Abstract
Mycobacterium tuberculosis EsxA and EsxB proteins are founding members of the WXG100 (WXG) protein family, characterized by their small size (∼100 amino acids) and conserved WXG amino acid motif. M. tuberculosis contains 11 tandem pairs of WXG genes; each gene pair is thought to be coexpressed to form a heterodimer. The precise role of these proteins in the biology of M. tuberculosis is unknown, but several of the heterodimers are secreted, which is important for virulence. However, WXG proteins are not simply virulence factors, since nonpathogenic mycobacteria also express and secrete these proteins. Here we show that three WXG heterodimers have structures and properties similar to those of the M. tuberculosis EsxBA (MtbEsxBA) heterodimer, regardless of their host species and apparent biological function. Biophysical studies indicate that the WXG proteins from M. tuberculosis (EsxG and EsxH), Mycobacterium smegmatis (EsxA and EsxB), and Corynebacterium diphtheriae (EsxA and EsxB) are heterodimers and fold into a predominately α-helical structure. An in vivo protein-protein interaction assay was modified to identify proteins that interact specifically with the native WXG100 heterodimer. MtbEsxA and MtbEsxB were fused into a single polypeptide, MtbEsxBA, to create a biomimetic bait for the native heterodimer. The MtbEsxBA bait showed specific association with several esx-1-encoded proteins and EspA, a virulence protein secreted by ESX-1. The MtbEsxBA fusion peptide was also utilized to identify residues in both EsxA and EsxB that are important for establishing protein interactions with Rv3871 and EspA. Together, the results are consistent with a model in which WXG proteins perform similar biological roles in virulent and nonvirulent species.
The WXG100 (WXG; pfam06013) proteins are a class of effector molecules found in gram-positive bacteria (26). WXG proteins are characterized by their small size (∼ 100 amino acids [aa]) and the presence of a WXG motif, or its structural equivalent, near the midpoint of their primary sequence (26). Bioinformatic analyses have shown that one WXG gene is frequently positioned near, or directly adjacent to, a second, related, WXG gene (14). The gene pairs characterized thus far encode proteins that associate to form 1:1 complexes (20, 31). The WXG proteins were once thought to be restricted to the mycobacteria, but homologues have now been detected in species of Bacillus, Listeria, Streptomyces, and Corynebacterium, among others, and the Pfam server lists >89 distinct WXG-encoding species and strains (10).
The identification of WXG proteins encoded by the pathogens Mycobacterium tuberculosis (15, 17, 19, 36), Mycobacterium marinum (13), and Staphylococcus aureus (5) has created significant interest in the proteins' biological activity. Nevertheless, these proteins are not a priori virulence factors (39), since organisms expressing WXG proteins are not necessarily capable of causing disease. In addition to pathogenesis, the WXG proteins are associated with processes as disparate as zinc homeostasis (24) and conjugal gene transfer (9, 11). A model for the mechanism(s) of action of these proteins that includes an explanation for their apparent functional versatility is at present lacking. One reason for this ambiguity may be the near-absence of studies comparing virulence-associated and non-virulence-associated WXG proteins, which is a goal of this study.
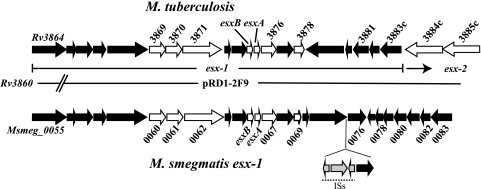
The M. tuberculosis secreted virulence factors EsxA (also called ESAT-6, or Rv3875) and EsxB (CFP-10; Rv3874) are the founding members of the WXG family, and M. tuberculosis derivatives defective in EsxA and EsxB are attenuated (17, 19, 36). The results of biochemical and structural studies indicate that EsxA and EsxB form a tightly associated heterodimer, EsxAB (25, 30, 31). The M. tuberculosis genome contains 23 WXG genes, named esxA to esxW, and the majority of these are expressed as tandem pairs (26). Of the pairs, five, including esxA and esxB, are contained within larger, highly conserved genetic loci, called esx-1 to esx-5 (Fig. 1). These loci have been the focus of much research, since mutants of esx-1 are attenuated, and esx-3 and esx-5 are necessary for in vitro growth of M. tuberculosis and M. marinum (1, 2, 32-34). The esx loci are proposed to encode secretory apparatuses dedicated to the secretion of their cognate WXG proteins (1).
FIG. 1.
Genetic map of the esx-1 loci of M. tuberculosis and M. smegmatis. The M. tuberculosis esx-1 genes discussed in the text are indicated by white arrows, as are their M. smegmatis homologues. The M. tuberculosis map also shows the Rv3884 and Rv3885 genes, which are part of the adjacent esx-2 locus. pRD1-2F9 is the cosmid that was used to create an esx-1-specific prey library. pRD1-2F9 includes the Rv3860 to Rv3885 genes, thus encompassing the entire esx-1 locus and part of esx-2. The four genes below the M. smegmatis map include defective insertion sequences (ISs) inserted into MSMEG_0075.
Although the majority of genes required for the secretion of the EsxAB heterodimer are encoded from within esx-1, additional non-esx-1 genes are necessary for secretion. In particular, one M. tuberculosis locus, esp, encodes three proteins essential for EsxAB secretion (12, 23). The first gene of the operon encodes a protein, EspA, that is cosecreted with EsxAB via the ESX-1 apparatus (12). Although no direct physical evidence has been presented, the inference from the interdependent cosecretion of the three proteins is that they likely form a complex, which is secreted by the ESX-1 apparatus. In this paper we provide the first genetic evidence that these three proteins interact.
The lack of a genetic assay for the study of ESX-1 activity in M. tuberculosis has hindered the identification of all of the protein components of the apparatus and all of the substrates that it secretes. However, the fast-growing, nonpathogenic organism Mycobacterium smegmatis has a conserved esx-1 locus that is essential for DNA transfer, and we have exploited this requirement for genetic studies (9). These analyses have shown that the M. smegmatis ESX-1 apparatus is functionally related to that of M. tuberculosis (11) and that M. smegmatis encodes non-esx-1 genes necessary for the secretion of the EsxAB heterodimer, including orthologues of EspA (9).
Here we have examined whether the secondary and quaternary structures of M. tuberculosis EsxA and EsxB are prototypical for other, functionally distinct and evolutionarily distant members of the WXG family (Fig. 2A). Comparisons were made to homologues encoded by M. smegmatis (esxA and esxB), Corynebacterium diphtheriae (esxA and esxB), and an additional non-virulence-related pair from M. tuberculosis (esxG and esxH, encoded from the esx-3 locus). Structural characterization of these proteins establishes that their secondary and quaternary structures are conserved, with each pair folding into a predominately α-helical structure and associating to form a heterodimer. We next devised and tested the utility of a novel strategy to identify proteins that interact specifically with these WXG heterodimers. This involved fusing EsxB and EsxA to create a biomimetic heterodimer for use in mycobacterial two-hybrid experiments. We reasoned that the use of this unique bait would allow the detection of proteins that interact with both components of the native heterodimer and that these proteins would normally go undetected in the conventional, single-protein two-hybrid screens. Indeed, using this approach, we identified novel protein partners of M. tuberculosis EsxBA (MtbEsxBA). We show for the first time that EspA proteins from M. tuberculosis and M. smegmatis interact with the EsxBA heterodimer (from both species) but not with EsxA or EsxB alone. We also provide evidence for promiscuity between the different M. tuberculosis ESX apparatuses by showing that EsxBA, encoded by esx-1, can interact with Esx proteins encoded by esx-2. Taken together, our studies suggest that the WXG proteins possess similar structures and properties, regardless of the host species and the apparent biological function.
FIG. 2.
Sequence alignment of WXG proteins characterized in this study and the strategy used to facilitate their expression. (A) Amino acid sequence alignment of four pairs of WXG proteins. Conserved sequences are in boldface, and the signature WXG motif is indicated with asterisks. Three residues in Rv3874 (EsxB) and a single residue in Rv3875 (EsxA) are underlined; they are the sites of amino acid substitutions discussed in the text that abrogate Rv3871 interactions. (B) (Bottom) Scheme for coexpression of tandemly arranged WXG genes. (Top) The ribbon cartoon (30) shows how the two monomers are freed from the expressed fusion protein by thrombin cleavage (scissors) at the peptide tether (balls and sticks).
MATERIALS AND METHODS
Enzymes and reagents.
Restriction and DNA-modifying enzymes were purchased from New England Biolabs, Inc. Restriction-grade thrombin was obtained from Novagen/EMD Biosciences, Inc. Genomic DNAs, extracted from heat-killed C. diphtheriae and M. tuberculosis strain H37RV, were kindly provided by Kimberlee Musser and Kathleen McDonough, respectively. M. smegmatis genomic DNA was isolated as described previously (18).
Vector construction.
The four WXG gene pairs were amplified by PCR using genomic DNA as a template. Each DNA fragment was used as a template for overlap extension (16), during which the two open reading frames were joined into a single gene; the DNA sequences of the resulting clones were verified. The 27-bp linker used for overlap extension encoded a thrombin-cleavable 9-mer, GLVPRGSTG. Open reading frames were fused in the native orientation for M. tuberculosis (esxHG, esxBA), and C. diphtheriae esxBA (6) but in the esxAB orientation for M. smegmatis. Fusion cassettes were cloned into pET22b (Novagen) in frame with the vector's C-terminal six-His tag, as an NdeI/XhoI fragment, or as an NdeI/NotI fragment for M. tuberculosis esxBA.
Overexpression and purification of WXG100 proteins as heterodimers.
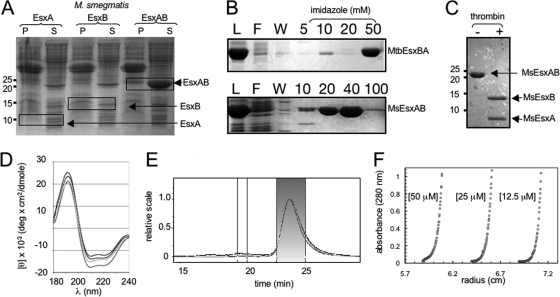
A 5-ml starter culture of BL21 Star (Invitrogen) cells carrying the expression plasmid was inoculated into 400 ml of broth, grown at 37°C to an optical density at 600 nm of 0.7, and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (3 × 10−4 M). Induced cells were incubated at 30°C for 4 h before they were harvested by centrifugation (6,000 × g, 10 min) and frozen at −70°C. Cell pellets were resuspended in 15 ml of a buffer containing urea (8 M), Tris-HCl (0.01 M), and NaH2PO4 (0.1 M) and were lysed by sonication with 10 to 15 bursts (10 s) at maximum power. The resulting lysate was cleared by centrifugation at 14,000 × g for 30 min and was applied to a 5-ml His-trap chelating column (GE Biosciences, Inc.). Fusion proteins eluted from the column at an imidazole molarity between 0.04 and 0.1 M (see Fig. 3B). Appropriate fractions were pooled and dialyzed into Tris buffer (0.02 M; pH 8) containing NaCl (0.05 M) at 4°C. Each of the four fusion proteins folded spontaneously during dialysis, as indicated by the circular dichroism (CD) spectra. Calculated extinction coefficients at 280 nm for the fusion proteins were 24,980 for MtbEsxBA, 32,430 for MtbEsxHG, 21,703 for M. smegmatis EsxAB (MsEsxAB), and 30,480 for C. diphtheriae EsxBA (CdEsxBA).
FIG. 3.
WXG protein pairs from different species have similar structural properties. (A) Enhanced overexpression in E. coli of the MsEsxAB fusion protein compared with the individual EsxA or EsxB protein. Protein extracts, soluble (S) and insoluble (P), collected from cells expressing MsEsxA, MsEsxB, or MsEsxAB, were separated on a 15% SDS-polyacrylamide gel, followed by staining with Fast Green (Sigma). The expected position for each protein is boxed, although EsxA and EsxB can be detected only by Western blot analysis. The sizes of protein standards (in kilodaltons) are given to the left of the gel. (B) Purification of the MtbEsxBA and MsEsxAB fusion proteins from E. coli using a 5-ml nickel-nitrilotriacetic acid affinity column and protein elution with imidazole. Protein samples of the load (L), the column flowthrough (F), the column wash (W), and imidazole elutions are indicated. (C) Proteolytic cleavage of purified MsEsxAB into MsEsxA and MsEsxB following overnight incubation with thrombin. Protein samples were separated on a 15% SDS-polyacrylamide gel and were then stained with Fast Green. (D) Overlay of CD spectra, obtained in aqueous solution, for MtbEsxBA (solid black), MsEsxAB (solid gray), MtbEsxHG (dashed line), and CdEsxBA (dotted line). (E and F) Representative quaternary-structure analyses of the complex formed from MtbEsxHG using SECMALLS (E) and AUC (F). (E) Data traces from the instrument's three in-line detectors, measuring the refractive index, light scattering, and UV absorbance (280 nm), are shown in arbitrary units. Molecular weight was determined for the major species using the data within the shaded area. (F) Three AUC equilibrium sedimentation profiles of equimolar mixtures of EsxG and EsxH at the indicated concentrations of total protein are shown. Absorbance values at 280 nm are plotted as a function of increasing radial position. See Table 1 for results.
Thrombin digestion.
Fusion proteins were cleaved site-specifically within the linker by digesting 1 mg of fusion protein with 1 μg of thrombin for 14 h at room temperature (Novagen) in Tris buffer (0.02 M; pH 8.4) containing NaCl (0.15 M) and CaCl2 (0.0025 M). The extent of digestion was >90%, as assessed by sodium dodecyl sulfate (SDS) gel electrophoresis.
SECMALLS.
For quaternary structure determination by size exclusion chromatography coupled to multiangle laser light scattering (SECMALLS), samples of proteins (200 μl) were dialyzed into Tris or HEPES buffer (0.01 M; pH 7.5) with 0.1 M NaCl and 5 mM dithiothreitol, filtered (pore size, 0.2 μm; Whatman Anotop syringe filter), and injected onto a Superdex 75 column (GE Healthcare) outfitted with an in-line 18-angle Wyatt Dawn Heleos laser light-scattering detector and a Wyatt Optilab rEX refractive index detector. Molecular masses were calculated from laser light-scattering data by using the ASTRA V (version 5.3.0.18) software package. The results were in reasonable agreement with results calculated by others (40), in which the ratio of light-scattering (LS) to refractive-index (RI) peak areas of the eluted solute is compared to a calibration curve comprising the LS/RI peak areas of RNase (14 kDa), ovalbumin (45 kDa), and bovine serum albumin (67 kDa).
CD spectroscopy.
CD spectra of samples containing equimolar mixtures of thrombin-cleaved proteins in phosphate buffer (0.05 M; pH 7.4) were acquired at 25°C using a JASCO J-715 spectropolarimeter equipped with a PTC-423S Peltier temperature control unit. Spectra were analyzed with JASCO CDPro software (version 1.53.01).
Analytical ultracentrifugation (AUC).
Sedimentation equilibrium studies were carried out using a Beckman XL-1 ultracentrifuge. Data from equilibrium experiments performed at 4 and 20°C, with rotor speeds of 16,000 rpm or 22,000 rpm, were collected after equilibrium was attained. Equilibrium was reached after ∼12 h, as indicated by examination of successive scans taken at 90-min intervals. Blank values for analysis of the centrifugation data were obtained from the absorbance in the reference cell, and from a reading of the absorbance in the sample cell after prolonged overspeed that removed all of the protein to the bottom of the cell. MtbEsxHG, MsEsxAB, and CdEsxBA were analyzed at each of three protein concentrations, 45, 22.5, and 11.5 × 10−3 M. MtbEsxBA was analyzed only at 45 ×10−3 M.
Protein-protein interaction studies.
Protein-protein interaction studies were performed as described previously using the mycobacterial protein fragment complementation (M-PFC) vectors pUAB300 and pUAB400 (35). We have consistently observed that, for reproducible and robust results, it is important to use colonies freshly transformed with prey or bait plasmids. To ensure uniform inoculation of plates, transformants were resuspended in 50 μl of broth before either spreading of a loopful, or spotting of 10 μl, of the cell suspension onto trimethoprim (Tp)-containing medium. Tp concentrations used in the medium ranged from 25 μg/ml to 75 μg/ml. Screens were generally performed using Tp at 25 μg/ml, but interactions were then confirmed at higher concentrations. In general, positive interactions (as scored by colony growth) were seen at all Tp concentrations, but the growth was less robust at 75 μg/ml. Individual prey clones of esx-1 genes were tested directly against a tripartite fusion of the M. tuberculosis genes esxB and esxA and the F[3] region of the murine dhfr gene. For construction of the esx-1 minilibrary, cosmid pRD1-2F9 (28), which encompasses Rv3860 to Rv3885, was partially digested with AciI. After digestion, DNA fragments were purified using Size Separation 400 spin columns (GE Healthcare, formerly Amersham) according to the supplier's instructions. Following purification, the DNA was ligated with either pUAB400 or pUAB300, which had been previously digested with ClaI. The ligation products were transformed into EP-Max 10B T1-resistant competent cells (Bio-Rad). The resulting colonies (5.34 × 105) were scraped off the plates, and plasmid DNA was extracted. This esx-1 minilibrary was then electroporated into mc2155 expressing the relevant bait protein, which was plated directly onto a medium containing Tp at 25 μg/ml. Clones exhibiting positive interactions were purified, and the interaction was confirmed, before DNA sequence analysis was performed to identify the gene encoding the interacting protein.
Forced misincorporation mutagenesis of MtbesxBA.
The esxBA gene fusion was subjected to random mutagenesis using the GeneMorph II random mutagenesis kit (Stratagene). The mutation frequency was controlled by manipulating the template concentration and the number of cycles during PCR; a final concentration of 10 ng/μl of DNA was used in the reaction, which ran for 23 cycles. The mutagenized PCR products were gel purified and digested with ClaI and MfeI and were then ligated into pUAB400. The ligation products were transformed into Mach1-T1 chemically competent Escherichia coli cells (Invitrogen) to generate a library of 3.2 ×103 transformants. The transformant colonies were pooled, and plasmid DNA was isolated (Qiagen). To estimate the overall level of mutagenesis, plasmid DNAs from 10 of the original transformants were isolated and subjected to DNA sequencing; 75% of the clones had one to two mutations in esxBA. The library was transformed into M. smegmatis containing pUAB300 expressing an Rv3871-dihydrofolate reductase (DHFR)-prey fusion. Kmr Hygr transformants were then screened on a Tp-containing medium to identify noninteracting (Tps) clones. Tps clones were restreaked to confirm their mutant phenotype. To identify the mutations, the esxBA gene was PCR amplified from genomic DNA, and the product was used directly for DNA sequence analysis. Before subsequent screens against other prey constructs, the Rv3871-pUAB300 plasmid was cured from the strain by passaging without selection.
RESULTS
Improved expression of WXG100 proteins by covalent linking.
Previous attempts to overexpress EsxA or EsxB proteins of M. tuberculosis individually in E. coli were hampered by technical difficulties (poor expression, formation of inclusion bodies, and proteolysis), which resulted in low yields of protein (31). We experienced similar difficulties with the M. smegmatis homologues and therefore elected to express the two proteins together by fusing the respective genes into a single open reading frame, which we hypothesized would facilitate folding and dimerization without aggregation (Fig. 2B). The resulting polypeptide included a thrombin-sensitive 9-mer (GLVPRGSTG) as the linker, and a six-His tag located at the C terminus for ease of purification. Model building using the nuclear magnetic resonance (NMR) structure (30) of the M. tuberculosis EsxAB heterodimer suggested that a 9-mer was sufficient to connect the disordered C terminus of EsxB to the equally disordered N terminus of EsxA, while still allowing the two halves to interact. Four WXG gene pairs were tested: M. tuberculosis esxBA (to encode MtbEsxBA), M. tuberculosis esxHG (MtbEsxHG), M. smegmatis esxBA (these genes were cloned in their nonnative orientation to encode MsEsxAB), and C. diphtheriae esxBA (CdEsxBA). The gene fusion approach was successful in producing robust overexpression of all four proteins (Fig. 3A; also data not shown). High yields of soluble, recombinant protein (ranging from 25 to 125 mg protein/liter of cells [Fig. 3B]) were achieved for each fusion protein following application of the cell lysate to a Ni2+ affinity resin. This fusion approach should dramatically simplify structural and biochemical studies of related protein pairs from the WXG family.
CD spectroscopy confirms that the purified proteins are folded into similar α-helical structures.
A number of approaches were used to confirm that all four overexpressed fusion proteins were folded in a native conformation. Each fusion protein was subjected to thrombin digestion, which cleaved the engineered protein linker between the two fused proteins into the two constituent proteins (Fig. 3C). The digested products remained soluble and stable at 4°C for months, suggesting that the heterodimers were properly folded.
As a more direct test of folding, and as a prelude to subsequent in vivo functional assays, covalently fused and thrombin-cleaved protein pairs were analyzed by CD spectroscopy. The CD spectra for the four pairs are virtually superimposable (Fig. 3D) (this is also true for uncleaved proteins [data not shown]). The spectrum for each pair indicates a folded structure, with the positions of the extrema and the associated amplitudes characteristic of a predominantly α-helical structure, consistent with the four-helix structure of the MtbEsxAB heterodimer (30).
Conserved secondary and quaternary structures for WXG100 protein dimers.
The molecular weight of each purified, thrombin-digested protein pair was determined by AUC and SECMALLS. The results from the two analyses were in excellent agreement, indicating that stable heterodimers formed between each protein pair (Table 1 and Fig. 3E and F). No higher-order multimers were detected in either of these assays. The heterodimerization of the M. tuberculosis protein pairs (EsxBA and EsxHG) is in agreement with the findings from genetic and biochemical studies, which have shown that the two pairs of proteins interact and form stable heterodimers (20, 21, 31, 37). Thus, our CD results for the four WXG100 protein pairs, combined with the AUC and SECMALLS data, suggest that the NMR structure of MtbEsxAB is an excellent prototypical model for all other WXG heterodimers.
TABLE 1.
WXG100 protein pairs associate stably as heterodimers
The MtbEsxBA fusion mediates robust protein-protein interactions.
Two-hybrid systems used for the detection of protein-protein interactions in vivo traditionally use a single protein, or a segment of a protein, as the bait for a second protein segment (the prey). A drawback to this approach is the assumption that the single segment of the protein—in an isolated, nonnative context—will fold properly and will thereby interact with a protein partner. It also assumes that a prey protein interacts only with a single protein, and not with multiple proteins within a complex. This is particularly problematic in two-hybrid analyses conducted in a nonnative system, where the expressed bait cannot associate with its usual partners. Thus, some protein-protein interacting partners are likely to be missed in standard two-hybrid systems. Since our data indicated the EsxBA fusion protein was folded in its native state, we reasoned that it could be used as a protein bait, which would obviate some of the drawbacks mentioned above and would therefore facilitate the development of a more comprehensive mycobacterial protein-protein interaction network.
A DHFR based two-hybrid system adapted for use with mycobacteria has been described previously (35). In this M-PFC system, interactions between test proteins (bait and prey) allow the functional reconstitution of DHFR and result in Tp resistance (Tpr). This genetic screen, with MtbEsxB as the bait, identified and verified interacting protein partners that included FtsQ, ClpC1, Rv2240, and ESX-1 components EsxA and Rv3871 (35).
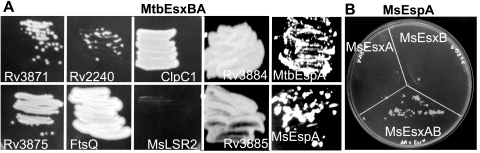
A tripartite-bait protein, consisting of the fused M. tuberculosis EsxB and EsxA proteins joined to the F[3] domain of DHFR, was expressed; the feasibility of the tripartite bait was then tested in pairwise combinations with known EsxB-interacting proteins (Fig. 4A). In all cases, Tpr colonies resulted, indicating maintenance of protein-protein interactions, while no Tpr colonies were obtained with control plasmids expressing no prey (see Fig. 5) or a noninteracting prey (MsLSR2). The ability of the MtbEsxBA fusion-protein to interact with the same prey as EsxB (32; also data not shown) supports our hypothesis that MtbEsxBA is folded in its native heterodimeric state and confirms our experimental rationale: the MtbEsxBA fusion protein acts as a biomimetic of the natural MtbEsxBA heterodimeric protein.
FIG. 4.
Protein-protein interactions mediated by virulence factors EsxAB and EspA. (A) Mycobacterial two-hybrid analyses indicate that the MtbEsxBA heterodimeric bait interacts with M. tuberculosis prey proteins previously shown to interact with MtbEsxB (32), which we confirmed independently (data not shown). MsLSR2 is a negative control encoding a histone-like protein cloned from M. smegmatis (K. Nguyen, unpublished data). Rv3871, Rv2240c, ClpC1, and FtsQ all interact with MtbEsxBA. The plates shown contain Tp at 50 μg/ml. Protein interactions between MtbEsxAB and MtbEspA, and with the M. smegmatis homologue MsEspA, are also shown. Two esx-2-encoded proteins, Rv3884 and Rv3885, which were identified by screening a library of clones encompassing esx-1 and part of esx-2, also interact with MtbEsxBA (Table 2). (B) MsEspA interacts specifically with MsEsxAB but not with MsEsxB or MsEsxA alone.
FIG. 5.
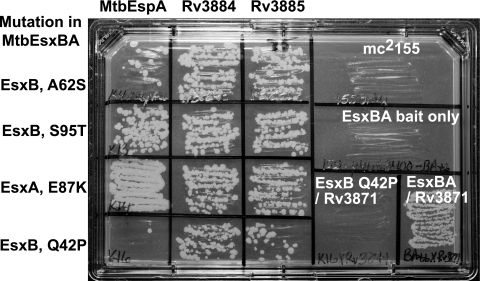
Abrogation of specific protein-protein interactions by mutations in the EsxBA fusion protein. Results of a two-hybrid analysis comparing interactions between the four different mutant MtbEsxBA fusion proteins discussed in the text (listed on the left) and three different prey proteins (MtbEspA, Rv3884, and Rv3885). (Right, top and center) Lack of growth on trimethoprim (75 μg/ml) medium of the host strain, mc2155, with and without the MtbEsxBA-pUAB400 vector. (Right, bottom) Control growth obtained when the MtbEsxBA bait is coexpressed with the Rv3871 prey and the effect of the Q42P mutation in EsxB. This mutation also disrupts interaction with EspA but not with Rv3884 or Rv3885 (compare growth in the bottom left panels). In contrast, the EsxB(S95T) and EsxA(E87K) mutants, which are defective in Rv3871 interaction (see Fig. S1 in the supplemental material), retain their abilities to interact with EspA (center left panels).
The MtbEsxBA fusion interacts with EspA.
MtbEspA is an M. tuberculosis protein that is not encoded from within the esx-1 locus but is required for EsxBA secretion (12, 23). EspA is secreted in an EsxBA-dependent manner, and it has been proposed that they are cosecreted as a complex (12, 23). Earlier efforts to detect protein interactions between EspA and the individual EsxA and EsxB proteins using yeast two-hybrid studies were not successful (23, 35), nor were EspA interactions detected using EsxB as the bait in the M-PFC system against an M. tuberculosis genome prey library (35). We hypothesized that the inability to detect protein interactions with EspA was due either to EspA interacting with both proteins in the heterodimer or to the EsxB baits not being folded properly. We therefore examined the ability of EspA to interact with the biomimetic MtbEsxBA bait. We constructed two dhfr fusion derivatives of MtbespA, one encoding the full-length protein (392 aa), and one encoding the N-terminal 100 aa (this latter segment is predicted to fold similarly to WXG proteins). Tpr colonies were observed in cells expressing both EsxBA and the N-terminal WXG-domain, indicative of protein-protein interactions (Fig. 4A and 5). However, no interactions were observed between the EsxBA fusion and the full-length EspA protein (data not shown), perhaps because conformational constraints imposed by the significantly larger size of the full-length protein prevent productive protein-protein interactions that would allow the reconstitution of DHFR.
In recent studies from this laboratory, we have identified a gene that encodes an M. smegmatis EspA orthologue (MsEspA), which was identified by genetic analyses that linked it to DNA transfer genes and by bioinformatic studies suggesting it was an orthologue of MtbEspA (9). Using the tripartite two-hybrid approach, we observed a positive interaction between MsEsxAB and MsEspA, confirming a direct interaction between these proteins; in contrast, no interactions were detected between MsEspA and either MsEsxA or MsEsxB (Fig. 4B). Together these results indicate that an MsEsxAB-EspA interaction requires substantial protein-protein contacts, which are provided only by the MsEsxAB heterodimer.
We also examined the conservation, between pathogenic and nonpathogenic species, of functional protein-protein interactions. MsEspA was shown to interact with the MtbEsxBA bait but not with the individual MtbEsxA or MtbEsxB protein (Fig. 4A and data not shown). Cross-species interactions were also observed between Rv3871 and MsEsxAB and between their respective homologues MSMEG0062 and MtbEsxBA. In each case, interactions with the individual EsxA and EsxB protein, as opposed to the heterodimer, either were not detected or resulted only in weak growth on Tp-containing medium (data not shown). The cross-species interactions suggest that the conservation of these proteins extends beyond amino acid homology to include conservation of critical protein-protein interactions.
Exploiting MtbEsxAB to identify novel ESX protein interactions.
To identify other novel MtbEsxBA interactions, we constructed an M-PFC library derived from a cosmid DNA encompassing Rv3861 to Rv3885 (29), which includes the entire M. tuberculosis esx-1 locus (Rv3864 to Rv3883 [Fig. 1]). This library was composed of 5.34 × 105 independent transformants. To verify the veracity of the library, it was introduced into M. smegmatis cells containing an integrated vector expressing the MtbEsxB-DHFR bait, and Tpr colonies were selected. Consistent with previous studies using both mycobacterial and yeast two-hybrid systems, known EsxB-interacting clones encoding Rv3875 (EsxA), Rv3874 (EsxB), and segments of Rv3871, an FtsK/SpoEIIIE-like protein essential for ESX-1 activity, were identified (7, 35-37). Two novel interactions between EsxB and the uncharacterized alanine-rich proteins Rv3876 and Rv3878 were also detected; their identification is presumably a consequence of screening against a smaller, more defined esx-1-based library rather than a complete genomic library (Table 2).
TABLE 2.
Comparison of M. tuberculosis ESX proteins identified using either MtbEsxB or MtbEsxBA as an interacting bait with a library of prey clones generated from the esx-1 locus
| Protein | Isolationa with the following protein bait: |
Segment of prey proteinb interacting with: |
||
|---|---|---|---|---|
| MtbEsxB | MtbEsxBA | MtbEsxB | MtbEsxBA | |
| Rv3869 | N | Y | 72-342 | |
| Rv3870 | Y | Y | 62-747 | 62-332 |
| Rv3871 | Y | Y | 1-210 | 1-261 |
| EsxB (Rv3874, Cfp-10) | Y | N | 47-100 | |
| EsxA (Rv3875, Esat-6) | Y | N | 9-95 | |
| Rv3876 | Y | Y | 1-81 | 35-186 |
| Rv3878 | Y | Yc | 76-280 | 76-280 |
| Rv3884 | N | Y | 522-619 | |
| Rv3885 | N | Y | 138-211 | |
N, no (the M. tuberculosis protein was not isolated in the screen); Y, yes (prey clones expressing the indicated protein were isolated).
Numbers are amino acid positions. The segments indicated are the largest region identified by overlapping clones.
Rv3878 was also identified using MsEsxBA as the bait.
Once the complexity of the library was verified, it was introduced into M. smegmatis cells expressing the MtbEsxBA-DHFR bait. Importantly, the tripartite bait identified interactions with Rv3871, Rv3876, and Rv3878, as observed when EsxB only was used (Table 2). The MtbEsxBA interactions were located within the N-terminal 261 aa of Rv3871, while overlapping clones of both Rv3876 and Rv3878 identified interactions mediated by regions spanning aa 35 to 186 and 76 to 280, respectively; these are the same protein segments that were shown to interact with EsxB. Remarkably, in an independent screen using the M. smegmatis EsxAB fusion as the bait with the same library, an interacting clone expressing the same region of Rv3878 was also isolated. This latter observation reinforces the results described above and is consistent with the conservation of both protein sequence and protein-protein interactions across species. The specificity of these interactions was underscored by the fact that no interactions were detected between MtbEsxBA and either EsxB or EsxA (0/27 Tpr clones), while more than half of the interactions identified with MtbEsxB as the bait were with EsxA (19/33 Tpr clones).
Three novel interactions that were not identified by using just MtbEsxB as the bait were detected by using the MtbEsxBA bait. The first was with Rv3869, for which three independent and overlapping clones that spanned Rv3869 residues 72 to 342 were isolated. Rv3869 has no assigned ESX-1 function, although it is required for secretion of the EsxBA heterodimer in both M. tuberculosis and M. smegmatis, and it is predicted to be localized within the membrane via its N terminus. Interactions with both Rv3884 and Rv3885 were also detected. These two proteins are encoded from the esx-2 locus, which lies immediately adjacent to esx-1 in the M. tuberculosis genome; the Rv3884 and Rv3885 genes were, therefore, part of the library (Fig. 1). No function has been assigned to either Rv3884 or Rv3885; however, Rv3884 shares 30% identity at the amino acid level with the recently characterized ATPase Rv3868, which is an essential component of the ESX-1 apparatus (22). This is the first observation of an interaction between ESX-2 and ESX-1 proteins. Multiple clones expressing segments of the Rv3884 and Rv3885 genes were isolated, providing additional confidence in the protein interactions. Positive interactions were also observed using clones expressing the C-terminal half of each protein, while clones expressing the full-length proteins failed to grow on Tp-containing medium.
Exploiting the biomimetic fusion to identify key residues necessary for protein-protein interactions.
To further demonstrate the specificity of the interactions observed, and to provide insight into the key residues required for interaction, we used the MtbEsxBA fusion to screen for mutants that no longer interacted with Rv3871. An earlier study examining interactions with Rv3871 had used a targeted mutagenesis approach with just EsxB in a yeast two-hybrid system and had shown that the C-terminal tail of EsxB was important for interaction with Rv3871 and secretion (7). Here, error-prone PCR was used to introduce mutations into the MtbEsxBA gene fusion. Full-length PCR products were purified and cloned directly into the pUAB400 two-hybrid vector, which generated a library of 3.2 × 103 independent clones. DNA sequencing of 10 transformants showed that 75% of the clones contained at least one to two mutations in the esxBA gene. The mutant library was introduced into M. smegmatis containing the pUAB300-Rv3871 bait, and transformants were screened for lack of interaction (Tps). Approximately 7% (23/384) of the transformants were Tps, and these were picked and retested before the sequence of esxBA was determined. Consistent with the phenotype, many of the mutant genes contained multiple mutations and/or stop codons. Here we will focus on just four single point mutations to emphasize the specificity and utility of the MtbEsxBA fusion (Fig. 5). Three mutations were in esxB (A62S, Q42P, and S95T), and one was in esxA (E87K). Notably, Q42P is adjacent to the WXG motif of EsxB, and the S95T and E87K mutations are in the unstructured C-terminal tails of EsxB and EsxA, respectively (30). The abrogation of interaction with Rv3871 by single point mutations (Fig. 5; see also Fig. S1 in the supplemental material) indicated that the interactions we observed with the fusion protein were not due to nonspecific “sticky” interactions mediated by a misfolded MtbEsxBA protein. Moreover, this is the first description of a mutation in EsxA that disrupts interaction with Rv3871 (previously assumed to be mediated by EsxB).
We took advantage of the mutant EsxBA proteins to determine the role of the mutated residues in determining interaction specificity. The four mutants were screened against a panel of three prey proteins (EspA, Rv3884, and Rv3885) that we have demonstrated here interact specifically with the MtbEsxBA bait (Fig. 5). All four mutant proteins exhibited robust interactions with Rv3884 and Rv3885, demonstrating that the proteins must be natively folded and that the mutated residues do not contribute to interactions with either Rv3884 or Rv3885. The EsxB(S95T) and EsxA(E87K) mutant proteins also interacted with EspA, indicating that their defect is specific to Rv3871. This was also the case for EsxB(A62S), although we observed weaker growth on Tp-containing medium when this protein was coexpressed with EspA, in contrast to more-robust growth with the other two mutant proteins. However, the EsxB(Q42P) mutation abolished interaction with EspA, indicating that this amino acid contributes to both Rv3871 and EspA interactions. Together these results demonstrate the specificity of the EsxBA protein-protein interactions we observed and also the potential of using a biomimetic-heterodimeric bait in interaction screens.
DISCUSSION
Proteins of the WXG-family have received considerable attention because of their association with pathogenesis and their potential use as vaccine targets and as biomarkers for detection of infections (4, 26). Mycobacteria possess multiple copies of genes expressing these proteins, several of which are known to play important, but still poorly defined, roles in pathogenesis. The discovery that homologues are encoded by nonpathogenic species has made defining possible functional roles for the WXG proteins yet more problematic. Do the proteins mediate similar functions in pathogenic and nonpathogenic species? Our laboratory's interest in the role of these proteins in conjugal DNA transfer in M. smegmatis (9, 11) prompted us to examine their structural characteristics as a first step toward determining their biological role(s).
Conserved secondary and quaternary structures of virulence-associated and non-virulence-associated WXG100 proteins.
The present work describes the development of a two-hybrid system to identify novel protein-protein interactions with the heterodimeric EsxAB proteins, demonstrating that protein-protein interactions were conserved between species. Exploitation of the fusion of EsxA and EsxB into a single molecule also circumvented problems of low protein expression, protein insolubility, and stability. The high yields of protein now attainable are sufficient for both crystallographic and biochemical studies; we pursued the latter to demonstrate that four distinct pairs of WXG proteins, from three different species, exhibited similar secondary and quaternary structures.
M. tuberculosis EsxA and EsxB are factors required for the virulence of this pathogen and the most intensely studied members of the WXG family. M. tuberculosis EsxA is speculated to promote pathogenesis by (i) subverting host defenses through direct interactions with Toll-like receptor II (27) and (ii) by perturbing the membranes of acidic compartments of infected cells in which the pathogen is transiently contained (38). The results of biochemical and structural studies indicate that EsxA and EsxB form a tightly associated heterodimer, EsxAB (25, 30, 31). The strength of interaction of EsxA and EsxB has been measured by calorimetry and Trp fluorescence titration, both of which indicate that the Kd (dissociation constant) for the EsxAB complex is in the 10−9 M range (25, 31). The NMR solution structure of the heterodimer shows that EsxA and EsxB adopt a similar helix-loop-helix structure, with the canonical WXG motif located in the loop of each protein (30).
Despite the specialized host interactions associated with EsxA and EsxB, we find that their solution properties closely match those of the non-virulence-associated WXG proteins: MtbEsxHG, MsEsxAB, and a pair, CdEsxBA, encoded by an evolutionarily distant species (Fig. 3D and E). All four protein pairs (studied as both protein fusions and cleaved proteins) were predominantly helical in nature and were observed to exist as a single heterodimeric species by two independent molecular-weight sizing experiments. Thus, our data indicate that adaptations required for virulence functions of the WXG proteins do not include gross structural changes. Structure-function comparisons at a higher resolution may help us to understand the diverse roles evident for the WXG proteins. However, in light of the structural similarities described here, it seems more likely that the heterodimers, MtbEsxBA, MtbEsxHG, MsEsxAB, and CdEsxBA, will function by a related mechanism.
Exploitation of an EsxBA protein fusion for two-hybrid studies.
The lack of a genetic assay for the study of ESX-1 activity in a pathogen has hindered the identification of all the protein components of the apparatus and the substrates it secretes. One alternative approach is the two-hybrid screen, which was successfully adapted for use in mycobacteria to identify proteins interacting with EsxB (35). Despite this success, there are likely disadvantages associated with using EsxB or EsxA individually. EsxA and EsxB are each minimally structured when expressed in the absence of the other (25, 30, 31), a property that will undoubtedly influence the strength and specificity of protein-protein interactions. Moreover, if only one member of the pair is used as a bait, proteins that interact with EsxA and EsxB simultaneously (i.e., with the native EsxBA heterodimer) are less likely to be identified. Here, by using a fusion of two proteins, we describe a new twist to the traditional method of using a single protein, or protein segment, as a biological bait. We believe that the utilization of a fusion protein for protein-protein interaction studies has broad applications for the study of proteins that exist as dimers and for those proteins that carry out a biological function within a protein complex; our strategy allows the bait protein to present a native protein interface to prey proteins that recognize multiple proteins or that recognize structural folds induced upon dimerization. The EsxBA heterodimer bait identified novel protein interactions. These new interactions could be due either to increased stability of the bait or to the presence of native protein surfaces on the heterodimeric bait, which are absent on the more artifactual surfaces of the EsxA and EsxB baits used to date.
A protein prey library, generated from a cosmid encompassing the M. tuberculosis esx-1 region, allowed the identification of interactions between MtbEsxBA and Rv3869, Rv3884, or Rv3885 (Fig. 4A and 5; also data not shown). These interactions were not observed when the bait was EsxB alone, nor had they been detected in a previous genomewide screen (35). The Rv3884 and Rv3885 genes are actually within esx-2, which is located immediately adjacent to the 3′ end of esx-1 in M. tuberculosis, with Rv3884 and Rv3885 being the two most esx-1-proximal genes. Interaction with ESX-2 proteins was unexpected; the five M. tuberculosis esx loci have generally been assumed to mediate nonoverlapping functions. Perhaps these two ESX-2 proteins have maintained their ability to interact with EsxBA yet have diverged so as to acquire ESX-2-specific functions. The conservation of ESX1-ESX2 interactions supports the proposed evolutionary link between esx-1 and esx-2; duplication of esx1 resulted in the formation of esx3 and then esx2 (14).
The EsxBA bait was also used to determine residues responsible for interactions with Rv3871. In a very elegant series of experiments, targeted mutagenesis had been used to demonstrate that the seven terminal amino acids of EsxB (Cfp-10) were necessary and sufficient for ESX-1-mediated secretion via Rv3871 (7). In contrast, the approach described here allowed the interaction contributions of both EsxB and EsxA proteins to be examined without bias. A prediction of our screen was that mutations within the C-terminal 7 aa of EsxB would be isolated. Thus, it was satisfying to identify the S95T mutation of EsxB, which specifically abrogated interaction with Rv3871. An S95A substitution at this location had been shown to reduce Rv3871 interactions by 50% in a yeast two-hybrid screen but not to affect EsxA-EsxB dimerization (7). The other single mutations we described have not been described previously and map in both proteins. Site-directed mutagenesis had been used to examine the contributions of conserved residues of EsxA to secretion and virulence (3), but not Rv3871 interactions. The EsxA(E87K) mutation had the same phenotype as EsxB(S95T) in that it specifically prevented interaction with Rv3871. This is a particularly interesting finding, since previous studies have focused on how EsxB interacts with Rv3871 and have led to the proposal that only EsxB determines ESX-1-mediated secretion. EsxA(E87) is a highly conserved residue located in the unstructured C terminus of EsxA (30). This fact, combined with the phenotype of the E87K mutation, suggests that the C terminus of EsxA also contributes to the interaction with Rv3871, perhaps in a fashion similar to that of the C terminus of EsxB. The isolation of the EsxA mutant also underlines the benefit of our two-hybrid screen. We assume that the inability of previous studies to detect EsxA-Rv3871 interactions is due to the limitations of using either EsxB or EsxA, rather than the heterodimeric EsxBA bait.
Finally, we used the heterodimeric bait to demonstrate direct protein-protein interactions between the EsxBA and EspA proteins from both M. tuberculosis and M. smegmatis, interactions inferred from studies showing cosecretion of the M. tuberculosis proteins. The protein interactions shown here allow us to predict that MsEspA will be cosecreted with MsEsxAB. Notably, the EsxAB-EspA protein interactions were conserved across species. We also observed cross-species protein interactions between MSMEG0062 and the MtbEsxBA heterodimer (and their respective homologues, Rv3871 and MsEsxAB), indicating that these protein-protein interactions serve similar and important functional roles in each species.
Evidence of functional conservation of ESX-1 proteins has previously been inferred from the ability of the M. tuberculosis esx-1 locus to complement M. smegmatis transfer mutants (11) and from the ability of the M. smegmatis ESX-1 apparatus to secrete heterologously expressed EsxA and EsxB of M. tuberculosis (8). These observations are consistent with the idea that WXG proteins from different species retain a common function. One possibility is that EsxA and EsxB are not directly secreted but are instead structural subunits of the type VII apparatus; this role could be the unifying function of the WXG proteins (26). Alternatively, WXG proteins might serve as generic chaperones, escorting the “true,” function-specifying effectors, via the type VII apparatus (9). Attempts to determine the mode(s) of action of the WXG proteins, and to provide a more detailed model for the type VII apparatus, will be the subjects of forthcoming studies that will build on these biochemical and genetic observations.
Supplementary Material
Acknowledgments
We thank W. R. Jacobs for pRD1-2F9, J. Jaeger for help in modeling the thrombin linker in EsxAB, A. Verschoor for comments on the manuscript, the Wadsworth Center Biochemistry Core for advice and guidance in using CD spectroscopy, and the Molecular Genetics Core for DNA sequence analysis.
B.C. and K.N. were partly supported by NIH training grant 1T32AI05542901A1 and by NIH grants AI42308 and AI0666 (awarded to K.M.D.). A.C. was supported by AI0666. K.V. was supported by an NSF Research Experiences for Undergraduates fellowship. D.K.C. was supported by NIH training grant T32AI07041. A.J.C.S. was supported by NIH grants AI068928 and AI058131.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdallah, A. M., N. C. Gey van Pittius, P. A. Champion, J. Cox, J. Luirink, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, and W. Bitter. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883-891. [DOI] [PubMed] [Google Scholar]
- 2.Abdallah, A. M., T. Verboom, F. Hannes, M. Safi, M. Strong, D. Eisenberg, R. J. Musters, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, J. Luirink, and W. Bitter. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62:667-679. [DOI] [PubMed] [Google Scholar]
- 3.Brodin, P., M. I. de Jonge, L. Majlessi, C. Leclerc, M. Nilges, S. T. Cole, and R. Brosch. 2005. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J. Biol. Chem. 280:33953-33959. [DOI] [PubMed] [Google Scholar]
- 4.Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. [DOI] [PubMed] [Google Scholar]
- 5.Burts, M. L., W. A. Williams, K. DeBord, and D. M. Missiakas. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U. S. A. 102:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerdeño-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. G. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632-1636. [DOI] [PubMed] [Google Scholar]
- 8.Converse, S. E., and J. S. Cox. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 187:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coros, A., B. Callahan, E. Battaglioli, and K. M. Derbyshire. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 69:794-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U. S. A. 101:12598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortune, S. M., A. Jaeger, D. A. Sarracino, M. R. Chase, C. M. Sassetti, D. R. Sherman, B. R. Bloom, and E. J. Rubin. 2005. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 102:10676-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677-1693. [DOI] [PubMed] [Google Scholar]
- 14.Gey Van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guinn, K. M., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528-535. [PubMed] [Google Scholar]
- 17.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guérin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. U. S. A. 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guérin attenuation. J. Infect. Dis. 187:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lightbody, K. L., D. Ilghari, L. C. Waters, G. Carey, M. A. Bailey, R. A. Williamson, P. S. Renshaw, and M. D. Carr. 2008. Molecular features governing the stability and specificity of functional complex formation by M. tuberculosis CFP-10/ESAT-6 family proteins. J. Biol. Chem. 283:17681-17690. [DOI] [PubMed] [Google Scholar]
- 21.Lightbody, K. L., P. S. Renshaw, M. L. Collins, R. L. Wright, D. M. Hunt, S. V. Gordon, R. G. Hewinson, R. S. Buxton, R. A. Williamson, and M. D. Carr. 2004. Characterisation of complex formation between members of the Mycobacterium tuberculosis complex CFP-10/ESAT-6 protein family: towards an understanding of the rules governing complex formation and thereby functional flexibility. FEMS Microbiol. Lett. 238:255-262. [DOI] [PubMed] [Google Scholar]
- 22.Luthra, A., A. Mahmood, A. Arora, and R. Ramachandran. 2008. Characterization of Rv3868, an essential hypothetical protein of the ESX-1 secretion system in Mycobacterium tuberculosis. J. Biol. Chem. 283:36532-36541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGurn, J. A., S. Raghavan, S. A. Stanley, and J. S. Cox. 2005. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol. Microbiol. 57:1653-1663. [DOI] [PubMed] [Google Scholar]
- 24.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189:730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meher, A. K., N. C. Bal, K. V. Chary, and A. Arora. 2006. Mycobacterium tuberculosis H37Rv ESAT-6-CFP-10 complex formation confers thermodynamic and biochemical stability. FEBS J. 273:1445-1462. [DOI] [PubMed] [Google Scholar]
- 26.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 27.Pathak, S. K., S. Basu, K. K. Basu, A. Banerjee, S. Pathak, A. Bhattacharyya, T. Kaisho, M. Kundu, and J. Basu. 2007. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat. Immunol. 8:610-618. [DOI] [PubMed] [Google Scholar]
- 28.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 29.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533-539. [DOI] [PubMed] [Google Scholar]
- 30.Renshaw, P. S., K. L. Lightbody, V. Veverka, F. W. Muskett, G. Kelly, T. A. Frenkiel, S. V. Gordon, R. G. Hewinson, B. Burke, J. Norman, R. A. Williamson, and M. D. Carr. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24:2491-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, R. G. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 277:21598-21603. [DOI] [PubMed] [Google Scholar]
- 32.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 33.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeone, R., D. Bottai, and R. Brosch. 2009. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr. Opin. Microbiol. 12:4-10. [DOI] [PubMed] [Google Scholar]
- 35.Singh, A., D. Mai, A. Kumar, and A. J. Steyn. 2006. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U. S. A. 103:11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. U. S. A. 100:13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teutschbein, J., G. Schumann, U. Mollmann, S. Grabley, S. T. Cole, and T. Munder. 2009. A protein linkage map of the ESAT-6 secretion system 1 (ESX-1) of Mycobacterium tuberculosis. Microbiol. Res. 164:253-259. [DOI] [PubMed] [Google Scholar]
- 38.van der Wel, N., D. Hava, D. Houben, D. Fluitsma, M. van Zon, J. Pierson, M. Brenner, and P. J. Peters. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287-1298. [DOI] [PubMed] [Google Scholar]
- 39.Way, S. S., and C. B. Wilson. 2005. The Mycobacterium tuberculosis ESAT-6 homologue in Listeria monocytogenes is dispensable for growth in vitro and in vivo. Infect. Immun. 73:6151-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen, J., T. Arakawa, and J. S. Philo. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240:155-166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.