Abstract
Salmonella has evolved several strategies to counteract intracellular microbicidal agents like reactive oxygen and nitrogen species. However, it is not yet clear how Salmonella escapes lysosomal degradation. Some studies have demonstrated that Salmonella can inhibit phagolysosomal fusion, whereas other reports have shown that the Salmonella-containing vacuole (SCV) fuses/interacts with lysosomes. Here, we have addressed this issue from a different perspective by investigating if the infected host cell has a sufficient quantity of lysosomes to target Salmonella. Our results suggest that SCVs divide along with Salmonella, resulting in a single bacterium per SCV. As a consequence, the SCV load per cell increases with the division of Salmonella inside the host cell. This demands more investment from the host cell to counteract Salmonella. Interestingly, we observed that Salmonella infection decreases the number of acidic lysosomes inside the host cell both in vitro and in vivo. These events potentially result in a condition in which an infected cell is left with insufficient acidic lysosomes to target the increasing number of SCVs, which favors the survival and proliferation of Salmonella inside the host cell.
Pathogenic bacteria belonging to the genus Salmonella cause a spectrum of diseases ranging from mild gastroenteritis to life-threatening systemic diseases, like typhoid fever in humans and animals of economic importance. Being an intracellular pathogen, Salmonella has evolved strategies to avoid intracellular microbicidal agents, like reactive oxygen and nitrogen species and antimicrobial peptides (8, 13, 41).
Lysosomes, membrane-bound organelles containing acid hydrolases, constitute an important intracellular defense strategy of a eukaryotic cell. They form the terminal degradative compartment of the endocytic pathway. It is vital for an intracellular pathogen to evade lysosomal degradation in order to colonize a eukaryotic cell.
Different intracellular pathogens have evolved a variety of mechanisms to avoid lysosomal degradation (12). For example, Mycobacterium stalls the maturation of its vacuole at an early endosomal level (9), Escherichia coli modulates the trafficking of its vacuole to avoid fusion with lysosomes (26), and Shigella and Listeria escape from phagosomes and enter the cytoplasm (19, 33), whereas amastigote Leishmania can survive in the harsh environment of lysosomes (2). However, the mechanism by which Salmonella evades lysosomal degradation is not clearly understood. Salmonella thrives inside a specialized intracellular compartment termed the Salmonella-containing vacuole (SCV). The biogenesis of the SCV involves sequential interactions with the endocytic pathway (39). Many elegant studies have demonstrated that Salmonella blocks the fusion of the SCV with terminal acidic lysosomes (4, 16, 22, 25). Nevertheless, there are some reports that show that the SCV fuses/interacts actively with lysosomes (7, 10, 34). Thus, there is uncertainty in this matter in the case of Salmonella (39). In this study, we addressed this problem from a different perspective by investigating if there is a sufficient quantity of lysosomes inside the host cell to target Salmonella. Our results demonstrate that Salmonella tackles the lysosomal degradation problem in an elegant manner by causing an imbalance in the ratio of SCVs to acidic lysosomes.
MATERIALS AND METHODS
Bacterial strains.
Salmonella enterica serovar Typhimurium strain NCTC 12023 was used in all the experiments. Bacteria expressing green fluorescent protein (GFP) through pFPV25.1 were used for confocal laser scanning microscopy (CLSM). A ΔssaV strain was constructed in the same parental strain. Bacteria were routinely cultured in Luria broth (LB) at 37°C. Ampicillin (50 μg/ml) was used whenever required.
Eukaryotic cell culture.
RAW 264.7 cells (a mouse monocyte/macrophage-like cell line) and Intestine 407 cells (a human intestine epithelial cell line) were grown routinely in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum (Sigma) at 37°C in 5% CO2.
Infection experiments in cell culture.
Eukaryotic cells were infected as described previously (13). RAW 264.7 or Intestine 407 cells were seeded on coverslips in a 24-well plate (1 × 105 to 3 × 105 cells per well) for CLSM experiments, and they were seeded in six-well plates (7 × 105 to 15 × 105 cells per well) for transmission electron microscopy (TEM) experiments. After 12 to 24 h, these cells were infected with wild-type Salmonella at a multiplicity of infection (MOI) of 1:1 (for CLSM) or 50:1 (for TEM). RAW 264.7 cells were infected with bacteria from overnight cultures. The overnight cultures were diluted at a 1:33 ratio and grown for 3 h to late log phase prior to the infection of Intestine 407 cells. The plate was centrifuged at 1,000 rpm for 5 min and incubated at 37°C for 20 min. The cells were then washed with phosphate-buffered saline (PBS) to remove excess bacteria, and fresh medium containing 100 μg/ml of gentamicin was added. After 1 h, the medium was discarded, and the cells were again washed with PBS and incubated with fresh medium containing 25 μg/ml of gentamicin and incubated at 37°C until later time points. Sodium orthovanadate (SOV) was used to inhibit dynein (1, 17, 24). The cells were treated with 100 μM of SOV 3 h before infection, and it was maintained throughout the infection process till the end of the experiment. Intestine 407 cells were transfected either with pCS2p50 (a kind gift from Ron Vale) or pCS2 (vector control) using Lipofectamine (Invitrogen). After 48 h, the transfected cells were infected with Salmonella and processed for microscopy as described below. The fold intracellular multiplication of Salmonella was calculated by dividing the intracellular bacterial load at 10 h by the intracellular bacterial load at 2 h.
Confocal laser scanning microscopy.
CLSM was done as described previously (13). Infected cells were fixed with 3.5% paraformaldehyde (Sigma) for 20 min at the indicated time points. After being washed three times with PBS, the cells were incubated with specific antibody (Rab7 [Santa Cruz Biotechnology] and LAMP1 [DSHB]) diluted in blocking buffer (0.1% saponin, 2% bovine serum albumin [BSA], and 2% goat serum [all from Sigma] in PBS) for 1 h. The cells were then washed twice with PBS and incubated with appropriate fluorescent secondary antibodies (Dynova or Jackson Laboratory) diluted in the blocking buffer. Then, the cells were washed three times with PBS, and coverslips were mounted on a glass slide and observed under a confocal laser scanning microscope (Zeiss LSM Meta). To stain actin, fixed cells were incubated with phalloidin-Texas red (Molecular Probes) for 1 h. After 1 h, the cells were washed three times with PBS and the coverslips were mounted on a glass slide and observed under a confocal laser scanning microscope. To visualize lysosomes, infected (for 10 h) cells were washed in PBS, treated with 5 μg/ml of acridine orange (Sigma) for 5 min, and immediately observed under a confocal laser scanning microscope. Adobe Photoshop 7 was used to adjust the contrast and brightness of images.
SCVs were classified according to the number of bacteria per vacuole (a single bacterium per vacuole or multiple bacteria per vacuole) based on LAMP1 staining of SCVs inside RAW 264.7/Intestine 407 cells. Only those cells in which SCVs were clearly defined were included in the analysis. At least 50 infected cells were counted in each case.
Transmission electron microscopy.
Infected cells were washed three times with sodium phosphate buffer, gently scraped, and fixed with Karnowsky's fixative (4% paraformaldehyde [Sigma] plus 3% gluteraldehyde [Sigma] in 0.1 M sodium phosphate buffer) for 24 h. The fixed samples were washed with sodium phosphate buffer and incubated with 1% osmium tetroxide (TAAB) for 90 min. Then, the samples were again washed with sodium phosphate buffer and incubated with 70% ethanol for 1 h, followed by 90% ethanol for 30 min. After this step, samples were treated with 2% uranyl acetate (TAAB) in 95% ethanol for 30 min, followed by 100% ethanol for 1 h. Then, the samples were treated two times with propylene oxide (TAAB) for 10 to 15 min. After that, the samples were infiltrated with an araldite (TAAB) and propylene oxide mixture (1:1) overnight. The next morning, fresh araldite was added and incubated for 3 h. After 3 h, the samples were embedded in molds and kept at 60°C for 48 h. Then, the samples were sectioned using an ultramicrotome (Leica EM UC6) and observed under a transmission electron microscope (JEOL JEM-100CX II).
Flow cytometry.
RAW 264.7 cells were infected with Salmonella at different MOIs and incubated at 37°C until later time points. One hour before a specific time point, fresh medium containing LysoTracker-Green (LT) (Molecular Probes) at a 100 nM concentration was added and incubated for 30 min to 1 h. Then, the cells were washed twice with PBS, harvested by gentle scraping, and analyzed in a flow cytometer (excitation, 488 nm, and absorption, 530/30 nm [FL-1 channel] in a BD FACScan). In the acridine orange experiment, cells were stained with 5 μg/ml of acridine orange 5 min before flow cytometry (excitation, 488 nm, and absorption, ≥520 nm [BD LSR II system]). For autofluorescence, cells were analyzed without any staining in a BD LSR II system (excitation, 355 nm, and emission, 420/70 nm). The cells were treated with 10 μM of carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Sigma) to block the autofluorescence of mitochondria. The data were analyzed using the Winmdi program.
Animal experiments.
All procedures with animals were carried out in accordance with institutionally approved protocols. Eight-week-old BALB/c mice were infected intraperitoneally with Salmonella expressing GFP (104 bacteria/mouse). After 4 days of infection, the mice were sacrificed and their spleens were isolated. Splenocytes were extracted from the spleens by gently crushing the organs and lysing the red blood cells (RBCs) using RBC lysis buffer (0.1 M ammonium chloride, 1 mM potassium bicarbonate, and 1 mM EDTA in water). GFP-positive (infected) and GFP-negative (uninfected) splenocytes were sorted using a fluorescence-assisted cell sorter (Dako MoFlo). The lysosome-specific autofluorescence of the sorted cells was analyzed (excitation, 351 nm; emission, 450/65 nm). The cells were treated with 10 μM of CCCP (Sigma) to block the autofluorescence of mitochondria. The data were analyzed using the Summit V4.3 program.
RESULTS
As SCVs are the potential targets of lysosomes, it is important to know the load of SCVs in an infected host cell in order to understand if the number of lysosomes in a cell is sufficient to target the growing number of Salmonella bacteria. In this context, we examined whether multiple bacteria stay inside a single (or a few) large SCV resulting in single (or few) SCVs per cell or whether each bacterium has its own separate SCV, resulting in multiple SCVs per cell. Many reviews and studies of Salmonella or its vacuole depict the SCV as a big vacuole containing multiple bacteria (10, 15, 21, 28, 37, 39); however, electron micrographs of SCVs in some studies (imaged for other purposes) clearly indicate only one bacterium per vacuole (4, 23, 31). In order to clarify this, we looked at the number of bacteria residing inside an SCV, which in turn determines the load of SCVs (potential targets of lysosomes) per cell.
SCVs contain a single bacterium.
Using both confocal laser scanning microscopy and transmission electron microscopy, we observed that each bacterium was enclosed in a separate vacuole inside Salmonella-infected RAW 264.7 (murine macrophage-like) cells, resulting in multiple SCVs per cell (Fig. 1A and C; see Fig. S1 and S2 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). Salmonella, being rod shaped, is more likely to undergo transverse sectioning than longitudinal sectioning when ultrathin sections are taken. Therefore, Salmonella appears circular in electromicrographs. For confocal microscopy, the cells were infected at an MOI of 1:1 to avoid many bacteria infecting the same cell, and Rab7 (Fig. 1), LAMP 1, and actin (see Fig. S1 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html) were stained to mark the SCV. For electron microscopy, cells were infected at an MOI of 50:1 in order to increase the possibility of finding intracellular bacteria in ultrathin sections.
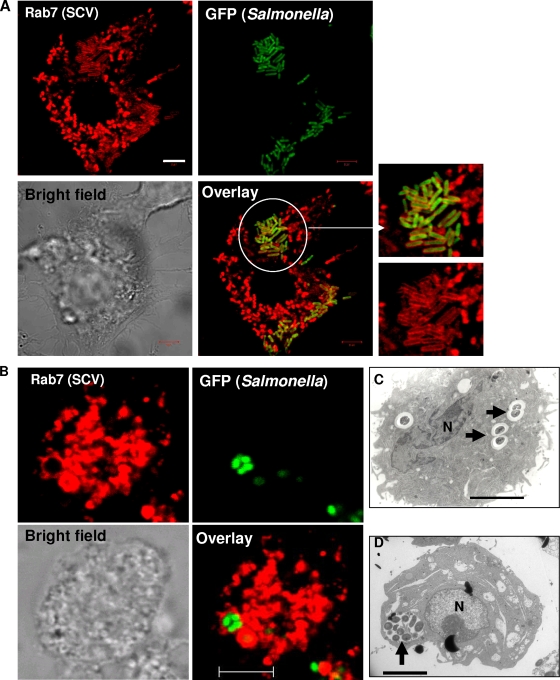
FIG. 1.
SCVs contain a single bacterium per vacuole. (A) Confocal laser scanning microscope images of RAW 264.7 cells infected with GFP-expressing Salmonella for 12 h and immunostained for Rab7 (red). The image shows many SCVs that have a single bacterium per vacuole. The white circle indicates the enlarged part. (B) Micrographs of an SCV containing multiple bacteria. RAW 264.7 cells were treated with 100 μM of sodium orthovanadate for 3 h, after which they were infected with GFP-expressing Salmonella for 12 h and immunostained for Rab7 (red), and the image was taken using a confocal laser scanning microscope. Sodium orthovanadate was maintained throughout the experiment. (C) Transmission electron microscope image of a RAW 264.7 cell infected with Salmonella for 12 h. The image shows SCVs that have a single bacterium per vacuole and also demonstrates the division of SCVs along with the bacteria (arrows). (D) Transmission electron microscope image of a RAW 264.7 cell treated with sodium orthovanadate (100 μM) and infected with Salmonella as described for panel B. The image shows an SCV that has multiple bacteria (arrow). N, nucleus. In all images, the scale bars correspond to 5 μm.
With LAMP1 as the SCV marker inside RAW 264.7 cells, we observed that 96.7% ± 0.7% of infected cells showed a single bacterium per SCV. The SCVs in the remaining infected cells (<4%) had three or more bacteria per vacuole (termed “multiple bacteria per SCV”). The scenario of a single bacterium per SCV was observed as early as 4 h (see Fig. S1B at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html) and as late as 24 h (see Fig. S1C at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html) after infection. The scenario of multiple bacteria per SCV was observed in less than 4% of infected cells in all cases.
SCV undergoes division inside the host cell.
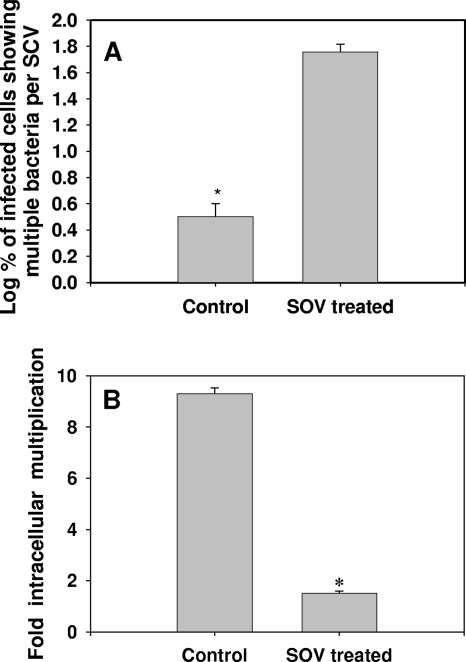
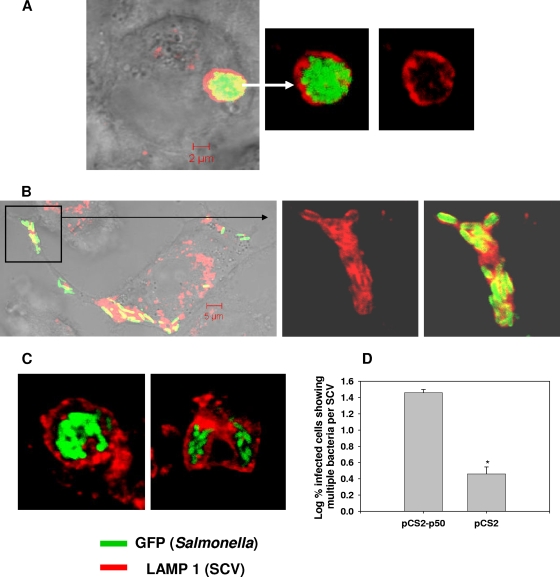
In an infected cell harboring many bacteria, the scenario of a single bacterium per SCV arises when the SCV divides. Electron micrographs of infected RAW 264.7 cells demonstrated the SCV division, along with the division of Salmonella bacteria residing within it (Fig. 1C; see Fig. S2 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). Similar events were observed in the livers of Salmonella-infected mice (see Fig. S3 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). What, then, is the mechanism of SCV division? We ruled out a role of the actin cytoskeleton and cholesterol in SCV division by using their respective pharmacological inhibitors—cytochalasin D for actin and lovastatin for cholesterol (data not shown). Then, we tried to get an answer to the question by studying the mechanism of mitochondrial division. SCVs are similar to mitochondria (endosymbiont bacteria present in eukaryotic cells) because the membrane of the SCV and the outer membrane of the mitochondrion are derived from the eukaryotic host cell and the inner membrane of the mitochondrion is prokaryotic in nature, like the Salmonella cell wall. A dynein motor molecule is involved in the division of the mitochondrial outer membrane by recruiting dynamin-related protein 1 (Drp1) onto the membrane (40). Interestingly, dynein is also reported to control the membrane dynamics of SCVs and to regulate the intracellular replication of Salmonella (18, 30). Therefore, we hypothesized that dynein might be involved in the division of SCVs. To test this hypothesis, we used a sodium orthovanadate and p50 (dynamitin) overexpression system to inhibit the function of dynein (5, 17). Inhibition of dynein using sodium orthovanadate (100 μM) resulted in a remarkable increase (3.3% ± 0.7% to 58% ± 7.6%) in the number of infected cells having multiple bacteria per SCV and a significant decrease in the proliferation of Salmonella (Fig. 1B and D and 2; see Fig. S4 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). A similar observation was made when dynein was inhibited by overexpressing p50 inside Intestine 407 cells (Fig. 3). Interestingly, only a fraction of bacteria were found to be enclosed in vacuoles inside Intestine 407 cells. Nonetheless, those SCVs had only one bacterium per vacuole. In addition, Drp 1 was found to colocalize with Salmonella inside RAW 264.7 cells, suggesting that the SCV probably uses mitochondrial division machinery for its division (see Fig. S5 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). More experiments are required to show the exact role of the mitochondrial division apparatus in the division of SCVs. Nonetheless, these results unequivocally demonstrate that the majority of the SCVs have only one bacterium per vacuole because of the division of SCVs.
FIG. 2.
A single bacterium per vacuole is important for the intracellular proliferation of Salmonella. (A) Percentages (log10) of infected RAW 264.7 cells that have SCVs containing more than three bacteria per vacuole (multiple bacteria per SCV). The graph represents the mean percentages obtained from three experiments. The error bars represent standard errors. Confocal laser scanning microscope images of at least 50 infected cells were analyzed in each case. (B) Fold multiplication of Salmonella inside RAW 264.7 cells. The fold multiplication was calculated by dividing the intracellular bacterial load at 10 h postinfection by the intracellular bacterial load at 2 h postinfection. The graph represents the mean values obtained from an experiment with samples in triplicate. The error bars represent standard errors. *, P < 0.05 (Student's t test).
FIG. 3.
Overexpression of p50 (dynamitin) inhibits the division of SCVs. Images acquired using a confocal laser scanning microscope show Intestine 407 cells transfected with pCS2p50 (A) or pCS2 (vector control) (B), followed by infection with Salmonella for 12 h. The enlarged part of the cell (different plane) is shown inside the box. (C) SCVs containing multiple bacteria per vacuole inside Intestine 407 cells transfected with pCS2-p50. Twelve hours after infection, cells were fixed and stained for LAMP1 (red). Overexpression of p50 inhibits SCV division, resulting in multiple bacteria per vacuole. (D) Percentages (log10) of infected RAW 264.7 cells that have SCVs containing more than three bacteria per vacuole (multiple bacteria per SCV). The graph represents mean percentages obtained from three experiments. The error bars represent standard errors. Confocal laser scanning microscope images of at least 50 infected cells were analyzed in each case. *, P < 0.05 (Student's t test).
Salmonella infection reduces the volume of the acidic compartment contributed by lysosomes inside the host cell.
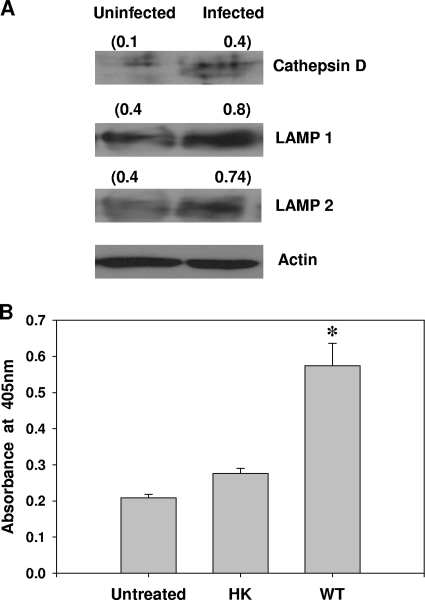
Next, we set out to investigate if the number of lysosomes present in the host cell is sufficient to target the overwhelming number of SCVs. For this purpose, we initially quantified some lysosomal proteins in infected cells. We observed a significant increase in LAMP1, LAMP2, and cathepsin D protein levels and also in acid phosphatase activity in Salmonella-infected RAW 264.7 cells (Fig. 4). As SCV harbors many lysosomal proteins, like vacuolar ATPase, LAMP1, LAMP2, cathepsin D, and acid phosphatase (14, 22, 28), quantification of lysosomal proteins or their functions may not provide the true picture of the state of lysosomes in Salmonella-infected cells. Therefore, we made use of the acidic nature of lysosomes; we stained infected cells with LT, a fluorophore that accumulates in acidic compartments of the cell. The fluorescence of LT can be measured using a flow cytometer. SCVs are not as acidic as lysosomes, and therefore, they do not accumulate LT. Hence, we used the fluorescence of LT to measure the volume of the acidic compartment of the host cell, to which lysosomes are the principal contributors.
FIG. 4.
Lysosomal proteins in Salmonella-infected RAW 264.7 cells. (A) Western blot analysis of cathepsin D, LAMP1, and LAMP2 expression in RAW 264.7 cells infected with Salmonella for 10 h. Two bands observed in the case of cathepsin D (infected sample) correspond to 54 kDa and 46 kDa. The numbers in parentheses are normalized values from densitometric analysis. (B) Acid phosphatase activity of RAW 264.7 cells infected with Salmonella for 10 h. The results are representative of three independent experiments. The error bars represent standard errors. The results are representative of three independent experiments (A) and two independent experiments (B). *, P < 0.01 (Student's t test). HK, heat-killed bacteria; WT, wild type.
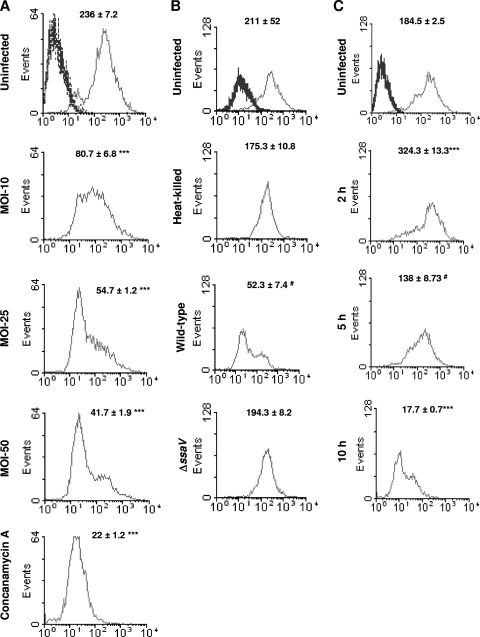
Interestingly, we observed an MOI-dependent decrease in the LT fluorescence of infected RAW 264.7 cells (Fig. 5A). Concanamycin A, a macrolide antibiotic known to increase the pH of lysosomes by inhibiting vacuolar ATPase, was used as a positive control (11). The type 3 secretion system (TTSS) encoded in Salmonella pathogenicity island 2 (SPI-2) is required for the proliferation of bacteria inside the host cell (38). Heat-killed bacteria and a ΔssaV strain (in which the TTSS encoded by SPI-2 is inactive) were unable to cause any significant change in the LT fluorescence in RAW 264.7 cells, indicating that proliferation of bacteria is essential to reduce the LT fluorescence in infected RAW 264.7 cells (Fig. 5B). Interestingly, Staphylococcus aureus, an extracellular pathogen, was unable to show a similar reduction in the LT fluorescence (see Fig. S6 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). We observed a significant increase in the LT fluorescence of infected cells at 2 h after infection, which might have been a host cell response to the invading pathogen. However, a decrease in the LT fluorescence started as early as 5 h after infection, which coincides with the beginning of multiplication of Salmonella inside macrophages (Fig. 5C) (39).
FIG. 5.
Salmonella infection reduces the volume of acidic compartments contributed by lysosomes in murine macrophages as inferred from LysoTracker-Green fluorescence. (A) Flow cytometric analysis of RAW 264.7 cells infected with Salmonella (at different MOIs for 10 h) and stained with LysoTracker-Green. Concanamycin A (50 nM), an inhibitor of vacuolar ATPases, was used as a positive control. (B) Flow cytometric analysis of RAW 264.7 cells infected with either wild-type, heat-killed, or ΔssaV strains of Salmonella (at an MOI of 50 for 10 h) and stained with LysoTracker-Green. (C) Flow cytometric analysis of RAW 264.7 cells infected (MOI, 50) with wild-type Salmonella for 2 h, 5 h, and 10 h and stained with LysoTracker-Green. Profiles of unstained cells are shown as heavy lines (left peaks) in panels A, B, and C. The x axis represents the fluorescence of LysoTracker-Green. The numbers represent means ± standard errors of mean fluorescence intensities obtained from three samples. The results are representative of at least two independent experiments. The statistical significance (Student's t test) of the difference compared to the corresponding uninfected samples are indicated (***, P < 0.001; #, P < 0.05).
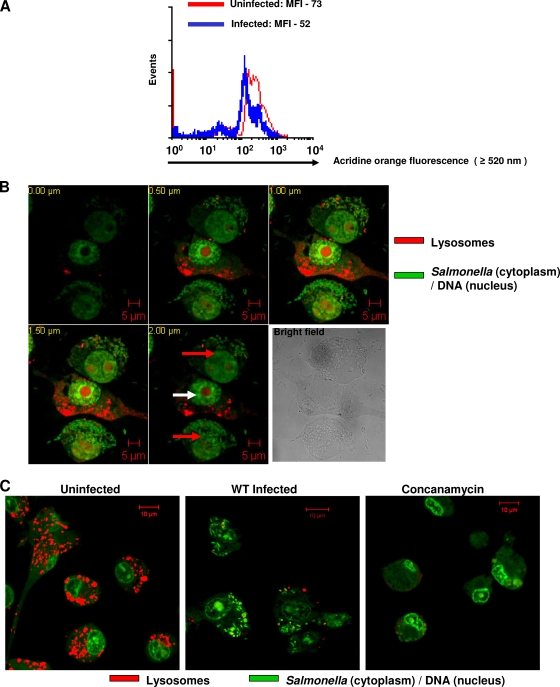
Acridine orange, a metachromatic fluorophore, has been extensively used to stain lysosomes (6, 29, 32, 42). We used acridine orange to confirm the results obtained using LT fluorescence. Flow cytometry demonstrated a significant decrease in the acridine orange fluorescence of RAW 264.7 cells upon Salmonella infection (Fig. 6A). In order to visualize lysosomes directly, we stained RAW 264.7 cells with acridine orange and observed them under a confocal laser scanning microscope. RAW 264.7 cells were infected with GFP-expressing Salmonella at an MOI of 10 to enable us to visualize both infected (containing many rod-shaped green bacteria) and uninfected cells in the same field. Acridine orange also stains nucleic acids. Nevertheless, lysosomes (discrete) could be easily distinguished from RNA (diffuse), and also, GFP-expressing bacteria (cytoplasm) could be easily distinguished from DNA (nucleus). We observed very few lysosomes stained with acridine orange inside RAW 264.7 cells infected with Salmonella compared to uninfected cells (Fig. 6B and C). Concanamycin A was used as a positive control. However, this decrease in lysosomal numbers was not observed in cells infected with the ΔssaV strain (see Fig. S7 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). These results clearly demonstrate that Salmonella infection reduces the volume of the acidic compartment in the host macrophages, to which lysosomes are the principal contributors.
FIG. 6.
Salmonella infection reduces the volume of acidic compartments contributed by lysosomes in murine macrophages as inferred from acridine orange fluorescence. (A) Flow cytometric analysis of RAW 264.7 cells infected with Salmonella (MOI, 50 for 10 h) and stained with acridine orange. The difference in MOIs is statistically significant (P < 0.05; Student's t test). MFI, mean fluorescence intensity. (B) Confocal laser scanning microscope images of RAW 264.7 cells infected with GFP-expressing Salmonella for 10 h and stained with acridine orange. Five different optical sections (0.5-μm interval) of three RAW 264.7 cells are shown. The upper and lower cells are infected (red arrows), whereas the middle cell is uninfected (white arrow). (C) Confocal laser scanning microscope images of RAW 264.7 cells stained with acridine orange. Representative images of cells either uninfected or infected with GFP-expressing Salmonella or treated with 50 nM of concanamycin A are shown. Rod-shaped GFP-expressing Salmonella bacteria can be seen inside infected cells in both panels B and C. The results are representative of at least two independent experiments. Scale bars, 5 μm (B) and 10 μm (C).
Salmonella reduces the lysosomal autofluorescence of the host cell.
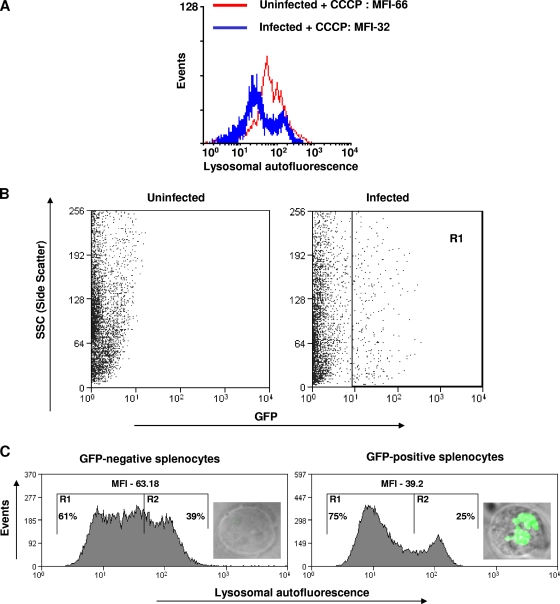
Lysosomes exhibit autofluorescence that originates from the lipofuscin pigment (excitation, 360 nm, and emission, 420 to 470 nm) (3). We used this property to quantify lysosomes by using flow cytometry. However, mitochondria also exhibit autofluorescence with spectral properties similar to those of lysosomal autofluorescence. Carbonyl cyanide m-chlorophenylhydrazone, a mitochondrial uncoupler, was used to block the autofluorescence of mitochondria (3). As expected, we observed a significant decrease in the lysosomal autofluorescence of RAW 264.7 cells upon infection for 10 h with Salmonella at an MOI of 10 (Fig. 7A). A similar result was observed at an MOI of 50 (data not shown). These results suggest that Salmonella infection decreases the number of lysosomes inside the host macrophages.
FIG. 7.
Salmonella reduces the lysosomal autofluorescence of host cells. (A) Flow cytometric analysis of lysosome-specific autofluorescence of RAW 264.7 cells infected with Salmonella (MOI, 10 for 10 h). Ten micromolar of carbonyl cyanide m-chlorophenylhydrazone was used to block the mitochondrial autofluorescence. The difference in MFI is statistically significant (P < 0.05; Student's t test). (B) Flow cytometry profile of splenocytes isolated from the spleens of either uninfected mice or mice infected intraperitoneally with GFP-expressing Salmonella (104 bacteria per mouse). GFP-positive cells (region R1) and GFP-negative cells were sorted using a fluorescence-assisted cell sorter. (C) Lysosome-specific autofluorescence of GFP-positive and GFP-negative splenocytes (20,000 each) isolated from a Salmonella-infected mouse. Ten micromoles of carbonyl cyanide m-chlorophenylhydrazone was used to block the mitochondrial autofluorescence. The difference in MFI is statistically significant (P < 0.05; Student's t test). The percentages of cells falling in R1 or R2 are shown. Representative images of GFP-positive (infected) and GFP-negative (uninfected) splenocytes are shown inside the respective graphs. The results are representative of two independent experiments.
Salmonella is known to cause cytotoxicity in host cells. Early, rapid cytotoxicity is caused by SPI-1 within 2 h of infection, and delayed cytotoxicity is SPI-2 dependent and begins after 12 h of infection (27). In our experiments, RAW 264.7 cells were infected with stationary-phase bacterial culture, in which SPI-1 is not induced, and the decrease in acidic lysosomes started at 5 h. Therefore, our observations are very unlikely to be due to cytotoxic effects of Salmonella. In order to rule out this possibility, we evaluated the cytotoxic effects of Salmonella. As expected, we observed minimal cell death (5 to 10%) in RAW 264.7 cells infected with Salmonella at an MOI of 50 (the maximum MOI used in this study) using both an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay and an acridine orange/ethidium bromide microscopic assay (35) (see Fig. S8 at http://mcbl.iisc.ernet.in/Welcome%20to%20MCBL/Faculty/Dipshikha/supplimentary.html). This much cytotoxicity cannot explain the extent of decrease in lysosomal fluorescence we have observed using various methods. Therefore, the effect of Salmonella on the lysosomes inferred via LT fluorescence, acridine orange fluorescence, and lysosomal autofluorescence is not due to the cytotoxic effect of Salmonella on the host cells.
Effect of Salmonella infection on lysosomes in vivo.
Next, we investigated the effect of Salmonella infection on lysosomes in vivo. Mice were intraperitoneally infected with GFP-expressing Salmonella (104 bacteria/mouse) and were sacrificed after 4 days of infection. Splenocytes infected with GFP-expressing Salmonella were sorted using a fluorescence-assisted cell sorter (Fig. 7B, R1 population). The presence of GFP-expressing Salmonella inside the sorted cells was confirmed by observation under a fluorescence microscope (Fig. 7C). Splenocytes were chosen for this analysis, as the spleen is one of the organs in which Salmonella proliferates within cells of monocytic lineage. We compared the autofluorescence of infected splenocytes with that of uninfected splenocytes, both isolated from the spleen of an infected mouse. Consistent with the in vitro results, we observed a significant decrease in the lysosomal autofluorescence of infected splenocytes (Fig. 7C). Carbonyl cyanide m-chlorophenylhydrazone was used to block the autofluorescence of mitochondria. This result demonstrates that Salmonella infection reduces the lysosomal autofluorescence of host cells in vivo.
Sodium orthovanadate partially rescues the Salmonella-induced decrease in acidic lysosomes inside RAW 264.7 cells.
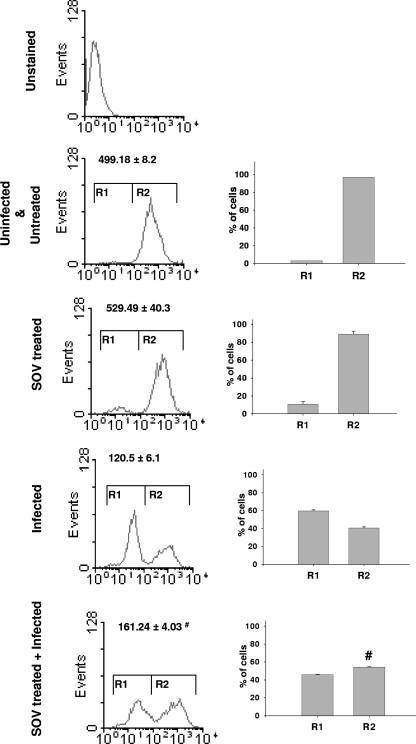
As mentioned above, the SCV is known to recruit many lysosomal proteins, like vacuolar-type ATPase, LAMP1, LAMP2, cathepsin D, and acid phosphatase (14, 22, 28). Probably because of this redistribution of lysosomal proteins to a growing number of SCVs, there is a shortage of raw materials for the biogenesis of acidic lysosomes inside the host cells infected with Salmonella. We next proceeded to test this hypothesis by investigating the number of acidic lysosomes in infected cells treated with SOV, where multiple bacteria cluster inside a single SCV. The lysosomal fluorescence levels of infected RAW 264.7 cells with and without SOV treatment were compared. Interestingly, we observed a partial increase (about 33%) in the lysosomal fluorescence in infected cells that were treated with SOV (Fig. 8). There was also a significant increase in the number of cells exhibiting high lysosomal fluorescence (Fig. 8, R2 population). The effect of SOV alone in the absence of infection was not statistically significant. This result suggests that the division of SCVs contributes to the depletion of acidic lysosomes inside RAW 264.7 cells.
FIG. 8.
Sodium orthovanadate treatment partially rescues depletion of acidic lysosomes by Salmonella. Shown is flow cytometric analysis of RAW 264.7 cells infected with Salmonella (at an MOI of 50 for 10 h) and stained with LysoTracker-Green. RAW 264.7 cells were treated with sodium orthovanadate (100 μM) as described in Materials and Methods and maintained throughout the experiment. The x axis represents the fluorescence of LysoTracker-Green. The numbers represent means ± standard errors of MFI obtained from three samples. The bars represent means and standard errors of the percentages of cells in the R1 and R2 populations. The results are representative of three independent experiments. The statistical significance (Student's t test) of the difference between infected samples with and without SOV treatment is shown (#, P < 0.05).
DISCUSSION
Pathogenic bacteria have evolved a variety of strategies to counteract the defense mechanisms of the host. Such strategies are vital for a bacterium to become a successful pathogen. Escaping lysosomal degradation is one such strategy that is very important for intracellular pathogens like Salmonella. The mechanism by which Salmonella escapes from lysosomal degradation is not clearly understood. In this study, we report a different perspective on this intriguing problem by analyzing the vacuolar and lysosomal loads inside infected cells.
Our results unequivocally demonstrate that a majority of the SCVs have only one bacterium per vacuole because of the division of SCVs, which causes an increase in the vacuolar load of the cell. Having a single bacterium per SCV could be advantageous to Salmonella in many ways. Exposure of Salmonella to microbicidal agents like lysosomes and the availability of nutrients to Salmonella are different in these two contrasting situations, i.e., multiple bacteria per SCV and a single bacterium per SCV. It is relatively difficult for the host cell to defend itself when there are many SCVs inside it; a host cell has to target each SCV separately with lysosomes, reactive oxygen and nitrogen intermediates, antimicrobial peptides, and other microbicidal agents. In contrast, a host cell has to target only a single or a few SCVs if many bacteria are clustered inside one or a few SCVs, which is advantageous for the host cell. In addition, in the case of a single bacterium per SCV, there is no competition for nutrients and each bacterium gets enough access to the SCV membrane to secrete effector proteins into the host cytoplasm using its type 3 secretion system. However, in the case of multiple bacteria per vacuole, bacteria have to compete with each other for nutrition inside the stringent environment of the SCV and the bacteria present at the center of the SCV might not get access to the SCV membrane to secrete their effector proteins into the host cytoplasm. Secretion of effector proteins by Salmonella into the host cytoplasm via its type 3 secretion system is essential for its survival and multiplication (20). The number of bacteria per SCV, which determines the SCV load per cell, thus appears to influence the survival and proliferation of Salmonella inside the host cell.
Our results obtained by flow cytometry and confocal laser scanning microscopy also demonstrate that Salmonella, on the other hand, reduces acidic lysosomes inside host macrophages, resulting in insufficient lysosomes for the growing number of SCVs. This is partially the result of SCV division, which increases the number of SCVs, causing redistribution of molecules, like vacuolar-type ATPase, LAMP1, LAMP2, cathepsin D, and acid phosphatase, that are required for lysosomal biogenesis. The remaining mechanisms involved in this extraordinary feat of Salmonella need to be investigated.
Our inference regarding the depletion of lysosomes inside Salmonella-infected macrophages was based on three different fluorescence sources. The fluorescence of acridine orange and LT depends on the pH of the lysosomes. However, the relation of lysosomal autofluorescence to the lysosomal pH is not known. Nevertheless, it is reasonable to conclude from our results that Salmonella reduces the quantity of acidic lysosomes inside the host cell. To our knowledge, there has been no report of any bacterial pathogen that affects the number of acidic lysosomes inside the host cells. Because this peculiar behavior of Salmonella reduces the microbicidal ability of the host cell, the host can potentially become susceptible to other intracellular and intravacuolar pathogens, like E. coli and Mycobacterium tuberculosis, and also to commensal bacteria.
Overall, our results unequivocally demonstrate that on one hand SCV undergoes division, increasing the number of vacuoles to be targeted by lysosomes and other microbicidal agents, and on the other hand, Salmonella decreases the number of acidic lysosomes in the infected host cells. As Salmonella proliferates, there is an imbalance in the ratio of the number of vacuoles to the number of acidic lysosomes that favors the bacteria. Probably because of this, earlier studies failed to find many SCV-lysosome fusion events and concluded that Salmonella blocks SCV-lysosome fusion. Actually, there are not enough lysosomes present in the infected host cells to fuse with all the SCVs. Nevertheless, in the initial period of infection, i.e., the first 4 to 5 h, when Salmonella does not multiply, it is crucial for Salmonella to block the SCV-lysosome fusion or to survive inside the harsh environment of the lysosomes, in case SCV-lysosome fusion takes place. As lysosomal biogenesis is a continuous process (36), Salmonella also has to avoid lysosomal degradation at later time points, which is done by causing an imbalance in the vacuole-to-lysosome ratio, as demonstrated in this study. Thus, Salmonella seems to use different mechanisms at different times to escape from lysosomal degradation.
Acknowledgments
This work was supported by the Department of Biotechnology (DBTO/197) and the Department of Atomic Energy (DAEO/119), the Director of IISc [Provision (2A) Tenth Plan (191/MCB)], the ICMR Center for Medical Microbiology, and DBT program support on Basic Biology of Microbial Pathogens. Infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST), and UGC (special assistance) is acknowledged. V.D.N. acknowledges C.S.I.R. for a fellowship.
We thank the flow cytometry facility and the confocal laser scanning microscope facility of IISc and the electron microscope facility of NIMHANS and IISc.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 26 October 2009.
REFERENCES
- 1.Abrahams, G. L., P. Muller, and M. Hensel. 2006. Functional dissection of SseF, a type III effector protein involved in positioning the salmonella-containing vacuole. Traffic 7:950-965. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, J., and K. Vickerman. 1975. Fusion of host cell secondary lysosomes with the parasitophorous vacuoles of Leishmania mexicana-infected macrophages. J. Protozool. 22:502-508. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, H., T. Baechi, M. Hoechl, and C. Richter. 1998. Autofluorescence of living cells. J. Microsc. 191:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., and F. Heffron. 1991. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 59:2232-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkhardt, J. K., C. J. Echeverri, T. Nilsson, and R. B. Vallee. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canonico, P. G., and J. W. Bird. 1969. The use of acridine orange as a lysosomal marker in rat skeletal muscle. J. Cell Biol. 43:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrol, M. E., P. S. Jackett, V. R. Aber, and D. B. Lowrie. 1979. Phagolysosome formation, cyclic adenosine 3′:5′-monophosphate and the fate of Salmonella typhimurium within mouse peritoneal macrophages. J. Gen. Microbiol. 110:421-429. [DOI] [PubMed] [Google Scholar]
- 8.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic, V., S. Singh, S. Master, J. Harris, E. Roberts, G. Kyei, A. Davis, S. de Haro, J. Naylor, H. H. Lee, and I. Vergne. 2006. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol. 8:719-727. [DOI] [PubMed] [Google Scholar]
- 10.Drecktrah, D., L. A. Knodler, D. Howe, and O. Steele-Mortimer. 2007. Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8:212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drose, S., K. U. Bindseil, E. J. Bowman, A. Siebers, A. Zeeck, and K. Altendorf. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32:3902-3906. [DOI] [PubMed] [Google Scholar]
- 12.Duclos, S., and M. Desjardins. 2000. Subversion of a young phagosome: the survival strategies of intracellular pathogens. Cell Microbiol. 2:365-377. [DOI] [PubMed] [Google Scholar]
- 13.Eswarappa, S. M., K. K. Panguluri, M. Hensel, and D. Chakravortty. 2008. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154:666-678. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-del Portillo, F., and B. B. Finlay. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol. 129:81-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-del Portillo, F., C. Nunez-Hernandez, B. Eisman, and J. Ramos-Vivas. 2008. Growth control in the Salmonella-containing vacuole. Curr. Opin. Microbiol. 11:46-52. [DOI] [PubMed] [Google Scholar]
- 16.Garvis, S. G., C. R. Beuzon, and D. W. Holden. 2001. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell Microbiol. 3:731-744. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons, I. R., M. P. Cosson, J. A. Evans, B. H. Gibbons, B. Houck, K. H. Martinson, W. S. Sale, and W. J. Tang. 1978. Potent inhibition of dynein adenosinetriphosphatase and of the motility of cilia and sperm flagella by vanadate. Proc. Natl. Acad. Sci. USA 75:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guignot, J., E. Caron, C. Beuzon, C. Bucci, J. Kagan, C. Roy, and D. W. Holden. 2004. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117:1033-1045. [DOI] [PubMed] [Google Scholar]
- 19.Hamon, M., H. Bierne, and P. Cossart. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423-434. [DOI] [PubMed] [Google Scholar]
- 20.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 21.Haraga, A., M. B. Ohlson, and S. I. Miller. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53-66. [DOI] [PubMed] [Google Scholar]
- 22.Hashim, S., K. Mukherjee, M. Raje, S. K. Basu, and A. Mukhopadhyay. 2000. Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J. Biol. Chem. 275:16281-16288. [DOI] [PubMed] [Google Scholar]
- 23.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishibashi, Y., and T. Arai. 1990. Specific inhibition of phagosome-lysosome fusion in murine macrophages mediated by Salmonella typhimurium infection. FEMS Microbiol. Immunol. 2:35-43. [DOI] [PubMed] [Google Scholar]
- 26.Kim, K. J., S. J. Elliott, F. Di Cello, M. F. Stins, and K. S. Kim. 2003. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol. 5:245-252. [DOI] [PubMed] [Google Scholar]
- 27.Knodler, L. A., and B. B. Finlay. 2001. Salmonella and apoptosis: to live or let die? Microbes Infect. 3:1321-1326. [DOI] [PubMed] [Google Scholar]
- 28.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 29.Kurz, D. J., S. Decary, Y. Hong, and J. D. Erusalimsky. 2000. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J. Cell Sci. 113:3613-3622. [DOI] [PubMed] [Google Scholar]
- 30.Marsman, M., I. Jordens, C. Kuijl, L. Janssen, and J. Neefjes. 2004. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol. Biol. Cell 15:2954-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriyama, Y., T. Takano, and S. Ohkuma. 1982. Acridine orange as a fluorescent probe for lysosomal proton pump. J. Biochem. 92:1333-1336. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa, M., and C. Sasakawa. 2006. Intracellular survival of Shigella. Cell Microbiol. 8:177-184. [DOI] [PubMed] [Google Scholar]
- 34.Oh, Y. K., C. Alpuche-Aranda, E. Berthiaume, T. Jinks, S. I. Miller, and J. A. Swanson. 1996. Rapid and complete fusion of macrophage lysosomes with phagosomes containing Salmonella typhimurium. Infect. Immun. 64:3877-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parks, D. R., V. M. Bryan, V. T. Oi, and L. A. Herzenberg. 1979. Antigen-specific identification and cloning of hybridomas with a fluorescence-activated cell sorter. Proc. Natl. Acad. Sci. USA 76:1962-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saftig, P., and J. Klumperman. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623-635. [DOI] [PubMed] [Google Scholar]
- 37.Sansonetti, P. J. 2004. War and peace at mucosal surfaces. Nat. Rev. Immunol. 4:953-964. [DOI] [PubMed] [Google Scholar]
- 38.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steele-Mortimer, O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varadi, A., L. I. Johnson-Cadwell, V. Cirulli, Y. Yoon, V. J. Allan, and G. A. Rutter. 2004. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J. Cell Sci. 117:4389-4400. [DOI] [PubMed] [Google Scholar]
- 41.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 42.Zelenin, A. V. 1966. Fluorescence microscopy of lysosomes and related structures in living cells. Nature 212:425-426. [DOI] [PubMed] [Google Scholar]