Abstract
The pneumococcus obtains its energy from the metabolism of host glycosides. Therefore, efficient degradation of host glycoproteins is integral to pneumococcal virulence. In search of novel pneumococcal glycosidases, we characterized the Streptococcus pneumoniae strain D39 protein encoded by SPD_0065 and found that this gene encodes a β-galactosidase. The SPD_0065 recombinant protein released galactose from desialylated fetuin, which was used here as a model of glycoproteins found in vivo. A pneumococcal mutant with a mutation in SPD_0065 showed diminished β-galactosidase activity, exhibited an extended lag period in mucin-containing defined medium, and cleaved significantly less galactose than the parental strain during growth on mucin. As pneumococcal β-galactosidase activity had been previously attributed solely to SPD_0562 (bgaA), we evaluated the contribution of SPD_0065 and SPD_0562 to total β-galactosidase activity. Mutation of either gene significantly reduced enzymatic activity, but β-galactosidase activity in the double mutant, although significantly less than that in either of the single mutants, was not completely abolished. The expression of SPD_0065 in S. pneumoniae grown in mucin-containing medium or tissues harvested from infected animals was significantly upregulated compared to that in pneumococci from glucose-containing medium. The SPD_0065 mutant strain was found to be attenuated in virulence in a manner specific to the host tissue.
Streptococcus pneumoniae is the leading cause of bacterial pneumonia, otitis media, bacterial meningitis, and septicemia (21). Furthermore, a high percentage of the population carries the bacterium in the nasopharynx, asymptomatically or as a prelude to disease (53). The upsurge in antibiotic-resistant strains and the search for new vaccines highlight the need to understand more completely the nature of pneumococcal virulence. A survey of pneumococcal virulence studies indicates a clear asymmetry in favor of research focusing on factors involved in attachment and damage to host tissues and in immune evasion (2, 11, 15, 22, 38). Although these studies have revealed useful information about pneumococcal virulence, there is a severe lack of knowledge on the basic physiology of S. pneumoniae, such that little information is available on how the pneumococcus fulfils its nutritional requirements for the generation of energy.
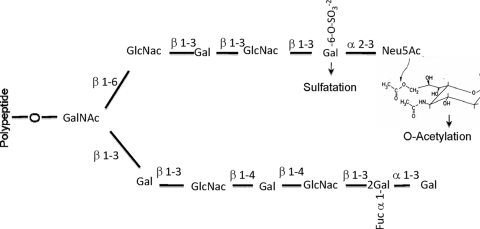
Carbohydrates are the principal energy sources for the pneumococcus, and these must be obtained exclusively from its host (16). Although the pneumococcus is known to utilize a variety of free sugars through at least 14 sugar utilization operons (10), in the upper respiratory tract the concentration of free sugars is low (39). However, monosaccharides are copious within the structures of O- and N-linked oligosaccharides of glycoproteins, which are found predominantly in the structures of mucins (Fig. 1) and in circulatory glycoproteins, respectively. Previous studies have shown that the sequential deglycosylation of N-linked glycan structures by pneumococcal glycosidases mediates in vitro growth of S. pneumoniae (5).
FIG. 1.
Schematic representation of the oligosaccharide structure of mucins. The carbohydrate chain is O-glycosidically linked to the apomucin through N-acetylgalactosamine (GalNAc). Galactose (Gal), N-acetylglucosamine (GlcNAc), sialic acid (Neu5Ac), sulfate (SO32−), and fucose (Fuc) residues are indicated. The structure of 9-O-acetylated sialic acid is shown as a structural formula (constructed from data presented in reference 49).
The apical surfaces of the respiratory tract epithelium are covered with mucus, which plays a vital role in the removal of pathogens and environmental toxins via the mucocilliary escalator (42). Paradoxically, mucus is a potentially rich source of carbon and nitrogen for bacteria with a sufficient array of glycosidase activities (43). Mucin is the major macromolecular compound of mucus, and the heavily glycosylated side chains of this glycoprotein may be utilized by bacteria as a carbohydrate source. Although the detailed structure of mucins in different organs exhibits variation, the majority of mucin glycans contain N-acetylgalactosamine, N-acetylglucosamine, galactose, fucose, sulfated sugars, and sialic acids in various proportions (Fig. 1) (41, 43). The acetylation of terminal sialic acids and sulfation of other monosaccharides, to a certain degree, protect carbohydrate side chains against the cleavage by microbial glycosidases (7).
Pneumococci degrade mucin for growth, and NanA is essential for the mucin degradation ability of the pathogen (52), presumably for removal of terminal sialic acid residues to expose the carbohydrate core of oligosaccharides to further degradation by other glycosidases. Recently it was also shown that NanB plays a role in the deglycosylation of human glycoconjugates by some pneumococcal strains (5). It has been demonstrated that mucin stimulates expression of the neuraminidase in the pneumococcus (49), thus enhancing its own degradation. Using isogenic nanA and nanB mutant strains, the involvement of neuraminidases in the pathology of pneumococcal otitis media, meningitis, and pneumonia has been demonstrated by us and others (30, 48). In addition to neuraminidases NanA (SPD_1504) and NanB (SPD_1499), BgaA (SPD_0562) and StrH (SPD0063), which catalyze the hydrolysis of terminal nonreducing β-d-galactose and N-acetyl-β-d-glucosamine residues, respectively, have been shown to be involved in the deglycosylation of human glycoconjugates and to contribute to pneumococcal colonization and pathogenesis of disease (24).
The roles of other glycosidases remain unknown, yet at least one-third of the pneumococcal genome encodes proteins involved in sugar transport, degradation, and processing, suggesting that carbohydrate metabolism has a central impact on pneumococcal biology (16, 27, 47). In the sequenced pneumococcal genome, several genes are annotated as sugar hydrolases (otherwise known as glycosidases); however, the importance of these in glycoprotein degradation is not known. In this study we explored the role in glycoprotein degradation and virulence of SPD_0065, annotated as β-galactosidase 3 in the D39 genome (27). During preparation of this paper, Jeong et al. (20) reported some properties of SPD_0065. Because pneumococcal β-galactosidase activity previously had been attributed exclusively to SPD_0562 (54), we investigated the contribution of this newly identified galactosidase, SPD_0065, to total β-galactosidase activity by constructing a pneumococcal strain with both genes mutated. Using an isogenic mutant, it was shown that this glycosidase is important in mucin degradation. The involvement of SPD_0065 in pneumococcal virulence was also tested using this mutant strain in a mouse model of pneumonia and bacteremia, where it was found that SPD_0065 is important primarily for survival in the nasopharynx.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptococcus pneumoniae serotype 2 strain D39 and serotype 6B strain IO11966, kindly provided by Brian Spratt (Imperial College, London), were used in this study. Routinely, pneumococci were grown in brain heart infusion (BHI) broth or on blood agar plates supplemented with 5% (vol/vol) defibrinated horse blood under microaerophilic conditions at 37°C. Where appropriate, spectinomycin (100 μg/ml) or kanamycin (250 μg/ml) was added to the culture medium.
Sicard's defined medium (44), with modifications, was also used for bacterial growth. The medium was modified to contain mucin as the sole carbon source, replacing glucose and bovine serum albumin (BSA). Porcine gastric mucin (PGM) (Sigma, Dorset, United Kingdom) was dissolved in water at a concentration of 10 mg/ml and dialyzed against water overnight at 4°C using a snakeskin dialysis membrane (molecular mass cutoff, 10 kDa; Pierce, Hoddesdon, United Kingdom). After freeze-drying, the mucin was dissolved in 10 mM potassium phosphate buffer (pH 7.0), which was autoclaved at 121°C for 15 min. This was then briefly centrifuged to remove insoluble residues and mixed with 5× concentrated Sicard's medium.
Construction of mutants.
To construct strain D39 mutants, the chromosomal regions encompassing SPD_0065 and SPD_0562 were amplified with the primers listed in Table 1 (those indicated with an “F” or “R” tag). The amplified products were incubated with Himar1 transposase (26) and plasmid pR412, which contains the mariner minitransposon conferring spectinomycin resistance (32). The in vitro-mutagenized DNA was then transformed into the pneumococcus using competence-stimulating peptide (1). Transformants were selected for spectinomycin resistance, and insertion of the resistance cassette was confirmed by PCR using transposon-specific primers, MP127 or MP128, with appropriate chromosomal primers. In addition, mutations were also confirmed by sequencing. A representative strain for each mutation was selected, and these were designated SPD0065M and SPD0562M. The SPD_0065 homolog was also mutated in S. pneumoniae serotype 6B. To do this, the mutated region was amplified from SPD0065M using SPD0065F and SPD0065R primers, and the amplified region was transformed into S. pneumoniae serotype 6B, as before. One of the transformants, designated serotype 6B/65 M, was chosen for further analysis.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ → 3′) | Target region in D39 pneumococcal genome (reference)d |
|---|---|---|
| SPD0065F | CTGCTAAACTGCCCAGAGATG | 65426-67226 |
| SPD0065R | GGATACGCAACGTAAGGGAA | |
| SPD0562F | CATGGCAAGGTTGAAACTCA | 580628-582435 |
| SPD0562R | GCCGTGAACGCTATAAGGCGC | |
| SPD0065CF | GCGAGATCTTGATGACACGATTTGAGATACGAGAT | NAa |
| SPD0065CF | GCGAGATCTTGTCATAAGTTTTCCCCCTTTATATGTT | |
| MALF | GCTTGAAAAGGAGTATACTT | NAb |
| PCEPR | AGGAGACATTCCTTCCGTATC | |
| MP127 | CCGGGGACTTATCAGCCAACC | pR410 and pR412 (32) |
| MP128 | TACTAGCGACGCCATCTATGTG | |
| SPD0709F | TCGTGTGGCTGCCAAGCGTG | 724517-724650 |
| SPD0709R | GGCTGATCCACCAGCTGAGTC | |
| SPD0065RTF | GGACCTCTTTGTAACAGGAA | 66449-66608 |
| SPD0065RTR | CATCTGCCAATTCCTTAGGA | |
| SPD0562RTF | CCTTAGAGCTAAGCATCCAA | 581545-581721 |
| SPD0562RTR | GTTGCTGTTTTCCCCCAA | |
| SPD0065IFF | TAAGGCCTCTGTCGACACACGATTTGAGATACGAGAc | 65847-67631 |
| SPD0065IFR | CAGAATTCGCAAGCTTTCATAAGTTTTCCCCCTTTA |
To make the double galactosidase mutant of strain D39, first a kanamycin-resistant mutant (SPD0562K) was constructed as described above, with the exception that plasmid pR410, which carries a kanamycin resistance cassette, was used. The mutated region in the spectinomycin-resistant SPD0065M strain was then PCR amplified with SPD0065F and SPD0065R primers, and the amplified region was transformed into SPD0562K. Transformants were selected in the presence of spectinomycin and kanamycin, and the presence of the mutation in each target was confirmed as described above. One transformant, designated SPD0065M/0562K, was selected for further study.
Complementation of SPD0065M.
To confirm that mutation of SPD_0065 introduced no polar effects, SPD0065M was complemented with an intact copy of the gene using pCEP, which is a nonreplicative plasmid that allows controlled gene expression following ectopic integration into the chromosome (13). SPD_0065 was amplified with the SPD0065CF and SPD0065CR primers, which incorporate BglII sites in the 5′ and 3′ ends of the gene, respectively (Table 1). The amplicons were digested with BglII, and the digested products were ligated with BamHI-digested pCEP. A sample of ligation mixture was directly transformed into SPD0065M as described previously (1), and the transformants were selected in the presence of spectinomycin and kanamycin (500 μg/ml). The successful introduction of the intact copy of the gene was confirmed by PCR using MALF and pCEPR primers (Table 1), whose recognition sites are localized immediately up- and downstream of the cloning site, respectively. The complemented strain was designated SPD0065C.
RNA extraction from bacterial cells and purification.
The extraction of RNA was done by the Trizol method using mid-log-phase cultures, as described previously (45). Before use, the RNA was treated with amplification grade DNase I (Qiagen, Crawley, United Kingdom) and subsequently purified with an RNeasy mini kit (Qiagen).
Extraction of pneumococcal RNA from infected tissues.
Outbred 9-week-old female MF1 mice (Harlan Olac, Bicester, United Kingdom) were intranasally infected with 50 μl phosphate-buffered saline (PBS) containing 1 × 106 passaged D39 pneumococci, as before (51, 53). When the mice became severely lethargic, they were anesthetized and blood was collected by cardiac puncture. After killing by cervical dislocation, the lungs and nasopharynx were removed and homogenized on ice in 10 ml of sterile PBS using a tissue homogenizer. To separate pneumococci from host cells, lung homogenates and blood samples were centrifuged at 900 × g for 6 min at 4°C. Supernatants were subsequently centrifuged at 15,500 × g for 2 min at 4°C, and the bacterial pellet was stored at −80°C until further processing. Prior to pelleting, 20 μl of homogenate was removed, serially diluted in PBS, and plated onto blood agar in order to enumerate pneumococci and to exclude the presence of contaminating microflora. RNA extraction and purification were done as described in the previous section.
Quantitative reverse transcription-PCR (RT-PCR).
First-strand cDNA synthesis was performed on approximately 1 μg of DNase-treated total RNA, immediately after isolation, using 200 U of SuperScript II reverse transcriptase (Invitrogen, Paisley, United Kingdom) at 42°C for 55 min and random hexamers (52). cDNA (15 ng) was amplified in a 20-μl reaction volume that contained 1× SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and 3 pmol of each primer (indicated with an “RTF” or “RTR” tag in Table 1). The transcription levels of specific genes were normalized to transcription of gyrB, which was amplified in parallel with SPD0709F and SP0709R primers. The results were analyzed by the comparative threshold cycle (CT) method (29).
Cloning, expression, and purification of recombinant enzymes.
The Talon Express bacterial expression and purification kit (Clontech, Saint-Germain-en-Laye, France) was used for the cloning, expression, and purification of polyhistidine-tagged proteins. Briefly, SPD_0065 was amplified by PCR with the SPD0065IFF and SPD0065IFR primers (Table 1). These primers incorporate 15 bases of flanking sequences that match the ends of the In-Fusion Ready pEcoli-Nterm 6XHN vector. The PCR conditions for amplification were as follows: 1 cycle of initial denaturation at 95°C for 45 s; 35 cycles of denaturation at 95°C for 45 s, annealing at 55°C for 60 s, and extension at 72°C for 2 min; and a final extension at 72°C for 3 min. The amplicons were mixed with prelinearized In-Fusion Ready pEcoli-Nterm 6XHN vector and In-Fusion Dry-Down mixture, which contains ligase and ligation buffer. The recombinant constructs were transformed into Fusion-Blue competent Escherichia coli, and two transformants were selected for sequencing. Once it was ensured that no mutational event had taken place, the construct DNA was transformed into the expression host E. coli BL21(DE3) (Novagen, Nottingham, United Kingdom). The bacterial culture for recombinant protein expression was maintained at 25°C on a shaking platform. The protein purification used Imac resin and nondenaturing conditions as instructed by the manufacturer (Clontech). The identity of the protein was verified by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometric analysis of tryptic digests of the products by PNACL at the University of Leicester.
Enzyme assays.
Sicard's medium supplemented with mucin (10 ml) was inoculated with approximately 1 × 107 CFU/ml pneumococci. After 6 h of incubation at 37°C, the bacteria were pelleted by centrifugation at 3,000 × g for 10 min. The pellet was resuspended in ice cold 50 mM Tris-HCl, pH 7.5. After freeze-thawing, lysates were prepared by sonication (51). The glycosidase assays used chromogenic or fluorogenic substrates and followed the previously described procedure (33). The protein concentration was determined by the Bradford assay, using BSA as the standard (4). The activities are expressed in units (μmol of p-nitrophenol or 4-methylumbelliferone produced per min) per mg of protein.
The pH optima of glycosidases in lysates of the wild-type strain were determined using 0.2 M sodium citrate buffer (pH 3.0 to 6.0), 0.2 M sodium phosphate buffer (pH 6.0 to 8.0), and 0.2 M Tris-HCl buffer (pH 7.5 to 8.9). The substrate specificity of the recombinant SPD_0065 was determined using chromogenic and fluorogenic substrates, as described above. Km and Vmax were assessed at the pH optimum of the enzyme using the Lineweaver-Burk method (28).
The recombinant enzyme was also tested for its activity in releasing neutral or N-acetylated sugars from PGM, fetuin (bovine; Sigma), and their desialylated forms. Glycoproteins were desialylated by two rounds of treatment with an excess of immobilized neuraminidase (from Clostridium perfringens; Sigma), essentially according to the manufacturer's instructions. The desialylated glycoproteins were dialyzed against 1 M sodium acetate buffer (pH 5.0) and used as substrates in enzyme assays. Assays contained glycoproteins at a concentration of 0.2 to 1.5 mg/ml, and approximately 10 μg of recombinant SPD_0065 was added. Samples were incubated for 18 h at 37°C prior to analysis of released monosaccharides, as described below.
Preparation of antisera against recombinant SPD_0065.
Ten-week-old female MFI outbred mice (Harlan Olac, United Kingdom) were injected with 25 μg of recombinant protein, 33 μl of Imject alum adjuvant (Perbio Science, Cramlington, United Kingdom), and 67 μl of PBS. The control group was administered only adjuvant and PBS. Injections were repeated three times at 2-week intervals. At 2 weeks after the last injection, mice were anesthetized with 5% (vol/vol) fluothane (Astra Zeneca, Macclesfield, United Kingdom) over oxygen (1.5 to 2 liters/min), and blood was collected by cardiac puncture. After the blood was left at room temperature for 1 hour to clot, the serum was recovered by centrifugation at 5,000 × g for 10 min and was stored at −80°C until needed.
Cellular localization of SPD_0065.
The parental D39 strain was grown in minimal medium supplemented with mucin until late exponential phase, and the bacteria were harvested by centrifugation at 3,000 × g for 10 min. The pellet was resuspended in 10 mM Tris-HCl-1 mM EDTA buffer (pH 8.0) containing 25% (wt/vol) sucrose and 12.5 mg/ml lysozyme. The suspension was incubated at 37°C until a clear lysate was obtained. The cell lysate was centrifuged at 3,000 × g for 5 min, and the supernatant containing the cell wall was kept. The pellet was then resuspended in 500 μl of PBS, sonicated, and centrifuged at 3,000 × g for 10 min. The supernatant and the pellet were kept for the analysis of cytoplasmic and membrane proteins, respectively. The quality of fractionation was determined by assessing activities of lactate dehydrogenase, which is known have an intracellular localization, and neuraminidase, which is localized on the cell wall, in different fractions as described previously (17, 52).
Pneumococcal cellular fractions were subjected to SDS-PAGE and were electrophoretically transferred to nylon membranes at a constant current of 250 mA for 1 h. The membrane was blocked in TBST (100 mM Tris, 0.15 M NaCl, 0.05% [vol/vol] Tween 20, pH 7.5) containing 5% (wt/vol) milk powder for 16 h and then incubated with 1:1,000 antiserum (see above) diluted in PBS (pH 7.0) for 2 h. After washing three times with TBST for 10 min, the filter was incubated with 1:2,000 anti-mouse IgG Fc-specific alkaline phosphatase (Sigma) diluted in PBS pH 7.0 for 1 h. Bound antibody was detected with an alkaline phosphatase detection kit (Sigma). The reaction was stopped by washing the membrane with distilled water.
Analysis of glycoprotein degradation.
The pneumococcal strains were grown in modified Sicard's medium without glucose and supplemented with 1% (wt/vol) mucin for 48 h. Cells were pelleted by centrifugation at 1,700 × g for 10 min, and the supernatants were retained and stored at −80°C until required. Controls for the analysis of monosaccharide removal from mucin due to the action of pneumococcal glycosidases comprised uninoculated, mucin-supplemented medium. Mucin-containing supernatants were dialyzed against distilled water, and residual sialic acids (N-acetylneuraminic and N-acetylglycolylneuraminic acids) and neutral/amino sugars (galactose, fucose, N-acetylglucosamine, and N-acetylgalactosamine) were released by acid hydrolysis, essentially as previously described (6). Sialic acids and neutral/amino sugars were analyzed in separate runs by high-pH anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using a DX500 high-pressure liquid chromatography (HPLC) system (Dionex, Camberley, Surrey, United Kingdom), as previously described (6). The release of monosaccharides from PGM, fetuin, and their desialylated forms by recombinant SPD_0065 was also monitored by HPAEC-PAD. Monosaccharide concentrations of mucin after growth of pneumococcal strains were related to the protein concentrations in individual samples to take into account sample losses or volume changes occurring during dialysis. The statistical significance of removal of mucin monosaccharides by S. pneumoniae strains was determined relative to uninoculated medium controls using the Student t test (P < 0.05).
In vivo virulence studies.
Ten-week-old female MFI outbred mice (Harlan Olac) were used for virulence testing. A standardized inoculum was prepared as described previously (51, 53). To determine the virulence of pneumococcal strains, mice were lightly anesthetized as described above and a 50-μl sample of PBS containing approximately 5 × 105 S. pneumoniae organisms was given dropwise into the nostrils. The inoculum dose was confirmed by viable counting on blood agar plates. Mice were monitored for disease signs (progressively starry coat, hunched posture, and lethargy) for 7 days (36), and those that reached the severely lethargic stage were considered to have reached the end point of the assay and were killed humanely. The time to this point was defined as “survival time.” Mice that were alive 7 days after infection were deemed to have survived the infection. To determine the development of bacteremia in each mouse, approximately 20 μl of venous blood was obtained from intranasally infected mice at predetermined time points after infection, and viable counts were determined as before. Survival times were analyzed by the Mann-Whitney U test.
Growth of pneumococci in the nasopharynx and lungs was also determined. For this, at predetermined time intervals following intranasal infection, set groups of mice were deeply anesthetized, and subsequently the mice were killed by cervical dislocation. The lungs and nasopharynx were transferred separately into 10 ml of sterile PBS, weighed, and then homogenized in a Stomacher-Lab blender (Seward Medical, London, United Kingdom) (51). Viable counts in homogenates were determined as described above. Data were analyzed by analysis of variance followed by the Bonferroni posttest. P values of <0.05 were considered statistically significant.
RESULTS
The choice of target was based on bioinformatic data. SPD_0065 is predicted to be a member of the glycosyl hydrolase family of proteins, which is a widespread group of enzymes that hydrolyze the glycosidic bond between monosaccharides or between a carbohydrate and a noncarbohydrate moiety (14). SPD_0065 is annotated as a β-galactosidase 3 gene. It is present in all sequenced pneumococcal genomes (16, 27, 47) and was identified as part of the core genomes of strains associated with colonization and invasive disease (37).
Enzyme assays.
In addition to neuraminidase, the activities of several other glycosidases are required to mediate glycoprotein degradation: these include β-galactosidase and N-acetylhexosaminidase (24). Initially, the pH optima of these enzymes were determined using crude lysates of the wild-type D39 strain, and these were found to be 6.5 and 5.0 for β-galactosidase and N-acetylglucosaminidase, respectively. When strain SPD0065M was tested for N-acetylglucosaminidase activity, it was found to have a level of activity (57.3 ± 13 U/mg; n = 6) similar to that of the wild type (52.1 ± 13 U/mg; n = 6) (P > 0.05), demonstrating that SPD_0065 does not code for an N-acetylglucosaminidase activity.
The β-galactosidase activity in SPD0065M, as measured using p-nitrophenyl (pNP)-β-d-galactopyranoside as a substrate, was significantly lower (10.5 ± 0.5 U/mg; n = 6) than that of strain D39 (17.1 ± 0.5 U/mg; n = 8) grown under the same conditions (P < 0.05) (Fig. 2). Reintroduction of an intact copy of SPD_0065 into SPD0065M (strain SPD0065C) restored the β-galactosidase activity to a level (19.0 ± 1.7 U/mg; n = 6) similar to that in the wild type. SPD0562M, which is mutated in a previously described pneumococcal β-galactosidase, BgaA (54), also showed a significant reduction in activity (9.3 ± 0.3 U/mg; n = 6) compared to D39 (P < 0.001).
FIG. 2.
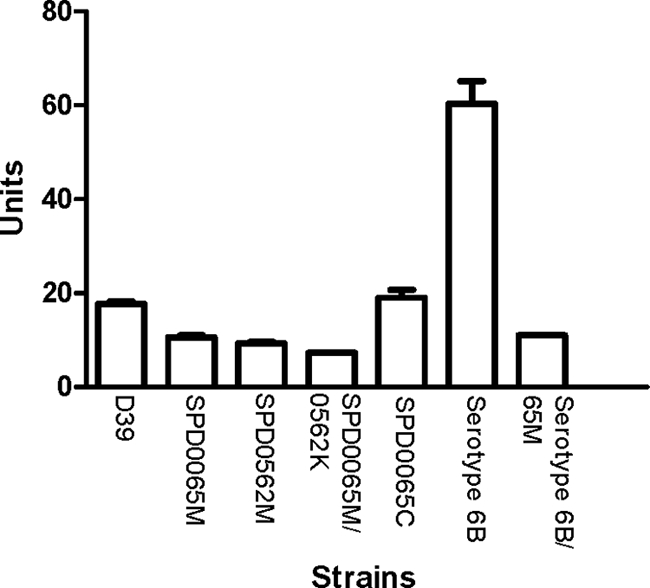
β-Galactosidase activities of pneumococcal strains. Pneumococcal cell lysates were prepared from the cultures containing PGM. The activities are expressed in units (nmol of p-nitrophenol produced per min) per mg protein. Each datum point is the mean of at least six individual measurements. Error bars indicate standard deviations.
Previously it was suggested that SPD_0562 is solely responsible for pneumococcal β-galactosidase activity (54); however, our results demonstrate otherwise. The double β-galactosidase mutant (SPD0065/0562K) had significantly less activity (7.3 ± 0.1 U/mg; n = 8) than D39 (P < 0.001) (Fig. 2). While there was no difference in activity between SPD0065M and SPD0562M (P > 0.05), both single mutants had more activity than the double mutant, (P < 0.001 for SPD0065M and P < 0.05 for SPD0562M). The residual activity in the double mutant suggests the existence of another pneumococcal hydrolase(s) that can utilize pNP-β-d-galactopyranoside as a substrate. Our results also showed that the absence of SPD_0065 is not compensated for by increased expression of SPD_0562 or vice versa (data not shown), indicating that the regulations of these genes are independent of each other.
In addition, we demonstrated that SPD_0065 expression is not limited to serotype 2 but is also produced by a serotype 6B strain. Total β-galactosidase activity in a serotype 6B strain (60.3 ± 4.9; n = 6) was significantly higher than in strain D39 (P < 0.05), and the mutation of the SPD_0065 homolog in the 6B strain resulted in significant reduction in total β-galactosidase activity relative to that in the parental strain (P < 0.01).
Gene expression.
The expression of SPD_0065 was measured in D39. Compared to growth in glucose, in mucin-containing Sicard's medium the mRNA level for SPD_0065 went up 189-fold (±11-fold; n = 3). In addition, the expression of this gene was measured in D39 recovered from intranasally infected mice. In bacteria from the nasopharynx, the SPD_0065 mRNA level, relative to that in bacteria cultured in vitro in glucose-containing medium, went up 2.2-fold, (±0.1-fold; n = 3), but the expression sharply increased in the lungs (35-fold ± 0.4-fold; n = 3) and blood (6.5-fold ± 1.1-fold; n = 3). The increased expression of this gene in mucin-containing medium and in vivo in the respiratory tract suggests a role for the β-galactosidase in glycoprotein degradation both in vivo and in vitro during growth on mucin.
Characterization of SPD_0065 recombinant protein.
It was decided to clone and express the protein coded by SPD_0065 for further characterization. Analysis by SDS-PAGE showed that SPD_0065 encoded a protein of approximately 69 kDa (data not shown), which is in line with the predicted molecular mass of the full-length polypeptide. The identity of the purified protein was verified by MALDI-TOF analysis (data not shown).
The substrate specificity of the recombinant protein was tested using various chromogenic and fluorogenic substrates (Table 2). It was found that the SPD_0065 protein had activity against pNP-β-d-galactopyranoside (1.03 ± 0.03 U/mg; n = 6). The observed activity of the protein was exclusively for this substrate, since the enzyme did not act on any other substrates tested (Table 2). Analysis of kinetic parameters showed that the β-galactosidase had a Km of 0.91 ± 0.29 mM and a Vmax of 5.88 ± 0.56 μmol/min/mg with pNP-β-d-galactopyranoside as substrate. These values are similar to those reported for the Streptococcus cremoris (Km, 0.38 mM) and Streptococcus thermophilus (Km, 0.25 mM) β-galactosidases (19, 40).
TABLE 2.
Substrate specificity of recombinant SPD0065 protein
| Substrate | Activitya |
|---|---|
| pNP-β-d-galactopyranoside | + |
| 4-Methylumbelliferyl-N-acetyl-α-d-glucosaminide | − |
| 4-Methylumbeliferyl-α-d-glucopyranoside | − |
| O-NP-α-d-galactopyranoside | − |
| pNP-α-d-glucoside-6-phosphate | − |
| pNP-α-d-mannose-6-phosphate | − |
| pNP-α-d-galactose-6-phosphate | − |
| pNP-β-d-glucose-6-phosphate | − |
Minus signs indicate the absence and plus signs show the presence of activity against individual substrates.
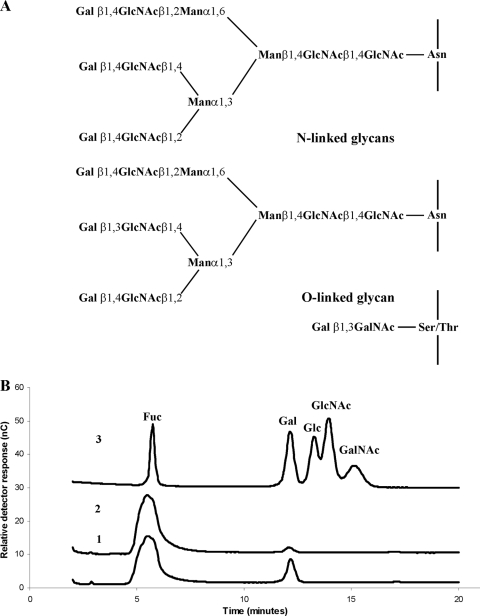
The ability of the recombinant β-galactosidase to release sugars from glycoproteins was examined using PGM and fetuin as substrates, either with or without prior treatment with immobilized neuraminidase because sialic acid removal has been shown to be a prerequisite for the activity of exoglycosidases, including the β-galactosidase BgaA (24). Fetuin, which is also known as α2-HS-glycoprotein in humans, is a serum glycoprotein. This was used as a substrate because the structural complexity of its oligosaccharides is substantially less than that for mucins (12) and the constituent sugars are subject to little modification by, for example, the sulfation that is extensive in many mucins and which is known to limit glycosidic activity. In addition, fetuin possesses galactose residues in both β1-4 and β1-3 linkages (Fig. 3A), suggesting that it would be a suitable substrate for the assay of galactosidases with unknown substrate specificities. Treatment of PGM with neuraminidase resulted in only approximately 10% removal of total sialic acids, perhaps reflecting O acetylation of the majority of these residues, which decreases their susceptibility to cleavage. Greater than 95% of sialic acid was removed from fetuin, however, after exposure to neuraminidase. This treatment resulted in a glycoprotein containing N- and O-linked glycans (Fig. 3A), the majority of which have reducing terminal galactose residues. Treatment of PGM (intact or partially desialylated) with recombinant SPD_0065 did not result in detectable release of any of the neutral or amino sugars which are found in the glycoproteins (data not shown), probably because galactose residues are blocked by the presence of other monosaccharides in this complex glycoprotein. We demonstrated, however, that recombinant SPD_0065 acted on desialylated fetuin, releasing galactose from the glycans (Fig. 3B). Activity of the enzyme against fully sialylated fetuin was minimal, with the release of only a trace amount of galactose (Fig. 3B), indicating that sialic acid removal from oligosaccharides to expose galactose was essential for the activity of the enzyme.
FIG. 3.
Release of galactose from fetuin by recombinant SPD_0065. (A) Glycan structures of desialylated fetuin (constructed from data presented in references 12 and 46 and references therein). (B) HPAEC-PAD separation and detection of monosaccharides. Traces 1 and 2 show monosaccharide release from desialylated and intact fetuin, respectively, following treatment with recombinant SPD_0065. Trace 3 shows the separation of monosaccharide standards (Fuc, fucose; Gal, galactose, Glc, glucose; GlcNAc, N-acetylglucosamine; GalNAc, N-acetylgalactosamine). Glucose, while not a component of PGM or fetuin oligosaccharides, was included in the standard mixture as it can be a contaminant introduced during processing of glycoproteins.
Mucin utilization.
As we were unable to show release of galactose from PGM by recombinant SPD_0065, probably because the sequential action of additional glycosidases was required to expose this monosaccharide, we cultured bacteria in Sicard's defined medium containing mucin as the sole carbon source. In this medium, SPD0065M grew with a lower growth rate during the exponential phase (0.5 ± 0.01/h; n = 6) than D39 (0.72 ± 0.02/h; n = 6) (P < 0.01) (Fig. 4). While strain D39 started to grow exponentially following a 6-h lag period with a net increase of 2.0 (±0.3) log10 CFU/ml over 6 h, SPD0065M exhibited an extended lag period of 12 h. A period of growth was then detected over 12 h, with a net increase of 1.7 (±0.3) log10 CFU/ml. However, the growth yields of the two strains were not significantly different (P > 0.05). The growth pattern and growth rate of the complemented mutant SPD0065C were similar to those of D39, indicating that the observed effect of the SPD_0065 mutation is not due to a polar effect (Fig. 4). There was no difference in the growth kinetics of the mutant and the parental strain in BHI or Sicard's medium containing glucose and BSA (data not shown).
FIG. 4.
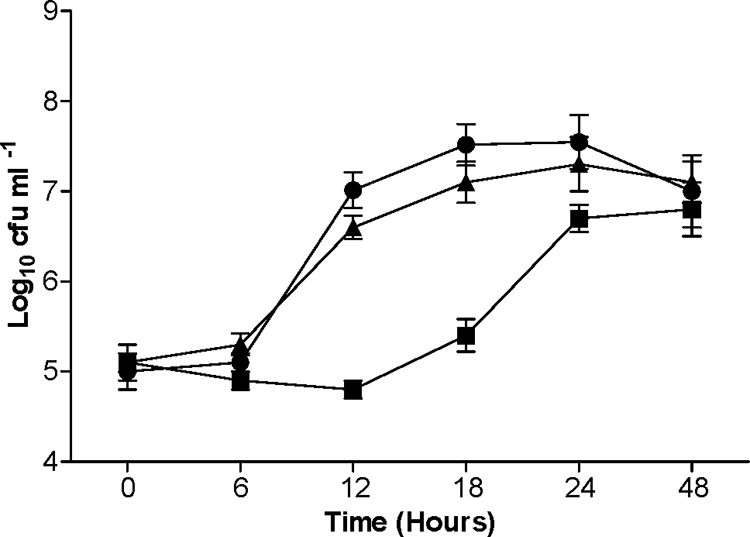
Growth kinetics of pneumococcal strains in defined medium with mucin as the sole carbon source. Growth was monitored over 48 h. Each datum point represents the mean of three to six independent experiments. Error bars indicate standard deviations. •, D39; ▪, SPD0065M; ▴, SPD0065C.
PGM is a heavily glycosylated protein, and our analysis of its composition showed that it was comprised of 7% (wt/vol) fucose, 44% (wt/vol) hexosamines (N-acetylglucosamine and N-acetylgalactosamine in a molar ratio of approximately 2:1), 14% (wt/vol) galactose, and 3% (wt/vol) neuraminic acids (N-acetylneuraminic acid and N-glycolylneuraminic acid in a molar ratio of approximately 6:1). After 48 h of growth of the wild-type D39 strain in mucin-containing Sicard's medium, approximately 40 to 50% of both N-acetyl- and N-glycolylneuraminic acid had been cleaved from the glycoprotein backbone. The galactose and N-acetylgalactosamine concentrations of PGM were reduced by approximately 30% and 35%, respectively. There was limited cleavage of fucose and N-acetylglucosamine following 48 h growth of bacteria, and the composition of PGM in the supernatants, with respect to these two sugars, did not differ significantly from that of uninoculated medium controls.
Strain SPD0065M, which lacked the putative galactosidase, released the same amount of sialic acids from PGM as the wild-type strain after 48 h of growth. Galactose cleavage was reduced from 30% in strain D39 to only 13% of the total in this mutant, and there was no significant removal of N-acetylgalactosamine, fucose, or N-acetylglucosamine from PGM by SPD0065M.
Determination of the cellular localization of SPD_0065.
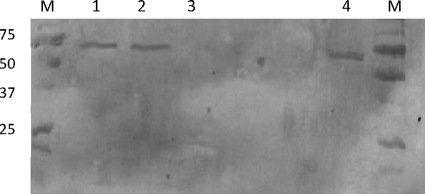
The cellular localization of the newly identified β-galactosidase (SPD_0065) was determined by immunoblotting using a polyclonal serum against the enzyme. Immune serum raised against the SPD_0065 protein reacted with the cell membrane and cell wall fractions, but no evidence for a cytoplasmic location was seen (Fig. 5).
FIG. 5.
Cellular localization of SPD_0065. Cellular fractions of S. pneumoniae, i.e., cell wall (lane 1), membrane (lane 2), and cytoplasm (lane 3), were separated on a 15% (wt/vol) SDS-polyacrylamide gel and capillary blotted to nylon membranes, and the localization of protein was determined using polyclonal antiserum specific for the enzyme. Lane 4, whole-cell lysate; lane M, protein markers with their molecular weights in thousands.
Virulence studies.
The median survival time of mice infected intranasally with SPD0065M (52 h ± 18 h; n = 10) was similar to that of the wild-type-infected cohort (48 h ± 15 h; n = 18) (P > 0.05). However, while bacteremia occurred sometime between 4 and 8 h after intranasal infection with the wild type, in the SPD0065M-infected mice it was delayed for a further 4 h. Once in the blood, SPD0065M grew at a rate similar to that of the wild type (data not shown). In addition, the progression of infection in the nasopharynx and lungs also was monitored (Fig. 6A and B). In the nasopharynx, SPD0065M colony counts (log10 1.75 ± 0.21, log10 1.99 ± 0.25, and log10 2.32 ± 0.47 [n = 10] for 12, 24, and 48 h postinfection, respectively) were less than those for D39 (log10 2.57 ± 0.09, log10 2.84 ± 0.11, and log10 3.43 ± 0.16; [n = 20] at 12, 24, and 48 h postinfection, respectively) (P < 0.01 for 12 and 24 h and P < 0.001 for 48 h). In the lungs, SPD0065M grew as well as the wild type at all time points (P > 0.05).
FIG. 6.
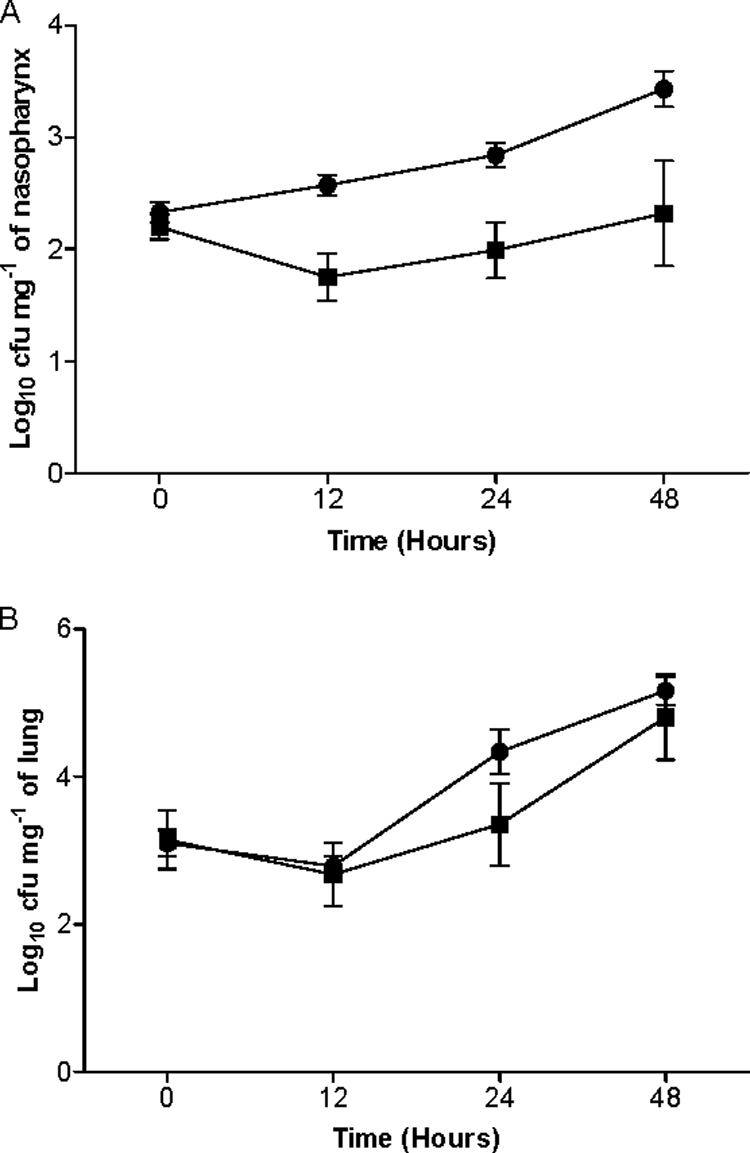
Growth of pneumococcal strains in nasopharynx (A) and lungs (B). For each time point, between 6 and 10 mice for SPD0065M (▪) and between 15 and 20 mice for D39 (•) were used. Error bars indicate standard errors of the means.
DISCUSSION
Given that carbohydrate is the principal pneumococcal energy source and at least one-third of the genome is devoted to sugar metabolism (16, 27, 47), we hypothesized that in addition to previously described pneumococcal glycosidases, there are likely to be others whose action is required to degrade host glycoconjugates, such as mucins. The search for novel and previously uncharacterized sugar hydrolases resulted in the identification of a β-galactosidase (SPD_0065) which is involved in the degradation of glycoproteins, including mucins.
SPD_0065 is a member of the glycosyl hydrolase family 35, which comprises enzymes with only β-galactosidase activity (14), and our analysis was consistent with this classification. The recombinant enzyme cleaved no synthetic substrates other than those with a β-linked galactose residue. During preparation of this paper, Jeong et al. (20) published a study in which the substrate specificity of SPD_0065 (BgaC) was investigated using a range of oligosaccharides. The enzyme was reported to have strict specificity for glycan structures terminating in Galβ1-3GlcNAc. In contrast to the previously characterized β-galactosidase, BgaA (SPD_0562), SPD_0065 was reported not to cleave galactose from Galβ1-4GlcNAc or to act on Galβ1-3GalNAc. Using desialylated fetuin, we showed that SPD_0065 also has the capability to act on intact glycoproteins. Fetuin contains oligosaccharides with terminal galactose in three different linkages (Fig. 4), and in light of the published study of the substrate specificity of the enzyme, it was concluded that SPD_0065 cleaved the glycan terminating in Galβ1-3GlcNAc. We demonstrated that, in common with many other pneumococcal glycosidases, the β-galactosidase did not act on fully sialylated oligosaccharides.
Approximately 85% of the total pneumococcal β-galactosidase activity in the TIGR4 strain, as measured using a synthetic substrate, was previously reported to be due to bgaA (54), the SPD_0562 homolog, which has activity against oligosaccharides containing the structure Galβ1-4GlcNAc (25). In a bgaA mutant, it was shown that BgaA-independent β-galactosidase activity is approximately sevenfold lower than that in the wild type of the same strain in the presence of lactose, which induces the activity of the enzyme. However, in strain D39, while SPD_0562 activity makes up 46%, SPD_0065 contributes 39% of the total activity. The residual activity in the double mutant presumably is due to the upregulation of other, currently unknown genes coding for β-galactosidase activity or at least those that can use pNP-β-d-galactopyranoside as a substrate. The discrepancy between the results of these different studies indicates that different pneumococcal strains probably use different combinations of enzymes.
β-Galactosidase activity in bacteria, other than S. pneumoniae, is encoded by multiple genes (35, 50), and they differ markedly with respect to substrate specificity, regulation, and cellular localization. The sizes of SPD_0065 and SPD_0562 differ substantially (69 and 247 kDa, respectively), and they share only 26% identity over 176 amino acids. SPD_0562 is classified within the glycosyl hydrolase family 2, the members of which have a broad range of enzymatic activity, including β-galactosidase (EC 3.2.1.23), β-glucuronidase (EC 3.2.1.31), and β-mannosidase (EC 3.2.1.25) activities. In different host niches multiple β-galactosidases with different substrate specificities may be required, as host glycoproteins exhibit variation in terms of tissue localization and glycosylation patterns (41). For example, in this study we observed that SPD_0065 is required mainly for nasopharyngeal colonization, in sharp contrast to the findings of King et al. (24), who reported that mutation of SPD_0562 (bgaA) did not attenuate nasopharyngeal colonization by D39. The discrepant results between the two studies indicate that although both pneumococcal galactosidases cleave galactose, due to their distinct substrate specificities, they are not functionally redundant.
The study by Jeong et al. (20) did not investigate the role of BgaC in growth of S. pneumoniae on glycoproteins. Here we show that SPD0065M is attenuated in its growth in mucin-containing defined medium and exhibits a reduced ability to cleave galactose and N-acetylgalactosamine from the glycoprotein relative to the wild-type strain. The reduced removal of galactose from oligosaccharides terminating in Galβ1-3GlcNAc (Fig. 1) by the mutant strain probably prevents the action of O-glycanase, which acts on the N-acetylgalactosamine-containing core disaccharide (31). The results also showed the complex nature of pneumococcal mucin degradation. Pneumococcal neuraminidases release N-glycolylneuraminic acid as well as N-acetylneuraminic acid. The recalcitrance of PGM to complete degradation by pneumococcal glycosidases is likely due to its complex structure, sulfation, and acetylation of some sugar residues. There was no release of fucose by the wild-type strain, which may be because the pneumococcus lacks a functional fucosidase, or the fucose residues in PGM may be masked by other sugars. Alternatively, the enzyme may be inducible, and the inducing residues are not unmasked during bacterial growth. There was also no removal of N-acetylglucosamine from PGM during growth of strain D39. This is surprising given that S. pneumoniae has a well-characterized N-acetylglucosaminidase (StrH) that acts on N-linked glycoproteins (24). The ability of StrH to cleave N-acetylglucosamine in O-linked oligosaccharides, as occur in mucins, has not been determined, however, and these residues could be masked by other sugars that are not cleaved during bacterial growth.
Our results indicated a cell surface localization for SPD_0065; however, the anchoring mechanism requires further investigation. Many glycosidases known to be involved in glycoprotein degradation, including NanA, BgaA, and StrH, are anchored in the cell wall in streptococci via the LPXTG motif (3). However, SPD_0065 does not contain this motif, nor does it have the carboxy-terminal choline-binding repeats or the transmembrane domains of other surface proteins (3). Despite the apparent lack of motifs for a cell surface localization for this enzyme, cell envelope-associated proteins can be released as a result of bacterial lysis and reassociate with the cell envelope (8).
Our analyses indicated that the expression of SPD_0065 both is repressed in the presence of glucose and is induced when mucin or in vivo glycoconjugates are present. A comprehensive characterization of the regulation of SPD_0065 expression in the presence of different sugars was beyond the scope of the present study. However, in Gram-positive bacteria the regulatory protein catabolite control protein A (CcpA) is primarily implicated in carbon catabolite repression (CCR) (18). CcpA has previously been shown to play a role in the regulation of expression of the β-galactosidase BgaA (SPD_0065) (23). The genes or operons regulated by CcpA bear an operator sequence known as the catabolite-repressible element (cre), to which CcpA binds. cre can be found either in the promoter region or in the coding sequence of the target gene, and it is known to have a consensus sequence of WWTGNAARCGNWWWCAWW (R for G or A, W for A or T, and N for any base) (34). We identified a perfect match to the cre consensus sequence in 37 bases upstream of the SPD_0065 start codon (ATGAAAGCGCAAACT). Therefore, it is plausible that this gene is regulated by CcpA.
As expected, our data on SPD_0065 confirm a number of key findings in the study by Jeong et al. (20). There are, however, some important differences between the two studies. Here we demonstrated that mutation of SPD_0065 does not cause a polar effect. This is particularly important because SPD_0065 is the first gene of a predicted operon (9). We also demonstrated that the synthesis of this enzyme is not limited to serotype 2 but that it also is produced by a serotype 6B strain. In contrast to Jeong et al. (20), who reported that the mutation of SPD_0065 increases nasopharyngeal colonization and that onset of bacteremia occurs earlier in the mutant-infected cohort than with wild-type infection, we observed that the mutation of SPD_0065 delays bacteremia and attenuates pneumococcal growth in the nasopharynx. These differences may be attributed to the mouse strain and the infective dose used. For example, while Jeong et al. used CD1 mice and 2 × 107 CFU/mouse in 10 μl, in the present study MF1 mice were infected with 5 × 105 CFU/mouse in 50 μl.
Due to the large genome polymorphism in the pneumococcal population (37), the contribution of this enzyme to virulence may vary for different serotypes or even for isolates of the same serotype. Consequently, the results presented here are limited to the strains that were investigated. Overall, these results demonstrate a fundamental link between pneumococcal glycosidase production and mucin degradation, which may account for reduction in virulence of SPD0065M. This emphasizes the connection between nutrient acquisition in the host and pathogenesis and goes some way toward furthering our understanding of the biological importance of the wide array of currently uncharacterized pneumococcal glycosidases extant in the genome.
Acknowledgments
This study was supported by a Wellcome Trust grant (078763/Z/05/Z) to H.Y. and P.W.A.
We are grateful to Brian Spratt (Imperial College, London, United Kingdom) for providing the S. pneumoniae serotype 6 strain, to Jack Thompson (National Institutes of Health, Bethesda, MD) for providing fluorogenic and chromogenic substrates, and to Marc Prudhomme (Centre National de la Recherche Scientifique, Toulouse, France) for his kind gift of plasmids for mariner mutagenesis.
Editor: A. Camilli
Footnotes
Published ahead of print on 19 October 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Alloing, G., C. Granadel, D. A. Morrison, and J. P. Claverys. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 2.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, S., and S. Hammerschmidt. 2006. Versatility of pneumococcal surface proteins. Microbiology 152:295-303. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byers, H. L., E. Tarelli, K. A. Homer, and D. Beighton. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469-479. [DOI] [PubMed] [Google Scholar]
- 7.Corfield, A. P., S. A. Wagner, J. R. Clamp, M. S. Kriaris, and L. C. Hoskins. 1992. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desvaux, M., E. Dumas, I. Chafsey, and M. Hebraud. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1-15. [DOI] [PubMed] [Google Scholar]
- 9.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 29:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giammarinaro, P., and J. C. Paton. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 70:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, E. D., G. Adelt, J. U. Baenziger, S. Wilson, and H. Van Halbeek. 1988. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-Megahertz 1H NMR spectroscopy. J. Biol. Chem. 263:18253-18268. [PubMed] [Google Scholar]
- 13.Guiral, S., V. Henard, M. H. Laaberki, C. Granadel, M. Prudhomme, B. Martin, and J. P. Claverys. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152:343-349. [DOI] [PubMed] [Google Scholar]
- 14.Henrissat, B., and A. Romeu. 1995. Families, superfamilies and subfamilies of glycosyl hydrolases. Biochem. J. 311:350-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirst, R. A., H. Yesilkaya, E. Clitheroe, A. Rutman, N. Dufty, T. J. Mitchell, C. O'Callaghan, and P. W. Andrew. 2002. Sensitivities of human monocytes and epithelial cells to pneumolysin are different. Infect. Immun. 70:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell, B. F., S. McCune, and R. Schaffer. 1979. Lactate-to-pyruvate or pyruvate-to-lactate assay for lactate dehydrogenase: a re-examination. Clin. Chem. 25:269-272. [PubMed] [Google Scholar]
- 18.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagota, S., M. V. Ramana Rao, and S. M. Dutta. 1981. β-Galactosidase of Streptococcus cremoris H. J. Food Sci. 46:161-168. [Google Scholar]
- 20.Jeong, J. K., O. Kwon, Y. M. Lee, D. B. Oh, J. M. Lee, S. Kim, E. H. Kim, T. N. Le, D. K. Rhee, and H. A. Kang. 2009. Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface β-galactosidase with specific hydrolysis activity for the Galβ1-3GlcNAc moiety of oligosaccharides. J. Bacteriol. 191:3011-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadioglu, A., and P. W. Andrew. 2004. The innate immune response to pneumococcal lung infection: the untold story. Trends Immunol. 25:143-149. [DOI] [PubMed] [Google Scholar]
- 22.Kadioglu, A., N. A. Gingles, K. Grattan, A. Kerr, T. J. Mitchell, and P. W. Andrew. 2000. Host cellular immune response to pneumococcal lung infection in mice. Infect. Immun. 68:492-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman, G. E., and J. Yother. 2007. CcpA-dependent and -independent control of β-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J. Bacteriol. 189:5183-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59:961-974. [DOI] [PubMed] [Google Scholar]
- 25.Kojima, K., M. Iwamori, S. Takasaki, K. Kubushiro, S. Nozawa, R. Iizuka, and Y. Nagai. 1987. Diplococcal β-galactosidase with a specificity reacting to β1-4 linkage but not to β1-3 linkage as a useful exoglycosidase for the structural elucidation of glycolipids. Anal. Biochem. 165:465-469. [DOI] [PubMed] [Google Scholar]
- 26.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 27.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lineweaver, H., and D. Burk. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56:658-666. [Google Scholar]
- 29.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 30.Manco, S., F. Hernon, H. Yesilkaya, J. C. Paton, P. W. Andrew, and A. Kadioglu. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 74:4014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marion, C., D. H. Limoli, G. S. Bobulsky, J. L. Abraham, A. M. Burnaugh, and S. J. King. 2009. Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect. Immun. 77:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller, P. L., F. Jorgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morton, D. B. 1985. Pain and laboratory animals. Nature 317:106. [DOI] [PubMed] [Google Scholar]
- 37.Obert, C., J. Sublett, D. Kaushal, E. Hinojosa, T. Barton, E. I. Tuomanen, and C. J. Orihuela. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74:4766-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661-1669. [DOI] [PubMed] [Google Scholar]
- 39.Philips, B. J., J. X. Meguer, J. Redman, and E. H. Baker. 2003. Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intensive Care Med. 29:2204-2210. [DOI] [PubMed] [Google Scholar]
- 40.Rao, M. V. R., and S. M. Dutta. 1981. Purification and properties of β-galactosidase from Streptococcus thermophilus. J. Food Sci. 46:1419-1423. [Google Scholar]
- 41.Rose, M. C., and J. A. Voynow. 2006. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 86:245-278. [DOI] [PubMed] [Google Scholar]
- 42.Sheehan, J. K., P. S. Richardson, D. C. Fung, M. Howard, and D. J. Thornton. 1995. Analysis of respiratory mucus glycoproteins in asthma: a detailed study from a patient who died in status asthmaticus. Am. J. Respir. Cell Mol. Biol. 13:748-756. [DOI] [PubMed] [Google Scholar]
- 43.Sheehan, J. K., D. J. Thornton, M. Somerville, and I. Carlstedt. 1991. Mucin structure. The structure and heterogeneity of respiratory mucus glycoproteins. Am. Rev. Respir. Dis. 144:S4-S9. [DOI] [PubMed] [Google Scholar]
- 44.Sicard, A. M. 1964. A new synthetic medium for Diplococcus pneumoniae, and its use for the study of reciprocal transformations at the amiA locus. Genetics 50:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 46.Takasaki, S., and A. Kobata. 1986. Asparagine-linked sugar chains of fetuin: occurrence of tetrasialyl triantennary sugar chains containing the Galβ1→3GlcNAc sequence. Biochemistry 25:5709-5715. [DOI] [PubMed] [Google Scholar]
- 47.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 48.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiggins, R., S. J. Hicks, P. W. Soothill, M. R. Millar, and A. P. Corfield. 2001. Mucinases and sialidases: their role in the pathogenesis of sexually transmitted infections in the female genital tract. Sex. Transm. Infect. 77:402-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yanahira, S., T. Kobayashi, T. Suguri, M. Nakakoshi, S. Miura, H. Ishikawa, and I. Nakajima. 1995. Formation of oligosaccharides from lactose by Bacillus circulans β-galactosidase. Biosci. Biotechnol. Biochem. 59:1021-1026. [DOI] [PubMed] [Google Scholar]
- 51.Yesilkaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Andrew. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yesilkaya, H., S. Manco, A. Kadioglu, V. S. Terra, and P. W. Andrew. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278:231-235. [DOI] [PubMed] [Google Scholar]
- 53.Yesilkaya, H., S. Soma-Haddrick, S. J. Crennell, and P. W. Andrew. 2006. Identification of amino acids essential for catalytic activity of pneumococcal neuraminidase A. Res. Microbiol. 157:569-574. [DOI] [PubMed] [Google Scholar]
- 54.Zahner, D., and R. Hakenbeck. 2000. The Streptococcus pneumoniae β-galactosidase is a surface protein. J. Bacteriol. 182:5919-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]