Abstract
Enhancer binding proteins (EBPs) control the temporal expression of fruiting body development-associated genes in Myxococcus xanthus. Eleven previously uncharacterized EBP genes were inactivated. Six EBP gene mutations produced minor but reproducible defects in fruiting body development. One EBP gene mutation that affected A-motility produced strong developmental defects.
When the intracellular starvation signal (p)ppGpp accumulates (10, 22, 23, 28), the deltaproteobacterium Myxococcus xanthus forms a biofilm containing a mat of peripheral rod cells and multicellular structures called fruiting bodies (29). Cells that aggregate into fruiting bodies differentiate into dormant and stress-resistant spores, while the peripheral rods outside these structures fail to sporulate (25). Fruiting body development is accompanied by large-scale changes in gene expression, and enhancer binding proteins (EBPs) form a regulatory cascade that controls the sequential expression of many developmental genes (N. B. Caberoy, K. M. Giglio, G. Suen, and A. G. Garza, submitted for publication). EBPs are transcriptional activators that work in conjunction with σ54-RNA polymerase; EBPs help σ54-RNA polymerase form a transcription-competent open promoter complex (33). To date, 17 EBPs that perform a variety of developmental functions have been linked to the formation of mature fruiting bodies (2, 6-9, 13, 14, 17, 30, 32).
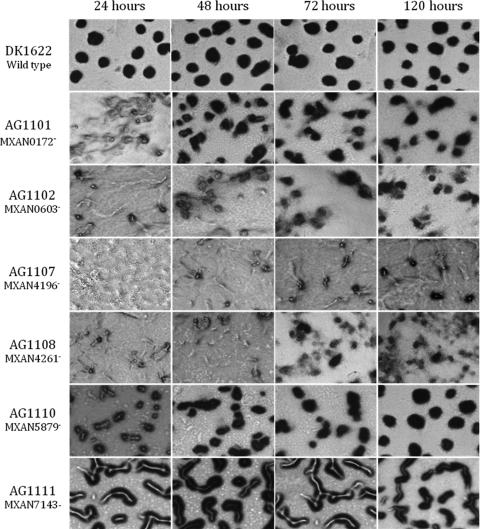
Eleven M. xanthus genes that code for EBPs have yet to be characterized. Here, we examined whether these uncharacterized EBP genes are important for fruiting body development. Insertions in the chromosomal copies of the EBP genes in wild-type strain DK1622 were created and confirmed as previously described (2). (Tables 1 and 2 show the bacterial strains, plasmids, and primers used in this study.) Subsequently, EBP mutant cells and wild-type cells were placed on 1.5% agar plates containing TPM starvation buffer (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, and 8 mM MgSO4) to monitor the progress of fruiting body development and to determine sporulation efficiencies. Six of the EBP mutants exhibited relatively weak developmental defects (Table 3 and Fig. 1). The MXAN0172, MXAN5879, and MXAN7143 mutants had wild-type sporulation efficiencies, but they exhibited fruiting body formation defects. In particular, fruiting body formation in the MXAN0172 and MXAN5879 mutants was delayed, and the MXAN7143 mutant failed to produce fruiting bodies with characteristic shapes. Fruiting body formation in the MXAN0603 and MXAN4261 mutants was both delayed and incomplete, and their sporulation efficiencies were reduced about 1.5- to 1.8-fold compared to that of wild-type cells. Finally, the MXAN0907 mutant produced normal-looking fruiting bodies (data not shown), but its sporulation efficiency was reduced about 2.2-fold compared to that of wild-type cells.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| AG1101 | DK1622 pKG01::MXAN0172 | This study |
| AG1102 | DK1622 pKG02::MXAN0603 | This study |
| AG1103 | DK1622 pKG03::MXAN0907 | This study |
| AG1104 | DK1622 pKG04::MXAN1189 | This study |
| AG1105 | DK1622 pKG05::MXAN1565 | This study |
| AG1106 | DK1622 pKG06::MXAN3555 | This study |
| AG1107 | DK1622 pKG07::MXAN4196 | This study |
| AG1108 | DK1622 pKG08::MXAN4261 | This study |
| AG1109 | DK1622 pKG09::MXAN4977 | This study |
| AG1110 | DK1622 pKG10::MXAN5879 | This study |
| AG1111 | DK1622 pKG11::MXAN7143 | This study |
| AG1112 | DK1253 pKG05::MXAN4196 | This study |
| AG1113 | DK1218 pKG05::MXAN4196 | This study |
| DK1218 | A-motility defect | 12 |
| DK1253 | S-motility defect | 12 |
| DK1622 | Wild-type motility and development | 15 |
| DK2161 | A-motility and S-motility defects | 12 |
| Plasmids | ||
| pCR2.1-TOPO | Kanr | Invitrogen |
| pKG01 | Kanr pCR2.1-TOPO containing a 455-bp MXAN0172 fragment | This study |
| pKG02 | Kanr pCR2.1-TOPO containing a 501-bp MXAN0603 fragment | This study |
| pKG03 | Kanr pCR2.1-TOPO containing a 548-bp MXAN0907 fragment | This study |
| pKG04 | Kanr pCR2.1-TOPO containing a 350-bp MXAN1189 fragment | This study |
| pKG05 | Kanr pCR2.1-TOPO containing a 404-bp MXAN1565 fragment | This study |
| pKG06 | Kanr pCR2.1-TOPO containing a 477-bp MXAN3555 fragment | This study |
| pKG07 | Kanr pCR2.1-TOPO containing a 502-bp MXAN4196 fragment | This study |
| pKG08 | Kanr pCR2.1-TOPO containing a 541-bp MXAN4261 fragment | This study |
| pKG09 | Kanr pCR2.1-TOPO containing a 485-bp MXAN4977 fragment | This study |
| pKG10 | Kanr pCR2.1-TOPO containing a 502-bp MXAN5897 fragment | This study |
| pKG11 | Kanr pCR2.1-TOPO containing a 350-bp MXAN7143 fragment | This study |
TABLE 2.
Primers used in this study
| Primer | Locus tag or gene | Sequence | Amplicon size (bp) |
|---|---|---|---|
| 3558 up | MXAN0172 | 5′-CGCTGCATTCGATGACTGCTC-3′ | |
| 3558 down | MXAN0172 | 5′-GCGAGCGAAGAAGGAGACGAA-3′ | 455 |
| 1181a | MXAN0603 | 5′-CGTCATCGTCACCGGCGAGTCC-3′ | |
| 1181b | MXAN0603 | 5′-GTGAGCTGCCGGACGAAGTGCC-3′ | 501 |
| mx2756-fwd | MXAN0907 | 5′-AGCGAGCTGCCCGTGCTGGTGTGC-3′ | |
| mx2756-rev | MXAN0907 | 5′-GCGGACAGCTCCATCTCCTCACGG-3′ | 548 |
| 1156a | MXAN1565 | 5′-CCTTCGTCACGCTCAACTGCGC-3′ | |
| 1156b | MXAN1565 | 5′-GAGGAAGGCGCACAACTGCGGC-3′ | 404 |
| 980a | MXAN1189 | 5′-GGCTCGTCGCCGTCAACTGCG-3′ | |
| 980b | MXAN1189 | 5′-CTGGAGAGGCATCACGTTGAGG-3′ | 350 |
| 1930 up | MXAN3555 | 5′-GGAGCTCATCGCCACCGCGCT-3′ | |
| 1930 down | MXAN3555 | 5′-TGGCGTGCTTGGCCACGAAGT-3′ | 477 |
| 3656 up | MXAN4196 | 5′-GCAGGCCACGGTGCTGCTGGT-3′ | |
| 3656 down | MXAN4196 | 5′-GCGCAGCAGCAGCTCCGACAA-3′ | 502 |
| 939a | MXAN4261 | 5′-CGATGCGGAACCTCTACGAGC-3′ | |
| 939b | MXAN4261 | 5′-GTGAAGTGCTCCACCAACAAGG-3′ | 541 |
| mx4346-fwd | MXAN4977 | 5′-CTGGCGAGAATGGGACGGGGAAGG-3′ | |
| mx4346-rev | MXAN4977 | 5′-CACAGGTGGGCGCACTGATTGAGG-3′ | 485 |
| 6911 up | MXAN5879 | 5′-CATCGCCGCCTCATCCATGAC-3′ | |
| 6911 down | MXAN5879 | 5′-GTCCGGGGACAGGCCGGATAC-3′ | 502 |
| 1254a | MXAN7143 | 5′-GGTGCGGCGGCTCATCGAGCG-3′ | |
| 1254b | MXAN7143 | 5′-AGCCCACCGGATGCAGCTCGC-3′ | 350 |
TABLE 3.
Developmental phenotypes of wild-type and EBP gene mutant strainsa
| Strain (genotype) | Fruiting body formationb | Fruiting body spores (% of wild type)c |
|---|---|---|
| DK1622 (wild type) | + | 100.0 ± 19.0 |
| AG1101 (MXAN0172) | +/− | 68.5 ± 8.5 |
| AG1102 (MXAN0603) | +/− | 54.3 ± 6.4d |
| AG1103 (MXAN0907) | + | 46.3 ± 10.0d |
| AG1104 (MXAN1565) | + | 100.1 ± 9.5 |
| AG1105 (MXAN3077) | + | 111.0 ± 11.1 |
| AG1106 (MXAN3555) | + | 85.9 ± 13.5 |
| AG1107 (MXAN4196) | − | <0.01d |
| AG1108 (MXAN4261) | +/− | 65.4 ± 3.8d |
| AG1109 (MXAN4977) | + | 102.1 ± 2.3 |
| AG1110 (MXAN5879) | +/− | 72.2 ± 11.9 |
| AG1111 (MXAN7143) | +/− | 116.0 ± 5.6 |
Cells were placed on TPM agar and allowed to develop for 5 days. Development was monitored visually using phase-contrast microscopy.
Symbols: +, produced normal-looking fruiting bodies; −, failed to produce normal-looking fruiting bodies; +/−, produced normal-looking fruiting bodies but aggregation was delayed.
Spore assays were performed three times for each strain. The mean values ± standard deviations for the spore assays are shown as percentages of DK1622 (wild type). The number of spores produced by wild-type cells ranged from 1.12 × 107 to 1.90 × 107. Values were determined by transferring sonication- and heat-resistant spores to CTTYE agar plates, incubating the plates for 5 days, and counting the number of colonies that arose from the spores.
Variances compared to wild type were found to be significant using a two-tailed t test (α = 0.05).
FIG. 1.
Development of EBP gene mutants on TPM agar plates. Wild-type and mutant cells were placed on TPM starvation agar, and the progress of fruiting body development was monitored for 5 days using phase-contrast microscopy. Photographs were taken at 24, 48, 72, and 120 h poststarvation using a total magnification of ×40.
We scanned the sequences of the EBP gene loci (5) and our findings suggest that three (MXAN0172, MXAN0907, and MXAN7143) out of the six insertions that yielded relatively weak developmental phenotypes have the potential to be polar. The genes located immediately downstream of MXAN0172, MXAN0907, and MXAN7143 are MXAN0171, MXAN0906, and MXAN7142, respectively. Using quantitative PCR analysis (26), we found no obvious signs that the three insertions in question are polar; we detected wild-type levels of MXAN0171, MXAN0906, and MXAN7142 expression in the MXAN0172, MXAN0907, and MXAN7143 mutants, respectively (data not shown).
One EBP mutant, MXAN4196, showed strong defects in fruiting body formation and sporulation (Table 3 and Fig. 1). This mutant failed to form normal-looking fruiting bodies, even when it was given 5 days to develop. Furthermore, the MXAN4196 mutant produced no viable spores. On the basis of the M. xanthus genome sequence (5), MXAN4196 is the last gene in an operon that contains two genes. This finding indicates that the insertion in MXAN4196 is unlikely to have polar effects. Because the MXAN4196 mutant has strong defects in fruiting body development, we chose to analyze it further.
M. xanthus cells use gliding motility to aggregate into multicellular fruiting bodies, and many EBPs that are important for fruiting body development have been linked to gliding motility (19). To determine whether the MXAN4196 mutant has a gliding motility defect, we used swarm expansion assays (16). MXAN4196 mutant cells and wild-type cells were placed on CTTYE (1.0% Casitone, 0.5% yeast extract, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4, and 8 mM MgSO4) plates containing 0.4% or 1.5% agar, and colony diameters were determined after 3 days of incubation at 32°C. The mean diameters of MXAN4196 mutant colonies on 0.4% and 1.5% agar plates were 64.1% (±5.6% [standard deviation]) and 44.9% (±5.3%) of wild-type colonies, respectively. These results indicate that the MXAN4196 mutant has a gliding motility defect.
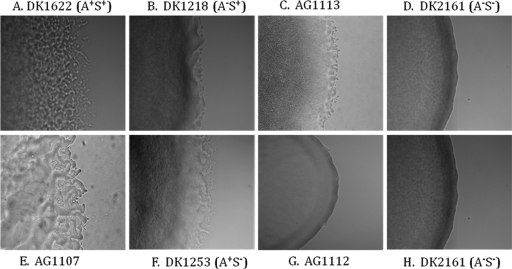
Mutants defective for either A-motility (A− S+ cells) or S-motility (A+ S− cells) swarm at a reduced rate, while mutants that are defective for both types of motility (A− S− cells) have a nonswarming phenotype and smooth colony edges (12). To determine whether the MXAN4196 insertion causes a defect in the A- or S-motility system, it was introduced into A− S+ (DK1218) and A+ S− (DK1253) mutant strains, and the colony edges of the double mutants were examined using phase-contrast microscopy (Fig. 2). When the MXAN4196 insertion was introduced into the DK1218 (A− S+) recipient, the colony edge was similar to that of wild-type cells carrying the same insertion. When the insertion was introduced into the DK1253 (A+ S−) background, we detected a smooth colony edge that was similar to that of the nonswarming A− S− double mutant DK2161. These findings indicate that the MXAN4196 insertion causes a defect in A-motility.
FIG. 2.
Colony edge morphologies produced by the MXAN4196 insertion. Colony edge morphologies produced by A+ S+ strain DK1622 (A), A− S+ strain DK1218 (B), A+ S− strain DK1253 (F), and A− S− strain DK2161 (D and H) are shown. The MXAN4196 insertion was introduced into strain DK1622 to generate strain AG1107 (E), into strain DK1218 to generate strain AG1113 (C) and into strain DK1253 to generate strain AG1112 (G). Colony edges were observed after 3 days of growth on CTTYE agar using phase-contrast microscopy (40× magnification).
Since sporulation takes place inside fruiting bodies and the MXAN4196 insertion disrupts A-motility and fruiting body formation, we examined whether this insertion has a direct effect on sporulation by performing glycerol spore assays (21). When glycerol is added to a nutrient broth culture, rod-shaped vegetative cells undergo a rapid and synchronous conversion into spores, bypassing many of the early events that are required for production of fruiting body spores by directly activating at least part of the sporulation program (4). Interestingly, the MXAN4196 mutant produced no viable glycerol spores in our assays (data not shown). Our interpretation of this result is that MXAN4196 plays a direct and important role in the M. xanthus sporulation process.
In this study, we identified six EBP mutants that have relatively minor defects in fruiting body development and one EBP mutant (MXAN4196) that has strong developmental defects. The MXAN4196 mutant fails to produce normal-looking fruiting bodies, and it fails to produce viable spores during development. Our data indicate that MXAN416 is an A-motility mutant. Although the mechanism of M. xanthus A-motility is not well understood, two models have been proposed: one model suggests that A-motility is powered by slime extrusion from the cell poles (31), and the other model suggests that A-motility is powered by motors associated with focal adhesion complexes (24). The A-motility system is known to require a complex network of more than 30 genes (reviewed in reference 11). Mutations in most A-motility genes have little or no effect on the formation of spore-filled fruiting bodies. Mutations that do produce developmental phenotypes seem to primarily affect sporulation. At this point, it is unclear whether the A-motility defect of the MXAN4196 mutant contributes to its developmental phenotype. However, we can state that the MXAN4196 mutant has a particularly strong developmental defect for an A-motility mutant. The MXAN4196 mutant also has a strong defect in glycerol-induced sporulation. This is a rather unique phenotype for an A-motility mutant, but we are aware of one other A-motility mutant that has such a defect, the EBP gene mutant nla24 (2, 20). Gliding motility is not required for glycerol-induced sporulation, suggesting that the MXAN4196 protein plays a critical role in sporulation that is distinct from its role in A-motility. Since EBPs regulate transcription at σ54 promoters, we looked for σ54 promoter signature sequences upstream of operons containing A-motility genes and operons containing sporulation-specific genes. As shown in Table 4, we found five A-motility gene operons and three sporulation gene operons that have putative σ54 promoters. This finding suggests that MXAN4196 might play a direct role in the regulation of both A-motility genes and sporulation genes. The goal of future work will be to determine whether any of these operons are under direct transcriptional control of this EBP.
TABLE 4.
Operons containing genes with putative σ54 promoters
| Gene type | First gene in operon | Relevant gene | No. of genes in operon | σ54 promoter sequencea | Reference |
|---|---|---|---|---|---|
| Genes known to be required for A motility | MXAN2991 | aglZ | 1 | TGGCAAC-N4-CTGCT | 34 |
| MXAN3502 | agmI | 2 | TGGGGCG-N4-TTGCC | 35 | |
| MXAN4799 | agmC | 2 | TGACAGA-N4-TTTCA | 35 | |
| MXAN5818 | agmR | 2 | TGGCACA-N4-GTGCT | 35 | |
| MXAN5820 | agmM | 1 | TGGCCCT-N4-CTGCT | 35 | |
| Genes known to be required for sporulation | MXAN2269 | mspA | 1 | TGGCCTA-N4-GTGCT | 3 |
| MXAN3225 | exo | 5 | TGGCACA-N4-CTGCT | 21 | |
| MXAN5432 | tps | 2 | TGGGGCA-N4-TTGCT | 18 |
The putative promoter regions of operons containing A-motility and sporulation genes were analyzed using the M. xanthus genome sequence (5) and PromScan (http://molbiol-tools.ca/promscan/), a bioinformatics tool that was specifically developed to identify σ54-RNA polymerase binding sites in the sequences of bacterial DNA. To be designated a σ54 promoter, there had to be a potential binding site for σ54-RNA polymerase and a potential EBP binding site, which is a tandem repeat of at least 7 bp (27). On the basis of tests done with known promoter sequences and intragenic sequences, we estimated that our analysis had a false-positive rate of about 4% and a false-negative rate of about 23%. The −12 and −24 regions of the putative σ54 promoters are shown. Bold, underlined nucleotides are those that match nucleotides in the σ54 consensus sequence, which is TGGCACG-N4-TTGC(T/A) (1).
Acknowledgments
This work was supported by National Science Foundation grant 0717653 to A. Garza.
Footnotes
Published ahead of print on 6 November 2009.
REFERENCES
- 1.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl, J. L., F. K. Tengra, D. Dutton, J. Yan, T. M. Andacht, L. Coyne, V. Windell, and A. G. Garza. 2007. Identification of major sporulation proteins of Myxococcus xanthus using a proteomic approach. J. Bacteriol. 189:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin, M., and S. M. Gibson. 1964. A system for studying microbial morphogenesis: rapid formation of microcysts in Myxococcus xanthus. Science 146:243-244. [DOI] [PubMed] [Google Scholar]
- 5.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronewold, T. M., and D. Kaiser. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, D., Y. Wu, and H. B. Kaplan. 2000. Identification and characterization of genes required for early Myxococcus xanthus gene expression. J. Bacteriol. 182:4564-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hager, E., H. Tse, and R. E. Gill. 2001. Identification and characterization of spdR mutations that bypass the BsgA protease-dependent regulation of developmental gene expression in Myxococcus xanthus. Mol. Microbiol. 39:765-780. [DOI] [PubMed] [Google Scholar]
- 10.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartzell, P., W. Shi, and P. Youderian. 2008. Gliding motility of Myxococcus xanthus, p. 103-122. In D. E. Whitworth (ed.), Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
- 12.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 172:177-191. [Google Scholar]
- 13.Jakobsen, J. S., L. Jelsbak, R. D. Welch, C. Cummings, B. Goldman, E. Stark, S. Slater, and D. Kaiser. 2004. σ54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 186:4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelsbak, L., M. Givskov, and D. Kaiser. 2005. Enhancer-binding proteins with a forkhead-associated domain and the σ54 regulon in Myxococcus xanthus fruiting body development. Proc. Natl. Acad. Sci. U. S. A. 102:3010-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser, D., and C. Crosby. 1983. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 3:227-245. [Google Scholar]
- 17.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komano, T., T. Furuichi, M. Teintze, M. Inouye, and S. Inouye. 1984. Effects of deletion of the gene for the development-specific protein S on differentiation in Myxococcus xanthus. J. Bacteriol. 158:1195-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroos, L., and S. Inouye. 2008. Transcriptional regulatory mechanisms during Myxococcus xanthus development, p. 149-168. In D. E. Whitworth (ed.), Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
- 20.Lancero, H., N. B. Caberoy, S. Castañeda, Y. Li, A. Lu, D. Dutton, X. Y. Duan, H. B. Kaplan, W. Shi, and A. G. Garza. 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous and social motilities. Microbiology 150:4085-4093. [DOI] [PubMed] [Google Scholar]
- 21.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxocococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manoil, C., and D. Kaiser. 1980. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J. Bacteriol. 141:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manoil, C., and D. Kaiser. 1980. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J. Bacteriol. 141:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossa, F., M. E. Diodati, N. B. Caberoy, K. M. Giglio, M. Edmonds, M. Singer, and A. G. Garza. 2007. The Myxococcus xanthus Nla4 protein is important for expression of stringent response-associated genes, ppGpp accumulation, and fruiting body development. J. Bacteriol. 189:8474-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rippe, K., N. Mucke, and A. Schulz. 1998. Association states of the transcription activator protein NtrC from E. coli determined by analytical centrifugation. J. Mol. Biol. 278:915-933. [DOI] [PubMed] [Google Scholar]
- 28.Singer, M., and D. Kaiser. 1995. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633-1644. [DOI] [PubMed] [Google Scholar]
- 29.Søgaard-Andersen, L. 2008. Contact-dependent signaling in Myxococcus xanthus: the function of the C-signal in fruiting body morphogenesis, p. 77-91. In D. E. Whitworth (ed.), Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
- 30.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 32.Wu, S. S., and D. Kaiser. 1997. Regulation of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:4138-4144. [DOI] [PubMed] [Google Scholar]
- 34.Yang, R., S. Bartle, R. Otto, A. Stassinopoulos, M. Rogers, L. Plamann, and P. Hartzell. 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 186:6168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youderian, P., N. Burke, D. J. White, and P. L. Hartzell. 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49:555-570. [DOI] [PubMed] [Google Scholar]