Abstract
More than 80 small regulatory RNAs (sRNAs) and 60 proteins of 16 to 50 amino acids (small proteins) are encoded in the Escherichia coli genome. The vast majority of the corresponding genes have no known function. We screened 125 DNA bar-coded mutants to identify novel cell envelope stress and acute acid shock phenotypes associated with deletions of genes coding for sRNAs and small proteins. Nine deletion mutants (ssrA, micA, ybaM, ryeF, yqcG, sroH, ybhT, yobF, and glmY) were sensitive to cell envelope stress and two were resistant (rybB and blr). Deletion mutants of genes coding for four small proteins (yqgB, mgrB, yobF, and yceO) were sensitive to acute acid stress. We confirmed each of these phenotypes in one-on-one competition assays against otherwise-wild-type lacZ mutant cells. A more detailed investigation of the SsrA phenotype suggests that ribosome release is critical for resistance to cell envelope stress. The bar-coded deletion collection we generated can be screened for sensitivity or resistance to virtually any stress condition.
Small regulatory RNAs (sRNAs) play critical regulatory roles in all domains of life. Numerous approaches have been taken to discover sRNA-encoding genes in bacteria (reviewed in references 1 and 26), including bioinformatic searches for conservation as well as promoter and Rho-independent terminator sequences in intergenic regions. sRNAs have also been detected directly by sequencing or microarray analysis, often after size selection or coimmunoprecipitation with RNA-binding proteins. Approximately 80 sRNAs have been identified in Escherichia coli. A few sRNAs bind proteins to effect a cellular response, but the vast majority of sRNAs characterized to date act by base pairing with mRNAs (reviewed in reference 52). sRNA base pairing with an mRNA can bring about any of a number of outcomes, including exposing or occluding a ribosome-binding site, increasing or decreasing mRNA stability, or terminating transcription.
Those sRNAs whose functions have been delineated regulate a wide array of physiological responses (reviewed in reference 52). For example, in E. coli, sRNAs are induced to promote translation of a stationary-phase-specific σ factor, to downregulate σ70-RNA polymerase activity at certain promoters in stationary phase, and to induce and repress genes in response to iron availability (52). In Vibrio species, sRNAs act to integrate quorum-sensing signals (52). Many Gram-negative bacteria also employ sRNAs to regulate the composition of outer membrane proteins (OMPs) within their cell envelopes (reviewed in references 16 and 50).
In work growing out of our screens for sRNAs, we have also initiated searches for unannotated genes encoding proteins between 16 and 50 amino acids in length (18). Approximately 60 genes have been shown to encode small proteins in E. coli (18). Very little is known about what the vast majority of small proteins do. However, the few whose functions have been elucidated act in a number of roles: as intercellular signals to regulate the onset of genetic competence in Gram-positive bacteria (7); as intracellular toxins (12) and antibiotics (22) in various bacteria; as kinase inhibitors in Bacillus subtilis (39).
sRNAs and small proteins of known function play diverse cellular roles, so how can those of unknown function be analyzed most efficiently? One approach is to uncover phenotypes associated with deletions of sRNA- and small protein-coding genes. The existence of a deletion phenotype indicates that an sRNA or small protein performs a biologically relevant function that is amenable to study in the laboratory. Aside from demonstrating the physiological relevance of the gene, the discovery of a deletion phenotype greatly facilitates the study of the corresponding sRNA or small protein by further genetic analysis. Biochemical and cytological approaches also are aided by knowledge of whether tagged or mutant derivatives complement a null phenotype.
Thus far, very little has been done to systematically associate deletion phenotypes with genes coding for bacterial sRNAs or small proteins. However, a number of studies have been undertaken to identify phenotypes tied to the absence of other genes in E. coli. Many of these investigations have made use of the Keio collection, a set of approximately 3,900 deletions of nonessential genes in E. coli which contains relatively few deletions of sRNA- and small protein-coding genes (2). This collection has been screened for mutants deficient in biofilm formation (31) and in resistance to various antibiotics (44). Two groups have also exploited bacterial conjugation to identify synthetically lethal interactions in a high-throughput manner (5, 47). Others have employed customized microarrays to analyze the Keio collection in batch competition experiments (42). In this approach (known as monitoring of gene knockouts [MGK]), every strain in the collection is mixed and subjected to mock and stress treatments. Individual strains are subsequently enumerated by quantifying DNAs amplified from the regions flanking every antibiotic resistance cassette on a custom microarray.
The yeast community has created a knockout collection of approximately 5,900 yeast genes (13). However, unlike the Keio collection, every strain in the yeast deletion collection contains two unique 20-mer DNA bar codes (13, 34). These bar codes enable the execution of parallel screens for deletion phenotypes in large-scale competition experiments using standardized microarrays. We have chosen this methodology to create a series of 125 DNA bar-coded deletion mutants in E. coli (Fig. 1). We employed this collection to identify deletion mutants of genes coding for sRNAs and small proteins that are sensitive or resistant to cell envelope stress or to acute acid stress, two conditions E. coli encounters during its life cycle as a pathogen or symbiont in higher eukaryotes (i.e., acid stress in the stomach and cell envelope stress in the intestine).
FIG. 1.
Diagram of bar-coded antibiotic resistance cassettes. Kanamycin resistance cassettes flanked by two unique 20-mer DNA bar code sequences (UP and DN) were generated by a two-step PCR process for each deleted gene. The bar-coded kanamycin resistance cassettes were incorporated at loci coding for sRNAs and small proteins by homologous recombination. For the analysis of the large-scale competition experiments, bar codes upstream and downstream of every kanamycin resistance cassette were amplified by means of common primer sequences (indicated by small black arrows) encoded within the regions bordering the UP and DN bar codes. The amplified bar codes were then hybridized to a DNA microarray to score each bar-coded deletion mutant within the population.
MATERIALS AND METHODS
Media and media supplements.
Luria-Bertani broth (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per liter) was prepared from a premixed stock (lot A08-23; Invitrogen). M63 minimal medium [15.2 mM (NH4)2SO4, 22.1 mM KH2PO4, 40.3 mM K2HPO4, 1 mM MgSO4, 3.30 μM FeSO4] was supplemented with 5% (wt/vol) sucrose, 0.2% (wt/vol) glycerol, 5 mg/liter vitamin B1, and 1 mg/liter biotin. When necessary, antibiotics were used at the following concentrations: kanamycin, 30 μg/ml; chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; carbenicillin, 100 μg/ml; tetracycline, 12.5 μg/ml. Isopropyl β-d-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) were used at final concentrations of 1 mM and 100 μg/ml, respectively.
Strains and oligonucleotides.
All strains are derivatives of the laboratory stock of E. coli K-12 MG1655. The strains and oligonucleotides used in the study are listed in Tables S1 and S2, respectively, of the supplemental material. Platinum Taq DNA Polymerase High Fidelity (Invitrogen) was employed in all PCRs.
Generation of bar-coded kanamycin resistance cassettes.
Bar-coded kanamycin resistance cassettes were generated by a two-step PCR process. First, P1 and P2 primers were used to amplify the kanamycin resistance cassette from pKD13 (8). The P1 and P2 primers contained common priming sequences, unique 20-mer “UP” and “DN” DNA bar codes specific to each locus being deleted and regions complementary to the kanamycin resistance cassette. P3 and P4 primers containing DNA sequences homologous to the regions flanking the locus to be deleted as well as DNA sequences complementary to the 5′ ends of the first PCR product were used to amplify the gel-purified first-round reaction products in a second-round PCR. The reaction products from the second-round PCR were incorporated into the chromosome by mini-λ-Red-mediated recombination (8, 56). The bar-coded kanamycin cassettes were moved to a fresh genetic background (wild-type E. coli K-12 MG1655 cells) by P1 transduction (45) and sequenced. For further characterization of the mutant strains, the kanamycin antibiotic resistance cassettes were excised from the chromosome by Flp-mediated recombination (6). In general, genes encoding sRNAs with mapped 5′ and 3′ ends and open reading frames (inclusive of the stop codon) encoding small proteins were deleted in their entirety. When possible, care was taken to avoid deleting flanking genes or their regulatory elements; however, intergenic regions containing sRNAs with unmapped 5′ and 3′ ends were deleted completely.
Generation of complementation constructs.
The counterselectable cat-sac cassette (24) was PCR amplified using primers ECH938 and ECH495, which also carried sequences homologous to regions upstream (123 to 177 bp before the start codon) and downstream (1 to 55 bp after the stop codon) of lacZ on the E. coli chromosome. The purified PCR product was used in conjunction with the mini-λ-Red system (8, 56) to replace lacZ with the cat-sac cassette and create GSO291.
Complementation constructs were generated by PCR amplifying the appropriate loci with primers that contained the lacZ-flanking regions described above. ssrA alleles were similarly integrated at lacZ after being amplified from pJW28 (ssrA+) (37) and pJW29 (ssrAO) (38) using primers ECH1218 and ECH1227. The mini-λ-Red system was employed to replace the cat-sac cassette with each complementation construct. A control lacZ deletion strain was created by using the primers ECH1012 and ECH1013 to PCR amplify two complementary oligonucleotides (ECH1007 and its complement) in which the upstream and downstream lacZ-flanking regions had been fused together. The resulting PCR product was used to replace the cat-sac cassette. Transformants were grown on M63 minimal medium supplemented with 5% sucrose to select for cells that had lost the cat-sac cassette. All complementation constructs were confirmed by sequencing.
Screening bar-coded deletion collection for novel phenotypes. (i) Cell growth.
Cells from each bar-coded deletion strain were inoculated separately into 50-ml conical tubes containing 5 ml of LB broth and grown for 16 h at 37°C with shaking (250 rpm).
For the cell envelope stress screen, the overnight cultures (optical density at 600 nm [OD600], ∼5.5) were pooled and used to inoculate 30 ml of prewarmed (37°C) LB broth at a dilution of 1:2,000. The culture was split into two 15-ml subcultures. Cell envelope stress was imposed in one subculture by adding SDS (final concentration, 0.025% [wt/vol]) and EDTA (pH 8.0; final concentration, 1 mM). Both 15-ml subcultures were incubated in a shaking (250 rpm) water bath at 37°C. A 1-ml aliquot of cells was harvested from the mock-treated subculture when the OD600 was between 0.280 and 0.400. A 1-ml aliquot of cells was collected from the cell envelope stress culture when the OD600 was between 0.280 and 0.400 and within 0.05 OD units of the OD600 achieved with the mock-treated cells at the time of their harvest.
For the acid shock screen, the overnight cultures were pooled and the OD600 of this pooled culture was determined (OD600, ∼5.5). Two 1-ml aliquots of the mixed culture were placed into 1.5-ml Eppendorf tubes. One aliquot was acidified to pH 1.8 with an aqueous solution of 37% (wt/vol) HCl. Both the mock-treated and acid-treated cells were incubated in a tabletop heating block at 37°C with shaking (1,400 rpm) for 10 min. Cells were subsequently washed three times with 1 ml of 1× phosphate-buffered saline (PBS) (pH 7.4). The mock-treated and acid-treated cells were inoculated (1:5,000) into separate 250-ml flasks each containing 30 ml of prewarmed (37°C) LB broth. Both cultures were incubated in a water bath (37°C, 250 rpm) until they achieved an OD600 within 0.2 OD units of the original mixed culture, at which point in time a 1-ml aliquot of cells was harvested.
(ii) Hybridization and scanning of microarrays.
UP and DN bar codes from each sample were quantified on the Genflex Tag 16K Array v2 (Affymetrix) (35). The methods summarized here have been described in greater detail by Pierce and colleagues (35). Recipes for making 12× morpholineethanesulfonic acid stock solution, 2× hybridization buffer, hybridization mix, wash A solution, wash B solution, and biotin staining solution as well as a step-by-step protocol for hybridizing DNA bar codes to the microarray can also be found in the supplemental material.
First, genomic DNA was prepared from each sample by using a Wizard genomic DNA purification kit (Promega). UP bar codes were PCR amplified using primers ECH361 and ECH427. DN bar codes were PCR amplified using primers ECH362 and ECH428. ECH427 and ECH428 were biotinylated at their 5′ ends. Approximately 0.2 μg of genomic DNA was used as a template in each reaction mixture.
Second, each microarray was filled with 140 μl of 1× hybridization buffer and incubated (42°C, 20 rpm) in an Affymetrix GeneArray hybridization oven for 10 min. The 1× hybridization buffer was subsequently removed from each microarray and replaced with a solution (previously boiled for 2 min and incubated on ice for 2 min) consisting of 30 μl each of the UP and DN bar code PCR products combined with 90 μl of hybridization mix. The arrays were then rotated at 20 rpm in the hybridization oven for 10 to 16 h at 42°C.
The following day, the hybridization mix was removed, and each microarray was washed twice with wash A solution (room temperature), six times with wash B solution (42°C), and once with wash A solution (room temperature). Then, the wash A solution was aspirated and replaced with biotin staining mix. Each microarray was rotated at 20 rpm for 10 min at 42°C. The arrays were washed six times with wash solution A (room temperature). The arrays were then filled with wash A (room temperature) and scanned at an emission wavelength of 560 nm with an Affymetrix GeneArray scanner.
(iii) Analysis of array data.
UP bar codes were analyzed separately from DN bar codes. GeneChip operating software (Affymetrix) was used to extract the arbitrary fluorescence values associated with each probe. Every bar code was queried by five probes on the microarray. The arbitrary fluorescence intensities associated with the individual probes in these quintets were averaged to yield a mean fluorescence intensity for each bar code. The background fluorescence intensity was determined by averaging the fluorescence intensities of probes associated with bar codes that were not present in any strain. The background fluorescence intensity was subtracted from the mean fluorescence intensity of each bar code. Bar codes with a background-corrected mean intensity of less than 200 arbitrary fluorescence units in the mock treatment sample were excluded from further analysis. One caveat to this approach is that the signal intensity observed for a bar code on the array does not scale in a linear manner with the actual concentration of the bar code in solution (34, 35). As a consequence, the difference in bar code concentrations between two samples tends to be underestimated in the final array analysis. As previously described, the remaining mean bar code fluorescence intensities were multiplied by a correction factor [e(0.00031 × mean bar code intensity)] to account for this effect (34, 35). The resulting corrected fluorescence intensity associated with each bar code in stress-treated cells was divided by its fluorescence intensity in mock-treated cells to obtain a relative abundance (RA) value. Both experiments were performed in triplicate, giving rise to three unique sets of UP and DN bar code RA values for each stress condition.
One-on-one competition assays.
Cells from strains to be tested were inoculated separately into 50-ml conical tubes containing 5 ml of LB broth and grown for 16 h at 37°C with shaking (250 rpm). An aliquot of the overnight cultures of each deletion mutant was mixed with an equal amount of the overnight culture of the ΔlacZ (NM601) cells.
For the cell envelope stress assays, each of the mixed cultures was used to inoculate 30 ml of prewarmed (37°C) LB broth at a dilution of 1:2,000. The 30-ml culture was split into two 15-ml subcultures. Cell envelope stress was imposed in one subculture by adding SDS (final concentration, 0.025% [wt/vol]) and EDTA (pH 8.0; final concentration, 1 mM). Both 15-ml subcultures were incubated in a shaking (250 rpm) water bath at 37°C. For the mock-treated subculture, cells were harvested when the OD600 was between 0.280 and 0.400. For the cell envelope stress subculture, cells were collected when the OD600 was between 0.280 and 0.400 and within 0.05 OD units of the OD600 achieved by the mock-treated cells at their time of harvest.
For the acid shock assays, two 1-ml aliquots of the mixed culture were placed into 1.5-ml Eppendorf tubes. One aliquot was acidified to pH 1.8 with an aqueous solution of 37% (wt/vol) HCl. Both the mock-treated and acid-treated cells were incubated in a tabletop heating block at 37°C with shaking (1,400 rpm) for 10 min. Cells were subsequently washed three times with 1 ml of 1× PBS (pH 7.4).
Aliquots of mock- and stress-treated subcultures were diluted appropriately and spread on LB agar plates containing IPTG and X-Gal. After overnight incubation at 37°C, the numbers of blue and white colonies arising from the mock treatment and the stress treatment were scored to obtain a competitive index (CI).
RESULTS
Generating bar-coded deletion strains.
The phenotyping of individual bacterial strains under numerous stress and growth conditions is time- and labor-intensive. The effort involved is compounded when hundreds or thousands of strains need to be screened simultaneously. An alternative methodology is to perform batch competition experiments in which all strains are mixed together and subjected to selective pressure. In this approach, conditions can be manipulated to select for extremely resistant strains. However, it is difficult to identify sensitive or moderately resistant mutants without a means to enumerate the number of cells corresponding to each strain within the population.
We incorporated unique 20-mer DNA sequences (bar codes) into a collection of 125 directed deletion mutants. These bar codes can be used with microarray analysis to allow the quantification of individual strains within large-scale competition experiments. At the time we performed the large-scale competition experiments described below, this collection contained 122 strains. A total of 47 of these strains were single deletion mutants of genes encoding sRNAs, and 50 strains were deletion mutants of genes encoding small proteins of 50 amino acids or less. Three additional strains were also created that were deleted for the repetitive sib and ldr loci (ΔsibABCDE, ΔldrABC, and ΔldrABCD). Thirteen strains were deleted for genes encoding proteins between 50 and 70 amino acids in length, and eight control strains were deleted for genes known to be required for survival under various stress conditions (e.g., smpA, gadE, trpA, uspA, uspB, uspD, uspE, and oxyR). One final strain was deleted for dppA, a target of the GcvB sRNA (49).
Homologous recombination was employed to replace the genes listed above with antibiotic resistance cassettes flanked by two unique bar codes (Fig. 1). Common priming sites were also incorporated upstream and downstream of each bar-coded antibiotic resistance cassette. Hence, all of the UP and DN bar codes in a population of cells could be amplified in two separate PCRs by using the UP and DN common primers in conjunction with primers designed to anneal to the antibiotic resistance cassette. One caveat of this strategy is that any phenotypes we uncovered could arise as a consequence of polarity effects imposed by the antibiotic resistance cassettes on downstream genes. To minimize this potential problem, the antibiotic resistance cassettes were excised from strains that were subjected to further analyses. The bar code and common primer sequences left behind after excision of the antibiotic resistance cassette were designed to limit cross-hybridization and should not give rise to any significant secondary structures that would affect downstream gene expression.
Strains sensitive to cell envelope stress.
Two pieces of information led us to hypothesize that it would be fruitful to screen our deletion collection for cell envelope stress phenotypes generated by exposure to SDS and EDTA. First, several sRNAs regulate the synthesis of OMPs in bacteria (reviewed in references 16 and 50). Second, it has been reported that up to 70% of the small proteins in E. coli are predicted to be membrane localized (18).
The bar-coded deletion collection was subjected to mock treatment and to cell envelope stress as described in Materials and Methods. UP and DN bar codes were amplified from the genomic DNAs of mock- and stress-treated cells and were hybridized to a microarray containing complementary probes. An RA value was obtained for each bar code by dividing its average stress treatment array intensity by its average mock treatment array intensity. An RA of 1 indicates that a deletion mutant has no phenotype. An RA of <1 indicates that a mutant is sensitive to the stress being imposed, while an RA of >1 indicates that a mutant is resistant to the stress.
A representative histogram plot of RA values obtained from one cell envelope stress screening experiment is shown in Fig. 2. The vast majority of the deletion mutants had an RA close to 1, indicating that they are wild type with respect to cell envelope stress. The cell envelope stress screening experiments were performed in triplicate. Due to the fact that each strain contains two unique bar codes, two independent RA measurements can be calculated for every deletion mutant within the population. Thus, six RA measurements were obtained for each deletion mutant. Although the RA values calculated for any particular deletion mutant differed across the three trials (see Table S3 in the supplemental material), the rank orders for the most sensitive and resistant strains were similar between experiments. A number of deletion mutants appeared repeatedly in the list of the twenty most sensitive strains (i.e., those with the lowest 20 RA values) (see Table S3); those that were among the 20 most sensitive strains in at least four of six RA measurements were analyzed further. As expected (41), the smpA deletion mutant was the most sensitive strain in every cell envelope stress experiment. Ten other deletion strains (ssrA, ybaM, micA, ryeF, yqcG, yobF, sroH ybhT, yqgB, and glmY) were also sensitive to growth in SDS and EDTA in at least four of the six measurements.
FIG. 2.
Most strains have no membrane stress phenotype under conditions of cell envelope stress. A representative histogram of RA values obtained from measurements of the fluorescence intensities of the DN tags in one experiment shows that the majority of strains have an RA value close to 1 (denoted by a solid black line). This indicates that they exhibit no phenotype under conditions of cell envelope stress. Sensitive strains have the lowest RA values, while resistant strains have the highest RA values.
Strains resistant to cell envelope stress.
The RA measurements were also analyzed for strains that might be resistant to cell envelope stress. As with the potentially sensitive strains, some deletion mutants appeared repeatedly among the most resistant strains (see Table S3 in the supplemental material). Deletion mutants of genes coding for one sRNA (rybB) and one small protein (blr) ranked with the five highest RA measurements at least four of six times and were analyzed further. The ybgT deletion mutant also appeared within this set, but its apparently intrinsic resistance to cell envelope stress is difficult to interpret and may be misleading given that ybgT cells grow very poorly on LB agar plates and in LB broth at 37°C (data not shown). As such, we did not analyze this potential phenotype further.
Although a combination of SDS and EDTA has been used previously to impose cell envelope stress (41), it should be noted that the phenotypes uncovered in the cell envelope stress screen may not have arisen as a consequence of cell envelope stress per se. Another possibility is that the cells are responding to the depletion of available divalent cations from the medium.
Verification of cell envelope stress phenotypes.
The phenotypes of putatively sensitive or resistant strains were verified in one-on-one competition assays with otherwise-wild-type lacZ mutants. In contrast to the large-scale screens, these experiments were conducted with deletion strains in which the antibiotic resistance cassettes incorporated at each deletion locus had been excised by Flp-mediated recombination. The one-on-one competition assays were conducted by mixing LacZ+ deletion mutant cells of interest (competitor strain) with otherwise-wild-type LacZ− cells (reference strain) and subjecting one-half of this mixture to a mock treatment and the other half to the cell envelope stress conditions described above. Cells from each sample were incubated on LB plates supplemented with IPTG and X-Gal. The numbers of blue and white colonies on these plates were scored. A CI was obtained by dividing the ratio of competitor cells to reference cells observed on the stress treatment plates by the ratio of competitor cells to reference cells observed on the mock treatment plates. Sensitive strains exhibit a CI of less than 1, while resistant strains have a CI greater than 1.
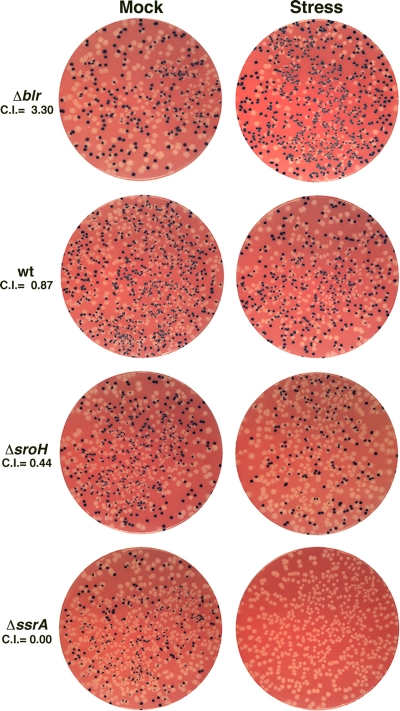
The results of four representative competition experiments are displayed in Fig. 3. The mock samples in each experiment contained blue and white cells in roughly equal proportions. The first panel shows that blr mutants are more resistant to growth in SDS and EDTA than wild-type cells, as evidenced by the increased ratio of blue to white cells after cell envelope stress treatment (CI, >1). When wild-type MG1655 was employed as a competitor strain, the ratio of blue to white cells remained unchanged after stress treatment, indicating that a deletion of lacZ does not affect E. coli fitness in either a positive or negative manner in this assay (Fig. 3 and Fig. 4). sroH and ssrA mutant cells exhibited increasingly severe sensitivity phenotypes, which was reflected in the decreasing ratios of blue to white cells after stress treatment (CI, <1).
FIG. 3.
Small-scale competition assays illustrate a range of phenotypes. Otherwise-wild-type LacZ− cells (NM601) were evaluated against one of four LacZ+ competitor strains, Δblr (GSO280), wild-type E. coli K-12 MG1655, ΔsroH (GSO278), or ΔssrA (GSO279), as described in Materials and Methods. The total numbers of blue and white colonies varied in the mock-treated samples, but the ratio of blue to white colonies was roughly 1:1 in all instances. For wild-type cells, this ratio was unchanged in the stress-treated sample. However, blr mutants were more resistant to cell envelope stress than lacZ mutants, as evidenced by the preponderance of blue colonies in the corresponding stress-treated samples. In contrast, sroH and ssrA mutant cells were sensitive to cell envelope stress, as shown by the reduced number of blue colonies relative to white colonies. The calculated CIs for these individual experiments are provided beneath each strain name.
FIG. 4.
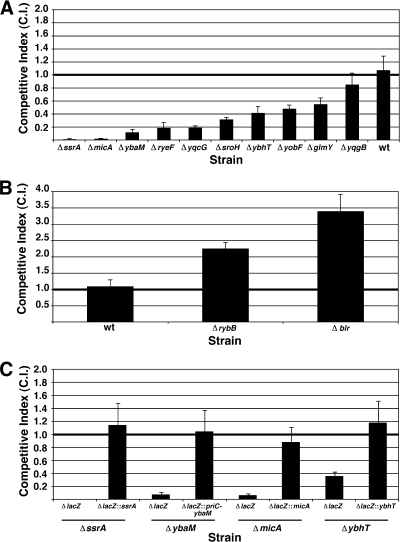
sRNA and small protein deletion mutants that were sensitive or resistant to cell envelope stress. (A and B) Competitor strains were grown in competition with LacZ− cells (NM601) under mock treatment conditions or conditions of cell envelope stress as described in Materials and Methods. A CI was calculated for each experiment; the CI values reported for all strains are the means of three trials, except for MG1655 (n = 4 trials). The error bars represent 1 standard deviation from the mean. Wild-type MG1655 cells did not exhibit a cell envelope stress phenotype and were employed as controls in both panels. (A) Cells mutant for ssrA (GSO279), ybaM (GSO283), micA (GSO271), ryeF (GSO277), or yqcG (GSO288), were sensitive to cell envelope stress. Cells mutant for sroH (GSO278), ybhT (GSO284), yobF (GSO287) or glmY (GSO269) exhibited more modest sensitivity to cell envelope stress, while yqgB (GSO289) deletion mutants were effectively wild type. (B) Cells mutant for blr (GSO280) or rybB (GSO276) were resistant to cell envelope stress. (C) Complemented LacZ− deletion mutants of ssrA (GSO298), ybaM (GSO299), micA (GSO297), and ybhT (GSO300) and uncomplemented LacZ− deletion mutants of ssrA (GSO294), ybaM (GSO295), micA (GSO293), and ybhT (GSO296) were evaluated against LacZ+ wild-type MG1655 cells as described in Materials and Methods.
One-on-one competition experiments were performed in triplicate with each of the putatively resistant or sensitive strains identified in the large-scale cell envelope stress assays (Fig. 4). After the ΔsmpA control strain (data not shown), ΔssrA and ΔmicA cells had the most severe cell envelope stress phenotypes (CI close to 0) (Fig. 4A). ybaM, ryeF, and yqcG deletion mutants were also very sensitive (CI values between 0.1 and 0.2) (Fig. 4A). sroH, ybhT, yobF, and glmY deletion mutants (CI values between 0.3 and 0.6) were only moderately sensitive. yqgB deletion mutants were not sensitive to cell envelope stress. In total, 9 of the 10 putatively sensitive strains exhibited significant phenotypes in one-on-one competition assays with LacZ− cells. Five of these nine strains were deleted for genes encoding sRNAs (ssrA, micA, ryeF, sroH, and glmY), three were deleted for genes encoding small proteins (yqcG, ybhT, and yobF), and one was deleted for a gene encoding a 53-amino-acid protein (ybaM). Finally, the resistance phenotypes exhibited by rybB and blr deletion mutants were also confirmed (CI values of 2.2 and 3.4, respectively) (Fig. 4B).
Complementation of select cell envelope stress phenotypes.
We examined whether the deleted gene was responsible for the phenotypes of the three deletion mutants most sensitive to cell envelope stress (ssrA, ybaM, and micA) as well as that of a more moderately sensitive deletion mutant (ybhT) by performing complementation experiments. To accomplish this, each of these genes was integrated under the control of its own promoter at the lacZ locus of the appropriate deletion mutant. The priC gene immediately upstream of ybaM was also included in the ybaM complementation construct. Each of these strains was subjected to cell envelope stress competition experiments against wild-type MG1655 cells. As shown in Fig. 4C, deletion mutations at ssrA, ybaM, micA, and ybhT could be complemented, as evidenced by the fact that all of the complemented strains exhibited CI values close to 1. We proceeded to further characterize the two strains with the most severe phenotypes, ΔssrA and ΔmicA.
SsrA function is required for cell envelope stress resistance.
ssrA encodes a specialized RNA (tmRNA) that frees stalled ribosomes from mRNA transcripts (reviewed in reference 10). During this process, a portion of SsrA that encodes a proteolysis tag is inserted into the ribosome concomitantly with displacement of the mRNA transcript. The tag is translated as the C terminus of the nascent polypeptide chain and targets the protein for degradation. Proteolysis of SsrA-tagged proteins is carried out primarily by the ClpXP protease (25). This is demonstrated by the fact that SsrA-tagged proteins accumulate and can be readily detected by immunoblot analyses in ΔclpX and ΔclpP cells, but not in deletion mutants of genes coding for other major cellular proteases (25).
To test if the freeing of stalled ribosomes from mRNAs and aborted polypeptides is sufficient for resistance or if both ribosome release and proteolysis tagging are required for cell envelope stress resistance, we determined the phenotype of ΔclpP cells. If the proteolysis of SsrA-tagged proteins is required for cell envelope stress resistance, then the major cellular protease required for carrying out this activity (ClpP) would be necessary for survival. However, in contrast to an ssrA deletion mutant (which is 100-fold or more sensitive to cell envelope stress), ΔclpP cells exhibit only a modest cell envelope stress phenotype (3- to 5-fold more sensitive) (Fig. 5). This result suggests that the proteolysis of SsrA-tagged proteins by ClpXP is at least partially dispensable with respect to cell envelope stress resistance and implies that ribosome release is the most critical aspect of the two SsrA functions.
FIG. 5.
Strains deficient in SsrA-mediated proteolysis are only moderately sensitive to cell envelope stress. Uncomplemented LacZ− deletion mutants of ssrA (GSO294) (n = 4) as well as deletion mutants of ssrA-deficient mutants complemented with either a wild-type allele of ssrA [ssrA+ (GSO301)] (n = 6) or an ochre codon mutant [ssrAO (GSO302)] (n = 5) were evaluated against LacZ+ wild-type MG1655 cells. A deletion mutant of clpP (GSO303) was also evaluated against LacZ− (NM601) cells (n = 3). A CI was calculated for each experiment. The error bars represent 1 standard deviation from the mean.
To further examine this possibility, ΔssrA cells were complemented with an allele (ssrAO) that is wild type for ribosome release but which contains a premature ochre stop codon that gives rise to a truncated tag with reduced affinity to the proteolysis machinery (54). As expected, the wild-type ssrA allele complements the SsrA phenotype (Fig. 5). Even though SsrAO is unable to target aborted polypeptides for proteolysis, the ssrAO allele also largely complements the SsrA phenotype, and cells containing SsrAO are phenotypically similar to ΔclpP cells (3- to 5-fold more sensitive) (Fig. 5). These results suggest that while SsrA-mediated proteolysis of aborted polypeptides is required to fully resist cell envelope stress, it is ribosome release that is primarily responsible for allowing E. coli to survive under these environmental conditions.
Roles of MicA and RybB in conferring resistance to cell envelope stress.
The outer membrane of a Gram-negative bacterium is studded with numerous β-barrel outer membrane proteins that contribute to its structural integrity and govern its permeability (3). Two signal transduction systems, σE and EnvZ-OmpR, employ sRNAs to downregulate OMP synthesis during periods of stress. The σE pathway is activated by misfolded OMPs that accumulate in the periplasm under conditions of cell envelope stress (51), and the EnvZ-OmpR system is responsive to high osmolarity (reviewed in reference 36). The sRNAs induced by σE and EnvZ-OmpR halt OMP synthesis by blocking ribosome binding to OMP-encoding mRNAs and promoting the degradation of the mRNAs (16). We were intrigued by the observation that a deletion mutant of one σE-regulated sRNA, MicA, was severely sensitive to cell envelope stress, while a deletion mutant of another σE-regulated sRNA, RybB, was resistant. We thus examined whether mutants of other OMP-regulating sRNAs exhibit cell envelope stress phenotypes that were missed in the large-scale screen.
In agreement with the results of the large-scale experiments, micC, micF, and cyaR deletion mutants did not show cell envelope stress phenotypes (Fig. 6). Individual omrA and omrB deletion mutants also displayed wild-type phenotypes with respect to cell envelope stress in the large-scale competition assay (see Table S3 in the supplemental material). Since omrA and omrB are functionally redundant and genetically linked (14, 15), we also tested cells that were doubly mutant for both genes in addition to deletion mutants of two other OMP-regulating sRNAs, RseX and IpeX, that were not initially included in our collection. None of these additional strains exhibited cell envelope stress phenotypes (Fig. 6).
FIG. 6.
Deletion mutants of most OMP-regulating sRNAs exhibit wild-type cell envelope stress phenotypes. Otherwise wild-type LacZ− mutant cells (NM601) were evaluated against one of nine LacZ+ competitor strains, ΔmicA (GSO271), ΔmicA ΔrybB (GSO290), ΔomrAB (GSO274), ΔmicC (GSO272), ΔmicF (GSO273), ΔrseX (GSO275), ΔipeX (GSO270), ΔcyaR (GSO268), and ΔrybB (GSO276), as described in Materials and Methods. Wild-type MG1655 cells did not exhibit a cell envelope stress phenotype and were employed as a control. A CI was calculated for each experiment; the CI values reported for each strain are the means of three trials, except for MG1655 (n = 4). The error bars represent 1 standard deviation from the mean.
Finally, to test whether RybB and MicA act in the same pathway, we constructed a strain that was doubly mutant for micA and rybB. If MicA and RybB were acting exclusively in the same genetic pathways, then the double mutant would be expected to exhibit a CI value close to the CI observed for one or the other single mutants. However, the double mutant exhibited an intermediate CI of 0.11, compared to the micA (CI, 0.013) and rybB (CI, 2.2) single mutants, and thus the two sRNAs most likely act independently of one another, possibly through different sets of mRNA targets (Fig. 6).
Acid stress screening experiments.
To examine the effects of another stress, the bar-coded deletion collection was subjected to mock treatment and to acid shock. The data arising from the acid shock experiments were analyzed as described above for the large-scale cell envelope stress experiments. As with the cell envelope stress experiments, the rank order of the most sensitive strains was roughly conserved in each of the three trials (see Table S4 in the supplemental material). A mutant deleted for a transcriptional activator of acid resistance genes in E. coli, gadE (27), appeared in the top 20 most sensitive strains all six times. Seven additional deletion mutants (yqgB, mgrB, yobF, yceO, ylcG, hokE, and ybgT) were among the most sensitive strains in at least four of six possible instances. No deletion mutants appeared to be resistant to acid shock.
Verification of acid shock sensitivity phenotypes.
We proceeded with one-on-one competition assays after verifying that lacZ deletion mutants were wild type with respect to acid sensitivity (Fig. 7) and that gadE cells were acid sensitive (CI, 0.0) (data not shown). One-on-one competition experiments were performed with the yqgB, mgrB, yobF, yceO, ylcG, and hokE mutant strains. As with the analyses of cell envelope stress phenotypes, we did not further analyze the slow-growing ybgT deletion mutant. The results in Fig. 7 demonstrate that the yqgB, mgrB, yobF, and yceO deletion mutants are all severely sensitive to acid stress (mean CI, ≤0.3), while ΔylcG and ΔhokE cells are not.
FIG. 7.
Four small protein deletion mutants were sensitive to acid stress. Otherwise-wild-type LacZ− mutant cells (NM601) were evaluated against one of seven LacZ+ competitor strains, ΔyqgB (GSO289), ΔmgrB (GSO282), ΔyobF (GSO287), ΔyceO (GSO285), ΔylcG (GSO286), ΔhokE (GSO281), and wild-type MG1655, as described in Materials and Methods. A CI was calculated for each experiment; the CI values reported for each strain are the means of three trials, except for ΔylcG (GSO286) and ΔhokE (GSO281) (n = 5 trials). The error bars represent 1 standard deviation from the mean.
DISCUSSION
We have created a collection of 125 DNA bar-coded mutants in E. coli. A total of 116 of these strains are deleted for genes encoding sRNAs and proteins of less than 70 amino acids, 1 strain is deleted for a known sRNA target, and the remaining 8 strains are deleted for genes known to be necessary for resistance to various stress conditions. We were able to detect an array of phenotypes of varied severities, ranging from mutants that were very sensitive to cell envelope stress or acid shock to moderately sensitive and resistant cells. Even deletion mutations that give rise to moderate phenotypes are of considerable value, since they can be combined with one another to identify redundant genetic pathways.
Importantly, we were able to identify subtle deletion phenotypes which would have remained undiscovered by more traditional methodologies. This is evidenced by the fact that none of the cell envelope stress sensitivity or resistance phenotypes are apparent when the corresponding mutant strains are incubated on LB agar plates containing 0.5% SDS and 1 mM EDTA (data not shown). This is in contrast to ΔsmpA cells (the control strain known to be sensitive to cell envelope stress), which are readily distinguished from wild-type cells on such medium (42).
Aside from the ability to detect subtle sensitivity and resistance phenotypes, the bar-coding approach we and one other group (38) have adapted for E. coli presents another advantage to traditional screening methodologies. Namely, the bar codes themselves, the microarray employed to detect them, and procedures to set up and analyze experiments have all been validated by the yeast community. Other groups have generated directed deletion mutants of almost every gene in E. coli (2), as well as some sRNA genes in E. coli (19) and Salmonella (32), but none of these collections incorporates DNA bar codes. In principle, MGK analysis could be employed to analyze our collection of bar-coded deletion mutants; however, at present, the chips employed in this methodology are not commercially available, have not been tested as extensively as the yeast bar code arrays used in our study (13, 34, 53), and would have to be custom designed for our application (41).
Identification of novel cell envelope stress phenotypes.
One-on-one competition experiments against otherwise-wild-type lacZ mutant cells confirmed that nine deletion mutants arising from large-scale screens were indeed sensitive to cell envelope stress, and two deletion mutants were resistant (Fig. 4). None of the cell envelope stress phenotypes we uncovered has been reported previously. Of particular note, we found that deletion mutants of two extensively studied genes, ssrA (10, 21) and micA (48), exhibited severe cell envelope stress phenotypes (Fig. 4A).
SsrA-mediated ribosome release is required for cell envelope stress resistance.
SsrA acts in conjunction with the SmpB protein to mediate trans-translation, a process that frees stalled ribosomes (reviewed in reference 10). In this process, SsrA exhibits two primary activities, protease tagging and ribosome release from mRNAs and aborted polypeptides. ssrA is essential in some bacteria (but not in E. coli) and is required for pathogenesis in Yersinia (10), survival of Salmonella enterica serovar Typhimurium in macrophages (10), and swimming motility in E. coli (23). Deletion mutants of ssrA also induce an elevated heat shock response in E. coli (30).
We are the first to show a cell envelope stress sensitivity phenotype associated with the deletion of ssrA. Furthermore, two strains that are deficient in the proteolysis of SsrA-tagged proteins, ΔclpP and ssrAO, show similar phenotypes and are only moderately sensitive to cell envelope stress (Fig. 5). This would imply that the more important activity of SsrA with respect to cell envelope stress resistance is ribosome release and not proteolysis tagging.
Many cell envelope proteins are cotranslationally secreted at the inner membrane (reviewed in reference 9). When nascent polypeptides misfold under conditions of cell envelope stress, it is conceivable that the activity of the secretory apparatus is inhibited, which would in turn halt translation. Without SsrA, these membrane-bound ribosomes would remain stalled and unavailable to participate in the response and possibly even contribute to cell envelope stress.
MicA and RybB have opposite effects on cell envelope stress resistance.
ΔmicA cells are almost as sensitive to cell envelope stress as ΔssrA cells (Fig. 4A). MicA is one of several E. coli sRNAs that repress OMP translation (48). The MicA phenotype we observed was striking, given that deletion mutants of seven other genes coding for OMP-regulating sRNAs (omrAB, micC, micF, ipeX, cyaR, and rseX) did not exhibit cell envelope stress phenotypes in our study (Fig. 6; see also Table S3 in the supplemental material), and the deletion mutant of one OMP-regulating sRNA (rybB) exhibited resistance (Fig. 6).
It is curious that ΔmicA cells are extremely sensitive to cell envelope stress, while ΔrybB cells are resistant. One might expect that ΔrybB cells are more resistant because they upregulate σE activity (46); however, this also occurs in micA mutants (see Fig. S1 in the supplemental material). Given that MicA and RybB target different sets of mRNAs in Salmonella (33), one plausible explanation for the RybB resistance phenotype is that the synthesis of an OMP(s) that makes the cell more resistant to cell envelope stress is upregulated in ΔrybB due to derepression but not in ΔmicA cells. Alternatively, RybB might normally upregulate genes that are detrimental to surviving cell envelope stress.
Identification of novel acid shock phenotypes.
Deletion mutants of four genes coding for small proteins (yobF, yceO, mgrB, and yqgB) were confirmed to exhibit novel acid sensitivity phenotypes (Fig. 7). mgrB exhibits regulation that is consistent with its acid-sensitive deletion phenotype. This gene was so named because it is regulated by the PhoQ-PhoP two-component system (20), which is responsive to low concentrations of Mg2+ (29). Expression of mgrB is also activated by the EvgS-EvgA two-component system in a PhoP-dependent manner (11). Although it is unclear what stimulates the sensor kinase EvgS in vivo, artificial activation of evgA has been reported to make exponentially growing E. coli cells acid resistant (28). The YqgB sensitivity phenotype could arise as a consequence of polarity effects on two downstream genes, speA and speB. SpeA and SpeB are required for the synthesis of polyamines (4), the presence of which has been shown to confer acid resistance to E. coli (40, 55). The yqgB gene contains an internal promoter that drives expression of speA and speB (43). Therefore, deleting yqgB could eliminate speAB expression and render the yqgB mutant cells acid sensitive. Another group has recently reported that ΔgcvB cells are acid sensitive (19). We did not observe this phenotype (see Table S4 in the supplemental material); however, we exposed cells to pH 1.8 for 10 min, while Jin et al. exposed cells to pH 2.0 for 30 min.
Overlap of phenotypic data with small-protein expression data.
A large number of small-protein-encoding genes are regulated by changes in growth or stress conditions (17). Deletion mutants of this set of stress-regulated small-protein-encoding genes did not exhibit cell envelope or acid shock phenotypes in the present study. This is perhaps not surprising, since the acid shock and cell envelope stress conditions employed in the two studies were slightly different. Additionally, two genes (yohP and yshB) that were shown to be induced in response to cell envelope stress were not included in our collection of bar-coded deletion mutants (17). However, a deletion mutant of yobF (which is posttranscriptionally induced by heat shock [17]) was moderately sensitive to cell envelope stress and severely sensitive to acute acid stress (Fig. 4A and 7). Absent any polarity effects on the downstream cspC gene, these data suggest that YobF warrants further investigation as a potential component of a generalized stress response pathway.
Future directions.
Until recently, the large numbers of sRNAs and small proteins encoded in the intergenic regions of bacterial genomes have been underappreciated. Advances in bioinformatic approaches, the development of densely tiled oligonucleotide microarrays, and cloning-based approaches coupled with DNA pyrosequencing technology are extending the list of sRNA and small protein genes of undefined function. Deletion mutants of these newly discovered genes can be readily added to our bar-coded collection and tested en masse under conditions of cell envelope stress, acid shock, and most any other stress condition (e.g., alkaline stress, ethanol stress, and heavy metal stress) for novel phenotypes. The phenotypes uncovered in these assays will facilitate genetic studies as well as the application of biochemical and cytological methodologies to further illuminate the roles sRNAs and small proteins play in the cell.
Supplementary Material
Acknowledgments
We thank S. Gottesman, D. Kearns, A. Typas, and members of the Storz laboratory (especially F. Fontaine and M. Hemm) for helpful discussions and comments.
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (E.C.H., J.L.A., and G.S.) and by the Pharmacology Research Associate Program at the National Institute of General Medical Sciences (E.C.H.).
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altuvia, S. 2007. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10:257-261. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191-214. [DOI] [PubMed] [Google Scholar]
- 4.Boyle, S. M., G. D. Markham, E. W. Hafner, J. M. Wright, H. Tabor, and C. W. Tabor. 1984. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (speA, speB, speC and metK). Gene 30:129-136. [DOI] [PubMed] [Google Scholar]
- 5.Butland, G., M. Babu, J. J. Diaz-Mejia, F. Bohdana, S. Phanse, B. Gold, W. Yang, J. Li, A. G. Gagarinova, O. Pogoutse, H. Mori, B. L. Wanner, H. Lo, J. Wasniewski, C. Christopolous, M. Ali, P. Venn, A. Safavi-Naini, N. Sourour, S. Caron, J. Y. Choi, L. Laigle, A. Nazarians-Armavil, A. Deshpande, S. Joe, K. A. Datsenko, N. Yamamoto, B. J. Andrews, C. Boone, H. Ding, B. Sheikh, G. Moreno-Hagelseib, J. F. Greenblatt, and A. Emili. 2008. eSGA: E. coli synthetic genetic array analysis. Nat. Methods 5:789-795. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 60:451-475. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driessen, A. J., and N. Nouwen. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643-667. [DOI] [PubMed] [Google Scholar]
- 10.Dulebohn, D., J. Choy, T. Sundermeier, N. Okan, and A. W. Karzai. 2007. trans-Translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 46:4681-4693. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi, Y., T. Okada, S. Minagawa, T. Oshima, H. Mori, K. Yamamoto, A. Ishihama, and R. Utsumi. 2004. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J. Bacteriol. 186:3006-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fozo, E. M., M. Kawano, F. Fontaine, Y. Kaya, K. S. Mendieta, K. L. Jones, A. Ocampo, K. E. Rudd, and G. Storz. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 70:1076-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 14.Guillier, M., and S. Gottesman. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 36:6781-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillier, M., and S. Gottesman. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59:231-247. [DOI] [PubMed] [Google Scholar]
- 16.Guillier, M., S. Gottesman, and G. Storz. 2006. Modulating the outer membrane with small RNAs. Genes Dev. 20:2338-2348. [DOI] [PubMed] [Google Scholar]
- 17.Hemm, M. R., B. J. Paul, J. Miranda-Rios, N. Soltanzad, and G. Storz. 2010. Small stress response proteins in Escherichia coli: proteins missed by classical proteomics studies. J. Bacteriol. 192:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemm, M. R., B. J. Paul, T. D. Schneider, G. Storz, and K. E. Rudd. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 70:1487-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, Y., R. M. Watt, A. Danchin, and J. D. Huang. 2009. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 22.Kleerebezem, M. 2004. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405-1414. [DOI] [PubMed] [Google Scholar]
- 23.Komine, Y., M. Kitabatake, T. Yokogawa, K. Nishikawa, and H. Inokuchi. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91:9223-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 25.Lies, M., and M. R. Maurizi. 2008. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J. Biol. Chem. 283:22918-22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livny, J., and M. K. Waldor. 2007. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 10:96-101. [DOI] [PubMed] [Google Scholar]
- 27.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 28.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 29.Monsieurs, P., S. De Keersmaecker, W. W. Navarre, M. W. Bader, F. De Smet, M. McClelland, F. C. Fang, B. De Moor, J. Vanderleyden, and K. Marchal. 2005. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J. Mol. Evol. 60:462-474. [DOI] [PubMed] [Google Scholar]
- 30.Munavar, H., Y. Zhou, and S. Gottesman. 2005. Analysis of the Escherichia coli Alp phenotype: heat shock induction in ssrA mutants. J. Bacteriol. 187:4739-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niba, E. T., Y. Naka, M. Nagase, H. Mori, and M. Kitakawa. 2007. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 14:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papenfort, K., V. Pfeiffer, S. Lucchini, A. Sonawane, J. C. Hinton, and J. Vogel. 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 68:890-906. [DOI] [PubMed] [Google Scholar]
- 33.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. Hinton, and J. Vogel. 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierce, S. E., R. W. Davis, C. Nislow, and G. Giaever. 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2:2958-2974. [DOI] [PubMed] [Google Scholar]
- 35.Pierce, S. E., E. L. Fung, D. F. Jaramillo, A. M. Chu, R. W. Davis, C. Nislow, and G. Giaever. 2006. A unique and universal molecular barcode array. Nat. Methods 3:601-603. [DOI] [PubMed] [Google Scholar]
- 36.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 37.Ranquet, C., and S. Gottesman. 2007. Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189:4872-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rooney, J. P., A. Patil, M. R. Zappala, D. S. Conklin, R. P. Cunningham, and T. J. Begley. 2008. A molecular bar-coded DNA repair resource for pooled toxicogenomic screens. DNA Repair (Amst.) 7:1855-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejewski, A. D. Grossman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13:689-701. [DOI] [PubMed] [Google Scholar]
- 40.Samartzidou, H., M. Mehrazin, Z. Xu, M. J. Benedik, and A. H. Delcour. 2003. Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J. Bacteriol. 185:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sklar, J. G., T. Wu, L. S. Gronenberg, J. C. Malinverni, D. Kahne, and T. J. Silhavy. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, L. K., M. J. Gomez, K. Y. Shatalin, H. Lee, and A. A. Neyfakh. 2007. Monitoring of gene knockouts: genome-wide profiling of conditionally essential genes. Genome Biol. 8:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szumanski, M. B., and S. M. Boyle. 1992. Influence of cyclic AMP, agmatine, and a novel protein encoded by a flanking gene on speB (agmatine ureohydrolase) in Escherichia coli. J. Bacteriol. 174:758-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamae, C., A. Liu, K. Kim, D. Sitz, J. Hong, E. Becket, A. Bui, P. Solaimani, K. P. Tran, H. Yang, and J. H. Miller. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190:5981-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomason, L. C., N. Costantino, and D. L. Court. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 1:1.17. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 189:4243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Typas, A., R. J. Nichols, D. A. Siegele, M. Shales, S. R. Collins, B. Lim, H. Braberg, N. Yamamoto, R. Takeuchi, B. L. Wanner, H. Mori, J. S. Weissman, N. J. Krogan, and C. A. Gross. 2008. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods 5:781-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19:2355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urbanowski, M. L., L. T. Stauffer, and G. V. Stauffer. 2000. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 37:856-868. [DOI] [PubMed] [Google Scholar]
- 50.Vogel, J., and K. Papenfort. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9:605-611. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 52.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 54.Withey, J., and D. Friedman. 1999. Analysis of the role of trans-translation in the requirement of tmRNA for λimmP22 growth in Escherichia coli. J. Bacteriol. 181:2148-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yohannes, E., A. E. Thurber, J. C. Wilks, D. P. Tate, and J. L. Slonczewski. 2005. Polyamine stress at high pH in Escherichia coli K-12. BMC Microbiol. 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.