Abstract
To investigate the Na+-driven flagellar motor of Vibrio alginolyticus, we attempted to isolate its C-ring structure. FliG but not FliM copurified with the basal bodies. FliM proteins may be easily dissociated from the basal body. We could detect FliG on the MS ring surface of the basal bodies.
The basal body, which is the part of the rotor, is composed of four rings and a rod that penetrates them. Three of these rings, the L, P, and MS rings, are embedded in the outer membrane, peptidoglycan layer and in the inner membrane, respectively (1), while the C-ring of Salmonella species is attached to the cytoplasmic side of the basal body (3). The C-ring is composed of the proteins FliG, FliM, and FliN (25), and genetic evidence indicates that the C-ring is important for flagellar assembly, torque generation, and regulation of rotational direction (33, 34). FliG, 26 molecules of which are incorporated into the motor, appears to be the protein that is most directly involved in torque generation (15). Mutational analysis suggests that electrostatic interactions between conserved charged residues in the C-terminal domain of FliG and the cytoplasmic domain of MotA are important in torque generation (14), although this may not be the case for the Na+-type motor of Vibrio alginolyticus (32, 35, 36). FliM interacts with the chemotactic signaling protein CheY in its phosphorylated form (CheY-P) to regulate rotational direction (30). It has been reported that 33 to 35 copies of FliM assemble into a ring structure (28, 29). FliN contributes mostly to forming the C-ring structure (37). The crystal structure of FliN revealed a hydrophobic patch formed by several well-conserved hydrophobic residues (2). Mutational analysis showed that this patch is important for flagellar assembly and rotational switching (23, 24). The association state of FliN in solution was studied by analytical ultracentrifugation, which provided clues to the higher-level organization of the protein. Thermotoga maritima FliN exists primarily as a dimer in solution, and T. maritima FliN and FliM together formed a stable FliM1-FliN4 complex (2). The spatial distribution of these proteins in the C-ring of Salmonella species was investigated using three-dimensional reconstitution analysis with electron microscopy (28). However, the correct positioning has still not been clarified.
The Na+-driven motor requires two additional proteins, MotX and MotY, for torque generation (19-21, 22). These proteins form a unique ring structure, the T ring, located below the LP ring in the polar flagellum of V. alginolyticus (9, 26). It has been suggested that MotX interacts with MotY and PomB (11, 27). Unlike peritrichously flagellated Escherichia coli and Salmonella species, V. alginolyticus has two different flagellar systems adapted for locomotion under different circumstances. A single, sheathed polar flagellum is used for motility in low-viscosity environments such as seawater (18). As described above, it is driven by a Na+-type motor. However, in high-viscosity environments, such as the mucus-coated surfaces of fish bodies, cells induce numerous unsheathed lateral flagella that have H+-driven motors (7, 8). We have been focusing on the Na+-driven polar flagellar motor, since there are certain advantages to studying its mechanism of torque generation over the H+-type motor: sodium motive force can be easily manipulated by controlling the Na+ concentration in the medium, and motor rotation can be specifically inhibited using phenamil (10). Moreover, its rotation rate is surprisingly high, up to 1,700 rps (compared to ∼200 rps and ∼300 rps for Salmonella species flagella and E. coli flagella, respectively) (12, 16, 17).
Although understanding the C-ring structure and function is essential for clarifying the mechanism of motor rotation, there is no information about the C-ring of the polar flagellar motor of Vibrio species or the flagella of any genus other than Salmonella. Since Vibrio species have all of the genes coding for C-ring components, we would expect its location to be on the cytoplasmic side of the MS ring, as in Salmonella species. In this study, we attempted to isolate the polar flagellar basal body with the C-ring attached and investigate whether it is organized similarly to the H+-driven flagellar motor of Salmonella enterica serovar Typhimurium.
Isolation of the Na+-driven flagellar basal bodies from V. alginolyticus.
The C-ring of species of Salmonella has been isolated with the flagellar basal body using a previously established mild isolation protocol (3). In the purification of the Na+-driven flagellar basal bodies, we used MotY, which is an essential protein for rotation and a component of the T ring, as the marker protein for the basal body. To detect the C-ring during isolation, FliG and FliM were used as the marker proteins. The proteins were detected by immunoblotting using anti-MotY (MotYB0079) (31), anti-FliG (see below), and anti-E. coli FliM (gifts from David Blair, University of Utah) antibodies. The antibody raised against FliG of V. alginolyticus was prepared as follows. FliG was expressed in BL21(DE3)/pLysS cells harboring pJN309, which encodes His6-tagged FliG (FliG-His6) in pET3d. Cells were collected and broken by using a French press. FliG-His6 was purified from the cytoplasmic fraction using Ni-nitrilotriacetic acid (NTA) agarose (Qiagen), a HiTrap Q column (GE Healthcare), and Superdex 200HR 10/30 (GE Healthcare). Purified FliG was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), stained with Coomassie blue R250, and excised for immunization. Rabbit anti-FliG antibody was produced by Biogate Co., Ltd. (Gifu, Japan).
We applied the Salmonella protocol to V. alginolyticus strain KK148, which produces multiple polar flagella (13), expecting to isolate more basal bodies from KK148 than from the wild-type strain used previously (26). However, we could not detect any FliG and FliM signals in the basal body fraction. When the basal body fraction was observed by electron microscopy, we could detect the basal body structure, but no structures could be seen on the cytoplasmic side of the MS ring where the C-ring is expected to be located (data not shown). In this protocol, which was optimized to isolate the C-ring of Salmonella, the C-ring of V. alginolyticus seems to have dissociated from the basal body. We therefore suspected that the detergent, Triton X-100, is not suitable for maintaining the association between the Vibrio C-ring structure and the basal body.
Screening for optimal solubilization conditions for the flagellar basal body.
To find the optimal conditions for isolating the C-ring of V. alginolyticus, 72 detergents were screened at several concentrations. Cells from 100-ml cultures were harvested, and spheroplasts were prepared as described above. The spheroplast solution (1 ml) was transferred to an Eppendorf tube, and spheroplasts were lysed by adding detergent to a final concentration of around 0.5 to 2% (wt/vol) to each tube (Hampton Research). After the solution became clear, MgSO4 was added to a 10 mM final concentration, the mixture was incubated on ice for 30 min, and EDTA was added to 5 mM (final). Thus, unlysed cells and cell debris were removed by centrifugation at 6,000 × g for 5 min, and the supernatant fraction was subjected to ultracentrifugation at 100,000 × g for 30 min. The pellet was resuspended in TE buffer (10 mM Tris-HCl [pH 8.0], 5 mM EDTA) and SDS sampling buffer. The FliG and FliM proteins were analyzed by immunoblotting. FliG signals were detected when the following detergents were used: 0.1% Triton X-100, 0.5% CHAPS (3-[(3-chloramidopropyl)-dimethylammonio]-1-propanesulfonate), 1% octyl glucoside, 0.1% Fos-choline 12, 0.5% sodium cholate, and 0.5% sodium deoxycholate. FliM, however, was not detected by using any of the tested detergents. SDS-PAGE and immunoblots of fractions of CHAPS-solubilized samples are shown in Fig. 1. Solubilization with 0.5% (wt/vol) CHAPS seemed to be the best condition for isolating basal bodies containing FliG. CHAPS also gave good results for purifying the stator complex composed of PomA and PomB in V. alginolyticus (5).
FIG. 1.
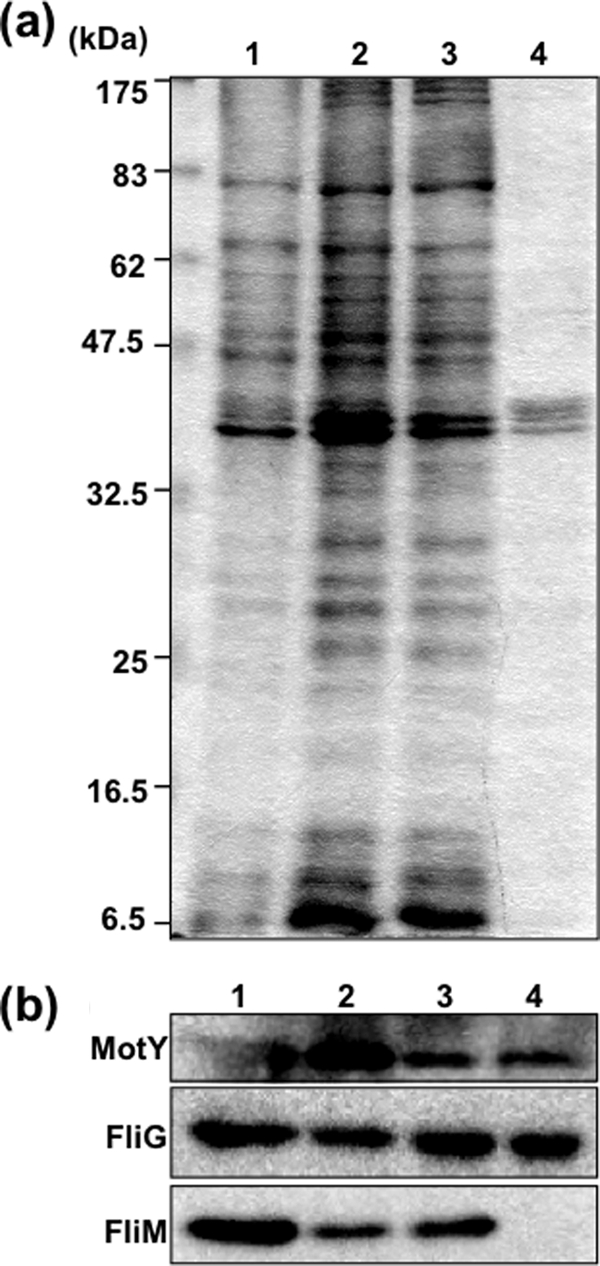
Isolation of V. alginolyticus basal bodies solubilized using CHAPS. (a) Coomassie blue-stained SDS-PAGE of proteins in the purification steps using CHAPS. Lane 1, whole-cell lysate. Lane 2, supernatant fraction from low-speed centrifugation. Lane 3, supernatant fraction from ultracentrifugation. Lane 4, precipitate fraction from ultracentrifugation. (b) The proteins in the purification steps were detected by immunoblotting using anti-MotY, anti-FliG, and anti-FliM antibodies. Fractionated samples were loaded in the same order as in panel a.
To improve the conditions used for isolating the C-ring, we examined the effects of several reagents on the isolation of FliG or FliM along with the basal bodies (data not shown). Prior to solubilization by 0.5% CHAPS, various reagents (KCl, NaCl, or glycerol) were added. FliM signals were not detected with the basal body fraction under any of the tested conditions. On the other hand, addition of 100 mM NaCl or 25% glycerol to the cell suspension resulted in stronger FliG signals.
FliG proteins associate with isolated Vibrio flagellar basal bodies.
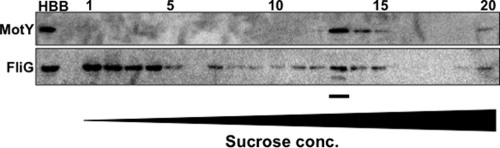
To confirm that FliG associates with the basal bodies, the basal body fraction obtained after solubilization by CHAPS with 100 mM NaCl was separated using sucrose density gradient centrifugation. To identify the fractions containing basal bodies, we used the T-ring protein MotY as the basal body marker. MotY was detected in fractions 13 to 15 (Fig. 2). FliG signals were detected in a broader range of fractions than MotY. FliG was detected in the upper fractions (1 to 5) and also in the fractions containing basal bodies (13 to 15) (Fig. 2). When the purified recombinant FliG proteins, which were prepared from BL21(DE3)/pLysS cells harboring FliG/pCold encoding His6-tagged FliG (His6-FliG) in pColdI, were fractionated in the same way, all FliG signals were detected in the upper fractions (data not shown).
FIG. 2.
Fractionation by sucrose density gradient centrifugation. The preparation of basal bodies was applied to a 20 to 60% (wt/wt) stepwise sucrose gradient in TEC (TE buffer containing 0.5% CHAPS). After centrifugation at 72,000 × g for 90 min at 4°C, the gradient was divided into 20 fractions from the top to the bottom. For observation by electron microscopy, basal bodies in some fractions were collected by ultracentrifugation at 100,000 × g for 30 min and the pellet was resuspended using TEC. The proteins in each fraction were separated by SDS-PAGE and detected by immunoblotting using anti-MotY and anti-FliG antibodies. HBB indicates the basal body fraction before the sucrose density gradient centrifugation. The underlined fraction was used for electron microscopy.
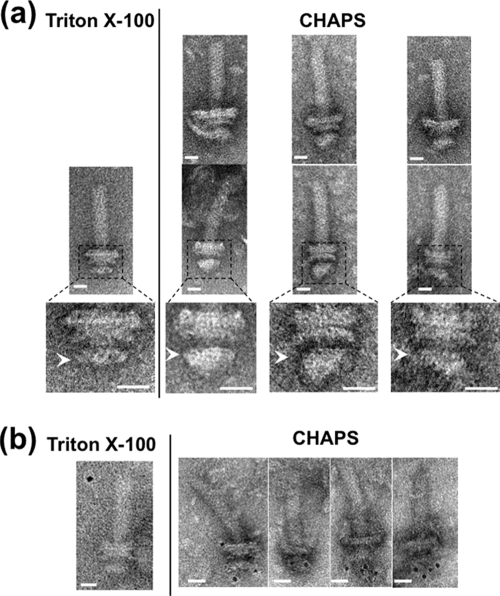
The fractions containing both MotY and FliG were observed by electron microscopy (Fig. 3). In these samples we could detect the basal bodies and an additional structure at the cytoplasmic side of the MS ring (Fig. 3a). We also identified a structural difference between isolated basal bodies solubilized by CHAPS and Triton X-100 (Fig. 3a, lower panels). When immunoelectron microscopic observation was performed using anti-FliG antibody followed by immunogold labeling, we could detect basal bodies with immunogold dots on the cytoplasmic side of the MS ring (Fig. 3b). This indicates that FliG assembled on the MS ring.
FIG. 3.
Electron micrographs of basal bodies isolated using CHAPS. (a) Comparison of electron micrographs of basal bodies isolated by CHAPS (right) and by Triton X-100 (left). Six representative images are shown for CHAPS-solubilized basal bodies in fraction 13 (Fig. 2) of sucrose density gradient (upper and middle). Enlarged images of the middle panel corresponding to the dotted frame are shown in the lower panels. Arrowheads indicate the MS rings. The samples containing flagellar structures were put on a hydrophilic-treated carbon-coated copper grid. The grids were washed with TE buffer, negatively stained with 3% uranyl acetate and observed with a JEM-1200EX or JEM-2010 electron microscope (JEOL, Japan). Bar, 20 nm. (b) Immunogold labeling of the basal bodies isolated by CHAPS (right) and by Triton X-100 (left) using anti-FliG antibody. 3 μl of the basal body suspension was mixed with 1 μl of buffer A (10 mM Tris, 5 mM EDTA, 600 mM NaCl at pH 8.0) and with 1 μl of the antibody solution diluted 20-fold with buffer B (10 mM Tris, 5 mM EDTA, 150 mM NaCl at pH 8.0). The mixture was kept on ice for 10 min. One μl of 5-nm colloidal gold-conjugated anti-rabbit IgG antibody diluted 2-fold with buffer C (10 mM Tris, 5 mM EDTA, 300 mM NaCl at pH 8.0) was then added to this mixture and it was left at room temperature for 30 min. Next, the mixture was put on a hydrophilic-treated carbon-coated copper grid. The grids were washed twice with buffer (10 mM Tris-HCl [pH 8.0], 5 mM EDTA), negatively stained with 3% uranyl acetate, and observed with a JEM-1200EX or JEM-2010 electron microscope (JEOL, Japan). Bar, 20 nm.
In this study, we attempted to isolate flagellar basal bodies from the Na+-driven polar flagellar motor of Vibrio alginolyticus, with the C-ring structure attached. All of the genes encoding C-ring components (fliG, fliM and fliN) identified in Salmonella exist in the genome of V. alginolyticus, and previous studies showed that GFP-fused FliG, which is functional, localized at the base of the single polar flagellum (4). Therefore the C-ring structure is believed to be located on the cytoplasmic side of the MS ring, and to have important roles in assembly, rotational switching and motor function, as has been shown in Salmonella and E. coli.
Stability of FliG with the basal body.
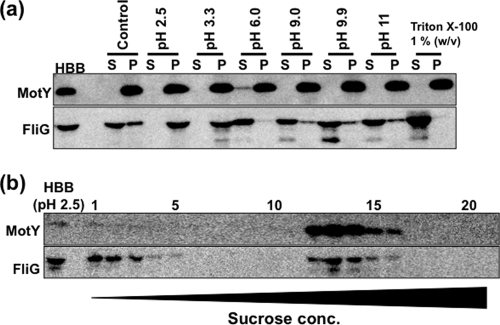
To examine the stability of FliG on the MS ring of the basal body, the isolated basal bodies were treated with acidic or alkaline buffer, or with Triton X-100. The treated samples were fractionated by ultracentrifugation (Fig. 4a). Following alkaline treatment (pHs of 9.0, 9.9, and 11), MotY and FliG signals were detected in both the supernatant and pellet fractions, whereas after acidic treatment (pHs of 2.5 and 3.3), MotY and FliG signals were detected only in the pellet fractions. Basal bodies treated at pH 2.5 were further fractionated by sucrose density gradient centrifugation. MotY and FliG signals were detected in the same fractions as the control samples without acid treatment (Fig. 4b). This may indicate that the interaction between FliG and the MS ring of Vibrio flagella is relatively stable under acidic conditions. Following Triton X-100 treatment, little or no FliG signal was detected in the pellet fractions (Fig. 4a).
FIG. 4.
The effect of pH on the association of FliG with basal bodies. (a) After isolating the basal bodies by solubilization using 0.5% CHAPS, they were treated with several pH buffers (pH 2.5 to 11) and 1% Triton X-100, or were left untreated (Control). After the basal bodies were separated by ultracentrifugation into the supernatant fraction (S) and the precipitate fraction (P), the proteins in the separated fractions as well as in the original fraction (HBB) were separated by SDS-PAGE and detected by immunoblotting using anti-MotY and anti-FliG antibodies. (b) The basal bodies treated with pH 2.5 (HBB) were separated by sucrose density gradient centrifugation. The proteins in each fraction were separated by SDS-PAGE and detected by immunoblotting using anti-MotY and anti-FliG antibodies.
The FliG signal was detected in the upper fractions following sucrose density gradient centrifugation, indicating that some FliG still dissociated from the basal body even in the improved protocol. Therefore, more delicate treatment and special caution are required to isolate the whole C-ring structure. The complete C-ring of Salmonella can be isolated and purified with the basal body. In this case, FliM could be dissociated from the basal body by treatment with acid (pH 4.5) or by CsCl density gradient centrifugation, but FliG dissociated only from the basal body when the pH was lowered to 2.5 (3). We found that FliG was still associated with the basal body of Vibrio isolates even when they were exposed to acidic buffer at pH 2.5. This suggests that the FliF-FliG interaction is retained under acidic conditions in Vibrio. Although we do not have any biochemical evidence for interactions between FliG, FliM, and FliN in Vibrio isolates, it is plausible that they form a C-ring structure as seen in Salmonella. In the present study, we isolated FliG proteins together with the Na+-driven flagellar basal bodies from V. alginolyticus for the first time. This protocol will be a very useful tool for analyzing various mutant FliG proteins and their interaction with the basal bodies. We hope to determine the structural features of the C-terminal region of FliG that is thought to associate with the stator proteins. For example, isolating basal bodies from V. alginolyticus cells that are expressing a functional FliG-GFP fusion (35) will contribute to structural analysis of the FliG region involved in flagellar rotation. Recently, we were able to demonstrate that the stator complex is dynamically assembled and disassembled in the plasma membrane depending on the flow of Na+ through the stator, but that the switch protein, FliG, did not show such dynamic behavior (6).
Acknowledgments
We appreciate T. Miyata and K. Namba for assistance in isolating the flagellar structures from S. enterica serovar Typhimurium and T. Gotoh for technical advice on electron microscopy.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan, the Japan Science and Technology Corporation (to M.H. and to S.K.), and from the Japan Society for the Promotion of Science (to H.T.).
Footnotes
Published ahead of print on 30 October 2009.
REFERENCES
- 1.Aizawa, S. I., G. E. Dean, C. J. Jones, R. M. Macnab, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, P. N., M. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka, H., Y. Sowa, S. Kojima, A. Ishijima, and M. Homma. 2007. Visualization of functional rotor proteins of the bacterial flagellar motor in the cell membrane. J. Mol. Biol. 367:692-701. [DOI] [PubMed] [Google Scholar]
- 5.Fukuoka, H., T. Yakushi, and M. Homma. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J. Bacteriol. 186:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuoka, H., T. Wada, S. Kojima, A. Ishijima, and M. Homma. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825-835. [DOI] [PubMed] [Google Scholar]
- 7.Kawagishi, I., M. Imagawa, Y. Imae, L. McCarter, and M. Homma. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20:693-699. [DOI] [PubMed] [Google Scholar]
- 8.Kawagishi, I., Y. Maekawa, T. Atsumi, M. Homma, and Y. Imae. 1995. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol. 177:5158-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima, S., A. Shinohara, H. Terashima, T. Yakushi, M. Sakuma, M. Homma, K. Namba, and K. Imada. 2008. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. USA 105:7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojima, S., T. Atsumi, K. Muramoto, S. Kudo, I. Kawagishi, and M. Homma. 1997. Vibrio alginolyticus mutants resistant to phenamil, a specific inhibitor of the sodium-driven flagellar motor. J. Mol. Biol. 265:310-318. [DOI] [PubMed] [Google Scholar]
- 11.Kojima, S., Y. Furukawa, H. Matsunami, T. Minamino, and K. Namba. 2008. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J. Bacteriol. 190:3314-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudo, S., Y. Magariyama, and S. Aizawa. 1990. Abrupt changes in flagellar rotation observed by laser dark-field microscopy. Nature 346:677-680. [DOI] [PubMed] [Google Scholar]
- 13.Kusumoto, A., K. Kamisaka, T. Yakushi, H. Terashima, A. Shinohara, and M. Homma. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J. Biochem. 139:113-121. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe, G., M. Meister, and H. C. Berg. 1987. Rapid rotation of flagellar bundles in swimming bacteria. Nature 325:637-640. [Google Scholar]
- 17.Magariyama, Y., S. Sugiyama, K. Muramoto, Y. Maekawa, I. Kawagishi, Y. Imae, and S. Kudo. 1994. Very fast flagellar rotation. Nature 371:752. [DOI] [PubMed] [Google Scholar]
- 18.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 19.McCarter, L. L. 1994. MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarter, L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okabe, M., T. Yakushi, Y. Asai, and M. Homma. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130:879-884. [DOI] [PubMed] [Google Scholar]
- 22.Okunishi, I., I. Kawagishi, and M. Homma. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, K., and D. F. Blair. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 188:2502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul, K., J. G. Harmon, and D. F. Blair. 2006. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J. Bacteriol. 188:5240-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 26.Terashima, H., H. Fukuoka, T. Yakushi, S. Kojima, and M. Homma. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 62:1170-1180. [DOI] [PubMed] [Google Scholar]
- 27.Terashima, H., S. Kojima, and M. Homma. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell. Mol. Biol. 270:39-85. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, D. R., N. R. Francis, C. Xu, and D. J. DeRosier. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C-ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yagasaki, J., M. Okabe, R. Kurebayashi, T. Yakushi, and M. Homma. 2006. Roles of the intramolecular disulfide bridge in MotX and MotY, the specific proteins for sodium-driven motors in Vibrio spp. J. Bacteriol. 188:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakushi, T., J. Yang, H. Fukuoka, M. Homma, and D. F. Blair. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 188:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi, S., S. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi, S., H. Fujita, A. Ishihara, S. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yorimitsu, T., A. Mimaki, T. Yakushi, and M. Homma. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 334:567-583. [DOI] [PubMed] [Google Scholar]
- 36.Yorimitsu, T., Y. Sowa, A. Ishijima, T. Yakushi, and M. Homma. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403-413. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, R., N. Pathak, H. Jaffe, T. S. Reese, and S. Khan. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195-208. [DOI] [PubMed] [Google Scholar]