burgdorferi 297 (3); BbCRASP-3 to -5 of B. burgdorferi LW2 and ZS7 (16, 17, 27); and BaCRASP-2, BaCRASP-4, and BaCRASP-5 of B. afzelii EB1 and FEM1 (24) are able to specifically bind human CFH. In addition, BbCRASP-3/ErpP, BbCRASP-4/ErpC, and BbCRASP-5/ErpA also exhibit significant affinities for CFHR1 (16, 17). Here, we identified three Erp proteins of diverse B. spielmanii isolates as being CFH- and CFHR1-binding proteins, displaying binding properties identical to those reported for BbCRASP-3/ErpP and BbCRASP-5/ErpA. These three Erp proteins are produced during all stages of mammalian infection (37, 38).
All infectious Lyme disease-associated spirochetes contain multiple, highly similar circular DNA elements of approximately 32 kb in size (cp32s) (46, 49, 50) that harbor one mono- or bicistronic erp locus. An individual bacterial cell can contain several different cp32 elements that each carries a distinct erp locus (9, 47). This general feature of Lyme disease spirochetes holds true also for B. spielmanii as Erp-encoding genes could be amplified using B. burgdorferi cp32-derived primers for PCR, and Southern blot analyses detected B. spielmanii erp genes on ∼30-kb plasmids (data not shown).
Although the identified B. spielmanii Erp alleles shared similar degrees of sequence identity (>92%), only three of the four analyzed proteins, Erp60, Erp61, and Erp62 bind CFH and CFHR1. Erp63 of isolate TIsar3 did not bind to any complement regulator. Sequence comparison (Fig. 4 and data not shown) excluded two short stretches of additional residues at the N terminus of Erp63 since Erp62 also contained those additional amino acids but bound both of the complement regulators. Site-directed mutagenesis suggested that a single amino acid residue of Erp63, R69, might be responsible for the inability of this protein to bind CFH and CFHR1. All the other identified B. spielmanii CFH-binding Erp proteins (Erp60, Erp61, and Erp62) contain a histidine at that position. It is of note that this residue is highly conserved within all known CFH-binding Erp proteins, except ErpC (strain B31), OspE (strain 297), and BaCRASP-4 (strain FEM1) (data not shown). Although a single amino acid exchange led to a complete loss of binding, other residues within the B. spielmanii Erp molecule probably also interact with CFH and CFHR1. Earlier studies from other investigators suggest that interaction between CFH and an Erp ortholog of B. burgdorferi strain 297, OspE, involves multiple lysine residues (1-3). Higher-order structures also appear to be involved (34, 36). Furthermore, two highly conserved amino acid residues at positions 61 and 82 within the OspE protein are associated with CFH binding (34, 36). It was suggested that mutations of these neutral amino acid residues (G61 and L82) might affect protein folding as both positions immediately precede predicted alpha-helical regions. Secondary structure predictions of the Erp60 protein revealed that H79 is also predicted to be located in proximity to an alpha helix. In addition, replacement of histidine by an arginine residue with an extended side chain may sterically hinder interactions between CFH/CFHR1 and Erp60. In this work, replacement of the negatively charged glutamine residue at position 77 with a neutral glycine residue did not affect binding of CFH or CFHR1. Mutations in two additional regions leading to a change of charge (glutamine at position 115 to glutamic acid and glutamic acid at position 131 to lysine) did not affect ligand binding. Mutagenesis studies of BbCRASP-3/ErpP and the OspE protein of B. burgdorferi strain 297 have shown that an intact C terminus is essential for binding of CFH (3, 25, 36). The 14 residues at the C termini of the analyzed B. spielmanii Erp proteins are nearly identical to those of BbCRASP-3/ErpP and OspE (data not shown). Ongoing efforts to crystallize Erp60 and related proteins may help explain these results.
All CFH- and CFHR1-binding Erp orthologs analyzed so far follow the same mode of interaction, binding to the C-terminal SCR domains 19 and 20 of both complement regulators (1, 16, 17, 25, 27). Accordingly, ligand blot analysis with deletion constructs of CFH suggested that interaction with Erp proteins of B. spielmanii is mediated by the C terminus of CFH. The importance of the C terminus of CFH in binding of diverse bacterial proteins has also been demonstrated for the B. hermsii CRASP-1, FhbA, and HcpA proteins of relapsing fever spirochetes (13, 23, 35, 42); the LenA/LfhA protein of Leptospira interrogans (52); PspC of Streptococcus pneumoniae (15); beta protein of Streptococcus agalactiae (5); Scl1 of Streptococcus pyogenes (10); PorA1 of Neisseria gonorrhoeae (39); and Tuf of Pseudomonas aeruginosa (31).
To understand the biological significance of CFH binding, we examined cofactor activity of CFH when bound to recombinant B. spielmanii Erp proteins. When bound to wild-type and most of the mutant Erp proteins, CFH promoted factor I-mediated cleavage of C3b, as indicated by the respective degradation products (Fig. 7). This finding implies that B. spielmanii spirochetes protect themselves from complement-mediated killing by the expression of at least two distinct classes of surface-exposed proteins, CspA (BsCRASP-1) (22) and Erps (BsCRASP-3), both of which independently bind functionally active CFH.
In conclusion, we characterized the CFH/CFHR1-binding Erp proteins of B. spielmanii and identified a single amino acid residue which is critical for the binding of host complement regulators. This study provides additional information on the molecular aspects of the interaction of Erp orthologs with CFH by Lyme disease spirochetes.
Acknowledgments
We thank Christa Hanssen-Hübner for skillful and expert technical assistance.
This work was funded by the Deutsche Forschungsgemeinschaft DFG, grant Kr3383/1-2 to P. Kraiczy and U.S. National Institutes of Health grant R01-AI44254 to B. Stevenson.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 26 October 2009.
REFERENCES
- 1.Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z.-Z. Cheng, T. S. Jokiranta, I. J. T. Seppala, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195-6201. [DOI] [PubMed] [Google Scholar]
- 2.Alitalo, A., T. Meri, P. Comstedt, L. Jeffery, J. Tornberg, T. Strandin, H. Lankinen, S. Bergström, M. Cinco, S. R. Vuppala, D. R. Akins, and S. Meri. 2005. Expression of complement factor H binding immunoevasion proteins in Borrelia garinii isolated from patients with neuroborreliosis. Eur. J. Immunol. 35:3043-3053. [DOI] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 4.Alitalo, A., T. Meri, L. Ramo, T. S. Jokiranta, T. Heikkila, I. J. T. Seppala, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Areschoug, T., M. Stalhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 277:12642-12648. [DOI] [PubMed] [Google Scholar]
- 6.Bacon, R. M., K. J. Kugeler, and P. S. Mead. 2008. Surveillance for Lyme disease—United States, 1992-2006. MMWR Surveill. Summ. 57:1-9. [PubMed] [Google Scholar]
- 7.Breitner-Ruddock, S., R. Würzner, J. Schulze, and V. Brade. 1997. Heterogeneity in the complement-dependent bacteriolysis within the species of Borrelia burgdorferi. Med. Microbiol. Immunol. 185:253-260. [DOI] [PubMed] [Google Scholar]
- 8.Brissette, C. A., K. Haupt, D. Barthel, A. E. Cooley, A. Bowman, C. Skerka, R. Wallich, P. F. Zipfel, P. Kraiczy, and B. Stevenson. 2009. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77:300-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 10.Caswell, C. C., R. Han, K. M. Hovis, P. Ciborowski, D. R. Keene, R. T. Marconi, and S. Lukomski. 2008. The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, factor H, and inhibits the alternative pathway of complement. Mol. Microbiol. 67:584-596. [DOI] [PubMed] [Google Scholar]
- 11.El Hage, N., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 13.Grosskinsky, S., M. Schott, C. Brenner, S. J. Cutler, P. Kraiczy, P. F. Zipfel, M. M. Simon, and R. Wallich. 2009. Borrelia recurrentis employs a novel multifunctional surface protein with anti-complement, anti-opsonic and invasive potential to escape innate immunity. PLoS ONE 4:e4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt, S., V. Agarwal, A. Kunert, S. Haelbich, C. Skerka, and P. F. Zipfel. 2007. The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J. Immunol. 178:5848-5858. [DOI] [PubMed] [Google Scholar]
- 16.Haupt, K., P. Kraiczy, R. Wallich, V. Brade, C. Skerka, and P. Zipfel. 2007. Binding of human factor H-related protein 1 to serum-resistant Borrelia burgdorferi is mediated by borrelial complement regulator-acquiring surface proteins. J. Infect. Dis. 196:124-133. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, K., P. Kraiczy, R. Wallich, V. Brade, C. Skerka, and P. F. Zipfel. 2008. FHR-1, an additional human plasma protein, binds to complement regulator-acquiring surface proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 298(Suppl. 1):287-291. [Google Scholar]
- 18.Hauser, U., G. Lehnert, and B. Wilske. 1999. Validity of interpretation criteria for standardized Western blots (immunoblots) for serodiagnosis of Lyme borreliosis based on sera collected throughout Europe. J. Clin. Microbiol. 37:2241-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinen, S., A. Hartmann, N. Lauer, U. Wiehl, H.-M. Dahse, S. Schirmer, K. Gropp, T. Enghardt, R. Wallich, S. Halbich, M. Mihlan, U. Schlotzer-Schrehardt, P. F. Zipfel, and C. Skerka. 2009. Factor H related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114:2439-2447. [DOI] [PubMed] [Google Scholar]
- 20.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 21.Herzberger, P., C. Siegel, C. Skerka, V. Fingerle, U. Schulte-Spechtel, A. van Dam, B. Wilske, V. Brade, P. F. Zipfel, R. Wallich, and P. Kraiczy. 2007. Human pathogenic Borrelia spielmanii sp. nov. resists complement-mediated killing by direct binding of immune regulators factor H and factor H-like protein 1. Infect. Immun. 75:4817-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzberger, P., C. Siegel, C. Skerka, V. Fingerle, U. Schulte-Spechtel, B. Wilske, V. Brade, P. F. Zipfel, R. Wallich, and P. Kraiczy. 2009. Identification and characterization of the factor H and FHL-1 binding complement regulator-acquiring surface protein 1 of the Lyme disease spirochete Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 299:141-154. [DOI] [PubMed] [Google Scholar]
- 23.Hovis, K. M., E. Tran, C. M. Sundy, E. Buckles, J. V. McDowell, and R. T. Marconi. 2006. Selective binding of Borrelia burgdorferi OspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 74:1967-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraiczy, P., K. Hartmann, J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, R. Wallich, and B. Stevenson. 2004. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi. Int. J. Med. Microbiol. 293:152-157. [DOI] [PubMed] [Google Scholar]
- 25.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement-inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 26.Kraiczy, P., K. P. Hunfeld, S. Breitner-Ruddock, R. Wurzner, G. Acker, and V. Brade. 2000. Comparison of two laboratory methods for the determination of serum resistance in Borrelia burgdorferi isolates. Immunobiology 201:406-419. [DOI] [PubMed] [Google Scholar]
- 27.Kraiczy, P., C. Skerka, V. Brade, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and Peter F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 29.Kühn, S., C. Skerka, and P. F. Zipfel. 1995. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J. Immunol. 155:5663-5670. [PubMed] [Google Scholar]
- 30.Kühn, S., and P. F. Zipfel. 1995. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene 162:225-229. [DOI] [PubMed] [Google Scholar]
- 31.Kunert, A., J. Losse, C. Gruszin, M. Huhn, K. Kaendler, S. Mikkat, D. Volke, R. Hoffmann, T. S. Jokiranta, H. Seeberger, U. Moellmann, J. Hellwage, and P. F. Zipfel. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J. Immunol. 179:2979-2988. [DOI] [PubMed] [Google Scholar]
- 32.Kurtenbach, K., H.-S. Sewell, N. H. Ogden, S. E. Randolph, and P. A. Nuttall. 1998. Serum complement sensitivity as a key factor in Lyme disease ecology. Infect. Immun. 66:1248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambris, J. D., D. Ricklin, and B. V. Geisbrecht. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDowell, J. V., J. Wolfgang, L. Senty, C. M. Sundy, M. J. Noto, and R. T. Marconi. 2004. Demonstration of the involvement of outer surface protein E coiled coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 173:7471-7480. [DOI] [PubMed] [Google Scholar]
- 35.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metts, M. S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection in mice. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. C., K. Narayan, B. Stevenson, and A. R. Pachner. 2005. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 39:27-33. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. C., and B. Stevenson. 2004. Increased expression of Borrelia burgdorferi factor H-binding surface proteins during transmission from ticks to mice. Int. J. Med. Microbiol. 293(Suppl. 37):120-125. [DOI] [PubMed] [Google Scholar]
- 39.Ngampasutadol, J., S. Ram, S. Gulati, S. Agarwal, C. Li, A. Visintin, B. Monks, G. Madico, and P. A. Rice. 2008. Human factor H interacts selectively with Neisseria gonorrhoeae and results in species-specific complement evasion. J. Immunol. 180:3426-3435. [DOI] [PubMed] [Google Scholar]
- 40.Pangburn, M. K., R. D. Schreiber, and H. J. Muller-Eberhard. 1977. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein beta1H for cleavage of C3b and C4b in solution. J. Exp. Med. 146:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prodinger, W. M., J. Hellwage, M. Spruth, M. P. Dierich, and P. F. Zipfel. 1998. The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochem. J. 331:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossmann, E., P. Kraiczy, P. Herzberger, C. Skerka, M. Kirschfink, M. M. Simon, P. F. Zipfel, and R. Wallich. 2007. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 178:7292-7301. [DOI] [PubMed] [Google Scholar]
- 43.Siegel, C., P. Herzberger, C. Skerka, V. Brade, V. Fingerle, U. Schulte-Spechtel, B. Wilske, P. F. Zipfel, R. Wallich, and P. Kraiczy. 2008. Binding of complement regulatory protein factor H enhances serum resistance of Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 298(Suppl. 1):292-294. [DOI] [PubMed] [Google Scholar]
- 44.Siegel, C., J. Schreiber, K. Haupt, C. Skerka, V. Brade, M. M. Simon, B. Stevenson, R. Wallich, P. F. Zipfel, and P. Kraiczy. 2008. Deciphering the ligand-binding sites in the Borrelia burgdorferi complement regulator-acquiring surface protein 2 required for interactions with the human immune regulators factor H and factor H-like protein 1. J. Biol. Chem. 283:34855-34863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson, B. 2002. Borrelia burgdorferi erp (ospE-related) gene sequences remain stable during mammalian infection. Infect. Immun. 70:5307-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson, B. 2001. Borrelia burgdorferi: a (somewhat) clonal bacterial species. Trends Microbiol. 9:471-472. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309-324. [DOI] [PubMed] [Google Scholar]
- 49.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, B., W. R. Zuckert, and D. R. Akins. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2:411-422. [PubMed] [Google Scholar]
- 51.van Dam, A. P., A. Oei, R. Jaspars, C. Fijen, B. Wilske, L. Spanjaard, and J. Dankert. 1997. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect. Immun. 65:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verma, A., J. Hellwage, S. Artiushin, P. F. Zipfel, P. Kraiczy, J. F. Timoney, and B. Stevenson. 2006. LfhA, a novel factor H-binding protein of Leptospira interrogans. Infect. Immun. 74:2659-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallich, R., J. Pattathu, V. Kitiratschky, C. Brenner, P. F. Zipfel, V. Brade, M. M. Simon, and P. Kraiczy. 2005. Identification and functional characterization of complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii. Infect. Immun. 73:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whaley, K., and S. Ruddy. 1976. Modulation of the alternative complement pathways by beta 1 H globulin. J. Exp. Med. 144:1147-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zipfel, P. F., T. S. Jokiranta, J. Hellwage, V. Koistinen, and S. Meri. 1999. The factor H protein family. Immunopharmacology 42:53-60. [DOI] [PubMed] [Google Scholar]