Abstract
Intracellular Salmonella enterica serovar Typhimurium deploys the Salmonella pathogenicity island 2 (SPI2)-encoded type III secretion system (T3SS) to modify host cell functions and accomplish intracellular replication. This virulence function is controlled by the two-component system SsrAB that regulates the expression of several operons in SPI2 and, in addition, a large number of genes for non-SPI2-encoded effector proteins. Here, we analyzed the relative expression levels of members of the SsrAB virulon. We used a novel reporter fusion strategy for single-copy chromosomal fusions, all done in an identical manner in order to enable direct quantitative comparison. We observed very high expression levels for sseJ and sifA; high expression levels for ssaG, steC, sseL, and sopD2; moderate expression levels for ssaB, sseA, sseG, sifB, pipB2, and sspH1; and low expression levels for sspH2, sseI, slrP, sseK1, sseK2, pipB, and gogB. The expression of the SsrAB virulon was highly dependent on the function of SsrB but also required EnvR/OmpZ. Deletion of PhoP, part of the global regulatory system PhoPQ, resulted in highly delayed expression of the SsrAB virulon under in vitro conditions; however, maximal expression was similar to that in a wild-type background. The expression levels of SsrAB-dependent genes in intracellular bacteria were in good agreement with in vitro analyses. We provide here a comprehensive and fully comparable analysis of the expression of genes in the SsrAB virulon. This information will be of interest for the selection of in vivo-activated promoters, for example, for rational design of recombinant vaccines.
Bacterial virulence requires the continuous adaptation of the pathogen to changing environments in the host and the fine-tuned regulation of the expression of virulence genes in response to environmental cues. Salmonella enterica is a food-borne, facultative, intracellular pathogen that can survive and colonize niches in mammalian hosts as diverse as the intestinal lumen, an intracellular niche referred to as the “Salmonella-containing vacuole” (SCV), and biofilms on gall stones (18). A virulence factor essential for the systemic phase of Salmonella infections and the intracellular life style of the pathogen is the Salmonella pathogenicity island 2-encoded type III secretion system (SPI2-T3SS) (25). The SPI2-T3SS is activated by Salmonella bacteria residing within the SCV and translocates a complex set of effector proteins into the host cell cytoplasm (25). Although the precise molecular functions of the individual effector proteins are only partially known, their combined actions allow the biogenesis of the SCV into a replication-permissive compartment and the modification of host cell processes, such as microtubule-dependent transport, activities of antimicrobial defense mechanisms, or antigen presentation (1, 3, 20, 41).
The expression of genes within SPI2, encoding the T3SS and several effectors, as well as that of a large number of loci outside of SPI2, encoding further effectors, is under the control of the SPI2-encoded two-component regulatory system SsrAB (11). The complex regulon of SsrAB-controlled virulence genes is referred to here as the “SsrAB virulon.” The SsrAB virulon is controlled by unknown signals that are sensed by SsrA and that activate the expression of multiple transcriptional units via the transcriptional regulator SsrB (10, 43). In addition to SsrAB, further global regulatory mechanisms also control the expression of the SsrAB virulon (13). They include the additional two-component systems PhoPQ (5, 11) and EnvZ/OmpR (14, 27) and the transcriptional regulator SlyA (29, 33).
The analysis of the SsrAB virulon is of central importance for an understanding of the pathogenesis of Salmonella infections. Furthermore, promoters of this virulon might also be useful for “inverted pathogenicity,” i.e., the use of virulence determinants for prevention and therapy of infectious diseases (6). Promoters of the SsrAB virulon appear to be ideal in vivo-induced promoters, since they can be used for the temporally and spatially controlled expression of heterologous antigens by live attenuated bacterial carrier strains. We have previously used PsseA of the SsrAB virulon to generate recombinant vaccines using attenuated Salmonella bacteria as carriers (21, 22). This promoter is active after the uptake of Salmonella by antigen-presenting cells (23).
The SsrAB virulon consists of a large number of distinct transcriptional units, since most effector proteins are encoded by individual loci outside of SPI2. Fifteen separate loci outside of SPI2 encode translocated effector proteins (reviewed in references 1 and 18), and further loci are coregulated and might encode further effectors (15, 45). Although there are indications that various effectors are expressed at different levels (32, 36, 40), a systematic comparative analysis of the expression levels has not yet been performed.
For a fully comparative analysis of the expression levels of members of a large regulon, the investigation of chromosomal reporter fusions appears most suitable in order to ensure that effects of global regulatory mechanisms, such as DNA topology or binding of nucleoid-associated proteins (NAP), like H-NS, are integrated. We recently devised a novel approach for the generation of reporter gene fusions (16). This method allows the rapid generation of reporter gene fusions in the original chromosomal context. Thus, reporter fusions can be generated in an identical manner for various genes to be compared, and the approach circumvents artifacts commonly associated with the use of episomal reporter constructs. The Red recombination-based reporter approach was recently used to analyze the regulatory cascade of genes in Salmonella pathogenicity island 4 (SPI4) (17).
Here, we describe the results of systematic comparative analyses of the expression levels and regulatory control of genes encoding effector proteins of the SPI2-T3SS. Using in vitro conditions for induction of the SsrAB virulon, as well as analyses of intracellular bacteria, we observed pronounced differences in the expression levels of genes encoding the SPI2-T3SS effector.
MATERIALS AND METHODS
Bacterial strains and generation of mutant and reporter fusion strains.
All strains used in this study are isogenic derivatives of Salmonella enterica serovar Typhimurium NCTC 12023. The characteristics of the strains are listed in Table 1. Deletions of genes encoding regulators were generated by the “one-step inactivation” approach as described previously (9). Reporter gene fusions with firefly luciferase were constructed using Red recombinase-mediated homologous recombination of genes cassettes as previously described (16). For the construction of reporter strains, a cassette containing luc aph was amplified from template plasmid p3121 using target gene-specific primer pairs listed in Table 2, and the product was introduced by electroporation into the Salmonella wild type (WT) harboring pKD46. The Red-mediated integration of the cassettes was confirmed by PCR and DNA sequencing. If required for the construction of reporter fusions in the background of mutations of regulatory genes, P22 transduction (31) was used to move a chromosomal locus with the respective reporter fusion.
TABLE 1.
Bacterial strains and plasmids used in this study
| Designation | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| NCTC12023 | Wild type | Laboratory collection |
| MvP531 | ΔssrB::aph | 17 |
| MvP532 | ΔssrB::FRT | 17 |
| MvP1343 | ΔphoP::FRT | This study |
| MvP1344 | ΔompR envZ::FRT | This study |
| MvP1345 | ΔslyA::FRT | This study |
| MvP896 | ΔsirA::FRT | 17 |
| MvP607 | sifA::luc aph | 16 |
| MvP1102 | sseA::luc aph | This study |
| MvP1103 | ssaB::luc aph | This study |
| MvP1104 | sifB::luc aph | This study |
| MvP1105 | sseG::luc aph | This study |
| MvP1106 | sseI::luc aph | This study |
| MvP1107 | sseJ::luc aph | This study |
| MvP1108 | pipB::luc aph | This study |
| MvP1406 | pipB2::luc aph | This study |
| MvP1109 | sseK1::luc aph | This study |
| MvP1110 | sseK2::luc aph | This study |
| MvP1111 | sspH1::luc aph | This study |
| MvP1112 | sspH2::luc aph | This study |
| MvP1113 | slrP::luc aph | This study |
| MvP1114 | gogB::luc aph | This study |
| MvP1234 | sseL::luc aph | This study |
| MvP1235 | steC::luc aph | This study |
| MvP1236 | ssaG::luc aph | This study |
| MvP1237 | sopD2::luc aph | This study |
| MvP1115 | sseA::luc aph ΔssrB | This study |
| MvP1116 | ssaB::luc aph ΔssrB | This study |
| MvP1117 | sifA::luc aph ΔssrB | This study |
| MvP1118 | sseG::luc aph ΔssrB | This study |
| MvP1119 | sseJ::luc aph ΔssrB | This study |
| MvP1120 | sifB::luc aph ΔssrB | This study |
| MvP1121 | sopD2::luc aph ΔssrB | This study |
| MvP1122 | slrP::luc aph ΔssrB | This study |
| MvP1123 | gogB::luc aph ΔssrB | This study |
| MvP1124 | ssaG::luc aph ΔssrB | This study |
| MvP1425 | steC::luc aph ΔssrB | This study |
| MvP1426 | sseL::luc aph ΔssrB | This study |
| MvP1427 | pipB2::luc aph ΔssrB | This study |
| MvP1428 | sspH1::luc aph ΔssrB | This study |
| MvP1429 | sspH2::luc aph ΔssrB | This study |
| MvP1430 | sseI::luc aph ΔssrB | This study |
| MvP1431 | sseK1::luc aph ΔssrB | This study |
| MvP1432 | sseK2::luc aph ΔssrB | This study |
| MvP1433 | pipB::luc aph ΔssrB | This study |
| MvP1238 | sifA::luc aph ΔphoP | This study |
| MvP1239 | sifA::luc aph ΔslyA | This study |
| MvP1240 | sifA::luc aph ΔompR/envZ | This study |
| MvP1241 | sseJ::luc aph ΔompR/envZ | This study |
| MvP1242 | sseJ::luc aph ΔphoP | This study |
| MvP1243 | sseJ::luc aph ΔslyA | This study |
| Plasmids | ||
| pKD46 | Vector Red expression | 9 |
| p3121 | Template plasmid for luc reporter fusions | 16 |
TABLE 2.
Oligonucleotides used in this study
| Designation | Sequencea |
|---|---|
| Red-mediated deletions | |
| PhoP-Del-For | 5′-AACGCTAGACTGTTCTTATTGTTAACACAAGGGAGAAGAGGTGTAGGCTGGAGCTGCTTC-3′ |
| PhoP-Del-Rev | 5′-GCGCAATTCAAAAAGATATCCTTGTCCGCGTACGGTGGTACATATGAATATCCTCCTTAG-3′ |
| OmpR-Del-For | 5′-TACAATTTGTTGCGAACCTTTGGGAGTACAGACAATGCAAGTGTAGGCTGGAGCTGCTTC-3′ |
| EnvZ-Del-Rev2 | 5′-GAGAAGAAAGGGAGGGTAATACCTCCCTTTCTTATGCCTCCATATGAATATCCTCCTTAG-3′ |
| SlyA-Del-For2 | 5′-AATCAGCATAATAACTTAGCAAGCTAATTATAAGGAGATGGTGTAGGCTGGAGCTGCTTC-3′ |
| SlyA-Del-Rev2 | 5′-TGTGGTCACATGGCCACACGTATGCCCCTGCACCTCAATCCATATGAATATCCTCCTTAG-3′ |
| luc reporter fusions | |
| SseA-Red-Rep-For | 5′-TTAGCACGTTAATTATCTATCGTGTATATGGAGGGGAATGatggaagacgccaaaaacataa-3′ |
| SseA-Red-Rep-Rev | 5′-CCCCATAAGATGTTTCCTGAAGACATTATGCTTTACCTTTTcgtgtaggctggagctgcttc-3′ |
| SsaB-Red-Luc-For | 5′-AATTTGATAGAAACTCCCATTTATGTCTGAGGAGGGATTCatggaagacgccaaaaacataa-3′ |
| SsaB-Red-Rep-Rev | 5′-TAGTAAAATTAAGATTAAACGTTTATTTACTACCATTTTAcgtgtaggctggagctgcttc-3′ |
| SsaG-Red-Luc-For | 5′-CCAGTATCCTTACGATGTATTTATTTTAAGGAAAAGCATTatggaagacgccaaaaacataa-3′ |
| SsaG-Red-Rep-Rev | 5′-TTCCAGCAGCAACCGTCGAACATCGTCGCTAATAACTTCAcgtgtaggctggagctgcttc-3′ |
| SseG-Red-Luc-For | 5′-CATTAATGGACAGTTCTGATCATACATCTCGGGGAGAACCatggaagacgccaaaaacataa-3′ |
| SseG-Red-Rep-Rev | 5′-AGAAAGCAATGAACATCCGGTATATACCTGAAAACGATTAcgtgtaggctggagctgcttc-3′ |
| SseI-Red-Luc-For | 5′-CGGACAGATACTATATGTAAATTTATAAAGGTTTTTTGTTatggaagacgccaaaaacataa-3′ |
| SseI-Red-Rep-Rev | 5′-ATTGACAGGGTTCTGACAGACGTCCTCCACGGTGCGCTTAcgtgtaggctggagctgcttc-3′ |
| SseL-Red-FLuc-For | 5′-CATATTCGCTACTTTCACTTACCAGGAAACAGAGCAAAatggaagacgccaaaaacataa-3′ |
| SseL-Red-Rep-Rev | 5′-CATTTCAGGATAAGAGCCTAATGGGATAGGCTCTAAGTACcgtgtaggctggagctgcttc-3′ |
| SifA-Red-Luc-For | 5′-ATTATGTAGTCATTTTTACTCCAGTATAAGTGAGATTAATatggaagacgccaaaaacataa-3′ |
| SifA-Red-Luc-Rev | 5′-ACCCTGAACGTGACGTCTGAGAAAGCGTCGTCTGATTTTAgtgtaggctggagctgcttc-3′ |
| SifB-Red-Luc-For | 5′-CCAGTAATGAAGTATCATATAATCACTTGTGGTCTACATTatggaagacgccaaaaacataa-3′ |
| SifB-Red-Rep-Rev | 5′-GCCAGGGGATTGTAAATCCATACTATTTATGGTGTGATCAcgtgtaggctggagctgcttc-3′ |
| SseK1-Red-Luc-For | 5′-TAAAATATGTAATGAAGTAAGTATGGAGCATTTAATTGTTatggaagacgccaaaaacataa-3′ |
| SseK1-Red-Rep-Rev | 5′-TTTTATGTATTCAATAGCATGATTATTGCCATTTCCGCTAcgtgtaggctggagctgcttc-3′ |
| SseK2-Red-Luc-For | 5′-AAATTAAGGTTAAAAACTGATAATTTAAGCGTGTAAAAATatggaagacgccaaaaacataa-3′ |
| SseK2-Red-Rep-Rev | 5′-GACGGTGGGAAGGGTGAGTAAAACAAGGCTATCATGATTAcgtgtaggctggagctgcttc-3′ |
| SspH1-Red-Luc-For | 5′-TTAATCTCTTTTCATTGTGCTGTAAATTAGGCAGTGGAATatggaagacgccaaaaacataa-3′ |
| SspH1-Red-Rep-Rev | 5′-ACCGCACCACATTCGCCTGGTGCGGTGAATATCGTGCTCAcgtgtaggctggagctgcttc-3′ |
| SspH2-Red-Luc-For | 5′-CGGACAGATACTATATGTAAATTTATAAAGGTTTTTTGTTatggaagacgccaaaaacataa-3′ |
| SspH2-Red-Rep-Rev | 5′-ATATCTTTGTCGCACCGCACCTCATTCACCTGGTGCATCAcgtgtaggctggagctgcttc-3′ |
| PipB-Red-Luc-For | 5′-CCTATAAGGAGTCGGCTCACTTCCATAAGAAGGAATCAAAatggaagacgccaaaaacataa-3′ |
| PipB-Red-Rep-Rev | 5′-CTGTTTGAATACTTCTTGTTTATAAAATCCCTTTATCTCGAcgtgtaggctggagctgcttc-3′ |
| PipB2-Red-Luc-For2 | 5′-TTTTATCATGCACTGTGTTGCTGTCTCTGGGAGAAAATATatggaagacgccaaaaacataa-3′ |
| PipB2-Red-Rep-Rev | 5′-TTTCACTATAAAATTCGTTAAAGAGTGTTTGTGTGCTTGTcgtgtaggctggagctgcttc-3′ |
| SlrP-Luc-For | 5′-TCTGTTACTTTAGGTTACGTTCAGATCAGGTAGGGAAAATatggaagacgccaaaaacataa-3′ |
| SlrP-Rep-Rev | 5′-AACAGGCTCTCTCCCTCTTCTGATAAACTGCGTTCAGCTAcgtgtaggctggagctgcttc-3′ |
| GogB-Red-Luc-For | 5′-TTGCCTGAATGAAAATAAATGTAATAATGATAGCTTGGTAatggaagacgccaaaaacataa-3′ |
| GogB-Red-Rep-Rev2 | 5′-CTATATATAAATATATTAATTGCATATTTTTTTAAAGTCAcgtgtaggctggagctgcttc-3′ |
| SteC-Red-Luc-For | 5′-GTGGTTACAGTAGTTACAAATCAGGTTTTTATATGTTTGCatggaagacgccaaaaacataa-3′ |
| SteC-Red-Rep-Rev | 5′-GCCCCCGGCGATTCGCAGAAAAGAACGGAACTAAATGCTAcgtgtaggctggagctgcttc-3′ |
Lowercase letters indicate sequences complementary to the template plasmid.
Growth conditions.
Bacterial strains were routinely cultured in LB broth or on LB agar plates. The media contained carbenicillin and/or kanamycin at concentrations of 50 μg/ml where required for the selection of mutant strains or to propagate plasmids. For reporter analyses under in vitro conditions, a fully defined “phosphate-carbon-nitrogen” (PCN) synthetic minimal medium was used, as previously described (10). Briefly, the composition of PCN medium was 80 mM MOPS (morpholinepropanesulfonic acid), pH 7.4, or 80 mM MES (morpholineethanesulfonic acid), pH 5.8, 4 mM Tricine, 0.4 mM FeCl3, 376 μM K2SO4, 50 mM NaCl, 15 mM NH4Cl, 1 mM MgSO4, 1 μM CaCl2, 0.4% glucose, and micronutrients as described previously (10). Phosphate buffer was adjusted as required, and PCN medium at pH 5.8 containing 0.4 mM inorganic phosphate (Pi) was used as the SPI2-inducing growth condition (30). For noninducing conditions, PCN medium containing 25 mM Pi, adjusted to pH 7.4, was used. Bacterial cultures were grown in borosilicate test tubes containing 3 ml of culture. Culture tubes were aerated by rotation in a roller drum (New Brunswick Scientific) in an incubator set to 37°C.
Reporter analyses.
The quantification of the reporter activities of bacterial strains grown in vitro or within the macrophage cell line RAW 264.7 was performed basically as described previously (16). Briefly, for analyses in vitro, various reporter strains were grown in noninducing PCN medium (pH 7.4, 1.0 mM Pi) overnight. Overnight cultures were diluted 1:100 in fresh medium for growth under inducing or noninducing conditions. Aliquots of the overnight cultures were collected, and bacterial cells were harvested by centrifugation for 5 min at 4,000 × g at 4°C. The bacteria were resuspended in PCN medium-0.4 mM Pi, pH 5.8, for SPI2-inducing conditions or PCN medium-25 mM Pi, pH 7.4, for noninducing conditions. At various times of culture, samples were taken, and aliquots equivalent to 250 μl of a culture with an optical density at 600 nm (OD600) of 1 were processed for luciferase assays. The bacteria were pelleted by centrifugation for 5 min at 22,000 × g in a microcentrifuge at 4°C, and the supernatant was removed. The bacterial pellet was resuspended in 250 μl of freshly prepared Luc lysis buffer. Samples were frozen at −20°C and allowed to thaw at room temperature (RT). After thawing, the samples were incubated for 15 min at RT with repeated mixing on a Vortex mixer. Aliquots of 25 μl of the lysate were transferred in triplicate into wells of white microtiter plates (no. 236108; Nunc, Wiesbaden, Germany), and 50 μl of the Luc reagent mixture was added. Light emission was integrated over 60 s using a luminometer (Ascent Fluroscan FL; Labsystems). The background activity was subtracted.
Macrophage infection and reporter analyses.
For the quantification of reporter activities of intracellular Salmonella, the murine macrophage cell line RAW 264.7 was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL, Grand Island, NY) containing 10% heat-inactivated fetal calf serum (Gibco-BRL, Grand Island, NY) at 37°C under 5% CO2. Prior to infection with Salmonella, the cells were washed gently with DMEM and removed from the flasks by scraping. The cells were suspended in six-well plates for experiments at a density of 2 × 106 cells per well and allowed to adhere for at least 12 h.
Bacterial strains were grown overnight in LB, and the OD600 of the cultures was adjusted to 0.2 in 1 ml of phosphate-buffered saline (PBS). Aliquots of this suspension were added to a well with macrophages in order to yield multiplicities of infection (MOI) of approximately 10 or 50. The plates were centrifuged at 500 × g for 5 min at RT to synchronize the infection and subsequently incubated at 37°C in an atmosphere of 5% CO2 for 15 min. Thereafter, the plates were washed thrice with DMEM and incubated with DMEM containing 100 μg ml−1 gentamicin for 1 h, followed by DMEM containing 10 μg ml−1 gentamicin for the rest of the experiment. At the indicated time points after infection, the supernatants were removed, and the cells were washed three times with PBS and lysed in 0.1% Triton X-100. An aliquot of the lysate was used to determine the number of viable intracellular bacteria by plating serial dilutions on agar plates and counting CFU. The samples were centrifuged for 5 min at 22,000 × g at 4°C to harvest the released bacteria. The pellets containing bacteria were resuspended in 250 μl of freshly prepared Luc lysis buffer and further processed for Luc assays as described above.
RESULTS
Generation of a collection of isogenic reporter strains for the SPI2 genes and effector proteins of the SPI2-T3SS.
The SsrAB virulon consists of promoters within the SPI2 locus and a large number of transcriptional units for effector proteins of the SPI2-T3SS located outside of SPI2. We compared the activities of promoters in SPI2, i.e., promoters for sseA, ssaB, and ssaG, and promoters for effectors outside of SPI2, i.e., gogB, pipB, pipB2, sifA, sifB, slrP, sopD2, sseI, sseJ, sseK1, sseK2, sseL, sspH1, sspH2, and steC. We also generated a fusion to the effector gene sseG. In contrast to all other fusions, sseG is the distal gene of a larger operon controlled by PsseA or an additional, so far unknown promoter downstream of PsseA. For some of the promoters in SPI2, experimental data on the transcriptional start points and RNA polymerase binding sites are available (43). For most other transcriptional units, predictions were performed by comparison of 5′ regions to consensus sequences of Escherichia coli promoters (19) to define putative control elements. In almost all sequences, −35-bp and −10-bp boxes with similarity to the promoter consensus and appropriate spacing between the boxes could be detected. A compilation of promoters analyzed in this study and the E. coli promoter consensus sequence are depicted in Fig. 1A and B, respectively.
FIG. 1.
Promoter regions of transcriptional units of the SsrAB virulon and schematic presentation of recombineering for generation of isogenic reporter strains in S. enterica serovar Typhimurium. (A) Compilation of 5′ regions of transcriptional units of the SsrAB regulon. The sequences were aligned with respect to experimentally confirmed or predicted start codons. The first three codons are in lowercase. The asterisks indicate promoters of ssaB, sseA, and ssaG that were defined by previous experimental work (43). The transcribed regions for these promoters are indicated by italics, and the −35-bp and −10-bp boxes are underlined. For other transcriptional units, the −35-bp and −10-bp boxes were predicted by comparison to the E. coli promoter consensus sequence (B) as defined by Hawley and McClure (19) and are indicated by underlining. The numbers indicate the spacing between −35-bp and −10-bp boxes in bp. (C) In order to generate chromosomal reporter fusions to genes within SPI2 or various loci that encode effector proteins of the SPI2-T3SS, a cassette containing luc as a reporter gene and aph as a selectable marker was recombined into various positions of the chromosome of S. enterica serovar Typhimurium. As shown for the example of sifA, all fusions were generated in an identical manner, i.e., by insertion of luc after the first codon of the genes under study.
In order to analyze the relative expression levels of various genes within SPI2 and the various SPI2 effector loci, we generated reporter strains that harbored luciferase reporter fusions in identical positions with respect to the start codon. In detail, the first codon of luciferase replaced the first codon of the open reading frame (ORF) under study, thus allowing the comparative analysis of the promoter strength and the quality of the ribosome binding site (RBS). A schematic representation of the approach is shown in Fig. 1C.
Comparison of the expression levels of the genes in the SPI2 virulon.
The various reporter strains were grown under in vitro conditions that were previously defined to induce the expression of genes in the SsrAB virulon. For this purpose, the fully defined synthetic PCN medium was used. We have shown previously that alteration of a single component of this medium can result in the strong induction of the SsrAB virulon. In detail, PCN medium with a slightly acidic pH of 5.8 resulted in rapid induction, while PCN medium at neutral pH with reduced amounts of Pi (0.4 mM) resulted in delayed induction of SsrAB-controlled genes (30).
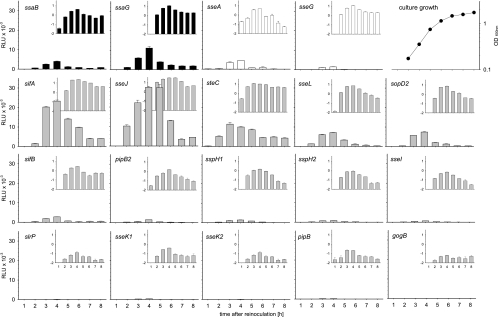
Analysis of the expression kinetics indicated that maximum luciferase activity was detectable around 4 h after the start of the subculture and coincided with the late exponential growth phase of the culture (Fig. 2). A decrease in reporter activity was observed a later time points that could have resulted from decreased expression and/or increased degradation of the reporter in cells entering the stationary growth phase. We did not observe major differences in the time points at which the maximal expression levels were reached for genes encoding structural components of the SPI2-T3SS, SPI2 effector genes within SPI2, or SPI2 effector genes outside of SPI2.
FIG. 2.
In vitro expression kinetics of reporter fusions for genes encoding SPI2 effectors or components of the SPI2-T3SS. Reporter strains representing different functional groups in the SsrAB virulon were grown in vitro under SPI2-inducing conditions (PCN medium-0.4 mM Pi, pH 5.8). Cells were harvested at various time points, and equal amounts of cells adjusted by OD600 were lysed for the quantification of Luc activities. Expression levels of reporter fusions to SPI2 genes encoding structural components of the T3SS or effector proteins are indicated in black or white histograms, respectively. Expression levels of reporter fusions to genes for effector proteins that are located outside of SPI2 are indicated by gray histograms. Luc activities are expressed with linear or logarithmic scaling in the large histograms and insets, respectively. A representative growth curve of S. enterica serovar Typhimurium in PCN-0.4 mM Pi, pH 5.8, minimal medium is shown on the upper right, and all reporter strains showed similar growth characteristics. The error bars indicate standard deviations.
The expression levels of the 19 reporter fusions were highly divergent. Very high expression levels (>20,000 relative light units [RLU]) were observed for fusions to the effector genes sseJ and sifA. Fusions to ssaG, steC, sseL, and sopD2 showed high expression levels (>5,000 RLU), while average expression levels (>1,000 RLU) were found for fusions to ssaB, sseA, sseG, sifB, pipB2, and sspH1. The expression levels of the other fusions, i.e., sspH2, sseI, slrP, sseK1, sseK2, pipB, and gogB, were low (<1,000 RLU). The expression of any of these reporter fusions under noninducing culture conditions was very low (data not shown).
Taken together, these data indicate high diversity in the expression levels of the various genes encoding the structural components and the effector proteins of the SPI2-T3SS.
Role of SsrB in control of the SsrAB virulon.
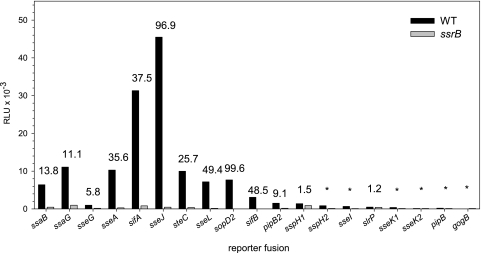
We next investigated the role of the SsrAB regulatory system in the expression of various reporter fusions to genes encoding structural components of the SPI2-T3SS and effector proteins (Fig. 3). The maximal expression levels of different fusions were compared in S. enterica serovar Typhimurium WT and ssrB deletion backgrounds. The expression of the majority of genes encoding effector proteins was strongly reduced in the absence of ssrB. We determined an almost 100-fold decrease in reporter activity for the sseJ or sopD2 fusion in an ssrB deletion background. The SsrB-dependent transcriptional induction of most genes was in the range of 25- to 50-fold. The smallest difference was observed for the reporter fusion to sseG, with about sixfold SsrB-dependent induction. In accordance with previous observations (32), the expression of the slrP reporter fusion was not affected by the presence or absence of SsrB. These data demonstrate that expression of genes in the SsrAB virulon is affected to different extents by the transcriptional activator SsrB.
FIG. 3.
Comparison of expression levels of SPI2 genes and genes encoding SPI2 effector proteins. The reporter fusions were analyzed in the background of the S. enterica serovar Typhimurium wild type or in ssrB strains. The reporter strains were grown under SPI2-inducing conditions, and the luciferase activity was determined at various time points as described for Fig. 2. The histogram shows the maximal expression levels for representative reporter fusions. The numbers indicate fold SsrB-dependent activation.
Effects of global regulatory systems on the expression of genes in the SsrAB virulon.
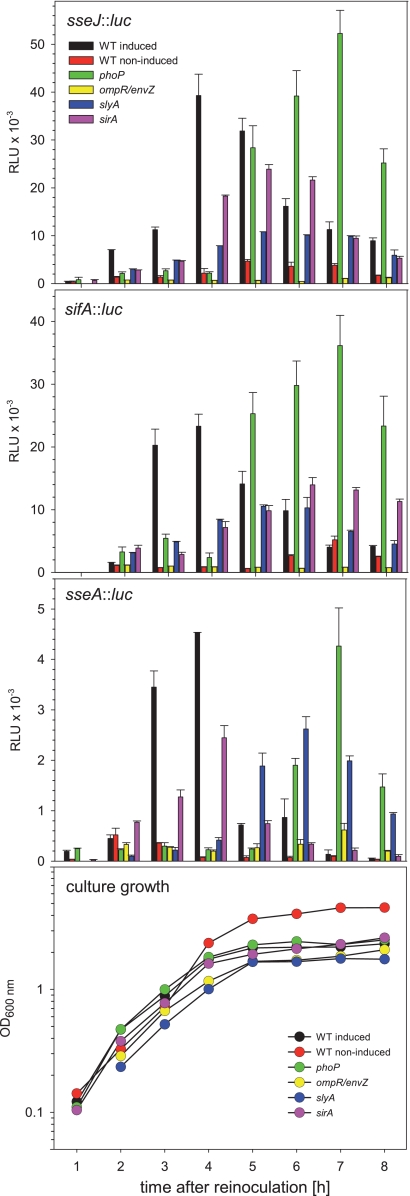
In addition to the SsrAB regulatory system, additional regulators that are involved in the control of the SsrAB virulon have been described. We generated strains to combine the reporter fusions to representative genes of the SsrAB regulon with mutations in genes for global regulators, i.e., ompR-envZ, phoP, sirA, or slyA. The deletions of the various regulators did not affect the overall growth of the reporter strains in PCN medium, and similar growth curves were observed for all strains (Fig. 4). Only during growth under noninducing conditions in PCN medium with neutral pH did the cultures reach higher cell densities.
FIG. 4.
Effects of global regulatory systems on the expression of the SsrAB virulon. The expression of representative genes of the SsrAB virulon was analyzed in strains defective in various regulatory systems, as indicated. Reporter fusions for sifA and sseJ were used to represent transcriptional units outside of SPI2, while sseA represents a transcriptional unit within SPI2. Reporter fusions were analyzed in the WT strain or in a phoP, ompR, slyA, or sirA deletion background. The strains were subcultured in noninducing minimal medium (PCN medium-25 mM Pi, pH 7.4; red) or under SPI2-inducing conditions (PCN medium-0.4 mM Pi, pH 5.8; all other histograms). Representative growth curves are shown in the bottom panel, and all reporter strains showed similar growth characteristics. Cells were harvested at various time points, and equal amounts of cells, as adjusted by OD600, were lysed for the quantification of Luc activities. The quantification of reporter activities was performed as described for Fig. 2, and means and standard deviations for triplicate assays are shown.
Quantification of reporter activities (Fig. 4) revealed that strains deficient in the two-component system OmpR/EnvZ showed the highest reduction in expression. Expression of sseJ::luc and sifA::luc fusions was highly reduced, and the luciferase activity was in the range of the background activity. Slightly higher expression levels were determined for the sseA::luc fusion in the ompR-envZ background.
The deletion of slyA, encoding a transcriptional regulator, had less severe effects on the expression of the reporter fusions. Compared to the WT background, the maximal level was reduced by a factor of 2 to 3. Further, the peak level of expression was delayed and was usually reached after 5 to 6 h of culture.
The PhoPQ two-component system is considered a global regulator of Salmonella adaptation to life inside multicellular hosts. The effects of the PhoPQ system on regulation of SPI2 genes have been described previously (5, 10). Our analyses of the expression kinetics in a phoP background indicated that expression of all three fusions was virtually absent at the time when the expression of the reporter fusions reached maximum in the WT background, i.e., 3 to 4 h after inoculation. Surprisingly, the analyses of the expression kinetics showed that the expression of fusions in the phoP background was highly delayed, with peak levels at 7 h of subculture. At this time, the expression level of the sifA::luc fusion even exceeded the peak activity of the WT strain at 3 to 4 h of culture, while the expression levels of sseJ and sseA fusions reached maxima similar to or even higher than those determined for fusions in the WT background.
SirA is part of the two-component system SirA/BarA that is considered a master regulator of enteropathogenesis (2). Due to this function, we did not anticipate a major effect of the sirA deletion on the expression of SPI2 reporter fusions. In fact, lack of SirA resulted in reduced expression of all three reporter fusions, with about 30 to 50% reduced maximal expression. The kinetics of expression of the sseA::luc fusion were similar in both the WT and sirA backgrounds, while peak expression of sseJ::luc and sifA::luc was delayed, with peak expression at 5 to 6 h of culture.
Taken together, our analyses with chromosomal reporter fusions supported an essential role of SsrB and OmpR/EnvZ for the expression of SPI2 genes under in vitro conditions mimicking the intracellular life of Salmonella and a limited contribution of SlyA and SirA. The defect of PhoP resulted in temporally deregulated expression of the SsrAB operon, since the peak of expression was shifted from the exponential growth phase to the stationary growth phase.
Analysis of intracellular expression levels.
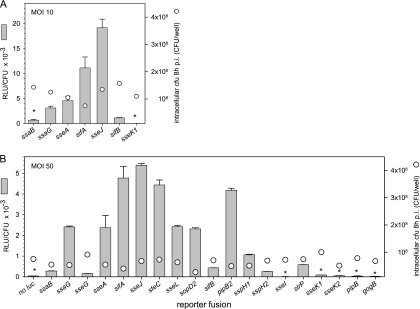
The previous experiments were performed using in vitro conditions for the activation of SsrAB virulon expression. In order to correlate these data with expression during the infection of host cells, we performed infections of macrophages with strains harboring reporter fusions. Representative reporter strains for T3SS genes or effector genes, as well as strains with high, average, or low expression levels under in vitro conditions, were selected for the infection of macrophages at an MOI of 10 (Fig. 5A). In order to compensate for the varying degrees of intracellular replication of different strains, the number of intracellular bacteria was determined and the luciferase activity per intracellular bacterium was calculated. The highest expression level was observed for sseJ, followed by sifA, sseA, and ssaG. Low levels of expression were observed for ssaB and sifB, and sseK1 expression was not detectable (Fig. 5A). In order to analyze the intracellular expression of the entire set of reporter strains and to determine the expression levels of weakly expressed genes, macrophages were infected with various reporter strains at an MOI of 50 (Fig. 5B). We observed the highest expression of sseJ, sifA, steC, and pipB2 by intracellular bacteria and high expression levels of sseA, ssaG, sseL, and sopD2. Low expression levels were detected for ssaB, sifB, sspH1, slrP, sseG, and sspH2, while the activities of the intracellular reporter strains for sseI, sseK1, sseK1, pipB, and gogB were at, or only slightly above, the detection limit.
FIG. 5.
Expression levels of genes of the SsrAB virulon in intracellular bacteria. RAW264.7 macrophages were infected at an MOI of 10 (A) or 50 (B) with various Salmonella strains harboring reporter gene fusions, as indicated, or the WT strain without the reporter (no luc). Noninternalized bacteria were killed by the addition of gentamicin. Infected host cells were lysed 8 h after infection (p.i.), intracellular bacteria were recovered, and the luciferase activities were quantified. In parallel, aliquots of the lysates were plated on agar plates for the determination of the number of intracellular bacteria (cfu). The numbers of intracellular bacteria recovered 8 h after infection are shown by circles. The expression of various genes of the SsrAB virulon was calculated as RLU per CFU. Means and standard deviations of triplicates are shown for an experiment representative of three independent assays. Reporter activities for several strains (indicated by asterisks) were close to the limit of detection of the assay.
Due to the insertion of the reporter cassette, some of the reporter strains had defects in the T3SS or lacked important effector proteins, such as SifA. However, at the time of sampling, i.e., 8 h after infection of RAW264.7 cells, only two- to threefold differences in the numbers of intracellular bacteria were observed (CFU are indicated by circles in Fig. 5). The reduction of intracellular replication was much more pronounced at 16 h after infection (data not shown). We also observed that intracellular expression of reporter fusions in the background of the ssrB strain was highly reduced, and values were in the range of the background (data not shown).
Overall, the expression levels of the intracellular reporter strains showed a clear correlation with the data for in vitro-grown bacteria (Fig. 2). This finding suggests that the conditions for the induction of the SsrAB virulon are well suited to mimic environmental conditions experienced by intracellular Salmonella during life within macrophages.
DISCUSSION
The genes encoding the SPI2-T3SS and the cognate effector proteins form a complex virulon that is controlled by the SsrAB two-component system (reviewed in reference 13). The analyses of the expression levels of such large virulons is difficult and may be hampered by different positions with respect to regulatory sequences in the case of randomly generated chromosomal reporter fusions. Episomal reporter fusions can be generated in a comparable manner but suffer from the absence of the chromosomal context of the gene under study. We have recently devised an approach to generate reporter fusions in the chromosome of Salmonella that can be identical with respect to the distance between the reporter gene and regulatory elements of the gene under study (16). Here, we applied this reporter approach to the comparative analysis of the elements of the SsrAB virulon. To our knowledge, this is the first report of fully comparable reporter fusions for genes of a complex virulon.
The comparison of the genes of the SsrAB virulon showed a large range of maximal expression levels, with the highest levels observed for some of the effectors encoded by genes outside of SPI2. The expression of some effectors was virtually undetectable, and further studies will be needed to determine if effector proteins, such as SseK1, SseK2, or GogB, accumulate in intracellular Salmonella bacteria in detectable amounts and are translocated into host cells. However, the expression levels of the effectors did not show a direct correlation with the role of the effector in the intracellular lifestyle of Salmonella. For example, sifA and sseJ both showed the highest expression levels. While an essential role of SifA in the formation of Salmonella-induced filaments (SIF) (42), the maintenance of the SCV, and systemic virulence in the murine model has been demonstrated (4), the role of SseJ is still unclear, and deletion of SseJ had no significant contribution to intracellular phenotypes or systemic virulence (34, 37). Interestingly, the detection of epitope-tagged translocated effectors by immunofluorescence analyses also indicated that effector proteins, like SseJ or SseF, are present in host cells in large amounts (26), while we have not been able to detect SseK1, SseK2, or GogB as translocated effectors (data not shown).
In addition to the regulation by the level of expression that was analyzed in this study, the amounts of SPI2-T3SS subunits and effector proteins are also affected by posttranscriptional mechanisms, e.g., by mRNA degradation, and on the posttranslational level, e.g., by proteolysis and secretion or translocation of a subset of the proteins. A comprehensive analysis of this complex regulatory network thus also requires the quantitative analysis of the proteins in the bacterial cell and in host cells after translocation.
The kinetics of expression under in vitro conditions were similar for all genes of the SsrAB virulon. We did not observe significant differences in the time points of maximal expression between genes encoding the structural components of the T3SS or effector proteins within SPI2 or loci outside of SPI2. We anticipated that the expression of the T3SS structural genes would precede the expression of the effector genes. However, the temporal resolution of luc reporter assays may not be sufficient to detect subtle differences in expression kinetics. The molecular functions of most effector proteins of the SPI2-T3SS are not known, and no temporal order of effector functions has been established to date. Studies of the effector proteins of the SPI1-T3SS indicate that the sequence of effector actions is not mediated by the sequential expression of various genes, but rather by differences in the half-lives of effectors after translocation (24) or by subtle differences in the kinetics of translocation (44).
As anticipated from previous studies, the transcriptional activator SsrB was essential for the expression of the members of the SsrAB virulon (7, 11, 14, 30). The analysis of the effects of various global regulatory systems revealed an important role of the OmpR/EnvZ system for the activation of the expression of members of the SsrAB virulon. These results are largely in line with previous studies (14, 27). A previous study (29) reported a role for SlyA in the regulation of the SsrAB virulon, but under the conditions applied in our assays, we observed only a minor contribution of SlyA to the regulation of SPI2 genes and other genes encoding effector proteins. The defect of the PhoPQ global regulatory system did not reduce the maximum expression level of the genes in the SsrAB virulon, but rather caused highly delayed expression. While reporter activities in the WT background reached a peak level at 3 to 4 h after the start of growth under in vitro conditions, maximal expression was observed at 6 to 7 h of culture in the phoP background. We propose that this delay prevents the timely expression of SPI2 virulence genes, resulting in defective intracellular virulence properties of Salmonella.
Salmonella is an interesting vehicle for the delivery of recombinant antigens for vaccination (reviewed in references 6 and 35). Attenuated strains of Salmonella can be used to deliver fusion proteins composed of effector proteins and heterologous antigens, and this approach has proven to be efficient for eliciting protective immunity against various infectious agents (22, 38, 39). One parameter for the optimization of recombinant Salmonella strains for vaccination is the use of optimized intracellular expression. Previous vaccination studies with recombinant Salmonella strains made use of the sseA promoter (22). This promoter belongs to the group of promoters with average expression levels, and we are currently investigating if the use of promoters with high or very high expression levels leads to improved performance in experimental vaccination (X. Xu, M. I. Husseiny, and M. Hensel, unpublished observations).
Previous microarray analyses (40) indicated a role for SsrAB in the control of the SsrAB virulon, similar to our observations. Under inducing conditions, the expression levels of genes in SPI2 and certain effectors were up to 100-fold higher in the WT background than in an ssrB mutant (40). Our results are only partially in accord with the quantification of the expression levels of Salmonella genes in vivo (36). This study identified sifA and sifB as the genes with the highest expression during systemic infection, followed by ssaG, sseA, pipB, and ssaB. Moderate expression was observed for ssaM, sopD2, sseJ, and pipB2. It is possible that the environmental stimuli applied in our study only partially resemble the complexity of the signals encountered by Salmonella during systemic virulence in the mammalian host. However, the study by Rollenhagen et al. (36) investigated promoter fragments randomly cloned in mid-copy-number vectors, while we compared the expression levels of chromosomal reporter fusions generated in an identical manner. One should be aware that detection limits still restrict the analyses of single-copy chromosomal reporters in the host tissue.
The effects of the DNA topology and global regulatory mechanisms by NAP such as H-NS (reviewed in reference 12) are more likely to be defined by analyses of chromosomal reporter fusions. In addition, in the global regulatory system with a role in the regulation of the SsrAB virulon, modulation of expression by NAP has been observed. These involve positive regulators of ssrAB or the SPI2 genes, such as Fis (28), and negative regulators, such as H-NS (43) and YdgT (8).
In this study, we performed comprehensive and comparative analyses of the expression levels of genes of the SsrAB virulon. The results can now be applied to the rational design of Salmonella-based live vaccines with in vivo-activated promoters of the SsrAB virulon.
Acknowledgments
This work was supported by grant HE1964 of the Deutsche Forschungsgemeinschaft to M.H. X.X. was supported in part by a fellowship from Gimex GmbH.
We thank Daniela Jäckel and Roman Gerlach for the construction of reporter strains and Deepak Chikkaballi for critical reading of the manuscript.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 26 October 2009.
REFERENCES
- 1.Abrahams, G. L., and M. Hensel. 2006. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell. Microbiol. 8:728-737. [DOI] [PubMed] [Google Scholar]
- 2.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 3.Bakowski, M. A., V. Braun, and J. H. Brumell. 2008. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9:2022-2031. [DOI] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. [DOI] [PubMed] [Google Scholar]
- 6.Cheminay, C., and M. Hensel. 2008. Rational design of Salmonella recombinant vaccines. Int. J. Med. Microbiol. 298:87-98. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 8.Coombes, B. K., M. E. Wickham, M. J. Lowden, N. F. Brown, and B. B. Finlay. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. USA 102:17460-17465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiwick, J., and M. Hensel. 1999. Regulation of virulence genes by environmental signals in Salmonella typhimurium. Electrophoresis 20:813-817. [DOI] [PubMed] [Google Scholar]
- 11.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 12.Dorman, C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157-161. [DOI] [PubMed] [Google Scholar]
- 13.Fass, E., and E. A. Groisman. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385-2396. [DOI] [PubMed] [Google Scholar]
- 15.Geddes, K., M. Worley, G. Niemann, and F. Heffron. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 73:6260-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlach, R. G., S. U. Hölzer, D. Jäckel, and M. Hensel. 2007. Rapid engineering of bacterial reporter gene fusions by using Red recombination. Appl. Environ. Microbiol. 73:4234-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach, R. G., D. Jäckel, N. Geymeier, and M. Hensel. 2007. Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect. Immun. 75:4697-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haraga, A., M. B. Ohlson, and S. I. Miller. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53-66. [DOI] [PubMed] [Google Scholar]
- 19.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry, T., J. P. Gorvel, and S. Meresse. 2006. Molecular motors hijacking by intracellular pathogens. Cell. Microbiol. 8:23-32. [DOI] [PubMed] [Google Scholar]
- 21.Husseiny, M. I., and M. Hensel. 2005. Evaluation of an intracellular-activated promoter for the generation of live Salmonella recombinant vaccines. Vaccine 23:2580-2590. [DOI] [PubMed] [Google Scholar]
- 22.Husseiny, M. I., F. Wartha, and M. Hensel. 2007. Recombinant vaccines based on translocated effector proteins of Salmonella pathogenicity island 2. Vaccine 25:185-193. [DOI] [PubMed] [Google Scholar]
- 23.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 24.Kubori, T., and J. E. Galan. 2003. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333-342. [DOI] [PubMed] [Google Scholar]
- 25.Kuhle, V., and M. Hensel. 2004. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61:2812-2826. [DOI] [PubMed] [Google Scholar]
- 26.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 27.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim, S., B. Kim, H. S. Choi, Y. Lee, and S. Ryu. 2006. Fis is required for proper regulation of ssaG expression in Salmonella enterica serovar Typhimurium. Microb. Pathog. 41:33-42. [DOI] [PubMed] [Google Scholar]
- 29.Linehan, S. A., A. Rytkonen, X. J. Yu, M. Liu, and D. W. Holden. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73:4354-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Löber, S., D. Jäckel, N. Kaiser, and M. Hensel. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435-447. [DOI] [PubMed] [Google Scholar]
- 31.Maloy, S. R., V. L. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarre, W. W., T. A. Halsey, D. Walthers, J. Frye, M. McClelland, J. L. Potter, L. J. Kenney, J. S. Gunn, F. C. Fang, and S. J. Libby. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492-508. [DOI] [PubMed] [Google Scholar]
- 34.Nawabi, P., D. M. Catron, and K. Haldar. 2008. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol. Microbiol. 68:173-185. [DOI] [PubMed] [Google Scholar]
- 35.Panthel, K., K. M. Meinel, V. E. Sevil Domenech, K. Trülzsch, and H. Rüssmann. 2008. Salmonella type III-mediated heterologous antigen delivery: a versatile oral vaccination strategy to induce cellular immunity against infectious agents and tumors. Int. J. Med. Microbiol. 298:99-103. [DOI] [PubMed] [Google Scholar]
- 36.Rollenhagen, C., M. Sorensen, K. Rizos, R. Hurvitz, and D. Bumann. 2004. Antigen selection based on expression levels during infection facilitates vaccine development for an intracellular pathogen. Proc. Natl. Acad. Sci. USA 101:8739-8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 38.Rüssmann, H., U. Gerdemann, E. I. Igwe, K. Panthel, J. Heesemann, S. Garbom, H. Wolf-Watz, and G. Geginat. 2003. Attenuated Yersinia pseudotuberculosis carrier vaccine for simultaneous antigen-specific CD4 and CD8 T-cell induction. Infect. Immun. 71:3463-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rüssmann, H., H. Shams, F. Poblete, Y. Fu, J. E. Galan, and R. O. Donis. 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science 281:565-568. [DOI] [PubMed] [Google Scholar]
- 40.Rytkonen, A., J. Poh, J. Garmendia, C. Boyle, A. Thompson, M. Liu, P. Freemont, J. C. Hinton, and D. W. Holden. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. USA 104:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele-Mortimer, O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 43.Walthers, D., R. K. Carroll, W. W. Navarre, S. J. Libby, F. C. Fang, and L. J. Kenney. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65:477-493. [DOI] [PubMed] [Google Scholar]
- 44.Winnen, B., M. C. Schlumberger, A. Sturm, K. Schupbach, S. Siebenmann, P. Jenny, and W. D. Hardt. 2008. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS ONE 3:e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]