Abstract
23S rRNA maturation in Bacillus subtilis is catalyzed by the recently characterized enzyme Mini-RNase-III. Mini-III is dispensable, however, and 23S rRNA is matured by other ribonucleases in strains lacking this enzyme. Here we show that these RNases are the 5′-to-3′ exoribonuclease RNase J1 and the 3′-to-5′ exoribonucleases, principally RNase PH and YhaM.
rRNA operons in bacteria are generally transcribed as 30S RNA precursors that are processed cotranscriptionally by RNase III (13, 16, 18). This yields 16S, 23S, and 5S rRNA precursors that are further matured by other ribonucleases, usually after ribosome assembly. Escherichia coli and Bacillus subtilis, two of the best-studied bacteria, have been shown to use organism-specific enzymes for each of the final steps of 16S, 23S, and 5S rRNA processing (2, 3, 15). While correct maturation of 16S rRNA can occur in E. coli in the absence of prior cleavage by RNase III, 23S rRNA maturation fails, and rRNAs with longer 5′ and 3′ extremities are incorporated into ribosomes (8, 9). In contrast, 23S rRNA maturation still occurs at the correct site and almost as efficiently in Bacillus subtilis strains lacking RNase III (7, 15). The final 23S rRNA maturation reaction in B. subtilis is catalyzed by Mini-III, which cleaves both sides of a double-stranded RNA processing stalk to produce mature 23S rRNA in one step (15). However, Mini-III is not essential, and a backup pathway of 23S rRNA maturation functions in the absence of Mini-III.
We have previously shown that the extremities of 23S rRNA in a Mini-III mutant (ΔmrnC) map close to the mature RNA ends found in wild-type cells. However, these extremities are highly heterogeneous; some are a few nucleotides (nt) shorter or longer at the 5′ end, and all are a few nucleotides longer at the 3′ end (15). The heterogeneity of the alternative 23S rRNA ends in the mrnC mutant suggests that they are matured by exoribonucleases. The most obvious candidate for the 5′ processing reaction is RNase J1, known to possess both endonucleolytic and 5′-to-3′ exonucleolytic activity (5, 12). To determine whether RNase J1 participates in 23S rRNA maturation in the absence of Mini-III, we first combined the Mini-III and RNase J1 mutations in a single strain. RNase J1 is essential in B. subtilis, so we used a strain (CCB084) in which the RNase J1 gene (rnjA) was placed under the control of a xylose-inducible promoter (Pxyl) (Table 1) (2). Competent CCB084 cells were transformed with chromosomal DNA isolated from strain SSB1044 (ΔmrnC) as described previously (15). The resulting double mutant (CCB166) was grown overnight in 2× yeast-tryptone (YT) medium supplemented with 2% xylose. The culture was pelleted and washed three times with 2× YT medium and inoculated into fresh medium at an optical density at 600 nm (OD600) of 0.02. Parallel cultures were grown in the presence of 2% xylose to induce rnjA expression or in 2% glucose to repress the xylose promoter. Cultures were harvested at an OD600 of 0.5, and total RNA was isolated as described previously (1).
TABLE 1.
B. subtilis mutant strains
| Strain | Genotypea | Reference |
|---|---|---|
| SSB1044 | mrnC::pMUTIN2 (mrnC::ery) | 15 |
| SSB1030 | pnp::cm | 17 |
| BG457 | rph::spc | 14 |
| BG393b | rph::cm rnr::spc | 17 |
| BG395b | rph::cm yhaM::pm | 17 |
| BG396 | rph::cm rnr::spc yhaM::pm | 1 |
| BG508c | pnp::kan rph::spc rnr::tc | 1 |
| CCB084 | amyE::pCT1[Pxyl-rnjA] Cm rnjA::spc | 2 |
| CCB166 | mrnC::ery amyE::pCT1[Pxyl-rnjA] Cm rnjA::spc | This work |
| CCB167 | mrnC::ery pnp::cm | This work |
| CCB168 | mrnC::ery rnr::spc | This work |
| CCB169 | mrnC::ery rph::spc | This work |
| CCB170 | mrnC::ery yhaM::pm | This work |
| CCB225 | mrnC::ery yhaM::pm rph::cm | This work |
| CCB171 | mrnC::ery rnr::spc rph::cm | This work |
| CCB172 | mrnC::ery rnr::spc rph::cm yhaM::pm | This work |
| CCB173 | mrnC::ery rnr::spc rph::cm yhaM::pm pnp::kan | This work |
Resistance markers are as follows: ery, erythromycin; cm, chloramphenicol; kan, kanamycin; pm, phleomycin; and spc, spectinomycin.
SSB1044 transformants with these DNAs were screened for Cms to obtain CCB168 and CCB170 double mutants only.
CCB172 transformants with DNA from strain BG508 were selected for Eryr, Pmr, Kanr, Cmr, Spcr, and Tcs.
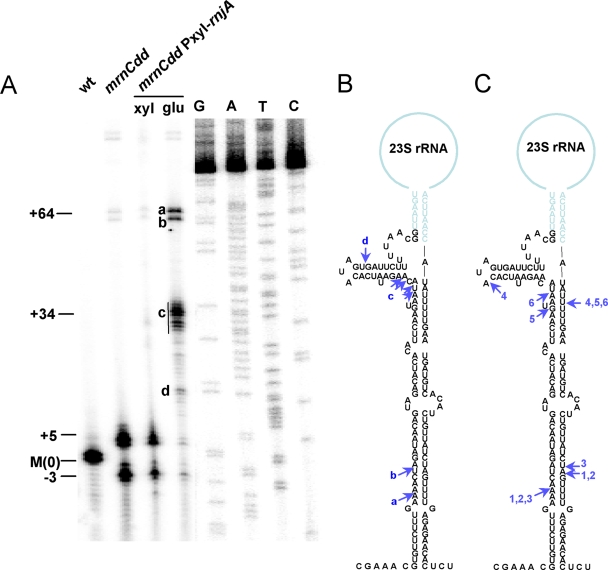
To determine whether RNase J1 played a role in the maturation of 23S rRNA in the absence of Mini-III, we performed primer extension analysis on total RNA in the conditional mutant strain, using a 32P-labeled primer (CC257) specific for 23S rRNA (15) and avian myeloblastosis virus (AMV) reverse transcriptase (Finnzymes). As shown in Fig. 1, depletion of RNase J1 in cells lacking Mini-III resulted in 23S rRNAs with significantly longer 5′ ends than in the Mini-III mutant alone. Clusters of cDNA products were observed with 5′ ends at positions +29 to +34 and at +64, the site of RNase III cleavage identified previously (15). A ladder of bands extended from the +34 position down to the −3 and +5 positions. We interpret this as evidence of 5′-to-3′ exonucleolytic action by RNase J1 (residual amounts are still present in isopropyl-β-d-thiogalactopyranoside [IPTG]-dependent constructs of RNase J1 [4], and we presume the same is true for the xylose-dependent construct used here). The product at the +34 position (band c) could be either the result of cleavage by an unknown endoribonuclease or a (residual) RNase J1 entry point in endonucleolytic mode. We also analyzed 23S rRNA extremities in RNase J1-depleted cells by circular reverse transcription-PCR (cRT-PCR) as described previously (15). The sequencing of cloned cRT-PCR products from the ΔmrnC strain depleted for RNase J1 confirmed the location of the 5′ ends mapped by primer extension and showed that the backup 3′ maturation pathway is not dependent on 5′ processing (Fig. 1C).
FIG. 1.
Maturation of the 5′ end of 23S rRNA in the absence of Mini-III. (A) Primer extension of total RNA with 32P-labeled oligonucleotide CC257. Strain CCB166 (mrnC Pxyl-rnjA) was grown in the presence of xylose (xyl) or glucose (glu). Bands characteristic of the mrnC mutant are indicated by −3, +5, and + 64, representing the number of nucleotides from the mature 5′ end. 5′ ends found in the mrnC Pxyl-rnjA strain grown in glucose are indicated (a to d). M(0), mature 23S rRNA 5′ end in wild-type cells. (B) Secondary structure of the 23S rRNA processing stalk showing 5′ ends (blue arrows) mapped by primer extension in the mrnC Pxyl-rnjA strain grown in the presence of glucose. (C) Secondary structure of the 23S rRNA processing stalk showing 5′ ends (blue arrows) mapped by sequencing of cRT-PCR products in the mrnC Pxyl-rnjA strain grown in the presence of glucose. Sequenced clones are labeled 1 to 6.
The implication of RNase J1 in the maturation of the 5′ end of 16S rRNA in B. subtilis (2) and in the secondary 5′-end processing pathway of 23S rRNA is very reminiscent of the pathways recently described in the nitrogen-fixing symbiotic alphaproteobacterium Sinorhizobium meliloti (10). Although this bacterium possesses both RNase J1 and RNase E, it uses RNase J1 for 5′-end maturation of both 16S and 23S rRNA; Mini-III is not encoded by the S. meliloti genome. We suspect, therefore, that the secondary 23S rRNA maturation pathway catalyzed by RNase J1 in B. subtilis is a vestige of an ancient 23S rRNA processing mechanism and that B. subtilis subsequently acquired Mini-III to more efficiently and rapidly mature both ends of 23S rRNA in a single step. We were unable to find culture conditions in which the RNase J1 pathway became the dominant pathway for 23S maturation, which would be manifested by 23S rRNA with alternate 5′ ends. Attempts to find conditions in which Mini-III might become limiting, such as a nutritional shift up from minimal to rich medium, a lag phase to exponential phase transition mimicking a shift out of starvation conditions, and a temperature shift up from 30° to 45°C, all of which are known to abruptly increase rRNA synthesis, all yielded 23S rRNA with 5′ ends characteristic of Mini-III cleavage (data not shown). Mini-III-derived extremities were also observed during growth in high concentrations of cysteine (mrnC is part of the T-box-regulated cysES operon that is likely turned down in the presence of cysteine [6]). Thus, if there is a selective pressure on B. subtilis to maintain the secondary pathway for 23S rRNA maturation, we were unable to identify it.
To identify the enzymes responsible for 3′-end maturation of 23S rRNA in the absence of Mini-III, we combined the mrnC mutation with mutations in genes encoding known 3′-to-5′ exoribonucleases. B. subtilis strain SSB1044 (ΔmrnC) competent cells were prepared and transformed with chromosomal DNA from the exoribonuclease mutants SSB1030, BG457, BG393, and BG395 (Table 1) to obtain mrnC double mutants with PNPase (pnp), RNase R (rnr), RNase PH (rph), and YhaM (yhaM) (yielding strains CCB167 to CCB170, respectively). Strains CCB225 (mrnC rph yhaM), CCB171 (mrnC rnr rph), and CCB172 (mrnC rnr rph yhaM) were obtained by transformation of BG395 (rph yhaM), BG393 (rnr rph), and BG396 (rnr rph yhaM) competent cells, respectively, with mrnC (SSB1044) chromosomal DNA. Strain CCB173 (mrnC rnr rph yhaM pnp) was created by transformation of CCB172 competent cells with BG508 chromosomal DNA and selection with the relevant antibiotics. The resulting strains were grown in 2× YT medium to an OD600 of 0.5, and total RNA was isolated.
We analyzed the 23S rRNA 5′ and 3′ extremities in the different multiple mutants by cRT-PCR, where the final PCR was performed with primers CC256 (15) and CC257, which was labeled with 6-carboxyfluorescein at the 5′ end to obtain fluorescently labeled products. The products were run on a 5% acrylamide-7 M urea gel and analyzed by Typhoon (GE Healthcare). Fluorescent DNA sequencing ladders were also generated to estimate the size of cRT-PCR products, using the same 6-carboxyfluorescein-labeled oligonucleotide. As expected, depletion of RNase J1 in the absence of Mini-III led to an accumulation of longer cRT-PCR products (Fig. 2A, compare lanes 3 and 4), confirming the results shown in Fig. 1. The ΔmrnC phenotype (Fig. 2A, lane 2) of two major cRT-PCR products, one shorter and one longer than the wild-type product (Fig. 2A, lane 1), was not altered in Mini-III mutants combined with PNPase, RNase R, or YhaM (Fig. 2A, compare lanes 5, 6, and 8, respectively, to lane 2). However, the mrnC rph double mutant yielded cRT-PCR products that were a few nucleotides longer in zone I than the mrnC mutant alone (Fig. 2A, compare lane 7 to lane 2, black arrowhead). This pattern was also seen in the triple mutant mrnC rnr rph (Fig. 2A, lane 9). These results suggest that RNase PH is involved in removing the last few nucleotides from the 3′ end of 23S rRNA in the absence of Mini-III. A longer precursor was evident in zone II in the triple mutant mrnC rph yhaM, suggesting that YhaM is also involved in this pathway (Fig. 2A, lane 10, white arrowhead). Interestingly, this precursor is a few nucleotides shorter in the quadruple and quintuple mutants, mrnC rnr rph yhaM and mrnC rnr rph yhaM pnp, than in the mrnC rph yhaM strain (Fig. 2A, compare lanes 11 and 12 to lane 10), suggesting that the presence of RNase R is slightly inhibitory to YhaM processing. Similar inhibitory effects of exoribonucleases on each other have been seen before, most notably the removal of poly(A) tails from the E. coli rpsO mRNA by RNase II that inhibits degradation by PNPase (11). Since we did not see an accumulation of cRT-PCR products corresponding to cleavage at the RNase III site (zone III) in the absence of all known 3′-to-5′ exonucleases, we assume that at least one other enzyme of this pathway remains at large.
FIG. 2.
Maturation of 3′ end of 23S rRNA in the absence of Mini-III. (A) Sequencing gel (5% acrylamide-7 M urea) showing fluorescently labeled cRT-PCR products. Major groups of bands for mrnC single and multiple 3′-to-5′ exoribonuclease mutants are indicated as zones I, II, and III. The RNase PH-dependent precursor is indicated by solid arrowheads, and the precursor observed upon further removal of YhaM is indicated by white arrowheads. (B) 23S rRNA processing stalk showing 5′ and 3′ ends of cloned cRT-PCR products excised from zones I and II of the mrnC rph mutant (blue arrows) and of the mrnC mutant (green arrows) (15). (C) Same as described for panel B but for the mrnC rnr rph yhaM mutant (blue arrows).
We mapped the 23S rRNA extremities in strains CCB167 (mrnC rph) (Fig. 2B), CCB172 (mrnC rnr rph yhaM) (Fig. 2C), and CCB173 (mrnC rnr rph yhaM pnp) (not shown) to compare them to the mrnC single mutant (Fig. 2B and C, green arrows) (15) and to confirm that the defects observed were in 3′ processing. cRT-PCR products corresponding to zones I and II were eluted from gels, cloned into pBluescript as described previously (15), and sequenced. The 3′ end of one of the cloned cRT-PCR products excised from zone II (of three clones sequenced) was 4 to 5 nt longer in the mrnC rph mutant than in the mrnC strain, confirming that the rph defect is at the 3′ end (Fig. 2B). We did not obtain any clone (three clones sequenced) from zone I with a longer 3′ end, presumably because of the relatively low abundance of this cRT-PCR product. The 3′ ends of the cloned cRT-PCR products from the quadruple mutant CCB172 were 4 to 7 nt (zone I) or 7 to 10 nt (zone II) longer than those observed in the mrnC background (Fig. 2C). This is strong evidence for the implication of RNase PH and YhaM in the secondary 23S rRNA 3′-trimming pathway. The cloned cRT-PCR products from the quintuple mutant CCB173 were similar to those identified in CCB172 (not shown), as was expected from data shown in Fig. 2A. Zone III corresponds to the site of RNase III cleavage (+64) and was the same for all three mutants (data not shown).
Our results suggest that RNase PH and YhaM play the most important roles in the secondary pathway of 23S rRNA 3′-end maturation. These enzymes remove nucleotides close to the mature 3′ end of 23S rRNA in the absence of Mini-III. While YhaM can be functionally replaced by RNase PH, the reverse is not true; the two enzymes are not, therefore, fully redundant. RNase PH has also been found to play a major role in removing the last few nucleotides of the 3′ extensions of one class of tRNAs in B. subtilis (those with an encoded CCA motif), while other 3′-to-5′ exonucleases play overlapping roles in this reaction (17). Like the tRNA 3′ maturation pathway, it is clear that at least one more enzyme is required to complete the puzzle. We are currently trying to identify this enzyme(s).
Acknowledgments
We thank members of the lab for helpful discussions and D. Bechhofer for exonuclease mutants.
This work was supported by funds from the CNRS (UPR 9073), Université Paris VII—Denis Diderot, and the Agence Nationale de la Recherche (ANR).
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Bechhofer, D. H., I. A. Oussenko, G. Deikus, S. Yao, N. Mathy, and C. Condon. 2008. Analysis of mRNA decay in Bacillus subtilis. Methods Enzymol. 447:259-276. [DOI] [PubMed] [Google Scholar]
- 2.Britton, R. A., T. Wen, L. Schaefer, O. Pellegrini, W. C. Uicker, N. Mathy, C. Tobin, R. Daou, J. Szyk, and C. Condon. 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 63:127-138. [DOI] [PubMed] [Google Scholar]
- 3.Condon, C., D. Brechemier-Baey, B. Beltchev, M. Grunberg-Manago, and H. Putzer. 2001. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function. RNA 7:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daou-Chabo, R., N. Mathy, L. Benard, and C. Condon. 2009. Ribosomes initiating translation of the hbs mRNA protect it from 5′-to-3′ exoribonucleolytic degradation by RNase J1. Mol. Microbiol. 71:1538-1550. [DOI] [PubMed] [Google Scholar]
- 5.Even, S., O. Pellegrini, L. Zig, V. Labas, J. Vinh, D. Brechemmier-Baey, and H. Putzer. 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33:2141-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagnon, Y., R. Breton, H. Putzer, M. Pelchat, M. Grunberg-Manago, and J. Lapointe. 1994. Clustering and co-transcription of the Bacillus subtilis genes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J. Biol. Chem. 269:7473-7482. [PubMed] [Google Scholar]
- 7.Herskovitz, M. A., and D. H. Bechhofer. 2000. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 38:1027-1033. [DOI] [PubMed] [Google Scholar]
- 8.King, T. C., and D. Schlessinger. 1983. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J. Biol. Chem. 258:12034-12042. [PubMed] [Google Scholar]
- 9.King, T. C., R. Sirdeshmukh, and D. Schlessinger. 1984. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc. Natl. Acad. Sci. USA 81:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madhugiri, R., and E. Evguenieva-Hackenberg. 2009. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett. 583:2339-2342. [DOI] [PubMed] [Google Scholar]
- 11.Marujo, P. E., E. Hajnsdorf, J. Le Derout, R. Andrade, C. M. Arraiano, and P. Regnier. 2000. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA 6:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathy, N., L. Benard, O. Pellegrini, R. Daou, T. Wen, and C. Condon. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681-692. [DOI] [PubMed] [Google Scholar]
- 13.Nikolaev, N., L. Silengo, and D. Schlessinger. 1973. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 70:3361-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oussenko, I. A., T. Abe, H. Ujiie, A. Muto, and D. H. Bechhofer. 2005. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 187:2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redko, Y., D. H. Bechhofer, and C. Condon. 2008. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 68:1096-1106. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 17.Wen, T., I. A. Oussenko, O. Pellegrini, D. H. Bechhofer, and C. Condon. 2005. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis. Nucleic Acids Res. 33:3636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zingales, B., and W. Colli. 1977. Ribosomal RNA genes in Bacillus subtilis. Evidence for a cotranscription mechanism. Biochim. Biophys. Acta 474:562-577. [DOI] [PubMed] [Google Scholar]