Abstract
To ensure survival in the host, bacteria have evolved strategies to acquire the essential element iron. In Neisseria gonorrhoeae, the ferric uptake regulator Fur regulates metabolism through transcriptional control of iron-responsive genes by binding conserved Fur box (FB) sequences in promoters during iron-replete growth. Our previous studies showed that Fur also controls the transcription of secondary regulators that may, in turn, control pathways important to pathogenesis, indicating an indirect role for Fur in controlling these downstream genes. To better define the iron-regulated cascade of transcriptional control, we combined three global strategies—temporal transcriptome analysis, genomewide in silico FB prediction, and Fur titration assays (FURTA)—to detect genomic regions able to bind Fur in vivo. The majority of the 300 iron-repressed genes were predicted to be of unknown function, followed by genes involved in iron metabolism, cell communication, and intermediary metabolism. The 107 iron-induced genes encoded hypothetical proteins or energy metabolism functions. We found 28 predicted FBs in FURTA-positive clones in the promoters and within the open reading frames of iron-repressed genes. We found lower levels of conservation at critical thymidine residues involved in Fur binding in the FB sequence logos of FURTA-positive clones with intragenic FBs than in the sequence logos generated from FURTA-positive promoter regions. In electrophoretic mobility shift assay studies, intragenic FBs bound Fur with a lower affinity than intergenic FBs. Our findings further indicate that transcription under iron stress is indirectly controlled by Fur through 12 potential secondary regulators.
The acquisition of iron is essential for bacterial pathogenesis. Because iron is insoluble in aqueous environments at neutral pH and has the potential to produce damaging free radicals, the mammalian host tightly sequesters iron (1, 51), lowering the level of free iron available to invading microorganisms to well below what is needed to survive (34). Since survival in the host and hence virulence is dependent on the acquisition of iron, pathogenic bacteria must be able to sense iron availability and globally regulate the transcription of iron acquisition genes. For many gram-positive and gram-negative organisms, including the sexually transmitted disease pathogen Neisseria gonorrhoeae, the crucial balance between acquiring enough iron to grow and avoiding the toxic effects of excess iron is controlled by the ferric uptake regulator (Fur) protein (12, 27, 49). Typically, Fur acts as a transcriptional repressor by binding as a dimer, along with ferrous iron as a corepressor, to regulatory Fur box (FB) sequences in the promoters of iron-regulated genes under iron-rich growth conditions. As intracellular iron stores are depleted, the Fur-Fe2+ dimer dissociates from the promoter, allowing the entry of RNA polymerase and subsequent transcription (22). Fur acts as a global regulator controlling not only the expression of iron acquisition and storage genes but also the expression of oxidative stress, energy metabolism, acid tolerance, and virulence genes (5, 6, 20, 34, 51, 57) and, as most recently reported, a small, iron-repressible regulatory RNA (15, 36-38).
Early studies of N. gonorrhoeae using a missense mutant confirmed that Fur regulated not only known iron acquisition genes but also a broad range of other genes (49). Later, in silico analysis by Sebastian et al. (43) confirmed by mobility shift assays that Fur was capable of binding, with various degrees of affinity, to the promoter regions of genes found in diverse metabolic pathways. Recently, we investigated the gonococcal transcriptional response to iron limitation at mid-log phase by using a pan-Neisseria microarray (16). The expression profiles of FA1090 grown under iron-replete and iron-depleted growth conditions, together with computational analysis of the promoter regions of iron-regulated genes, demonstrated that transcriptional changes in several functional categories were controlled by Fur, either directly or indirectly, through four secondary regulators (16). We determined that roughly 10% of the genes in the N. gonorrhoeae genome were responsive to iron, with 30% of those open reading frames (ORFs) regulated directly by Fur (16). Similar array reports have been made for the closely related pathogen Neisseria meningitidis grown in the absence and the presence of iron (25).
Fur has also been reported to activate gene expression (36, 38); thus, transcriptional regulation of the Fur regulon is likely to be a multifaceted process, leaving many questions as yet unanswered. In this report, we address the gonococcal response to iron and expand the scope of iron-responsive genes with a comprehensive approach by employing several global strategies, including temporal transcriptional analysis of iron-regulated gene expression using a custom gene-specific microarray; a genomewide bioinformatic approach to predict Fur binding sequences, including intragenic regions; and the screening of an FA1090 library using the Fur titration assay (FURTA) (47) for genomic regions capable of binding gonococcal Fur in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Strains and plasmids are listed in Table S1 in the supplemental material. Unless otherwise stated, the incubation temperature was 37°C. FA1090 was routinely cultivated from frozen stock to GCB agar (Becton Dickinson) supplemented with 2% IsoVitaleX (Becton Dickinson) under a 5% CO2 atmosphere. Escherichia coli isolates were grown on either LB agar (with or without ampicillin at 100 μg/ml, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] at 100 μg/ml, or isopropyl-β-d-thiogalactopyranoside [IPTG] at 50 μg/ml as needed), MacConkey agar (Becton Dickinson) supplemented with 60 μM ferric ammonium sulfate (FAS) (Sigma), MacConkey agar with 2,2′-dipyridyl (Sigma), or chemically defined medium with 100 μM or 10 μM FeNO3 (CDM-100 or CDM-10, respectively) (18, 41).
RNA isolation.
RNA was isolated from FA1090 grown under iron depleted and iron replete growth conditions as previously described (16). Cells were collected for RNA isolation at 1, 2, 3, and 4 h. RNA samples were then DNA digested on a column with RNase-free DNase (Qiagen) prior to quantification using the NanoDrop ND-1000 spectrophotometer (NanoDrop Products). RNA integrity was assessed using the Agilent 2100 bioanalyzer (Agilent Technologies).
cDNA labeling and array hybridization.
The custom Affymetrix GeneChip microarray MPAUTa1520274F has been described previously (23). RNA (10 μg) labeling and hybridization were done at the University of Iowa DNA Facility (http://dna-9.int-med.uiowa.edu/) as previously described (23). For each time point and growth condition, hybridizations were done on three separate biological replicates.
Microarray analysis.
Data files generated by GCOS were imported into GeneSpring GX, version 7.3.1 (Agilent Technologies) and were analyzed as described previously (23). Fold change was expressed as the ratio of the expression of an N. gonorrhoeae gene when the organism was grown under iron-depleted conditions to the expression of the same gene when the organism was grown under iron-replete conditions for the 1-, 2-, 3-, or 4-h time point. Genes with differential expression of ≥1.8-fold and a P value of 0.05 were selected. Hierarchical gene tree cluster analysis was carried out on array data from iron-depleted and iron-replete conditions using GeneSpring software (Agilent).
N. gonorrhoeae qRT-PCR.
Microarray fold changes were confirmed by quantitative reverse transcription PCR (qRT-PCR) with the same RNA samples used in the microarray experiments. cDNA was generated from 2 μg total RNA with random hexamers according to the instructions with the TaqMan reverse transcription reagent kit (Applied Biosystems). A negative reverse transcription reaction (minus the reverse transcriptase) was included for each sample in order to detect genomic DNA. Primers were designed with Primer Express software, version 1.5 (Perkin-Elmer Applied Biosystems) and are listed in Table S2 in the supplemental material. Real-time PCR amplification was performed on the ABI Prism 7500 sequence detection system (Applied Biosystems) using SYBR green PCR master mix (Applied Biosystems). Relative gene expression was quantified using the comparative threshold cycle 2−ΔΔCT method (user bulletin no. 2; Applied Biosystems) with porA (a constitutively expressed major outer membrane porin) as the endogenous reference. Validation experiments for all experimental primer pairs with the endogenous control primers were conducted to ensure that the efficiencies of the target amplification and the endogenous control were equal (user bulletin no. 2; Applied Biosystems). Each gene was assayed in duplicate, and the mean threshold cycle from three separate growth curves was used for comparisons.
Construction of an E. coli strain expressing gonococcal Fur.
The gonococcal Fur gene (NGO1779), including the 5′ promoter region containing the FB sequence, was PCR amplified (see Table S2 in the supplemental material) and inserted into the EZ-Tn5 pMOD transposon construction vector containing a multiple cloning site (MCS) flanked by 19-bp mosaic ends (EPICENTRE Biotechnologies), which had previously been digested with BamHI and end-polished with Klenow fragment (both from New England Biolabs). The resulting transposon, pDF1, was PCR amplified using the pMOD 〈MCS〉 FP-1 forward PCR primer and the pMOD 〈MCS〉 RP-1 reverse PCR primer (EPICENTRE Biotechnologies). The PCR-amplified transposon was then electroporated into E. coli DEC39 (strain H1780 carrying DH-001 EZ-Tn5::〈tfrA/DHFR-1〉, a custom transposome provided by EPICENTRE Biotechnologies) to generate E. coli DEC40. Briefly, the DH-001 transposon was constructed by ligation of a ∼2-kb DNA fragment containing the tfrA gene and control elements into the EZ-Tn::〈DHFR-1〉 transposon at the KpnII restriction site (EPICENTRE Biotechnologies). The transposon was released by enzyme digestion, and transposase was added to generate the custom transposome. The insertion site in E. coli DEC40 was mapped to 55 bp upstream of the ompN gene by rapid identification of EZ::TN transposon insertion sites (17, 32). White (Lac−) colonies from growth on MacConkey agar with 60 μM FAS were selected. To confirm that the Lac− phenotype did not arise from transposon insertion into the fiu::lacZ operon, Lac− colonies were plated onto MacConkey agar with the iron chelator 2,2-dipyridyl at 250 μM, which produced red (Lac-positive) colonies.
FURTA library construction and FURTA.
The FURTA was performed according to the work of Stojiljkovic et al. (47). Chromosomal DNA from FA1090 was randomly sheared by nebulization (IPI Medical Products) to produce 1.5- to 4-kb fragments that were end-repaired with a DNA Terminator kit (Lucigen). The blunt-ended DNA fragments were then ligated into plasmid pUC18 and electroporated into E. coli strain DH5α or XL1-Blue (Stratagene). A total of 34,000 individual white colonies comprising 24,000 DH5α transformants and 10,000 XL1-Blue transformants were combined into pools of 1,000. Plasmids from individual pools were isolated using a Qiagen Maxiprep plasmid kit, transformed into E. coli DEC40, and plated onto MacConkey agar containing 100 μg/ml ampicillin and 60 μM FAS. Lactose positive colonies were selected and restreaked onto MacConkey agar; then single colonies were inoculated into CDM-100 for overnight growth. Recombinant plasmids containing a Fur-regulated gene were obtained using a Qiagen Miniprep plasmid kit and were retransformed into E. coli DEC40 to confirm the FURTA-positive phenotype. Automated DNA sequencing of each recombinant plasmid was performed on the ABI 3730 DNA analyzer using universal M13 primers. The FA1090 chromosomal location for each FURTA-positive plasmid insert was determined by searching the NCBI database using blastn. E. coli DEC40 transformed with plasmid pUC18 alone and the Fur− E. coli DEC39 isolate, which constitutively produces the fiu::lacZ operon, served as negative and positive controls, respectively.
β-Galactosidase assays.
β-Galactosidase activity was quantified according to the work of Miller (40). E. coli DEC40 colonies harboring plasmids exhibiting lactose hydrolysis were first grown overnight in CDM-100 and then diluted 1:100 into fresh CDM-100; cultures were then harvested at mid-log phase (optical density at 600 nm, ∼0.60). Positive and negative controls were the same as those for the FURTA.
Gonococcal Fur cloning and expression.
Fur was cloned by amplifying the coding region of NGO1779 with primers containing NdeI and BamHI restriction enzyme sites on the 3′ and 5′ ends, respectively (see Table S2 in the supplemental material). The PCR product was then blunt-end cloned into pGEM-T Easy (Promega). Colonies were screened for the correct insert by sequencing on the ABI 3730 DNA analyzer using M13 forward and reverse primers. This construct was digested with NdeI and BamHI and was cloned into the pET15b vector (Novagen), generating pET15Fur.
Fur was expressed by transforming the pET15Fur construct into E. coli BLR(DE3)(pLysS) (Novagen). Overnight cultures grown in LB broth from frozen stocks were inoculated into 1 liter of LB broth supplemented with ampicillin (200 μg/ml). At an optical density at 600 nm of 0.8, Fur overexpression was induced by the addition of 1 mM IPTG, and the cells were grown for an additional 3 h. Cells were harvested by centrifugation at 3,000 × g (4°C) for 10 min and were immediately stored at −80°C overnight. Cells were resuspended in 100 ml of 10 mM 2-morpholino-ethanesulfonic acid (MES) (Research Organics)-150 mM NaCl (pH 6.5) (buffer A) with one tablet of Complete protease inhibitor cocktail (Roche). The cells were lysed by three passages through an EmulsiFlex-C5 high-pressure homogenizer (Avestin) at 15,000 lb/in2, and cell debris was removed by a 10-min centrifugation at 21,000 × g (4°C). His-tagged Fur was purified from the supernatant using a cobalt-chelating column (GE Healthcare). Bound proteins were eluted with 500 nM imidazole. Fractions containing Fur were treated with 2 U of biotinylated thrombin (Novagen) per mg of His-tagged Fur and were incubated at room temperature for 2 h. The biotinylated thrombin was removed according to the manufacturer's instructions (Novagen). Imidazole was removed from the filtrate using an Amicon Ultra-4 centrifugal filter unit (Millipore) with a nominal molecular weight limit of 10,000. The retentate from the Amicon filter was resuspended in 500 μl of 20 mM Tris-HCl-50 mM NaCl (pH 7.5). In order to remove any His-tagged Fur that was not cleaved, the retentate was incubated for 10 min at room temperature in 500 μl of a 50% nickel-nitrilotriacetic acid slurry (Qiagen). Following a 1-min centrifugation at 1,000 × g, the concentration of the purified Fur protein was determined by a bicinchoninic acid assay (Pierce). The identity of the purified Fur was confirmed by mass spectrometry at the Core Proteomic and Genomic Facility at the University of South Dakota.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed using Invitrogen's EMSA kit (catalog no. E-33075). DNA fragments including the FB sequence were PCR amplified (see Table S2 in the supplemental material for primers) and purified with a QIAprep kit (Qiagen). The DNA fragment sizes used in these EMSAs were as follows: for tbpA FB1, 141 bp; for tbpA FB2, 140 bp; for fetB, 139 bp; for the nonspecific DNA fragment (NGO1809), 141 bp. Varying concentrations of Fur were incubated for 20 min at room temperature with the respective DNA fragment at 25 nM in EMSA binding buffer (10 mM Tris-borate, 1 mM MgCl2, 100 μM MnCl2, 40 mM KCl, 5% glycerol, 0.1% Igepal, 1 mM dithiothreitol [pH 7.5]). Following incubation, 4 μl of EMSA loading buffer (EMSA kit) was added to each reaction mixture, which was then analyzed on a 4% nondenaturing polyacrylamide gel in cold (4°C) 1× running buffer (45 mM Tris base, 45 mM boric acid, 100 μM MnCl2) for 30 min at 200 V. The gels were stained in 1× SYBR green (EMSA kit) for 20 min and were visualized using the Kodak Gel Logic 1500 imaging system (Carestream Molecular Imaging). Band densities were determined with ImageJ, and GraphPad Prism, version 4, was used for analysis. The KD (equilibrium dissociation constant) of Fur binding to a given DNA fragment was defined as the concentration of Fur required for a 50% shift of the target DNA.
Bioinformatics.
HMMer (19) and Meta-MEME (26) were used to predict the locations of FB sequences in the N. gonorrhoeae FA1090 genome. Both programs were trained on experimentally derived Fur box sequences from N. gonorrhoeae, Pseudomonas aeruginosa, and E. coli (see Table S3 in the supplemental material). FURTA-positive FB sequences were compared in order to generate a sequence logo using WebLogo (11).
Statistical analysis.
Results were expressed as means ± standard errors of the means (SEM). Analysis of variance with a Newman-Keuls posttest was used for comparing groups. A P value of ≤0.05 was considered significant.
Microarray data accession number.
Microarray data were deposited at Gene Expression Omnibus (GEO) (http://www.ncbi.nih.gov/geo) under GEO series GSE16352.
RESULTS
N. gonorrhoeae iron-regulated transcriptional profile.
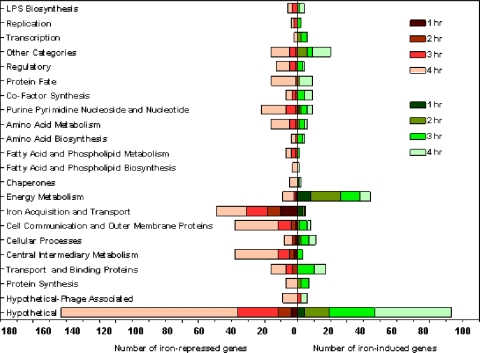
The effect of iron on the global gene expression of FA1090 was determined over a 4-h period by comparing the transcriptional profile of the organism grown under iron-depleted conditions to that for growth under iron-replete conditions. By 4 h, the transcription of 407 genes (20% of the FA1090 genome) was responding to iron availability, and of those genes with altered expression, 300 genes, or 75%, were upregulated during growth under iron-depleted conditions, while 107 (25%) were upregulated under iron-replete conditions. For a detailed summary of gene expression at all time points, see Table S4 in the supplemental material. When we classified the gene expression change according to functional categories, the greatest changes in the transcriptional profile under both iron-depleted and iron-replete growth conditions were those for genes encoding hypothetical proteins (Fig. 1).
FIG. 1.
Functional categories of iron-repressed and iron-induced genes. Fold changes in expression (expression under iron-depleted conditions/expression under iron-replete conditions) for FA1090 genes on the microarray were calculated at 1, 2, 3, and 4 h, and the genes were categorized according to biological function. Each bar represents the actual number of genes.
Response to iron-depleted growth conditions.
As would be expected, when iron was restricted, iron acquisition and storage genes were the most highly expressed, including exbBD, which are essential for TonB-dependent energy transduction, tonB, the receptor protein-encoding genes tbpAB and fetA, and the gene encoding the heme utilization protein (NGO1318). Of the three outer membrane TonB-dependent family genes (Tdf), tdfF, tdfG, and tdfH (54), we found tdfG (NGO0553) and tdfH (NGO0952) to be iron repressed. Turner et al. (54) found that TdfH did not act as a heme receptor, was not iron repressed as judged by protein expression, and lacked a FB (consistent with our data [see below]). These data led Turner et al. (54) to conclude that TdfH may function as a vitamin receptor or as a receptor for a trace metal. In contrast, our finding that tdfH is iron repressed could be due to differences in methodology. We did not see an increase in tdfH transcription in an iron-depleted growth medium until the stationary phase at the 4-h time point. Our findings that tdfH and tdfG are iron repressed suggest that Fur could be indirectly regulating these TonB-dependent receptors through a potential secondary regulator (Fig. 1; see also Table S4 in the supplemental material).
We also observed that iron repression controlled a number of cell communication and outer membrane proteins. Type IV pili and Opa are two major adhesin proteins that facilitate the anchoring of Neisseria to host cells, a process critical for colonization and the establishment of infection, through the binding of CD46 (complement receptor 3) and CEACAM on the human epithelium, respectively (56). The expression of all genes in the pilMNOPQ operon, which encode the components of the type IV secretion system, and that of the opaB and opaD genes, encoding Opa proteins, were iron repressed. The neisserial Maf family of glycolipid binding adhesins are not well characterized, but similar carbohydrate motifs present on host cells are capable of binding type IV pili of Pseudomonas aeruginosa (39). We found several genes in the Maf-like family of proteins, mafB3 (NGO1392), mafB4 (NGO1585), and mafB5 (NGO1971), to be iron repressed. Interestingly, we observed iron repression of app, encoding an adhesin penetration protein (NGO2105) recently described for N. meningitidis (45). App has been implicated in the regulation of adhesion to host cells in the early stages of colonization. Autocleavage of App may allow the release of bacteria to aid in the spread of the organism and reinfection (Fig. 1; see also Table S4 in the supplemental material).
One of the conserved hypothetical proteins (NGO0698) was of interest, since it was one of the two most highly expressed iron-regulated genes lacking an FB. The expression of NGO0698 at 4 h showed a 32-fold increase under iron-depleted growth conditions, a change similar to that of a known iron-repressible gene, fetA (see Table S4 in the supplemental material). NGO0698 is a putative DNA-binding protein showing 62% similarity to N. meningitidis NMB0830 and 48% similarity to rhuM, encoding a virulence factor identified in Salmonella enterica by using a Caenorhabditis elegans-based screen of a transposon library (48). The rhuM mutant was deficient in epithelial cell invasion and showed a reduction in polymorphonuclear leukocyte transepithelial migration assays (48). Further studies are needed to determine whether NGO0698 may play a similar role in gonococcal pathogenesis.
Response to iron-replete growth conditions.
In contrast to iron depletion, the addition of iron resulted predominantly in the upregulation of genes associated with energy metabolism (Fig. 1; see also Table S4 in the supplemental material), consistent with an increased growth rate in iron-containing medium (16). Subunits (sdhC and sdhA) of the sdhCDAB (NGO0923-to-NGO0920) gene cluster, encoding succinate dehydrogenase, were overexpressed under iron-replete growth conditions. The transcription of genes in the nuoA-to-nuoN operon (NGO1737 to NGO1751) encoding subunits of NADH dehydrogenase was also increased in FA1090 in response to high iron availability. The Nuo complex couples electron transfer from NADH to ubiquinone in the respiratory chain (proton motive force), leading to ATP production. The transcription of this operon has been reported to be iron induced in other organisms (13, 36, 55). Iron induced the transcription of a primary Na+ pump, encoded by nqrABCDEF (NGO1413 to NGO1418), that uses Na+ (sodium motive force) in place of H+ for additional ATP production. Sodium pumps are restricted to marine bacteria and selected pathogenic bacteria; they synthesize ATP and are important for solute uptake and motility (28).
Transcriptional regulators.
Lack of iron increased the levels of expression of several transcriptional regulators, including NGO1013 and NGO1116 (both putative phage repressors); NGO1813 (OxyR), a LysR family transcriptional repressor regulating oxidative stress responses (44, 52); MpeR (NGO0025), an AraC family regulator; NGO0718, an RpiR family regulator; and two regulators of PilE, RegF (NGO2130) and RegG (NGO2131) (see Table S4 in the supplemental material). The last observation is consistent with the effects of iron availability on the pilMNOPQ operon (noted above).
The expression of four transcriptional regulators was induced under iron-rich growth conditions: NmlR (NGO0602), a member of the MerR metal response regulator family; NGO1182, a putative nitrogen regulator protein; NGO1788, a putative translational regulator; and NGO1562, an ArsR family regulator (see Table S4 in the supplemental material).
Prediction of FB sequences in the gonococcal genome.
In order to predict the location of potential FB sequences, we trained HMMer and Meta-MEME programs using functional FB seed sequences (see Table S3 in the supplemental material) from organisms whose Fur proteins shared high degrees of identity and conservation with the gonococcal Fur protein (16). Rather than focus on promoter regions alone, we scanned both intergenic and intragenic sequences in the entire FA1090 genome. Our computational approach identified 163 putative FBs, with 36 of these putative FBs predicted by both programs. Some of these predictions overlapped; we found 102 predicted FBs upstream of 92 genes, with 10 genes having tandem FBs. Approximately 90% of the 102 upstream FB sequences were located within 300 bp of a translational start site. A total of 111 intragenic FBs were also predicted by HMMer and Meta-MEME. When this data set was reduced to compensate for overlapping predictions from the two methods, 85 FBs were found within the coding sequences of 82 genes, with 3 genes predicted to contain tandem FBs (see Tables S5 and S6 in the supplemental material).
FURTA.
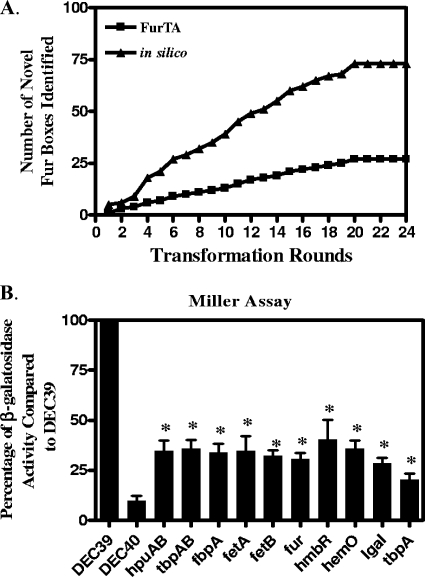
To test these in silico FB predictions, we screened an FA1090 genomic library for the ability to titrate gonococcal Fur in an E. coli background using the FURTA. Genomic regions on the transformed high-copy-number plasmids possessing sequences able to bind Fur will deplete the Fur bound to the chromosomally located fiu::lacZ fusion, allowing transcription and the production of a lactose-positive phenotype on MacConkey agar supplemented with ferric iron. The FURTA-positive clones were sequenced and compared to the in silico-predicted FBs in order to identify novel FB sequences that had not been identified in the previous transformation. After the 20th transformation round, the total number of FURTA-positive clones identified, 28, did not change, indicating that we had saturated the ability of the FURTA to detect FBs in the FA1090 genome (Fig. 2A).
FIG. 2.
(A) Effectiveness of the FURTA for the identification of novel gonococcal Fur box regions. A total of 34,000 clones from an FA1090 FURTA library were transformed into E. coli DEC40 in groups of 1,000 and were screened for the FURTA-positive phenotype as described in Materials and Methods. Following each transformation round, novel FB regions identified by the FURTA were compared to novel FB regions identified by in silico predictions. (B) Increased β-galactosidase activity in E. coli DEC40 FURTA-positive clones. Clones containing FB sequences were grown to mid-log phase in CDM-100 (iron-replete medium) and assessed for β-galactosidase activity. The activity of each individual clone is expressed as a percentage of the activity of E. coli DEC39, a strain constitutively expressing lacZ. The IgA1 and tbpA clones contain internal FB sequences. Data are means ± SEM and are representative of three separate experiments. Analysis of variance with a Newman-Keuls posttest was used. A P value of ≤0.05 was considered significant. Asterisks indicate results significantly different from that for E. coli DEC40 containing the negative-control plasmid pUC18.
We found predicted FB sequences in the promoter regions of 24 of the 28 FURTA-positive clones; the remaining 4 FURTA-positive clones were found in intragenic regions of the genome. Comparison of the FB prediction for the 24 FURTA-positive clones with microarray analysis showed that 14 genes were iron derepressed and 10 were iron induced (see Table S5 in the supplemental material). We found that the 24 FURTA-positive clones included several operons/genes previously reported to be regulated by iron in Neisseria: NGO0217 to NGO0215 (fbpABC), NGO1495-NGO1496 (tbpAB), NGO2093-NGO2092 (fetAB), NGO2109 (hpuB), NGO1318 (hemO/hmbR), NGO0553 (tdfG), and NGO1779 (fur) (8, 9, 16, 33, 42, 43, 49).
In addition, we found NGO0322, a hypothetical protein; NGO0108, a putative oxidoreductase; and NGO0554, an N. gonorrhoeae-specific protein upregulated during oxidative stress (46). NGO0114, a glutaredoxin, and NGO0652, a putative thioredoxin, were also iron repressed. The expression of NGO1751, which is involved in oxidative phosphorylation, a transposase (NGO1317), the ATP binding protein NGO2116, and NGO0904, a Fe-S oxidoreductase, was increased by growth under iron-replete conditions. NGO0711 (an alcohol dehydrogenase), NGO0076 (a putative phosphatase), and two hypothetical proteins (NGO0432 and NGO1430) were upregulated in the presence of iron by the 4-h time point. NGO1276 (aniA) and NGO1275 (norB), produced during anaerobic growth, were also upregulated (see Table S5 in the supplemental material) (29, 30). Isabella et al. (31) have shown that Fur indirectly activates NorB by preventing the binding of an ArsR regulator homologue that overlaps the FB found upstream of the promoter region of NorB.
The remaining 4 FURTA-positive clones out of the initial 28 had FB sequences located within the ORF. The transcription of NGO0024, encoding a putative FetB2 protein, immunoglobulin A1 (IgA1) protease (NGO0275), and NGO1013, a putative phage repressor, was upregulated under low-iron conditions by late log phase; this was confirmed by qRT-PCR (see Table S6 in the supplemental material). We cloned a region of DNA from TbpA (NGO1495), encoding a transferrin binding protein that was not detected in the FURTA but had two predicted internal FBs. The transformation of a single plasmid containing both tbpA intragenic FBs into E. coli DEC40 gave a weakly positive result by the FURTA, suggesting that these FBs barely reached the sensitivity threshold of the FURTA in agar plate cultures. When the plasmid containing both tbpA intragenic FBs was transformed into E. coli DEC40 and the transformant was assayed for β-galactosidase activity by the Miller assay, we observed iron-repressible β-galactosidase activity in this culture (Fig. 2B). This suggests that the FURTA has limited sensitivity in detecting FB activity in a plate assay.
Gonococcal Fur binding site.
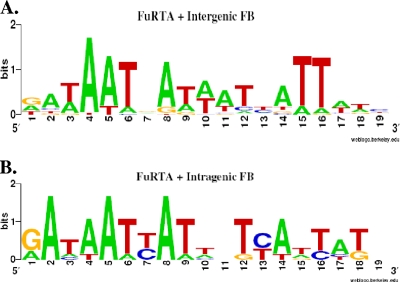
For other organisms, studies of the binding of Fur to DNA promoter regions and bioinformatic mining of the promoter regions of genes responding to iron have established a 19-bp sequence 5′-GATAATGATAATCATTATC-3′ as the functional target of Fur. The structure of this binding site has been variously interpreted as a 7-1-7 heptamer repeat or a combination of three 6-bp repeats (3, 16, 21, 25). Escolar et al. (21) have shown, using DNA footprinting, that in E. coli the strongest natural Fur binding site comprises two hexameric GATAAT direct repeats followed by a 6 bp inverted repeat, ATTATC. We generated a gonococcal FB sequence logo for comparison by including genes whose transcription was regulated by iron and that were able to bind Fur as determined by the FURTA. Figure 3A shows that this sequence logo is very similar to the arrangement reported for E. coli. The gonococcal sequence logo also supports the importance of the sixth residue in each of the two direct repeats (corresponding to nucleotides 6 and 12 in the sequence) and the fifth residue in the inverted repeat (nucleotide 15), which were reported by Escolar to be critical for Fur binding in E. coli (Fig. 3) (21). In the FURTA-positive genes with promoter-proximal FBs, thymidine is conserved and is found at a higher frequency than other residues. Intragenic FBs displayed the same arrangement of hexamers and a similar conservation of the thymidine residues (Fig. 3B). Nucleotides 15 and 16 in the sequence correlate to the two thymidines to which Tiss et al. (50) attribute the binding to the putative Fur DNA helix recognition region. The intragenic FB sequence logo, in contrast to the promoter-proximal FB sequence logo, showed a reduced frequency of thymidine residues able to bind Fur (Fig. 3), which might reduce the overall Fur binding affinity for these intragenic FBs. This would be consistent with the lower level of β-galactosidase activity observed for the intragenic tbpA FB, compared to the activity of the FURTA-positive clones.
FIG. 3.
FB logos. (A) FB sequence logo for FURTA-positive, iron-responsive genes. (B) FB sequence logo for FURTA-negative, iron-responsive genes. The bit score, or overall height, represents sequence conservation at a given position, while the height of each residue within each stack represents the frequency of that residue.
EMSAs for Fur binding.
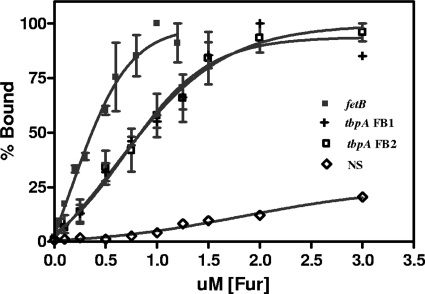
We used EMSAs to examine whether intragenic FBs bound Fur at a lower affinity than promoter-proximal FBs. The fetB gene has a predicted promoter-proximal FB sequence, is iron repressed, and bound Fur in the FURTA (see Table S5 in the supplemental material), as confirmed by β-galactosidase assays (Fig. 2B). As noted above, tbpA has two predicted intragenic FB sequences 216 bp apart (see Table S6). This gene, encoding the TonB-dependent transferrin receptor protein, is cotranscribed in an operon with tbpB, encoding a surface-exposed lipoprotein, as a bicistronic message under iron-stressed conditions (10, 42). The affinity of Fur for these predicted FB sequences was estimated by measuring the percentage of the target DNA fragment bound to Fur in the presence of increasing Fur concentrations. The Fur KD for the promoter-proximal fetB FB was estimated at 381 ± 1 nM (mean ± SEM) (Fig. 4). In contrast, the affinities of the tandem intragenic FB tbpA FB1 and tbpA FB2 were lower and similar to one another (KDs, 840 ± 182 nM and 855 ± 15 nM [means ± SEM], respectively) (Fig. 4). These data are consistent with the proposition that tbpA intragenic FBs bind Fur with a lower affinity than promoter-proximal FBs. Ronpirin et al. (42) demonstrated that, under iron-stressed conditions, the level of the TbpA transcript is twice that of the TbpB transcript. We observed a similar differential of the TbpB message from that of TbpA (see Table S4). Although we were able to identify a transcriptional start site for the entire tbpAB operon using primer extension analysis (data not shown), we could not detect a secondary transcriptional start site that would produce an mRNA encoding a full-length tbpA gene, separate from the tbpAB message (data not shown). Consequently, although the intragenic FBs found in tbpA bind Fur, the regulatory role of these low-affinity intragenic FBs in controlling the transcription of tbpA remains unclear.
FIG. 4.
Comparison of the relative binding affinity of gonococcal Fur for FBs located in the promoter region to that for FBs located within the coding region of TbpA. EMSAs were carried out by incubation of 25 nM concentrations of each respective DNA target containing a predicted FB sequence with increasing concentrations of Fur protein (for FetB, 0 nM, 50 nM, 100 nM, 200 nM, 300 nM, 500 nM, 600 nM, 800 nM, 1μM, and 1.2 μM Fur; for the TbpA and NS DNA fragments, 0 nM, 100 nM, 250 nM, 500 nM, 750 nM, 1 μM, 1.25 μM, 1.5 μM, 2 μM, and 3 μM Fur). DNA was visualized on a 4% nondenaturing polyacrylamide gel using SYBR green. Bound and unbound bands were measured to determine the percentage of Fur that was bound. The KD was defined as the concentration of Fur required for a 50% shift of the target DNA. NS is a nonspecific DNA fragment from NGO1809.
Cluster analysis of iron-regulated genes.
To examine whether groups of genes were being coregulated by iron availability, we analyzed the 4-h array data using a hierarchical gene tree, to include FURTA-positive and FURTA-negative genes with a predicted intergenic or intragenic FB. As shown in Fig. 5, the majority of the genes are iron repressed (clusters 1 to 3). The FURTA-positive genes clustered together, as did known iron-regulated operons, such as fetAB and tbpAB (Fig. 5, cluster 2). Since the FURTA-negative genes that were iron repressed, found in clusters 1 to 3, exhibit an expression pattern similar to that of FURTA-positive genes, we speculate that some may be capable of binding Fur, but at a lower affinity that was not detected in the plasmid pools used to screen the gonococcal genome, like the low-affinity FBs found within the tbpA coding sequence. Genes whose expression was upregulated in the presence of iron were largely FURTA negative (Fig. 5, cluster 4). The total number of predicted FBs found in the promoter-proximal regions of genes in all four gene clusters was only slightly higher than the number of predicted intragenic FBs found within the ORF, suggesting that intragenic FBs may play a regulatory role. Taken together, the results of this expression analysis indicate that Fur directly regulates FURTA-positive, iron-repressed genes and may indirectly affect the iron-induced expression of FURTA-negative genes in cluster 4 through secondary regulators.
FIG. 5.
Gene tree cluster of transcriptional changes in FURTA-positive and FURTA-negative genes with predicted FBs at 4 h. Red indicates an increase, and green indicates a decrease, in transcription levels under iron-depleted or iron-replete growth conditions.
DISCUSSION
Fur has been studied extensively as a classical repressor, binding along with iron to DNA sequences in the promoter regions of iron-responsive genes (22). In iron-poor environments, the Fur dimer is released from the FB, and transcription can ensue. Microarray profiling combined with focused bioinformatic mining of promoter regions has clearly shown that the cascade of iron-regulated gene expression by Fur affects several metabolic pathways (2, 4, 7, 14, 53, 58). More-recent evidence from transcriptome analysis demonstrates that Fur acts as a positive regulator of gene expression in several organisms (25, 53, 58). The mechanism is most likely indirect control of gene expression by Fur through repression of a repressor. In E. coli, the small RNA RyhB is repressed by Fur when iron is present (35, 37). As iron levels decrease, RyhB is transcribed and acts as a posttranscriptional repressor of several genes, including sodB, bacterioferritin A (bfrA), and bacterioferritin B (bfrB), by binding to complementary regions on the mRNAs with the help of the RNA chaperone Hfq, targeting the degradation of the mRNA. Small iron-regulated, Fur-dependent RNAs have been described for several organisms (37, 55), and most recently for N. meningitidis through a bioinformatics approach (38). NrrF (neisserial regulatory RNA responsive to iron) was found to control sdhA and sdhC, encoding subunits of succinate dehydrogenase, an iron-containing enzyme in the tricarboxylic acid cycle (38). An NrrF homologue in N. gonorrhoeae (intergenic space between NGO2002 and NGO2003) was identified by searching the FA1090 genome (38), and we also independently confirmed that report by other experimental methods (15). The gonococcal NrrF promoter, containing a predicted FB, bound Fur in the FURTA (see Table S5 in the supplemental material). Our array analysis showed that the genes whose expression was significantly upregulated under iron-rich conditions included genes known to be regulated in E. coli by RhyB: bfrA, bfrB, sodB, and genes encoding succinate dehydrogenase and NADH dehydrogenase subunits (36, 38, 55; see also Table S4 in the supplemental material).
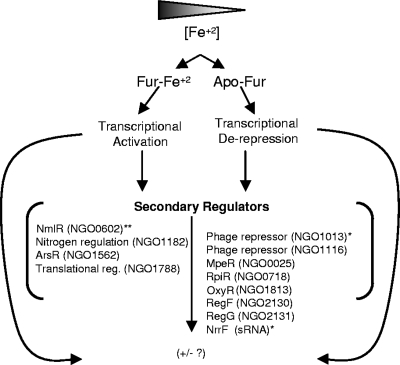
Consequently, we can propose a model of iron-regulated gene expression in the gonococcal Fur regulon (Fig. 6). We previously reported, using microarray analysis of iron-regulated gene expression, that Fur indirectly regulated genes through several secondary regulators: MpeR (NGO0025), an AraC-like regulator, and NGO1013, a putative phage repressor, each of which has a putative FB in the promoter region, as well as a heavy metal-responsive regulator, MerR (NGO0602), capable of responding to iron independently of Fur (16). Our bioinformatic search of intragenic regions located a putative FB within the ORF of NGO0602, suggesting that this transcriptional regulator is a secondary regulator. In the present study, MpeR (NGO0025) and NGO1013 were both iron derepressed by the mid-log to the late-log phase. MpeR has been shown by Folster and Shafer (24) to downregulate the expression of mtrF, encoding an auxilliary protein of the mtrCDE locus, an efflux pump conferring resistance to antibiotics, indicating that mtrCDE-mediated antibiotic resistance is a component of the gonococcal iron regulon.
FIG. 6.
Gonococcal Fur regulon, showing the proposed regulatory cascade of Fur and potential secondary transcriptional regulators. *, predicted FB and FURTA positive; **, predicted FB.
Supplementary Material
Acknowledgments
This work was supported with funds from the National Institute of Health, Centers of Biomedical Research Excellence (COBRE), grant 2 P20 RR-015564.
We thank Mark O'Brian for technical assistance with the EMSA and Rodney Tweten and Eileen Hotze for advice and support in purifying the Fur protein.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodríguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Angerer, A., and V. Braun. 1998. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 169:483-490. [DOI] [PubMed] [Google Scholar]
- 3.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 5.Belzer, C., B. A. M. van Schendel, E. J. Kuipers, J. G. Kusters, and A. H. M. van Vliet. 2007. Iron-responsive repression of urease expression in Helicobacter hepaticus is mediated by the transcriptional regulator Fur. Infect. Immun. 75:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulette, M. L., and S. M. Payne. 2007. Anaerobic regulation of Shigella flexneri virulence: ArcA regulates Fur and iron acquisition genes. J. Bacteriol. 189:6957-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderwood, C. M., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carson, S. D. B., P. E. Klebba, S. M. C. Newton, and P. F. Sparling. 1999. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 181:2895-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. J., P. F. Sparling, L. A. Lewis, D. W. Dyer, and C. Elkins. 1996. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect. Immun. 64:5008-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelissen, C. N. 2003. Transferrin-iron uptake by gram-negative bacteria. Front. Biosci. 8:d836-d847. [DOI] [PubMed] [Google Scholar]
- 11.Crooks, G. E., G. Hon, J.-M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081-1090. [DOI] [PubMed] [Google Scholar]
- 14.Deng, K., R. J. Blick, W. Liu, and E. J. Hansen. 2006. Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducey, T., L. Jackson, J. Orvis, and D. Dyer. 2009. Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microb. Pathog. 46:166-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducey, T. F., and D. W. Dyer. 2002. Rapid identification of EZ::TN transposon insertion sites in the genome of Neisseria gonorrhoeae. Epicentre Forum 9:6-7. [Google Scholar]
- 18.Dyer, D. W., E. P. West, and P. F. Sparling. 1987. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 20.Ernst, F. D., S. Bereswill, B. Waidner, J. Stoof, U. Mader, J. G. Kusters, E. J. Kuipers, M. Kist, A. H. M. van Vliet, and G. Homuth. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533-546. [DOI] [PubMed] [Google Scholar]
- 21.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 22.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folster, J. P., P. J. Johnson, L. Jackson, V. Dhulipali, D. W. Dyer, and W. M. Shafer. 2009. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J. Bacteriol. 191:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folster, J. P., and W. M. Shafer. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 187:3713-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundy, W. N., T. L. Bailey, C. P. Elkan, and M. E. Baker. 1997. Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 13:397-406. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 28.Häse, C. C., N. D. Fedorova, M. Y. Galperin, and P. A. Dibrov. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65:353-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Householder, T. C., W. A. Belli, S. Lissenden, J. A. Cole, and V. L. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Householder, T. C., E. M. Fozo, J. A. Cardinale, and V. L. Clark. 2000. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect. Immun. 68:5241-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isabella, V., L. F. Wright, K. Barth, J. M. Spence, S. Grogan, C. A. Genco, and V. L. Clark. 2008. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154:226-239. [DOI] [PubMed] [Google Scholar]
- 32.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28:1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- 33.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 34.Litwin, C. M., and C. M. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massé, E., H. Salvail, G. Desnoyers, and M. Arguin. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140-145. [DOI] [PubMed] [Google Scholar]
- 37.Massé, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellin, J. R., S. Goswami, S. Grogan, B. Tjaden, and C. A. Genco. 2007. A novel Fur- and iron-regulated small RNA, NrrF, is required for indirect Fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189:3686-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merz, A., and M. So. 2000. Interactions of pathogenic Neisseria with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Morse, S. A., and L. Bartenstein. 1980. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can. J. Microbiol. 26:13-20. [DOI] [PubMed] [Google Scholar]
- 42.Ronpirin, C., A. E. Jerse, and C. N. Cornelissen. 2001. Gonococcal genes encoding transferrin-binding proteins A and B are arranged in a bicistronic operon but are subject to differential expression. Infect. Immun. 69:6336-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal Fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seib, K. L., H.-J. Wu, S. P. Kidd, M. A. Apicella, M. P. Jennings, and A. G. McEwan. 2006. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol. Mol. Biol. Rev. 70:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serruto, D., J. Adu-Bobie, M. Scarselle, D. Veggi, M. Pizza, R. Rappuoli, and B. Arico. 2003. Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol. Microbiol. 48:323-334. [DOI] [PubMed] [Google Scholar]
- 46.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 48.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, C. E., and P. F. Sparling. 1996. Isolation and analysis of a Fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 178:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiss, A., O. Barre, I. Michaud-Soret, and E. Forest. 2005. Characterization of the DNA-binding site in the ferric uptake regulator protein from Escherichia coli by UV crosslinking and mass spectrometry. FEBS Lett. 579:5454-5460. [DOI] [PubMed] [Google Scholar]
- 51.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 52.Tseng, H.-J., A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsolis, R. M., A. J. Baumler, I. Stojiljkovic, and F. Heffron. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, P. C., C. E. Thomas, I. Stojiljkovic, C. Elkins, G. Kizel, D. A. A. Ala'Aldeen, and P. F. Sparling. 2001. Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 147:1277-1290. [DOI] [PubMed] [Google Scholar]
- 55.Vasil, M. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20:587-601. [DOI] [PubMed] [Google Scholar]
- 56.Virji, M. 2009. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat. Rev. Microbiol. 7:274-286. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, P., J. Sun, R. Geffers, and A.-P. Zeng. 2007. Functional characterization of the gene PA2384 in large-scale gene regulation in response to iron starvation in Pseudomonas aeruginosa. J. Biotechnol. 132:342-352. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, D., L. Qin, Y. Han, J. Qiu, Z. Chen, B. Li, Y. Song, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, and R. Yang. 2006. Global analysis of iron assimilation and Fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 258:9-17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.