Abstract
The development of molecular tools allowed light to be shed on several widespread genetic mechanisms aiming at limiting the effect of molecular damage on bacterial survival. For some bacterial taxa, there are limited tools in the genetic toolbox, which restricts the possibilities to investigate the molecular basis of their stress response. In that case, an alternative strategy is to study genetic variants of a strain under stress conditions. The comparative study of the genetic determinants responsible for their phenotypes, e.g., an improved tolerance to stress, offers precious clues on the molecular mechanisms effective in this bacterial taxon. We applied this approach and isolated two heat shock-tolerant strains derived from Bifidobacterium longum NCC2705. A global analysis of their transcriptomes revealed that the dnaK operon and the clpB gene were overexpressed in both heat shock-tolerant strains. We sequenced the hspR gene coding for the negative regulator of dnaK and clpB and found point mutations affecting protein domains likely responsible for the binding of the regulators to the promoter DNA. Complementation of the mutant strains by the wild-type regulator hspR restored its heat sensitivity and thus demonstrated that these mutations were responsible for the observed heat tolerance phenotype.
Over the last few decades, genetic analysis of the bacterial stress response has revealed a panoply of mechanisms protecting the bacterial cell from deleterious molecular damage (13). Mostly performed on model organisms like Escherichia coli, these experiments identified, sometimes fortuitously, a set of genetic components involved in the cellular stress response. Despite the strikingly high level of genetic conservation and the widespread nature of these genes, variations on the same theme were the rule. Differences exist even between closely related organisms (23, 33). The study of these mechanisms and their comparison were possible thanks to the genetic tools available for these model organisms. For some bacterial taxa, the genetic toolbox is rather limited, restricting the investigations of the molecular basis of the stress response to speculative comparisons with well-established model systems. In these cases, the “omics” technologies (genomics, transcriptomics, proteomics, and metabolomics) are frequently proposed to address questions in genetically less treatable organisms. Unfortunately, correlations are not causal relationships. Due to the large data sets generated by “omics” technologies, these approaches frequently raise more hypothesis than they can rule out. This may be circumvented by applying strategies to narrow down the complexity of the obtained “omics” data set. In that respect, comparison of parental strains with spontaneous mutants showing a defined phenotype is an interesting approach. Direct comparison of the global gene expression profiles between a mutant strain and the wild-type (wt) strain might lead directly to the genetic differences underlying the phenotypes. Additionally, the collection of data from analyses of several independent mutants allows identification of their common genetic determinants likely associated with the investigated phenotype. The few remaining genetic associations can then be tested by classical molecular approaches, when they are available for use to study this organism.
Some bacterial species, including Bifidobacterium longum, are known to have poor tolerance to temperature increase, oxygen, or desiccation (35). However, even in such a species, variations in stress tolerance have been documented (19). Robust stress-resistant mutants arise, sometimes spontaneously (9, 20, 29, 30). Specific stress-tolerant mutants might thus be obtained by natural selection. In this process, a strain is subjected to alternating periods of normal growth conditions and stress conditions. If stress-adapted mutants appear, they quickly outcompete the parental strain, facilitating their isolation. Hence, such an iterative natural evolution process allows us to quickly obtain genetic variants that are less sensitive to stress than the original bacteria.
The genetic response of Bifidobacterium longum to heat shock (HS) was recently documented in its complexity (27, 31). When doing a global transcriptomic analysis by microarrays, HS altered the transcription of 46% of the genes analyzed. The contributions of the various parts of this complex molecular response still needs to be separately evaluated. However, this is an arduous task due to the limited efficacy of the molecular tools available for this genus (11).
In this work, we proposed to untangle the complexity of the heat shock response in B. longum by combining an iterative natural evolution process, as described above, with a comparative global transcriptome analysis. Eventually, we identified mutations in a regulator gene and demonstrated by gene complementation its implication in the observed heat tolerance.
MATERIALS AND METHODS
HS assay and natural evolution process for heat shock tolerance.
Each cycle of selection consisted in applying a HS stress of 13 min to 5 ml of a culture grown overnight in MRS medium (Becton Dickinson AG, Basel, Switzerland) containing 0.05% (wt/vol) cysteine (MRS-Cys medium). HS was performed by successive dilution of the culture; the culture was diluted 10 times in 45 ml of preheated medium and placed in a heated water bath. Similarly, the temperature was rapidly decreased by diluting the 50 ml with 225 ml of medium at room temperature (RT). The resulting culture was grown for 16 h in anaerobiosis (AnaeroGen; Oxoid AG, Basel, Switzerland) at 37°C (until stationary phase) before being submitted to a new cycle of selection. Finally, clones were isolated by double streaking to generate pure cultures from colonies obtained after HS. The isolated clones were checked for a phenotype at least as good as the one of the corresponding population. For this HS assay, 1.5 ml of a culture grown overnight was diluted in 13.5 ml of prewarmed MRS-Cys medium and incubated 13 min, and then 100 μl was cooled down in a dilution microtiter plate. During the selection process and for the HS assays, cell counts were performed before and after HS. Plates were incubated 48 h at 37°C before counting. The loss in viability is expressed in log units after dividing the post-HS count by the pre-HS count.
Strain identification by PCR.
The primer pair oNCC2705-A/oNCC2705-B was used for the molecular identification of B. longum NCC2705 and NCC2705 derivative strains. The design of these PCR primers was based on the genome sequences of B. longum NCC2705 (32). Thirteen primer pairs, amplifying chromosomal sequences related to the position of mobile elements, were challenged against a reference collection of 24 different strains from the same species (data not shown). This approach was previously mentioned elsewhere (17). The primer pair oNCC2705-A/oNCC2705-B, which targeted the position of the insertion sequence ISBlo1b, generated a PCR product only with the NCC2705 strains. PCR mixtures (25 μl) contained 10 pmol of each primer, each nucleotide at a concentration of 50 μM, 1× reaction buffer, 1.25 U of AmpliTaq Gold (Applied Biosystems), and a tiny portion of colonies directly added to the PCR mix. The PCR program was as follows: (i) 5 min at 94°C; (ii) 30 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C; (iii) 7 min at 72°C. Finally, 12 μl of PCR product was loaded onto a 1.5% agarose gel and identified at 500 bp.
Bacterial growth and HS for transcriptomic study.
Bacteria were grown in MRS-Cys medium at 37°C. Precultures were incubated in anaerobic conditions, whereas the cultures to be used in the HS experiment were grown in 0.5-liter Sixfors fermentors (Infors AG, Bottmingen, Switzerland) with stirring (150 rpm) in a CO2 atmosphere. All fermentations were performed in duplicate. In mid-exponential-phase (optical density at 600 nm of 0.7), bacterial cultures were sampled before HS; 25-ml samples were centrifuged at RT for 5 min at 3,600 × g. Then, the bacterial pellet was immediately frozen in liquid nitrogen and stored at −80°C. In parallel, a 200-ml aliquot was centrifuged, and the pellets were suspended in 90 ml of MRS-Cys medium prewarmed to 50°C. The concentrated bacterial suspension was incubated 7 min at 50°C. A 10-ml sample was collected and centrifuged, and the pellet was immediately frozen in liquid nitrogen. To determine the loss in viability, cell counts were performed from the two samplings. For stationary-phase sampling, the optical density at 600 nm of batch cultures grown in 10-ml tubes was monitored for 16 h. The cells were then harvested as described above.
RNA extraction and hybridization on microarrays.
RNA extraction and quality checking were performed as previously described (26) except an additional DNase I treatment (Ambion) was performed before using the RNeasy kit (Qiagen). Agilent 60-mer oligonucleotide microarrays were designed with four or five probes per gene (Agilent Technologies Inc.). RNA labeling and cDNA synthesis were carried out using the 3DNA Array 900 MPX Genisphere kit (Genisphere Inc., Hatfield, PA) combined with the in situ hybridization kit plus (Agilent Technologies Inc.). The hybridization conditions and slide washing were performed as follows. To the 20 μl of cDNA (obtained with the Genisphere kit), 25 μl of the control target (Agilent), 60 μl of H2O, and 105 μl of Agilent buffer were added. After 10 min of incubation at 80°C and 15 min at 65°C, the reaction mixture was loaded on a prewarmed slide in the hybridization chamber and incubated for 16 h at 65°C, at 4 rpm in an Agilent oven. The cDNA hybridization mixture was washed for 10 min at 42°C with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.005% Triton X-100 and washed for 10 min at RT in 0.2× SSC and 0.00016% Triton X-100. The 3DNA hybridization was carried out as described in the Genisphere protocol except for the hybridization mixture volume, which was increased to 204 μl, and the washing was modified as follows: 10 min at 65°C in 2× SSC and 0.0016% Triton X-100, 5 min at RT in 2× SSC and 0.0016% Triton X-100, and 10 min at RT in 0.2× SSC and 0.00016% Triton X-100.
Microarray data analysis.
The slides were scanned at 10 μm with a Scanarray 4000 (Packard Biochip Technologies, Billerica, MA), and the data were extracted with Imagene 5.6 (Biodiscovery, El Segundo, CA). Data were treated with homemade scripts in Python language (www.python.org) and a local installation of the ArrayPipe web server (15). Probes showing a signal smaller than twice the standard deviation of the local background were considered without signal. Probes showing no signal or saturated signals in both channels were discarded from the analysis. Assuming an intensity-dependent variation in dye signal, (limma) loess global normalization was applied to the signal ratios. Having several probes per gene, we summarized the results for each gene as follow. Within each hybridization data set, the change in expression for the gene was given by the median of the corresponding probe values. The average intensity was calculated based on the same “selected” probes. Between hybridizations, the gene values are given by the mean of the gene values obtained from each hybridization. Results used in this study are based on four replicates, two biological replicates and two technical replicates. The data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE14628 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14628).
Gene amplification and sequencing.
Genomic DNA was extracted following the procedure previously described (12). Genes were amplified from genomic DNA with Expand long template PCR system (Roche, Germany) and sequenced on both strands by Fasteris (Fasteris SA, Geneva, Switzerland). Gene sequences were analyzed by BLAST (1).
HspR cloning and genetic complementation.
The hspR gene of B. longum NCC2705 and NCC2912 were amplified from genomic DNA with Expand high-fidelity PCR system (Roche, Germany) (primers listed in Table 1). The 600-bp amplicons of strain NCC2705 or NCC2912 were cloned into the XhoI and HindIII sites of the pGUSA plasmid (18) to obtain plasmids pDM12 and pDM13, respectively. Similar clonings in pGUSC (18) gave plasmids pDM14 and pDM15, respectively. The DNA sequences were verified by sequencing (Fasteris). Plasmids were transformed into strains NCC2705 and NCC2912. DNA manipulations, plasmid isolation, and transformation of E. coli and B. longum were performed as previously referred (18). High transcriptional levels were obtained with pDM12 and pDM13. Medium or low levels of transcription were obtained with pDM14 and pDM15 in the presence of glucose or raffinose, respectively.
TABLE 1.
Primers used in this study
| Primer | Sequencea | Purpose, use, or description |
|---|---|---|
| pHspR-F | TGCCCGATGTGAGATAACAA | hspR sequencing |
| pHspR-R | AAAAAGCCGGCGACTATCTC | hspR sequencing |
| pBL0516F1 | CCCGGGCTCGAGATGGCGCGGTTAGCCAACC | Cloning XhoI |
| pBL0516R1 | CCCGGGAAGCTTTCACCAACCCCACAGGACC | Cloning HindIII |
| pBL0516_F1_qPCR | CCAACATTCACCCCCAGACT | qPCR |
| pBL0516_R1_qPCR | TGCGCTGCGGACGAA | qPCR |
| pBL0520_F1_qPCR | TCATCAAGGGCGACCGTAAG | qPCR |
| pBL0520_R1_qPCR | TCATGATGCCACCCTTGGT | qPCR |
| pBL1250_F1_qPCR | GCGTCCTGAACGAAATCAAGA | qPCR |
| pBL1250_R1_qPCR | TGGTGTGAATCTCGTCGATGA | qPCR |
| pBL0118_F1_qPCR | TCTGCTGCCACCGAACTG | qPCR |
| pBL0118_R1_qPCR | CCCCAAATAATCTGGGCTTCA | qPCR |
| pBL0301_F1_qPCR | CAACCGCCGCGATCTTC | qPCR |
| pBL0301_R1_qPCR | CCAGCTGTGAAAGCAACGTATT | qPCR |
| oNCC2705_A | TCCAGATCATTTCCGATTCC | Strain-specific primer |
| oNCC2705_B | CGGCGTATTTCTATCGCATC | Strain-specific primer |
The XhoI and HindIII restriction sites used for cloning are underlined.
Gene expression measurement by qPCR.
Three biological replicates of cultures in stationary phase were analyzed by quantitative PCR (qPCR) as previously described (27). Results were normalized with the housekeeping genes BL0301 and BL0118. The primers designed with the Primer Express software (Applied Biosystems, Foster City, CA) are reported in Table 1. Results were analyzed by analysis of variance with three factors, complementation, biological replicate, and genes.
RESULTS
Natural selection of heat shock-tolerant mutants.
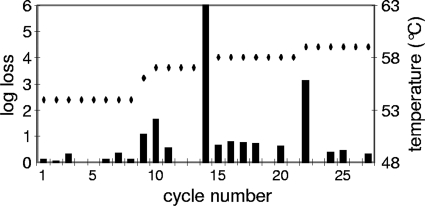
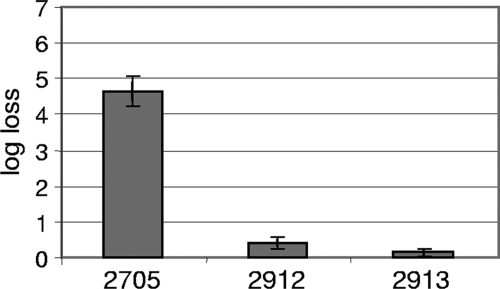
The HS tolerance of the B. longum strain NCC2705 was determined in liquid medium in stationary phase (data not shown). The sublethal temperature for a 13-min HS was measured as 53°C, and the lower limit of detection of our setup (6-log-unit loss) was reached at 62°C. Then, on the basis of these borderline cases, we performed an iterative selection of naturally occurring HS-resistant mutants. In order to do so, the bacteria were allowed to grow under normal conditions (nonharmful) until they reached stationary phase, when they were subjected to deleterious HS treatments. In serial passages starting at 54°C, the first batch was progressively brought to 59°C in a total of 27 cycles. Figure 1 shows that each temperature increase dramatically affects the bacterial viability. Then it rapidly returned back to a level close to the initial situation, indicating that the batch was rapidly enriched in HS-tolerant mutants for this temperature. From the population obtained after 27 cycles, we isolated a new strain, which we named NCC2912. With the second batch (data not shown), we started the selection at 60°C and observed a regular loss in viability of 5 log units at each cycle. After 10 cycles, the loss suddenly dropped close to zero, indicating that the population was considerably enriched in bacteria with improved resistance to HS. We then performed 10 additional cycles at 62°C, which first led to a loss in viability of 5 log units and then ended up with a steady 3-log-unit loss. A new strain, NCC2913, was isolated from this batch. In order to distinguish between a physiological adaptation and a genetic adaptation, more than 100 generations of subculturing were performed before submitting the strains to a new HS selection. The heat-tolerant phenotype of both strains was still observed (data not shown). This supports a genetic origin for this new trait, rather than a transient adaptation. The viability of strains NCC2912 and NCC2913 was subsequently tested at 59°C for 13 min and compared to the parent wt strain (Fig. 2). Both mutants showed an impressive survival improvement (4 to 5 log units) compared to the wt strain, with NCC2913 performing slightly better. To confirm the derivation of the two new strains from the original wt strain, they were tested by PCR with primers designed for NCC2705-specific genomic regions (see Materials and Methods for more details on this approach). Both strains yielded the expected PCR product, showing that they were NCC2705 derivatives (data not shown).
FIG. 1.
Cyclic selection of naturally occurring heat shock-tolerant mutants. The black bars represent the loss in viability at each heat shock treatment, expressed as a logarithm of the loss in viability (left-hand y axis). The limit of detection was reached at cycle 14. The black diamonds show the temperature of selection for each cycle (right-hand y axis).
FIG. 2.
Characterization of the newly isolated strains by measurement of their loss in viability after a 13-min heat shock at 59°C. The strains are shown in the figure without the NCC prefix. Strain NCC2705 is the wt strain. Strains NCC2912 and NCC2913 are independent heat shock-tolerant mutants. The bars show the mean values from 7 to 15 replicates (error bars show the 95% confidence intervals).
Transcriptomic characterization of the HS-tolerant derivative strains NCC2912 and NCC2913.
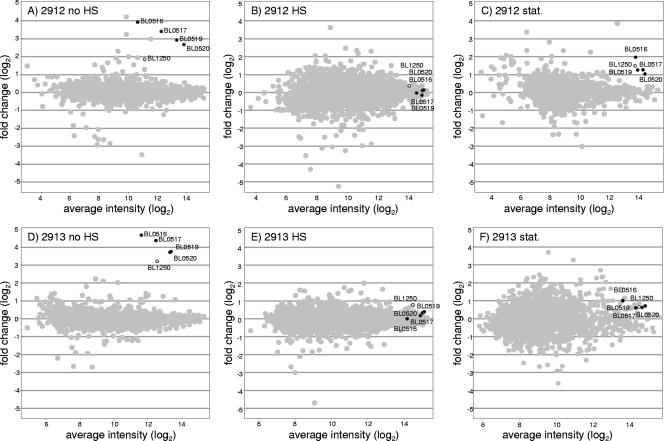
We hypothesized that the increased heat resistance of the new strains resulted from a difference in the expression of genes involved in the stress response, compared to the parental strain previously investigated (27). Whole-genome expression analysis of the mutants versus that of the wt NCC2705 would then focus on what make these strains more resistant to HS. In the documented HS study (27), a NCC2705 liquid culture was challenged in exponential phase at 50°C. In order to guarantee that the transcriptomic differences observed in the HS-tolerant mutants were relevant to their improved heat resistance, their phenotypes were validated in the same conditions: after 10 min of HS at 50°C, the viability of strain NCC2705 started an exponential decline and lost 2 log units after 50 min, whereas the viability of strains NCC2912 and NCC2913 remained unaffected (data not shown). For their transcriptomic analysis, the three strains were grown separately at 37°C in fermentors and harvested in mid-exponential phase with or without being subjected to HS, as previously described (27). Then, by microarray hybridizations, we compared the whole transcriptomes of these strains under two conditions, no HS and 7-min HS (Fig. 3A, B, D, and E).
FIG. 3.
Global transcriptome analysis of the heat shock-tolerant strains compared to the wt strain NCC2705. The change in expression versus average intensity is plotted in gray for each gene, except for the dnaK operon (black) and the clpB gene (white). Measurements were performed in exponential phase before (A and D) or during (B and E) heat shock treatment and in stationary phase (stat.) (C and F) for both mutants versus the wt strain. The strains are shown in the figure without the NCC prefix. The BL numbers are gene locus tags.
This comparative design, with the parental strain NCC2705 as a reference, allowed direct highlighting of the most striking difference observed in the transcriptomes of the mutants: the overexpression of the dnaK operon and the clpB gene. Figure 3A and D show the change in expression of all genes in the absence of HS. Among the few genes differentially transcribed, all the genes of the dnaK operon (BL0516 to BL0520) were overexpressed by at least 6-fold in strain NCC2912 and at least 13-fold in strain NCC2913. Similarly, the overexpression of the clpB gene was more pronounced in NCC2913 (9.3-fold) than in NCC2912 (3.6-fold). When measured after a 7-min HS, the transcription levels of these genes in the mutant strains and the wt strain proved to be similar, as shown in Fig. 3B and E. These two observations mean that in both mutant strains, the dnaK operon and the clpB gene are constitutively expressed at a level corresponding to the HS response of the wt strain.
Considering that we selected the HS-tolerant mutants in stationary phase, it seemed important to show that the mutants were also overexpressing dnaK and clpB in this growth phase. Thus, transcriptomes from cells harvested in stationary phase were analyzed to compare mutant strains NCC2912 and NCC2913 versus the wt strain NCC2705 (Fig. 3C and F). Although the upregulation of the dnaK operon and the clpB gene appeared lower than in the mid-exponential phase (approximately 1.5- to 4-fold instead of 4- to 25-fold), both mutants still expressed more of these chaperones than the wt strain. Overexpression of dnaK operon and clpB in strain NCC2912 was also documented by proteomics (14).
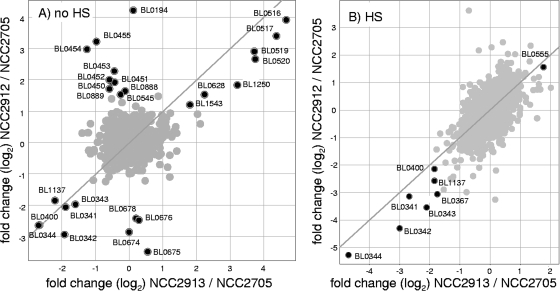
In addition to this convergence of dnaK-clpB overexpression between the two HS-tolerant mutants, other differences in gene expression exist compared to the wt strain transcriptome. Some are only observed in one of the mutants. Figure 4, which is based on the same data set as Fig. 3, compares the most pronounced up- and downregulated genes of the two strains in fermentor experiments, without HS (Fig. 4A) or with HS (Fig. 4B). In this unusual representation of transcriptomic data, the diagonal line clearly shows the positions of genes with similar transcript levels in both strains. In Fig. 4A and B, on the bottom left of the diagonal lines, a group of four genes organized in an operon (BL0341-BL0344) and coding for an ABC transporter is downregulated in both mutants, as well as a transcription regulator (BL1137) and a conserved hypothetical protein (BL0400). On the right part of the diagonal lines, the levels of transcription of an aspartate aminotransferase (BL0628) and an endo-beta-xylanase (BL1543) are slightly higher in both mutants and in both conditions. Observed below the left diagonal line in Fig. 4B, a phage-related transcription regulator (BL0367) is less transcribed in both mutants during HS. Strikingly, the only gene found more than threefold upregulated in both mutants upon stress conditions is htrA (BL0555), coding for a serine protease (Fig. 4B, right of the diagonal line). It is remarkable that, among 30 classical stress-related genes found in B. longum (27), the few showing an altered expression level in the HS-tolerant strains behave similarly in both mutants (the dnaK-clpB regulon and htrA). As shown in Fig. 4A, some alterations of transcription level are found in only one of the mutants. The putative fimbrial operon (BL0674-BL0678) is downregulated only in strain NCC2912. Two genes involved in potassium uptake (BL0194 and BL0545), a cluster of ABC transporter proteins associated with three membrane proteins (BL0450 to BL455), and a group of slightly upregulated ABC transporter components (BL0887 to BL0889) are upregulated only in strain NCC2912.
FIG. 4.
Comparison of the changes in gene expression obtained with the two heat shock-tolerant mutants versus the wt strain. The change in expression for one strain versus the change in expression for the other strain is plotted in gray for each gene, except for genes showing more than threefold differences of transcript level in at least one of the strains (A) or in the two strains (B), which are plotted in black. Measurements were performed in exponential phase before (A) or during (B) heat shock treatment. The diagonal line depicts the position of genes showing the identical change in expression in both strains.
Sequencing of the hspR gene in NCC2912 and NCC2913 mutants.
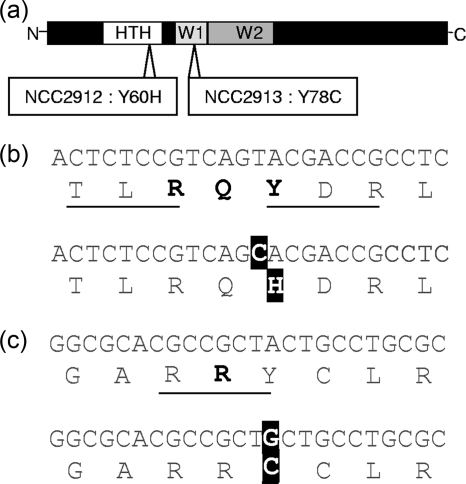
The coregulation of the dnaK operon and the clpB gene were previously documented in other members of the subclass Actinobacteridae (5) and proposed in B. longum (27). Considering the constitutive upregulation of these two features in our HS-tolerant strains, we hypothesized that some mutations affecting the transcriptional regulator HspR occurred in both mutants. Therefore, we PCR amplified the hspR gene of both strains and compared their whole sequences with the wt gene. In strains NCC2912 and NCC2913, we found point mutations modifying the amino acid sequence in the helix-turn-helix motif or the winged 1 motif of HspR, respectively (Fig. 5).
FIG. 5.
Positions of the mutations in the HspR regulator. (a) Schematic representation of the protein with the functional domains and the amino acid substitutions. N, N terminus; HTH, helix-turn-helix domain; W1 and W2, winged helix domains 1 and 2, respectively; C, C terminus. (b and c) Sequences of the mutations in strains NCC2912 (b) and NCC2913 (c). Underlined amino acids were conserved in Actinobacteridae. The boldface amino acids indicate that the residue interacts with the DNA molecule. Mutations in the DNA and protein sequences are indicated by white letters on black background.
Restoration of heat sensitivity by hspR complementation.
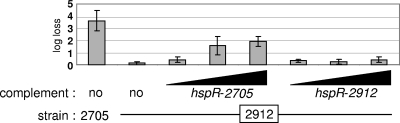
To demonstrate that the observed mutations in the hspR gene were responsible for the improved resistance to heat, we complemented the NCC2912 mutant with the wt hspR gene or mutated hspR from strain NCC2912. For each hspR version, the PCR-amplified gene was cloned into two vectors and under the control of previously characterized promoters (18). One promoter shows a constitutive strong expression level, whereas the second promoter has a mild to low expression level, depending on the carbon source of the medium (glucose or raffinose). All in all, three levels of gene expression were available for this complementation of mutant strain NCC2912 by the wt hspR gene or the NCC2912 mutated hspR gene. Similarly, we complemented the wt strain NCC2705 with the same constructions. The viability of all these complemented strains was measured after heat shock. Complementation with the empty vector was without effect (data not shown). Independently of the level of expression and the version of the hspR gene, strain NCC2705 always showed a higher sensitivity to heat stress when complemented. This sensitivity was slightly more pronounced for the wt HspR (data not shown). Complementation of the NCC2912 mutant by an increasing amount of wt HspR partially restored its sensitivity to heat in a dose-responsive way, whereas the mutated hspR showed only a weak effect, independently of its expression level (Fig. 6).
FIG. 6.
Loss in viability of the complemented strains after a 13-min heat shock at 59°C. The strains are shown in the figure without the NCC prefix (NCC2705 is the wt strain, and NCC2912 is the complemented heat shock-tolerant mutant). The black triangles depict an increased expression of the wt HspR (hspR from strain NCC2705 [hspR-2705]) or mutated HspR (hspR from strain NCC2912 [hspR-2912]) obtained by three transcriptional levels of promoters. The bars show the means from four or five replicates (error bars show the 95% confidence intervals).
Restoration of genetic negative control by hspR complementation.
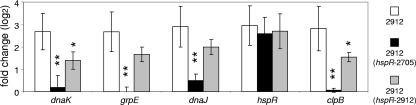
To demonstrate that the increased sensitivity to heat was linked to the genetic negative control brought by the hspR complementation, samples were harvested in stationary phase and prepared for transcriptomic analysis by qPCR (only the cultures complemented with the highest expression vector). Figure 7 shows the transcription levels for the genes in the dnaK operon and for the clpB gene of the mutant NCC2912, complemented or not, versus the wt NCC2705. In the mutant NCC2912, all the tested genes showed an increase of mRNA of more than 6.5-fold. When complemented with the wt hspR, all the genes tested except hspR were significantly downregulated to a level very close to that measured in the wt NCC2705. In the mutated hspR complementation, we observed an intermediate transcription level; the genes had a twofold downregulation compared to the mutant without complementation. In all cases, hspR mRNA was seen as highly present, which is not surprising, considering that the qPCR primers did not differentiate between mRNA from the chromosomal or plasmid copies.
FIG. 7.
Analysis by qPCR of the dnaK-clpB regulon transcription level in the complemented strains in stationary phase without heat shock. The bars show the means from three biological replicates of the mutant strain NCC2912 without complementation (2912), with the highest expression of the wt HspR (hspR from strain NCC2705 [hspR-2705]) or the mutated HspR (hspR from strain NCC2912 [hspR-2912]) (error bars show the 95% confidence intervals). Results are expressed in comparison with three biological replicates of the wt strain NCC2705. Significant differences from the value for the wt strain are indicated when the P value is lower than 0.05 (*) or 0.01 (**).
DISCUSSION
In light of the limited genetic tools available for Bifidobacterium species (11, 18), the isolation of heat shock-tolerant mutants represented a convenient way to unravel the genetic mechanisms underlying the stress response in this genus. We proposed to identify the most significant contributors of the genetic stress response by direct comparison between spontaneous resistant mutants and the original wt strain. Thus, the mechanisms truly contributing to the stress tolerance were identified only in the resistant mutants, whereas indirect effects of stress were observed in both parties being compared. We used a transcriptomic approach with microarrays to compare the strains. To complete our reductive approach, we hypothesized that the differences observed in two independent mutants are consequences of convergent evolution and are genetic mutations affecting key mechanisms of the HS response.
In order to obtain bacterial strains with better resistance to stress, the selection of natural variants was previously applied to bifidobacteria, e.g., for resistance to cholate (20), acid (9, 29), or bile (30). With the current work, we applied a similar approach that may be seen as an accelerated evolution of a bacterial strain: sequential selection of naturally occurring mutants showing an adaptation to specific physiological conditions. Growing the bacteria under normal (nonharmful) growth conditions between stress-induced selections allows the natural diversity of the bacterial culture to increase and for alternative stress-adapted solutions (“trajectories”) to appear and to persist (10). Another case deserving attention are survivors, which simply represent extreme cases of physiological adaptation to stress, instead of mutants (4). Their growth under nonharmful conditions allows them to behave exactly as the original culture and therefore to be eliminated at the next selection step. Better-adapted mutants rapidly outcompete the initial bacteria and are then easily isolated.
Such an iterative evolution process was previously used with Lactobacillus delbrueckii to improve survival to freezing (21). Our selection of heat shock-tolerant mutants similarly targeted a technological property. However, the genetic response of B. longum to HS is likely to confer a broader stress resistance, as shown by its numerous classical stress-associated genes induced by heat treatment (27, 31). The high complexity of the bifidobacterial HS response resembles a genetic stampede for survival. This observation is consistent with the stable temperature of the known ecological niche of B. longum, the mammalian gut (26).
During our evolution process, we applied two strategies, stepwise increased selective pressure or steady high selective pressure. In both cases, we obtained HS-tolerant mutants. However, when in the latter case we increased the selective pressure to 62°C, the isolation of a mutant showing a survival rate similar or better to that of the whole improved population was laborious (data not shown). Most of them were less resistant to HS than expected, although largely improved compared with the initial strain. This denoted higher population diversity and dynamic in this batch with harsher stress. This is an expected consequence of the nonharmful growth passages. Indeed, at high constraint, fitness gains obtained by beneficial mutations tend to represent a minor advantage (10). Due to the nonharmful growth passages, which further decrease the competitive advantage of the descendants showing higher fitness, several solutions to the selective pressure may coexist.
Analysis of the two HS-tolerant mutants showed convergent evolution. The genes coding for the chaperone proteins, DnaK, DnaJ, GrpE, the protease ClpB, as well as the transcriptional regulator HspR were found constitutively strongly upregulated. In fact, their level of expression corresponds to that observed in the wt strain after a HS induction, indicating an absence of negative regulation. Considering that the selection of the HS-tolerant mutants occurred in stationary phase, where an increased expression of the dnaK operon was observed (16), we verified that expression of these genes was also higher in the mutants in stationary phase. The relatively mild upregulation of the dnaK-clpB regulon that we observed by microarray analysis is explained by an artifact of measurement, which is frequent with this technology: the dynamic range of expression ratios tends to be lower for genes with high expression on both channels (34). The results obtained by qPCR confirmed this hypothesis: in the mutant NCC2912, all the genes tested show an increase of mRNA of more than 6.5-fold, instead of 2- to 3-fold as measured by microarrays. A comparison of the proteomes of strain NCC2912 and strain NCC2705 in stationary phase (14) undermines the hypothesis that the improved fitness of the mutant results from the accumulation of the DnaK and ClpB proteins during exponential phase. However, in the cellular machinery, DnaK is associated with the synthesis of a significant fraction of protein molecules (7). Consequently, growth in the presence of a constant high level of this chaperone may considerably influence the folding quality of many proteins and therefore their resistance to stress.
Apart from expression of the dnaK operon and clpB gene, other differences were observed in the mutants. Surprisingly, eight of them were clearly shared by the two independent mutants in exponential phase. For most of these differences, e.g., a complete ABC transporter operon, it is most likely an adaptive effect. Indeed, genetic adaptations may be accompanied by the selection of other mutations to fine-tune the level of expression of proteins indirectly affected by the initial mutation (3). Another noticeable convergent upregulation concerned htrA after 7 min of HS. HtrA is a widely distributed serine protease (25), which is strongly induced by HS in B. longum (27, 31). Again, its upregulation may be an indirect effect of the dnaK-clpB modulation, which could interfere with the expression of htrA and its proteolytic processing normally undergone during HS in B. longum (31). In any event, in this case, it would clearly contribute to the fitness of the mutants. Interestingly, other proteins whose expression was strongly promoted by HS in bifidobacteria, e.g., the Hsp20 chaperone (27, 36), were not upregulated in the two mutants. This shows that a high level of expression during HS does not necessarily entail that the corresponding protein plays a crucial role in the bacterial stress response.
In Streptomyces coelicolor, HspR is a regulator of HS gene expression which has been experimentally shown to be involved in the regulation of the dnaK operon and clpB (5, 6). The product of hspR, which is part of the dnaK operon, binds to the promoter of the dnaK operon and clpB to negatively regulate their expression. On the basis of the crystal structures of other MerR family members (24), a HspR model was previously proposed for several members of the subclass Actinobacteridae, including B. longum (37). HspR contains a DNA-binding motif belonging to the winged helix-turn-helix family and forms active multimers. Since the whole dnaK and clpB machinery of S. coelicolor has well-conserved homologues in bifidobacteria (37, 38) and the HS response in B. longum was consistent with a similar coregulation of dnaK and clpB by HspR (27), our microarray results prompted us to search for mutations impairing the functioning of HspR in our mutants. In both cases, we found one point mutation affecting tyrosine residues of the DNA-binding domain. Both tyrosines were well-conserved in Actinobacteridae, and for one of the two tyrosines, putatively involved in the direct interaction with the DNA molecule (24, 37).
To show to which extent the dnaK and clpB upregulation was responsible for HS tolerance through the failure of HspR in DNA binding, we genetically complemented the NCC2912 mutant by controlled expression of the wt hspR gene or the mutated (NCC2912) hspR gene on a plasmid vector. At a high level of expression, the mutated HspR variant caused partial repression of the dnaK-clpB regulon. Thus, the mutated regulator retains weak activity, despite its mutation in the DNA-binding domain. In the HS tolerance assay, this remaining activity is reflected in a hardly detectable decrease in survival of the complemented mutant. Following the wt hspR complementation, we expected that the potential multimerization of the wt protein with the cellular mutated HspR would have titrated its efficacy. However, the wt HspR fully restored the negative control on dnaK and clpB. Strikingly, when complemented with the wt hspR gene, the mutant still survived better than the wt strain. To explain this improved survival, we might consider the possibility that the HspR regulation was not restored to its full complexity. Though the quantity of plasmid-expressed wt HspR might be sufficient to exert its genetic control in the absence of stress, its expression is not regulated by HS. During HS, the mutated HspR produced by the cellular dnaK operon might multimerize with the wt HspR and thus weaken the normally loop-controlled negative regulation of the operon. However, it is most likely that other genetic modifications exist in the NCC2912 mutant that could explain the remaining HS tolerance. Since this mutant was obtained by stepwise HS temperature increases, which were clearly followed by progressive HS tolerance increases, it is expected that this hspR mutation is not the only stress-related genetic modification.
In Campylobacter jejuni (2) and Helicobacter pylori (28), HspR inactivation was shown to affect the motility of the bacteria and consequently their ability to invade humans. Therefore, it would be interesting to study whether the inactivation of HspR in our mutants would also affect their interaction with the host. Notably, one of our mutants showed a downregulation of genes similar to those involved in fimbria biogenesis of Actinomyces naeslundii and Actinomyces viscosus (17, 32). We also showed in this work that the hspR mutants express more DnaK chaperone protein than the wt strain does. According to a recent publication, this protein is present in large amounts on the cell surface and may be responsible for the activation of the plasminogen system of the host (8). This effect would be more pronounced with our DnaK-overexpressing mutants. Heat shock proteins are also known for their immunomodulator properties. In Mycobacterium tuberculosis, DnaK was reported for its immunosuppressive properties (22), which also suggests a new potential of our mutant strains for health-promoting applications.
Acknowledgments
We thank Enea Rezzonico for fruitful discussions on the initial idea of this work and on the results of the transcriptomic analysis.
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, M. T., L. Brondsted, B. M. Pearson, F. Mulholland, M. Parker, C. Pin, J. M. Wells, and H. Ingmer. 2005. Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151:905-915. [DOI] [PubMed] [Google Scholar]
- 3.Babu, M. M., and L. Aravind. 2006. Adaptive evolution by optimizing expression levels in different environments. Trends Microbiol. 14:11-14. [DOI] [PubMed] [Google Scholar]
- 4.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 5.Bucca, G., A. M. Brassington, G. Hotchkiss, V. Mersinias, and C. P. Smith. 2003. Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol. Microbiol. 50:153-166. [DOI] [PubMed] [Google Scholar]
- 6.Bucca, G., Z. Hindle, and C. P. Smith. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J. Bacteriol. 179:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukau, B., E. Deuerling, C. Pfund, and E. A. Craig. 2000. Getting newly synthesized proteins into shape. Cell 101:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Candela, M., S. Bergmann, M. Vici, B. Vitali, S. Turroni, B. J. Eikmanns, S. Hammerschmidt, and P. Brigidi. 2007. Binding of human plasminogen to Bifidobacterium. J. Bacteriol. 189:5929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collado, M. C., and Y. Sanz. 2007. Induction of acid resistance in Bifidobacterium: a mechanism for improving desirable traits of potentially probiotic strains. J. Appl. Microbiol. 103:1147-1157. [DOI] [PubMed] [Google Scholar]
- 10.Elena, S. F., and R. E. Lenski. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457-469. [DOI] [PubMed] [Google Scholar]
- 11.Fisseha, M., and F. Arigoni. 2005. Beyond genome sequences: approaches to genome-wide analysis of gut bacteria, p. 97-128. In G. W. Tannock (ed.), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 12.Germond, J. E., O. Mamin, and B. Mollet. 2002. Species specific identification of nine human Bifidobacterium spp. in feces. Syst. Appl. Microbiol. 25:536-543. [DOI] [PubMed] [Google Scholar]
- 13.Gross, C. A. 1996. Function and regulation of the heat shock protein, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 14.Guillaume, E., B. Berger, M. Affolter, and M. Kussmann. 2009. Label-free quantitative proteomics of two Bifidobacterium longum strains. J. Proteomics 72:771-784. [DOI] [PubMed] [Google Scholar]
- 15.Hokamp, K., F. M. Roche, M. Acab, M. E. Rousseau, B. Kuo, D. Goode, D. Aeschliman, J. Bryan, L. A. Babiuk, R. E. Hancock, and F. S. Brinkman. 2004. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 32:W457-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klijn, A. 2005. Physiological and molecular characterisation of stress response in Bifidobacterium longum NCC2705. Ph.D. thesis. University College Cork, Cork, Ireland.
- 17.Klijn, A., A. Mercenier, and F. Arigoni. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491-509. [DOI] [PubMed] [Google Scholar]
- 18.Klijn, A., D. Moine, M. Delley, A. Mercenier, F. Arigoni, and R. D. Pridmore. 2006. Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl. Environ. Microbiol. 72:7401-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian, W. C., H. C. Hsiao, and C. C. Chou. 2002. Survival of bifidobacteria after spray-drying. Int. J. Food Microbiol. 74:79-86. [DOI] [PubMed] [Google Scholar]
- 20.Margolles, A., L. Garcia, B. Sanchez, M. Gueimonde, and C. G. los Reyes-Gavilan. 2003. Characterisation of a Bifidobacterium strain with acquired resistance to cholate—a preliminary study. Int. J. Food Microbiol. 82:191-198. [DOI] [PubMed] [Google Scholar]
- 21.Monnet, C., C. Beal, and G. Corrieu. 2003. Improvement of the resistance of Lactobacillus delbrueckii ssp bulgaricus to freezing by natural selection. J. Dairy Sci. 86:3048-3053. [DOI] [PubMed] [Google Scholar]
- 22.Motta, A., C. Schmitz, L. Rodrigues, F. Ribeiro, C. Teixeira, T. Detanico, C. Bonan, H. Zwickey, and C. Bonorino. 2007. Mycobacterium tuberculosis heat shock protein 70 impairs maturation of dendritic cells from bone marrow precursors, induces interleukin-10 production and inhibits T-cell proliferation in vitro. Immunology 121:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Newberry, K. J., and R. G. Brennan. 2004. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 279:20356-20362. [DOI] [PubMed] [Google Scholar]
- 25.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 26.Parche, S., M. Beleut, E. Rezzonico, D. Jacobs, F. Arigoni, F. Titgemeyer, and I. Jankovic. 2006. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188:1260-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezzonico, E., S. Lariani, C. Barretto, G. Cuanoud, G. Giliberti, M. Delley, F. Arigoni, and G. Pessi. 2007. Global transcriptome analysis of the heat shock response of Bifidobacterium longum. FEMS Microbiol. Lett. 271:136-145. [DOI] [PubMed] [Google Scholar]
- 28.Roncarati, D., A. Danielli, G. Spohn, I. Delany, and V. Scarlato. 2007. Transcriptional regulation of stress response and motility functions in Helicobacter pylori is mediated by HspR and HrcA. J. Bacteriol. 189:7234-7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez, B., M. C. Champomier-Verges, M. del Carmen Collado, P. Anglade, F. Baraige, Y. Sanz, C. G. de Los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2007. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 73:6450-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez, B., M. C. Champomier-Verges, B. Stuer-Lauridsen, P. Ruas-Madiedo, P. Anglade, F. Baraige, C. G. de Los Reyes-Gavilan, E. Johansen, M. Zagorec, and A. Margolles. 2007. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl. Environ. Microbiol. 73:6757-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savijoki, K., A. Suokko, A. Palva, L. Valmu, N. Kalkkinen, and P. Varmanen. 2005. Effect of heat shock and bile salts on protein synthesis of Bifidobacterium longum revealed by [35S]methionine labelling and two-dimensional gel electrophoresis. FEMS Microbiol. Lett. 248:207-215. [DOI] [PubMed] [Google Scholar]
- 32.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal, R., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Sharov, V., K. Y. Kwong, B. Frank, E. Chen, J. Hasseman, R. Gaspard, Y. Yu, I. Yang, and J. Quackenbush. 2004. The limits of log-ratios. BMC Biotechnol. 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2005. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J. Appl. Microbiol. 99:493-501. [DOI] [PubMed] [Google Scholar]
- 36.Ventura, M., C. Canchaya, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2007. Molecular characterization of hsp20, encoding a small heat shock protein of Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ventura, M., J. G. Kenny, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2005. The clpB gene of Bifidobacterium breve UCC 2003: transcriptional analysis and first insights into stress induction. Microbiology 151:2861-2872. [DOI] [PubMed] [Google Scholar]
- 38.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]