Abstract
Listeria monocytogenes is an intracellular bacterial pathogen that invades epithelial cells by subverting two cellular receptors, E-cadherin and Met. We recently identified type II phosphatidylinositol 4-kinases α and β (PI4KIIα and PI4KIIβ) as being required for bacterial entry downstream of Met. In this work, we investigated whether tetraspanins CD9, CD63, and CD81, which figure among the few described molecular partners of PI4KIIα, function as molecular adaptors recruiting PI4KIIα to the bacterial entry site. We observed by fluorescence microscopy that CD9, CD63, and CD81 are expressed and detected at the cellular surface and also within intracellular compartments, particularly in the case of CD63. In resting cells, colocalization of tetraspanins and PI4KIIα is detectable only in restricted areas of the perinuclear region. Upon infection with Listeria, endogenous CD9, CD63, and CD81 were recruited to the bacterial entry site but did not colocalize strictly with endogenous PI4KIIα. Live-cell imaging confirmed that tetraspanins and PI4KIIα do not follow the same recruitment dynamics to the Listeria entry site. Depletion of CD9, CD63, and CD81 levels by small interfering RNA demonstrated that CD81 is required for bacterial internalization, identifying for the first time a role for a member of the tetraspanin family in the entry of Listeria into target cells. Moreover, depletion of CD81 inhibits the recruitment of PI4KIIα but not that of the Met receptor to the bacterial entry site, suggesting that CD81 may act as a membrane organizer required for the integrity of signaling events occurring at Listeria entry sites.
Listeria monocytogenes is a food-borne bacterial pathogen responsible for listeriosis, a disease affecting mainly immunocompromised individuals and characterized by gastroenteritis, abortion in pregnant women, and meningitis in newborns (9). Listeria is able to induce its internalization into nonphagocytic epithelial cells by interacting with two cellular receptors, E-cadherin (the ligand of the bacterial surface protein internalin) and the hepatocyte growth factor receptor Met (the ligand of InlB) (15). Activation of Met by InlB leads to the recruitment of the ubiquitin ligase Cbl, the clathrin-dependent internalization of the receptor, and also the Gab1-mediated activation of a phosphatidylinositol 3-kinase (PI3K)-dependent pathway involved in the reorganization of the actin cytoskeleton around the bacterial entry site (20). Recently, we have identified a role for type II PI4K α and β isoforms (PI4KIIα and PI4KIIβ) during Listeria entry (17). At this stage, it is not known which signaling cascade(s) is activated downstream of the PI4KIIs during Listeria entry and how these lipid kinases are recruited to the bacterial entry site upon Met activation.
To date, very few molecules have been identified as molecular partners of the PI4KIIs. Members of the tetraspanin family, including CD9, CD63, and CD81, have been found in complexes with PI4KIIα (26). Tetraspanins are transmembrane molecules known to associate with one another and with many membrane molecules, including integrins, forming molecular microdomains (the tetraspan web) which are thought to function as specialized signaling platforms (4, 11). In this context, it has been proposed that tetraspanins can recruit PI4KIIα to specific membrane locations to influence phosphoinositide-dependent signaling. In the present study, we investigated the possible role of tetraspanins as molecular adaptors for PI4KIIα at the sites of Listeria entry into HeLa cells.
MATERIALS AND METHODS
Cells and bacteria.
HeLa cells (ATCC CCL-2) were grown as recommended by the ATCC in modified Eagle medium supplemented with 10% fetal calf serum and 2 mM glutamine (GIBCO). L. monocytogenes EGD strain BUG 600 (a wild-type bacterium of serovar 1/2a) and mutant L. monocytogenes EGD strain BUG 1047 (a BUG 600 mutant carrying an inlB gene deletion) were grown in brain heart infusion. L. monocytogenes EGD strain BUG 1641 is a variant of BUG 600 expressing the InlB protein covalently bound to the bacterial cell wall (5), and it was grown in brain heart infusion supplemented with 5 μg/ml of erythromycin.
Antibodies and reagents.
Polyclonal purified anti-PI4KIIα antibodies were generated in our lab by immunizing rabbits with the peptide Met-Asp-Glu-Thr-Ser-Pro-Leu-Val-Ser-Pro-Leu-Val-Ser-Pro-Glu-Arg-Ala-Gln-Pro-Pro-Asp-Tyr-Thr-Cys. Mouse monoclonal anti-CD9, anti-CD63, and anti-CD81 were described previously (7). Rabbit polyclonal anti-Met C-12 antibodies were purchased from Santa Cruz. Alexa Fluor 488-, 546-, and 647-conjugated goat anti-rabbit and goat anti-mouse antibodies were purchased from Molecular Probes (Invitrogen). Recombinant InlB was purified by ion-exchange chromatography as described previously (16).
Transfection with DNA and siRNA.
A DNA construct expressing PI4KIIα-green fluorescent protein (GFP) was kindly supplied by T. Balla (NIH, Bethesda, MD), and a construct expressing CD63-GFP was supplied by G. Griffiths (University of Oxford); CD9 and CD81 genes were cloned into the pEGFP-C2 vector by using standard PCR techniques. HeLa cells grown to 50% confluence on six-well plates containing 22- by 22-mm glass coverslips were transfected overnight with 1 μg of each DNA construct per well by using JetPei according to the instructions of the manufacturer (Ozyme); infections were performed the next day. For experiments with small interfering RNA (siRNA), 5 × 104 HeLa cells plated onto six-well plates were transfected with RNA oligonucleotides from Ambion specific for the inactivation of CD9 (identification no. 10309), CD63 (identification no. 13018), and CD81 (identification no. 14407) and a negative control (catalog no. AM4631) by using DharmaFECT1 according to the instructions of the manufacturer (Thermo Scientific); infections were performed 72 h after transfection.
Cell infection and immunofluorescence.
One colony of L. monocytogenes BUG 1641 was grown overnight in a shaking device, and the bacterial culture was diluted the next day until an optical density of 0.9 at 900 nm was reached. Bacteria were resuspended in serum-free modified Eagle medium at a multiplicity of infection of 50 bacteria per cell, and the suspension was centrifuged over HeLa cells at 1,000 rpm for 2 min at room temperature. Cells were then incubated for different time periods at 37°C. Infected monolayers were then washed once with phosphate-buffered saline (PBS) supplemented with 0.5% bovine serum albumin, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 in PBS supplemented with 0.5% bovine serum albumin for 4 min, blocked with 0.5% bovine serum albumin for 15 min, and incubated with primary antibodies for 30 min and with secondary antibodies for 30 min. Cells were mounted onto slides with Fluoromount G (Interchim, France) and analyzed with an Axiovert 200 M microscope (Zeiss) equipped with a CoolSNAP HQ camera (Photometrics), and image acquisition was performed with MetaMorph software (Universal Imaging Corporation). For the acetone permeabilization experiments, infected monolayers were washed once with PBS and incubated with acetone for 20 min at −20°C before being incubated with primary antibodies.
Immunoprecipitations.
HeLa cells grown to confluence in six-well plates were washed with cold PBS and then lysed with 100 μl per well of lysis buffer containing 50 mM Tris, 150 mM NaCl, and 1% Brij 98 supplemented with Complete protease inhibitors (Amersham). Lysates were incubated for 30 min at 4°C and centrifuged for 15 min at 13,000 rpm on a cold tabletop Eppendorf centrifuge, pellets were discarded, and supernatants were incubated with 50 μl of Sepharose-protein A beads (Amersham) for 30 min at 4°C on a shaking wheel. Afterwards, beads were centrifuged and discarded, and the precleared lysates were incubated overnight with 1 μg of anti-PI4KIIα antibodies. The next day, PI4KIIα-containing complexes were recovered by incubation with Sepharose-protein A beads and separated by SDS-10% PAGE under nonreducing conditions. Proteins were transferred onto nitrocellulose membranes, and Western blotting was performed using anti-CD9, anti-CD63, anti-CD81, and anti-PI4KIIα antibodies.
Gentamicin survival assay.
HeLa cells and L. monocytogenes BUG 600 or BUG 1641 were grown as described above. Bacteria were resuspended in serum-free modified Eagle medium at a multiplicity of infection of 50 bacteria per cell, and cells were inoculated for 1 h. Then cells were washed once and incubated for 1 h in modified Eagle medium supplemented with 10% fetal calf serum and 20 μg/ml gentamicin. Cells were finally disrupted in sterile distilled water, and serial dilutions were plated onto brain heart infusion agar plates for CFU counting the next day.
Time lapse microscopy.
Images were acquired with a motorized inverted fluorescence microscope (Axiovert 200 M; Carl Zeiss MicroImaging Inc.) equipped with a temperature-controlled stage by using 100× lens objectives (Carl Zeiss, Inc.). Fluorescent illumination was driven by an ultrahigh-speed wavelength switcher, Lambda DG-4 (Sutter Instrument), equipped with a 175-W xenon arc lamp and excitation filters for cyan fluorescent protein (CFP) and GFP (Chroma Technology). Emission filters were selected using a high-speed Lambda 10 filter wheel (Sutter Instrument). Images were acquired with exposure times between 100 and 500 ms with a cooled, digital, charge-coupled device camera (CoolSNAP HQ; Photometrics). All devices were controlled by MetaMorph imaging system software (Universal Imaging).
FRET stoichiometry measurement.
PI4KIIα-CFP and CD9-yellow fluorescent protein (YFP) were obtained by GFP replacement in constructs furnished by T. Balla (NIH, Bethesda, MD) and E. Rubinstein and C. Boucheix (INSERM, U602, Villejuif, France). Cells were cotransfected with the PI4KIIα-CFP and CD9-YFP plasmids and stimulated with 5 nM InlB or infected with L. monocytogenes BUG 1641, and fluorescent resonance energy transfer (FRET) measurement was done as described previously (12); images were collected every 15 s using our time lapse microscopy equipment.
RESULTS
Expression and distribution of the tetraspanins CD9, CD63, and CD81 in HeLa cells.
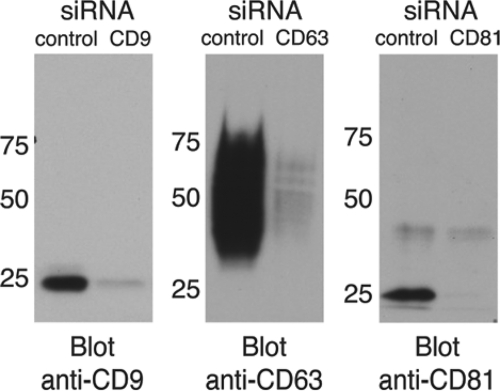
By Western blot analysis, we were able to detect the expression of endogenous CD9, CD63, and CD81 in HeLa cells (Fig. 1). CD9 was detected as a major band at 24 kDa and a minor band at 26 kDa, while CD81 appeared as a single clear band at around 26 kDa. CD63 was always detected as a protein smear at around 40 to 60 kDa due to its high levels of glycosylation. Using immunofluorescence, we investigated the distributions of these tetraspanins. In nonpermeabilized cells, we observed punctuated distribution of these molecules at the cell surface, with particular enrichment at sites of cell-to-cell contact, in line with previous observations (see Fig. S1A in the supplemental material) (4, 11). Intracellular enrichment was detected in the perinuclear region of cells following permeabilization, and this pattern was particularly remarkable in the case of CD63 (see Fig. S1B in the supplemental material), consistent with the reported accumulation of CD63 in late endosomal compartments. These results indicate that CD9, CD63, and CD81 are expressed in HeLa cells and detected both at the cellular surface and in enriched intracellular pools in the perinuclear region.
FIG. 1.
Expression of CD9, CD63, and CD81 in HeLa cells. Cell extracts from wild-type cells or cells treated with siRNAs were run on 10% polyacrylamide gels, samples were transferred onto nitrocellulose membranes, and Western blot analyses were performed under nonreducing conditions using specific anti-CD9, anti-CD63, or anti-CD81 antibodies. Left lanes show the detection of CD9 as a major band of 24 kDa, and a secondary minor band can be observed at 26 kDa. CD63 is detected as a smear due to the high levels of glycosylation of the protein. CD81 is detected as a band of 26 kDa. Right lanes show the absence of protein expression after inactivation with specific siRNAs.
Distribution of PI4KIIα and tetraspanins in HeLa cells.
We next investigated the distribution of PI4KIIα in relationship to the tetraspanins CD9, CD63, and CD81 in resting HeLa cells. Endogenous PI4KIIα was circumscribed mainly to the perinuclear region of cells, consistent with its described function at the trans-Golgi network and also in late endosomal compartments (see Fig. S2 in the supplemental material) (18, 24). In this region, we also detected partial colocalization of tetraspanins and PI4KIIα, CD63 being the molecule which presented the most distinct overlap with PI4KIIα (see Fig. S2 in the supplemental material). No accumulation of PI4KIIα was observed at cell-to-cell contact sites on the plasma membrane where tetraspanins were enriched (see Fig. S2 in the supplemental material). These results suggest that in resting HeLa cells, with the exception of a partial overlap in the perinuclear space area, no major colocalization of the tetraspanins CD9, CD63, and CD81 with PI4KIIα is observed.
CD9, CD63, and CD81 are recruited to the Listeria entry site.
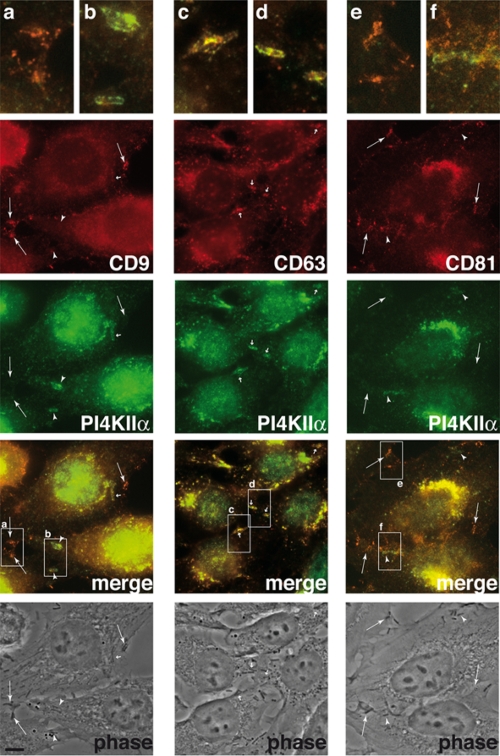
Since we hypothesized that tetraspanins could help target PI4KIIα to the site of Listeria entry into target cells, the potential colocalization of endogenous CD9, CD63, and CD81 with endogenous PI4KIIα upon bacterial infection was analyzed. For imaging experiments, we used the Listeria strain BUG 1641, which expresses the InlB protein covalently linked to the bacterial cell wall, fostering the activation of the InlB receptor Met and allowing the visualization of otherwise rare molecular events. As shown in Fig. 2, recruitment of CD9, CD63, and CD81 was detected at sites of bacterial entry, demonstrating for the first time the recruitment of members of the tetraspanin family to the Listeria entry site. In the same visual fields, we were able to observe recruitment of PI4KIIα by invading Listeria, but corecruitment with CD9 was detected only occasionally. In the case of CD63, we observed partial corecruitment of the tetraspanin and PI4KIIα, since the labeling of these two markers seemed to be distributed independently around bacteria (Fig. 2). Corecruitment of PI4KIIα and CD81 was not observed. Similar observations were obtained using different permeabilization methods, including acetone treatment (data not shown), which extracts smaller amounts of tetraspanins from membranes than Triton X-100 treatment (3). InlB-deficient Listeria (BUG 1047) bacteria are not able to interact with any of the tetraspanins, confirming the role of the Met signaling pathway in the redistribution of these molecules during bacterial entry (data not shown). These results demonstrate that tetraspanins CD9, CD63, and CD81 are recruited to the Listeria entry site but do not systematically colocalize with PI4KIIα.
FIG. 2.
Distribution of endogenous CD9, CD63, CD81, and PI4KIIα in HeLa cells infected with Listeria. Cells were infected for 10 min with L. monocytogenes strain BUG 1641; cells were then fixed and processed for immunofluorescence analyses using anti-CD9, anti-CD63, or anti-CD81 antibodies and anti-PI4KIIα immune serum. The colocalization of Listeria with tetraspanins is indicated by large arrows, the colocalization of Listeria with PI4KIIα is indicated by arrowheads, and the colocalization of Listeria with both markers is indicated by small arrows. Boxes labeled a to f in the merge panels have been enlarged at the top of the figure. Bar, 3 μm; phase, phase-contrast micrograph.
Live imaging of tetraspanin and PI4KIIα recruitment upon Listeria infection.
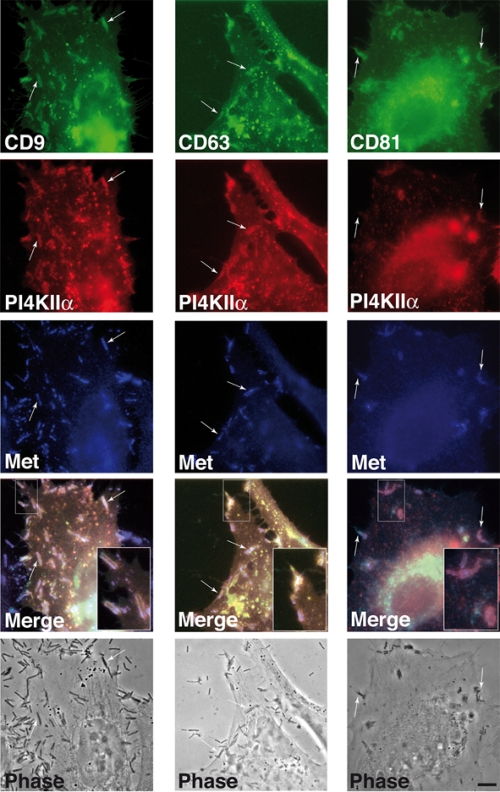
To better observe their recruitment to the bacterial entry site, tetraspanins and PI4KIIα were then overexpressed in HeLa cells by cotransfection of cells with constructs coding for CD9, CD63, and CD81 fused to GFP and PI4KIIα fused to CFP; after infection with Listeria, cells were fixed and labeled for the detection of Met, the cellular receptor for the Listeria invasion protein InlB. As shown in Fig. 3, recruitment of CD9-GFP was frequently associated with Listeria at sites of bacterial entry, where enrichment with Met and PI4KIIα-CFP was also observed. CD63-GFP and CD81-GFP were less frequently associated with bacterial entry foci, but when present, both molecules colocalized with Met and with PI4KIIα-CFP (Fig. 3). These results indicate that overexpression of tetraspanins and PI4KIIα may force these molecules to interact during Listeria internalization, suggesting that interactions can be very transient and difficult to capture in work with the endogenous proteins.
FIG. 3.
Distribution of Met, CD9, CD63, CD81, and PI4KIIα in HeLa cells infected with Listeria. Cells were transfected with CD9-GFP, CD63-GFP, CD81-GFP, and PI4KIIα-CFP constructs for 24 h, and then cells were infected for 10 min with wild-type L. monocytogenes strain BUG 1641; cells were then fixed and processed for immunofluorescence analyses using anti-Met antibodies. The colocalization of Met, CD9, PI4KIIα, and Listeria and of Met, CD63, PI4KIIα, and Listeria is observed (arrows). Boxes in the merge panels have been enlarged as insets. Bar, 3 μm.
To then examine the corecruitment of overexpressed CD9, CD63, CD81, and PI4KIIα to sites of Listeria internalization, we performed live-cell imaging analyses. We observed that the tetraspanins and the lipid kinase were concomitantly recruited by individual bacteria but that the dynamics of their recruitment were not identical (see Fig. S3 to S5 and Movies S1 to S5 in the supplemental material). The appearance of CD9-GFP labeling precedes PI4KIIα-CFP enrichment, and CD9-GFP labeling is located in membrane extensions that are not labeled by the lipid kinase during the first contacts between bacteria and target cells (see Fig. S3 and Movies S1 to S3 in the supplemental material); throughout internalization, CD9-GFP and PI4KIIα-CFP follow highly dynamic but not identical redistribution patterns around bacteria. A similar result was observed when cells were transfected with constructs encoding CD63-GFP and PI4KIIα-CFP (see Fig. S4 and Movie S4 in the supplemental material): interestingly, in these doubly transfected cells, we observed individual vesicles positive for both CD63-GFP and PI4KIIα-CFP which docked repeatedly with the invading Listeria but the distribution of the two fluorescent markers did not follow identical reorganization patterns. Concerning the dynamics of CD81-GFP and PI4KIIα-CFP (see Fig. S5 and Movie S5 in the supplemental material), an extremely transient accumulation of CD81-GFP is detected at bacterial invasion sites, but afterwards, the recruitment of PI4KIIα-CFP proceeds independently of interactions with CD81-GFP-positive structures. These results indicate that overexpressed tetraspanins and PI4KIIα are recruited by Listeria at different stages of the infection process.
Tetraspanins and PI4KIIα do not directly interact in HeLa cells.
Since our results suggested that tetraspanins are not associated in the same molecular complex with PI4KIIα while being corecruited by Listeria during bacterial entry, we analyzed the possible interaction of tetraspanins and PI4KIIα by immunoprecipitation. We first verified that our anti-PI4KIIα antibody was able to immunoprecipitate PI4KIIα itself (data not shown). When we immunoprecipitated the lipid kinase and investigated with anti-CD9, anti-CD63, or anti-CD81 antibodies, we were not able to detect any interaction between the tetraspanins and PI4KIIα in resting cells or in cells stimulated with 2.5 nM InlB for 2 min (data not shown). We then investigated by FRET the potential interaction between CD9-YFP and PI4KIIα-CFP in cells transfected to express these fluorescent constructs: no positive FRET signal was detected in cells stimulated with soluble InlB or infected with wild-type Listeria (data not shown). These results strongly suggest that in HeLa cells, there is no direct interaction between tetraspanins and PI4KIIα.
Assessment of tetraspanin function during Listeria entry.
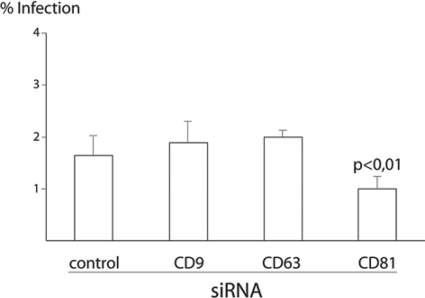
Despite the fact that we were not able to detect any interaction between tetraspanins and PI4KIIα, we clearly observed the recruitment of endogenous CD9, CD63, and CD81 to the Listeria internalization sites on HeLa cells. In order to determine if tetraspanins can contribute functionally to bacterial internalization, we depleted CD9, CD63, and CD81 levels with siRNA (Fig. 1) and performed a gentamicin invasion assay using Listeria BUG 1641. While inactivation of CD9 or CD63 did not modify bacterial entry into HeLa cells, inactivation of CD81 significantly reduced the levels of Listeria invasion (Fig. 4). Similar results were obtained with the wild-type BUG 600 Listeria strain (data not shown). This result demonstrates that CD81 is a novel molecular player required for the InlB-dependent entry of Listeria into target cells.
FIG. 4.
Inactivation of CD81 inhibits the entry of Listeria into HeLa cells. Cells were treated with specific siRNA against CD9, CD63, or CD81 for 72 h, and then cells were infected with L. monocytogenes strain BUG 1641 for 1 h. Bacteria were washed, and cells were incubated in the presence of gentamicin for 1 h; afterwards, cells were lysed, and bacteria that survived the gentamicin treatment were distributed on brain heart infusion plates for counting of the CFU 24 h later. Results are expressed as the percentage of the inoculum that survived the gentamicin treatment (the data shown are representative of results from four independent experiments). Analysis for statistical significance was performed using Student's t test.
Distribution of Met and PI4KIIα upon tetraspanin inactivation during Listeria entry.
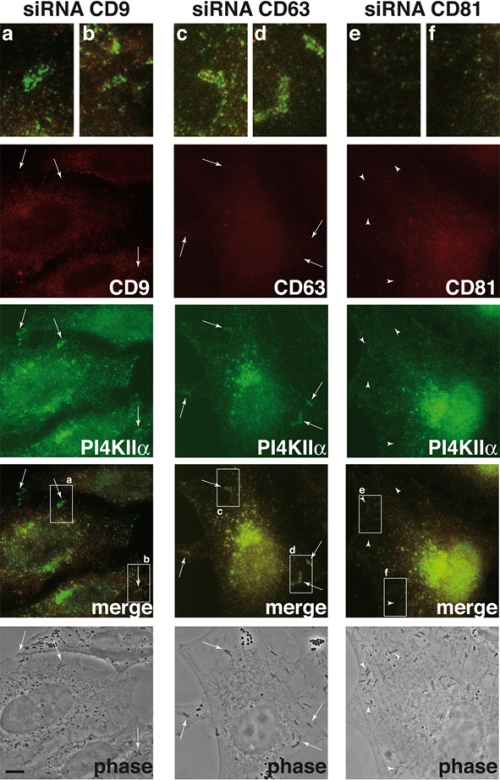
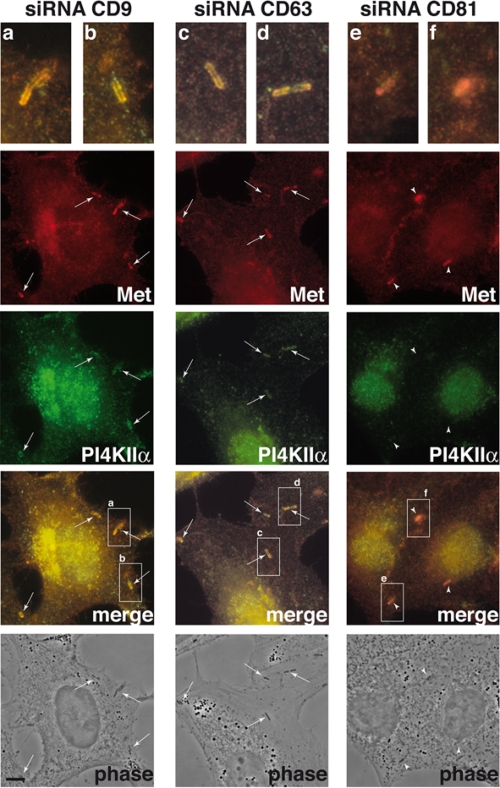
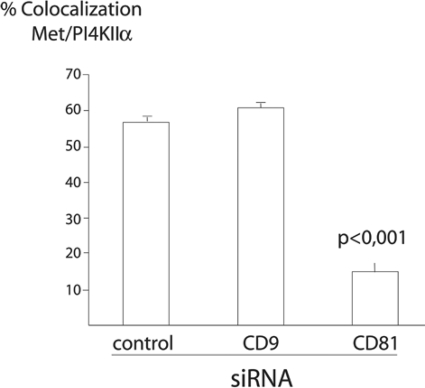
Having shown that depletion of tetraspanin levels by siRNA has different effects on the entry of Listeria, we investigated whether tetraspanin inactivation could also have differential roles in the recruitment of PI4KIIα to the Listeria invasion sites. In Fig. 5, we show that depletion of CD9 or CD63 does not affect the recruitment of PI4KIIα to bacterial entry sites but that CD81 depletion hampers the bacterium-induced recruitment of PI4KIIα. We investigated if Met recruitment was also abolished by depletion of CD81: as shown in Fig. 6, depletion of CD9, CD63, and CD81 does not prevent the colocalization of Met with invading bacteria. We took advantage of this observation to quantify the reduction of PI4KIIα recruitment to Listeria entry sites in cells treated with CD81-specific siRNA: as shown in Fig. 7, for the bacterial population colocalizing with Met, inactivation of CD81 (but not of CD9) significantly reduces the recruitment of PI4KIIα. These results suggest that CD81 may act as a surface membrane organizer upstream of PI4KIIα, coordinating the interaction of different molecules required for Listeria invasion downstream of the InlB receptor Met.
FIG. 5.
Distribution of PI4KIIα in Listeria-infected cells in which CD9, CD63, or CD81 has been inactivated. Cells were treated with specific siRNA against CD9, CD63, or CD81 for 72 h, and then cells were infected with L. monocytogenes strain BUG 1641 for 10 min; bacteria were washed, and cells were fixed, permeabilized with Triton X-100, and treated for immunofluorescence analyses using anti-CD9, anti-CD63, anti-CD81, and anti-PI4KIIα antibodies. Recruitment of PI4KIIα is still detected in cells in which CD9 or CD63 is inactivated but not in cells in which CD81 is inactivated. Arrows indicate bacteria that colocalize with PI4KIIα in cells with CD9 or CD63 inactivated; arrowheads indicate bacteria that do not colocalize with PI4KIIα in cells with CD81 inactivated. Boxes labeled a to f in the merge panels have been enlarged at the top of the figure. Bar, 3 μm.
FIG. 6.
Distribution of Met and PI4KIIα in Listeria-infected cells in which CD9, CD63, or CD81 has been inactivated. Cells were treated with specific siRNA against CD9, CD63, or CD81 for 72 h, and then cells were infected with L. monocytogenes strain BUG 1641 for 10 min; bacteria were washed, and cells were fixed, permeabilized with Triton X-100, and treated for immunofluorescence analyses using anti-Met and anti-PI4KIIα antibodies. Recruitment of Met by Listeria is detected in all cases. As in Fig. 5, bacterial colocalization with PI4KIIα is still detected in cells with CD9 or CD63 inactivated but not in cells with CD81 inactivated. Arrows indicate bacteria that colocalize with Met and PI4KIIα in cells with CD9 or CD63 inactivated; arrowheads indicate bacteria that colocalize with Met but do not colocalize with PI4KIIα in cells with CD81 inactivated. Boxes labeled a to f in the merge panels have been enlarged at the top of the figure. Bar, 3 μm.
FIG. 7.
Inactivation of CD81 reduces PI4KIIα recruitment to invading Listeria bacteria. Cells were treated with specific siRNA against CD9 or CD81 for 72 h, and then cells were infected with L. monocytogenes strain BUG 1641 and processed for immunofluorescence analyses as described in the legend to Fig. 6. Samples were analyzed by fluorescence microscopy, and bacteria which showed colocalization with the receptor Met were scored for the recruitment of PI4KIIα. As shown, inactivation of CD81 reduces the recruitment of PI4KIIα to invading bacteria interacting with Met. Results shown are the averages of results from three different infection experiments, and in each experiment, counts of more than 500 bacteria positive for Met were performed. Analysis for statistical significance was performed using Student's t test.
DISCUSSION
In the present work, we investigated the potential function of tetraspanins as possible molecular adaptors for the recruitment of PI4KIIα to Listeria internalization sites in target cells. To our knowledge, tetraspanins CD9, CD63, and CD81 have been identified among the only molecules which can directly interact with PI4KIIα (26). We detected endogenous CD9, CD63, and CD81 at the surfaces of resting HeLa cells; tetraspanins were also present in an intracellular perinuclear compartment in which we observed partial colocalization with PI4KIIα. Upon the infection of cells with Listeria, we documented for the first time the recruitment of tetraspanins to the bacterial entry site: interestingly, only CD63 was corecruited with the PI4KIIα. Overexpression of tetraspanins and PI4KIIα triggered their corecruitment by invading bacteria, but live-cell imaging demonstrated that tetraspanins and type II PI4Kα do not follow the same reorganization dynamics around Listeria. Moreover, by immunoprecipitation or FRET analyses, we did not detect any interaction between tetraspanins and PI4KIIα. Strikingly, siRNA inactivation of CD81 abolished the recruitment of PI4KIIα and the invasion of HeLa cells by Listeria without inhibiting the recruitment of Met, highlighting CD81 as the first tetraspanin known to play a role during the entry of Listeria into target cells.
Our imaging and biochemical results contrast with previous data suggesting that CD9, CD63, and CD81 were associated in complexes with PI4KIIα (2). The differences may be due to the specificities of the cell lines used, since previous studies were performed with B-cell, T-cell, and erythroleukemic cell lines, as well as fribrosarcoma cell lines (2, 25, 26), and not HeLa cells. In addition, despite the clear recruitment of CD9 to the Listeria entry site, inactivation of CD9 (and also of CD63) does not affect the internalization of Listeria into HeLa cells. It is possible that these tetraspanins are passively recruited or play redundant roles with other molecules including other tetraspanins which remain to be characterized.
The fact that CD81 plays a role in the entry of Listeria into target cells is highly interesting, taking into account that this molecule has been found to be required for the infectivity of hepatitis C virus (HCV) and of Plasmodium species (the agent of malaria) in hepatocytes (6, 23). Many similarities exist in the invasion pathways of these different infectious agents. Hepatocytes are important target cells for the establishment of disease in all three cases (6, 10, 23). Heparan sulfate has been described as serving as the initial docking site for HCV attachment in hepatocytes (1), and it also provides the signal to Plasmodium to stop migrating and productively invade cells (8); this glycosaminoglycan has been shown previously to induce the detachment of InlB from the Listeria cell wall (13) and plays an important role during both Listeria entry and the physiological activation of Met by its natural ligand, the hepatocyte growth factor (14).
Moreover, cholesterol is critical for replication, secretion, and entry of HCV into target cells (27), it contributes to the organization of CD81-enriched microdomains required for the infectivity of Plasmodium (22), and it is critical for the entry of Listeria into target cells by both the InlA and InlB entry pathways (19). It is interesting that cholesterol depletion does not preclude Met recruitment to the Listeria entry site but interferes with signaling downstream of the type I PI3K (21); in the case of CD81 inactivation, we also observed Met recruitment by invading Listeria but recruitment of PI4KIIα was inhibited. It would be important to determine whether for Listeria, as for Plasmodium, cholesterol plays a role in entry, organizing the distribution of CD81 at sites of bacterial entry, and to determine which are the events downstream of Met signaling which are inhibited by CD81 inactivation, precluding the recruitment of PI4KIIα and potentially inhibiting other signaling steps of the cascades already revealed to be required for Listeria entry into target cells.
Supplementary Material
Acknowledgments
We thank T. Balla and G. Griffiths for kindly supplying plasmids. We thank V. Villiers for help in the generation of anti-PI4KIIα antibodies.
This work received financial support from the Pasteur Institute, INSERM, INRA, and ERA-NET Pathogenomics, and ANR grant no. 05-MIME-013-01. P.C. is an international scholar from the Howard Hughes Institute.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 9 November 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Barth, H., E. K. Schnober, F. Zhang, R. J. Linhardt, E. Depla, B. Boson, F. L. Cosset, A. H. Patel, H. E. Blum, and T. F. Baumert. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berditchevski, F., K. F. Tolias, K. Wong, C. L. Carpenter, and M. E. Hemler. 1997. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272:2595-2598. [DOI] [PubMed] [Google Scholar]
- 3.Berditchevski, F., M. M. Zutter, and M. E. Hemler. 1996. Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol. Biol. Cell 7:193-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell. Mol. Life Sci. 58:1189-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, L., F. Nato, B. Payrastre, J. C. Mazie, and P. Cossart. 1999. The 213-amino-acid leucine-rich repeat region of the Listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol. 34:10-23. [DOI] [PubMed] [Google Scholar]
- 6.Brazzoli, M., A. Bianchi, S. Filippini, A. Weiner, Q. Zhu, M. Pizza, and S. Crotta. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charrin, S., S. Manie, C. Thiele, M. Billard, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33:2479-2489. [DOI] [PubMed] [Google Scholar]
- 8.Coppi, A., R. Tewari, J. R. Bishop, B. L. Bennett, R. Lawrence, J. D. Esko, O. Billker, and P. Sinnis. 2007. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe 2:316-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cossart, P., and A. Toledo-Arana. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10:1041-1050. [DOI] [PubMed] [Google Scholar]
- 10.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 11.Hemler, M. E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801-811. [DOI] [PubMed] [Google Scholar]
- 12.Hoppe, A., K. Christensen, and J. A. Swanson. 2002. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 83:3652-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonquieres, R., J. Pizarro-Cerda, and P. Cossart. 2001. Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol. Microbiol. 42:955-965. [DOI] [PubMed] [Google Scholar]
- 14.Kemp, L. E., B. Mulloy, and E. Gherardi. 2006. Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors. Biochem. Soc. Trans. 34:414-417. [DOI] [PubMed] [Google Scholar]
- 15.Pizarro-Cerda, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124:715-727. [DOI] [PubMed] [Google Scholar]
- 16.Pizarro-Cerda, J., M. Lecuit, and P. Cossart. 2001. Measuring and analysing invasion of mammalian cells by bacterial pathogens: the Listeria monocytogenes system. Methods Microbiol. 31:161-177. [Google Scholar]
- 17.Pizarro-Cerda, J., B. Payrastre, Y. J. Wang, E. Veiga, H. L. Yin, and P. Cossart. 2007. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell. Microbiol. 9:2381-2390. [DOI] [PubMed] [Google Scholar]
- 18.Salazar, G., B. Craige, B. H. Wainer, J. Guo, P. De Camilli, and V. Faundez. 2005. Phosphatidylinositol-4-kinase type II alpha is a component of adaptor protein-3-derived vesicles. Mol. Biol. Cell 16:3692-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seveau, S., H. Bierne, S. Giroux, M. C. Prevost, and P. Cossart. 2004. Role of lipid rafts in E-cadherin- and HGF-R/Met-mediated entry of Listeria monocytogenes into host cells. J. Cell Biol. 166:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seveau, S., J. Pizarro-Cerda, and P. Cossart. 2007. Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect. 9:1167-1175. [DOI] [PubMed] [Google Scholar]
- 21.Seveau, S., T. N. Tham, B. Payrastre, A. D. Hoppe, J. A. Swanson, and P. Cossart. 2007. A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell. Microbiol. 9:790-803. [DOI] [PubMed] [Google Scholar]
- 22.Silvie, O., S. Charrin, M. Billard, J. F. Franetich, K. L. Clark, G. J. van Gemert, R. W. Sauerwein, F. Dautry, C. Boucheix, D. Mazier, and E. Rubinstein. 2006. Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 119:1992-2002. [DOI] [PubMed] [Google Scholar]
- 23.Silvie, O., E. Rubinstein, J. F. Franetich, M. Prenant, E. Belnoue, L. Renia, L. Hannoun, W. Eling, S. Levy, C. Boucheix, and D. Mazier. 2003. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 9:93-96. [DOI] [PubMed] [Google Scholar]
- 24.Wang, Y. J., J. Wang, H. Q. Sun, M. Martinez, Y. X. Sun, E. Macia, T. Kirchhausen, J. P. Albanesi, M. G. Roth, and H. L. Yin. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114:299-310. [DOI] [PubMed] [Google Scholar]
- 25.Yauch, R. L., F. Berditchevski, M. B. Harler, J. Reichner, and M. E. Hemler. 1998. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell 9:2751-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yauch, R. L., and M. E. Hemler. 2000. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4-kinase. Biochem. J. 351(Pt. 3):629-637. [PMC free article] [PubMed] [Google Scholar]
- 27.Ye, J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.