Abstract
In Salmonella enterica, the CobT enzyme activates the lower ligand base during the assembly of the nucleotide loop of adenosylcobalamin (AdoCbl) and other cobamides. Previously, mutational analysis identified a class of alleles (class M) that failed to restore AdoCbl biosynthesis during intragenic complementation studies. To learn why class M cobT mutations were deleterious, we determined the nature of three class M cobT alleles and performed in vivo and in vitro functional analyses guided by available structural data on the wild-type CobT (CobTWT) enzyme. We analyzed the effects of the variants CobT(G257D), CobT(G171D), CobT(G320D), and CobT(C160A). The latter was not a class M variant but was of interest because of the potential role of a disulfide bond between residues C160 and C256 in CobT activity. Substitutions G171D, G257D, and G320D had profound negative effects on the catalytic efficiency of the enzyme. The C160A substitution rendered the enzyme fivefold less efficient than CobTWT. The CobT(G320D) protein was unstable, and results of structure-guided site-directed mutagenesis suggest that either variants CobT(G257D) and CobT(G171D) have less affinity for 5,6-dimethylbenzimidazole (DMB) or access of DMB to the active site is restricted in these variant proteins. The reported lack of intragenic complementation among class M cobT alleles is caused in some cases by unstable proteins, and in others it may be caused by the formation of dimers between two mutant CobT proteins with residual activity that is so low that the resulting CobT dimer cannot synthesize sufficient product to keep up with even the lowest demand for AdoCbl.
Cobalamin (Cbl, also known as B12) is a structurally complex cyclic tetrapyrrole with a cobalt ion coordinated by equatorial bonds with pyrrolic nitrogen atoms and is unique among cyclic tetrapyrroles (e.g., heme, chlorophylls, coenzyme F430) in that it has an upper and a lower axial ligand (Fig. 1). The coenzymic form of Cbl is known as adenosylcobalamin (AdoCbl) or coenzyme B12.
FIG. 1.
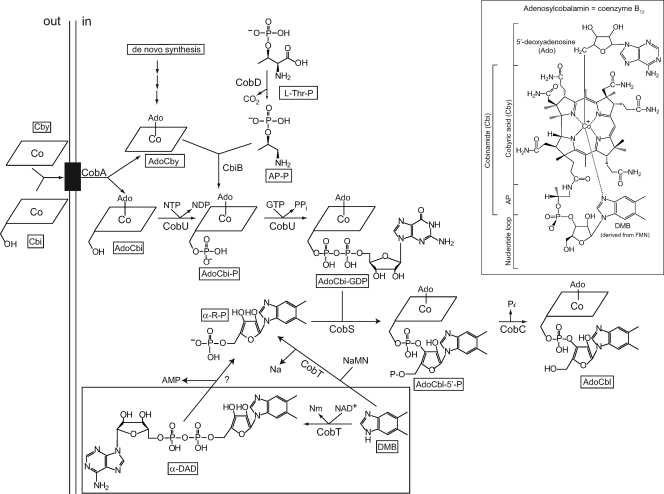
Role of CobT in the late steps of AdoCbl biosynthesis. This branch of the AdoCbl biosynthetic pathway is known as the NLA pathway. The black box in the inner membrane represents the corrinoid-specific ABC transporter BtuCD. The inset shows the chemical structure of AdoCbl; the coring ring in the scheme is represented by the rhomboid cartoon with the Co ion in the middle.
Some bacteria and archaea synthesize AdoCbl de novo or from preformed precursors such as cobyric acid (Cby) or cobinamide (Cbi) (Fig. 1) (11, 32). The enterobacterium Salmonella enterica serovar Typhimurium LT2 (hereafter referred to as S. Typhimurium) synthesizes the corrin ring of AdoCbl de novo only under anoxic conditions (15). Although oxygen blocks de novo corrin ring biosynthesis in this bacterium, it does not block the assembly of adenosylcobalamin (AdoCbl, coenzyme B12) if cells are provided with preformed, incomplete corrinoids such as Cbi or Cby (11, 13-15).
The late steps in AdoCbl biosynthesis can be divided into two different branches that comprise the nucleotide loop assembly (NLA) pathway (19). One of the branches of the NLA pathway activates the lower ligand base, while the other one activates adenosylcobinamide (AdoCbi) to AdoCbi-GDP (Fig. 1).
In this paper, we focus on the activation of 5,6-dimethylbenzimidazole (DMB), the lower ligand base of AdoCbl. There are two ways in which DMB can be activated. In both cases, the CobT enzyme (EC 2.4.2.21) catalyzes the reaction, but the product of the reaction varies, depending on whether the cosubstrate of CobT is NAD+ or its precursor nicotinate mononucleotide (NaMN). If NaMN is the substrate, CobT synthesizes α-ribazole-phosphate (α-RP) (reaction: DMB + NaMN → α-RP + Na) (30). If NAD+ substitutes for NaMN, CobT synthesizes α-DMB adenine dinucleotide (α-DAD) (reaction: DMB + NAD+ → α-DAD + Nm) (18). α-RP is a good substrate for the AdoCbl-5′-phosphate (AdoCbl-P) synthase (CobS; EC 2.7.8.26) enzyme (19, 34), suggesting that an unidentified enzyme cleaves α-DAD into α-RP and AMP (reaction: α-DAD → α-RP + AMP). Although it is possible that CobS may use α-DAD as a substrate, to date, data are not available to support this idea. The AdoCbl-P phosphatase (CobC; EC 3.1.3.73) enzyme catalyzes the last step of the NLA pathway to yield AdoCbl (Fig. 1) (34).
Early genetic studies identified four classes of cobT alleles, namely, classes J, K, L, and M (12); class M was an intriguing class of mutations because they did not display intragenic complementation (12). Here we identify the nature of class M cobT mutations, report the initial in vitro and in vivo characterization of class M CobT variant proteins, and propose structural explanations for the observed deficiencies in CobT activity caused by class M mutations.
MATERIALS AND METHODS
Genetic techniques. (i) Strains and growth conditions.
The genotypes of the strains and plasmids used in this study are listed in Table 1. Strains were grown in lysogeny broth (LB) (3, 4) rich medium or in no-carbon essential (NCE) minimal medium (2). NCE-glycerol medium contained glycerol (30 mM), MgSO4 (1 mM), and dicyanocobinamide [(CN)2Cbi; 15 nM] or cyanocobalamin (CNCbl; 15 nM). NCE-ethanolamine medium contained ethanolamine hydrochloride (30 mM), MgSO4 (1 mM), dicyanocobinamide [(CN)2Cbi; 150 nM) or cyanocobalamin (CNCbl; 150 nM), and NH4Cl (30 mM). When added, the concentration of DMB in the medium was 150 or 600 μM and l-methionine (l-Met) was present at 0.5 mM. When used, kanamycin or ampicillin was present at 50 μg/ml or 100 μg/ml, respectively. All chemicals were of high purity and were purchased from Sigma.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype | Protein encoded | Source or reference |
|---|---|---|---|
| LT2 | Wild type | Laboratory collection | |
| LT2 derivatives | |||
| JE2017 | cobA367::Tn10d(tet+)b/pGP1-2 T7 rpo+(ts) kan+ | Laboratory collection | |
| JE6583 | metE205 ara-9 | K. Sanderson | |
| JE6583 derivatives | |||
| JE2423 | recA1 cobT109::MudI1734c | ||
| JE2607 | recA1 cobT109::MudI1734 cobB1176:: Tn10d(tet+) | ||
| Plasmids | |||
| pT7-5 | Cloning vector, cat+ | 29 | |
| pSU18 | Cloning vector, cat+ | 20 | |
| pSU18 derivatives | |||
| pCOBT10 | cobT+ | CobTWT | |
| pCOBT18 | cobT298 | CobT(G320D) | |
| pCOBT22 | cobT1275 | CobT(C160A) | |
| pCOBT28 | cobT296 | CobT(G171D) | |
| pCOBT32 | cobT304 | CobT(G257D) | |
| pCOBT38 | cobT1278 | CobT(G257A) | |
| pT7-5 derivatives | |||
| pJO27 | cobT+ | CobTWT | 31 |
| pCOBT20 | cobT298 | CobT(G320D) | |
| pCOBT25 | cobT1275 | CobT(C160A) | |
| pCOBT30 | cobT296 | CobT(G171D) | |
| pCOBT33 | cobT304 | CobT(G257D) | |
| pCOBT39 | cobT1278 | CobT(G257A) | |
| pCOBT69 | cobT1293 | CobT(G257D P12G) | |
| pCOBT70 | cobT1294 | CobT(G320D H289A) | |
| pCOBT71 | cobT1334 | CobT(P12G) | |
| pCOBT72 | cobT1335 | CobT(H289A) | |
| pCOBT73 | cobT1336 | CobT(M325A) | |
| pCOBT75 | cobT1276 | CobT(G257N) | |
| pCOBT76 | cobT1277 | CobT(G171D M325A) | |
| pCOBT77 | cobT1341 | CobT(G257D Y284F) | |
| pCOBT79 | cobT1342 | CobT(G171N) |
(ii) In vivo assessment of CobT function.
The functionality of variant CobT proteins was assessed in vivo by their ability to restore growth of a cobT cobB strain (JE2607) under conditions that demanded Cbl-dependent methionine synthesis or growth of a cobT cobB+ strain (JE2423) under conditions that demanded the use of ethanolamine as a carbon and energy source. The cobB+ allele was retained in JE2423 because the lack of CobB causes a phenotype unrelated to AdoCbl biosynthesis (26). The above-mentioned difference in genetic background between strains JE2423 and JE2607 did not affect the interpretation of the data.
To calculate the generation time, we used the equation g = n/t, where g is the generation time, n is the number of generations, and t is the time it took to reach the final cell population. The number of generations (n) was calculated by using the equation Nt = N02n, where Nt and N0 are the final and initial optical densities (ODs) of the culture, respectively.
(iii) Cobalamin-dependent methionine biosynthesis.
Plasmids encoding wild-type or mutant cobT alleles were introduced into strain JE2607 (cobT cobB recA) by using previously described transformation protocols (24). Resulting strains were grown overnight in 2 ml of LB containing ampicillin. Cells were harvested by centrifugation at room temperature at 18,000 × g for 2 min using a Beckman-Coulter microcentrifuge and resuspended in 2 ml of sterile saline. A 10-μl sample of washed cells (∼2 × 107 CFU) was used to inoculate 190 μl of fresh NCE minimal medium containing glycerol as a carbon and energy source. Growth was monitored in a 96-well microtiter dish (Becton Dickinson) using an ELx808 high-throughput spectrophotometer (BioTek Instruments). The temperature in the plate chamber was maintained at 37°C, and OD readings were taken every 15 min. To maintain aeration, plates were shaken for 500 s prior to each reading.
(iv) AdoCbl-dependent ethanolamine catabolism.
Plasmids carrying wild-type or mutant cobT alleles were moved into strain JE2423 (cobT recA) by transformation. Cultures were grown overnight in LB medium containing ampicillin, and cells were centrifuged and resuspended as described above. A 100-μl sample of cell suspension (∼2 × 108 CFU) was used to inoculate 5 ml of NCE-ethanolamine medium in test tubes (16 by 150 mm). Cultures were grown at 37°C with shaking (∼150 rpm), and OD at 650 nm readings were taken every hour for up to 18 h using a Spectronic 20D spectrophotometer (Milton Roy). Each point of the growth curves is the average of the behavior of at least two cultures of each strain.
(v) Random mutagenesis.
To introduce substitutions into CobT(G171D) that would compensate for the effect of the G171D substitution, plasmid pCOBT30 was randomly mutagenized using Stratagene's XL1-Red competent cells in accordance with the manufacturer's instructions. Mutagenized plasmids were purified from 25 ml of LB using the PureYield Plasmid Midiprep system (Promega). A Bio-Rad Gene Pulser was used at settings of 2.0V, 200 Ω, and 25 mF to electroporate ∼2 μg of mutagenized plasmid DNA into 140 μl of JE2607 (cobB cobT recA) electrocompetent cells. Cells were recovered at 37°C for 1 h, plated onto LB supplemented with ampicillin, and grown overnight at 37°C; this procedure yielded approximately 500 colonies per plate. Ampicillin-resistant clones were screened for growth on minimal NCE medium supplemented with glycerol and (CN)2Cbi with or without DMB (10 μM). Cells that grew under these conditions were streaked onto the above-described medium containing DMB (10 μM), and their Cob phenotype was reassessed. The cobT allele in the plasmids isolated from cells that grew on NCE medium supplemented with glycerol, (CN)2Cbi, and DMB was sequenced by using primers specific to cobT and to the pT7 vector.
Recombinant DNA techniques. (i) General plasmid construction strategy.
The construction of plasmids pCOBT10 and pJ027 has been described previously (31). Plasmid pCOBT10 contains the cobT+ allele cloned into vector pSU18 (20), and plasmid pJO27 carries the cobT+ allele cloned into vector pT7-5 (29). Point mutations were introduced into the cobT+ gene on plasmid pCOBT10 by PCR as described previously (10) or by using Stratagene's QuikChange XL site-directed mutagenesis kit in accordance with the manufacturer's instructions. Each cobT allele was sequenced to verify the presence of the intended mutation. DNA sequencing was performed by using nonradioactive BigDye protocols (ABI Prism), and the reaction products were resolved and analyzed at the University of Wisconsin—Madison Biotechnology Center. Mutagenic primers used in PCR-based site-directed mutagenesis protocols were obtained from Integrated DNA Technologies. For detailed descriptions of the construction of the plasmids used in this work, see the supplemental material. All complementation studies were performed with cobT alleles cloned into pT7-5.
(ii) Sequence analysis of cobT chromosomal mutants.
Template DNA used for sequencing was generated by PCR amplification directly from the chromosome. Chromosomal cobT alleles were amplified by using a Perkin-Elmer model 2400 Gene Amp PCR thermocycler. To do this, cells from a single bacterial colony were resuspended in 100 μl of sterile, double-distilled H2O (ddH2O) and boiled at 100°C for 5 min, and debris was removed by centrifugation for 1 min at 18,000 × g in a Beckman-Coulter microcentrifuge. A 5-μl sample of the supernatant was used in a 100-μl PCR mixture that contained 2 U of Pfu DNA polymerase (Promega), 50 pmol of each primer, 10 μl of a 2 mM deoxynucleoside triphosphate mixture (Promega), 10 μl of 10× Pfu polymerase buffer, and ddH2O. The PCR amplification conditions were as follows: dwell, 94°C for 1 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1.1 min, followed by 10 min at 72°C. Reaction mixtures were run on 1% Tris-borate-EDTA buffer (pH 8) agarose gels (22). The amplified product was eluted from the agarose using a QIAquick gel extraction kit (Qiagen) and resuspended in 40 μl of ddH2O. A 5-μl sample was used as the template in sequencing reactions. Amplification products were purified using AutoSeq G-50 spin columns (Pharmacia Biotech). Reactions were resolved and analyzed by the DNA sequencing facility at the University of Wisconsin Biotechnology Center. Primers for PCR amplification and sequencing were obtained from Genosys.
Biochemical techniques. (i) Protein overproduction and purification.
Variant CobT proteins were overproduced as described for the wild-type protein (30). Plasmids encoding mutant cobT alleles under the control of the T7 promoter of vector pT7-5 were moved by transformation into strain JE2017 and overexpressed by using a temperature shift procedure (28). The reported protocol for the isolation of wild-type CobT (CobTWT) protein was modified for the use of an ÄKTA explorer fast protein liquid chromatography system (Amersham Pharmacia). Cell extracts (CFE) were obtained by using a French press operating at 1.034 × 104 kPa. Two 5-ml Amersham Pharmacia HiTrap Phenyl Sepharose FF (FastFlow) prepacked columns were equilibrated with buffer A (Tris-HCl buffer [50 mM, pH 7.5, 4°C] containing ethylene glycol [1.8 M] and ammonium sulfate [10%, wt/vol, 4°C]). Approximately 75 mg of CFE protein was loaded onto the columns flowing at a rate of 2 ml of buffer A/min. After loading was complete, the column was washed with 20 ml of buffer A. A 100-ml linear gradient that simultaneously reduced the concentration of ammonium sulfate to zero and increased the concentration of ethylene glycol to 8.9 M in buffer A was used to elute CobT proteins, whose elution off the column was monitored at 215 nm; the presence of CobT proteins in fractions was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (16) by using 12% polyacrylamide gels. Proteins were visualized after staining with Coomassie blue R-250 dye as described previously (25). Fractions containing CobT protein were pooled and quantified by using a Bio-Rad protein assay kit (Bio-Rad Laboratories), which is based on the Bradford dye-binding procedure (5).
After the Phenyl Sepharose purification step, Centricon-Plus 70 centrifugal filter devices (Amicon) were used to concentrate protein and exchange the buffer with Tris-HCl (25 mM, pH 7.5, 4°C) containing 1.4 M glycerol. The proteins were further purified by using two 5-ml Amersham Pharmacia Cibracon Blue columns in tandem equilibrated with Tris-HCl (25 mM, pH 7.5, 4°C) containing 1.4 M glycerol. Approximately 15 mg of protein was loaded onto the columns at a rate of 2 ml/min. After loading, the column was washed with 20 ml of the same equilibration buffer. A 100-ml linear gradient of NaCl (0 to 1 M) in the equilibration buffer was used to elute CobT proteins off the Cibacron Blue column. Prior to visualization by SDS-PAGE, fractions containing the CobT(G320D), CobT(G171D), and CobT(G257D) proteins were concentrated by using a Microcon YM-10 centrifugal filter device (Millipore). CobT-containing fractions were pooled, and the amount of protein in the sample was quantified. If needed, protein was further concentrated by using a Centricon-Plus 70 concentrator until it reached at least 1 mg/ml. The CobTWT, CobT(C160A), and CobT(G257A) proteins were stored at −80°C in Tris-HCl (50 mM, pH 7.5, 4°C) containing glycerol (3 M) until used. The CobT(G320D), CobT(G171D), and CobT(G257D) proteins were concentrated and stored at 4°C for immediate use. Protein purity was determined by gel densitometry by using the TotalLab v2005 software by Nonlinear Dynamics.
(ii) Preparation of extracts of strains carrying chromosomal cobT mutant alleles.
Full-density cultures of wild-type and mutant cobT strains were grown in 500-ml sidearm flasks containing 100 ml of medium. Cell density was monitored by using a Klett colorimeter (Manostat). Under the conditions used, full density was reached at ∼200 Klett units. Cells were concentrated at 7,000 × g for 10 min using a Sorvall RC-5B refrigerated centrifuge equipped with an SS-34 rotor (Dupont). Cell pellets were resuspended in 0.9 ml of ice-cold 50 mM Tris-HCl (pH 7.5, 4°C) and sonicated (microtip, setting 3, 50% duty) for 5 min at 4°C using a model 550 Sonic Dismembrator (Fisher Scientific). Sonication was stopped after 2.5 min, and the CFE were incubated for 2 min at 4°C to minimize protein denaturation by heat. Cell debris was removed by centrifugation at 44,000 × g for 1 h. Protein concentrations of the resulting extracts were determined as described above. Samples used for Western blot analysis were stored at −20°C overnight.
(iii) Western blot analysis of genome-encoded CobT proteins.
Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane (Immobilon-P transfer membrane; Millipore) for 1 h at 100 V. After transfer, membranes were blocked using a solution of 1× TBS (Tris-HCl [20 mM, pH 7.5, 25°C], NaCl [137 mM], Tween [0.1%, vol/vol]) containing low-fat powdered milk (5%, wt/vol) for at least 1 h at room temperature. After two rinses, membranes were incubated with primary antibodies for 1 h. Rabbit polyclonal CobT and CobU antibodies were obtained through the animal facility of the School of Medicine at the University of Wisconsin—Madison. Antiserum was diluted 1/50,000 and 1/10,000, respectively, in 1× TBS. After two rinses, a 15-min wash and a 5-min wash with 1× TBS, membranes were incubated with a secondary antibody, a 1/20,000 to 1/100,000 dilution of horseradish peroxidase-conjugated donkey anti-rabbit antibody (Pierce), for 1 h. Membranes were rinsed (same sequence as above), followed by treatment with ECL detection reagents (Amersham Life Science). ECL detection reagents were used for enhanced chemiluminescence detection of the horseradish peroxidase-coupled secondary antibody. Membranes treated with ECL detection reagents were exposed to X-ray film (Kodak BioMax AXR) for 1 or 5 min and developed.
(iv) Synthesis of [14C,C-2]DMB.
The chemical synthesis of DMB from 4,5-dimethyl-1,2-phenylenediamine (DMPDA) and phosphorylated aldoses has been reported (17). We modified the reported conditions to generate radiolabeled DMB. We synthesized [14C,C-2]DMB from [14C]HCHO (specific radioactivity, 57 mCi/mmol; American Radiolabeled Chemicals) and DMPDA. The reaction mixture (225 μl) contained morpholineethanesulfonic acid buffer (12.5 μmol, pH 8.0), DMPDA (0.3 μmol), and [14C]HCHO (2.5 μmol). The reaction mixture was placed in a 1.5-ml microcentrifuge tube (Eppendorf), which was incubated overnight with gentle shaking at 50°C. Radiolabeled DMB was purified by high-pressure liquid chromatography as described previously (17). Purified [14C,C-2]DMB was dried under vacuum using a SpeedVac concentrator (Savant). The dried material was solubilized in 1 ml of methanol, yielding a solution of radiolabeled DMB with a specific radioactivity of 43.6 mCi/mmol.
(v) In vitro NaMN:DMB phosphoribosyltransferase activity assay.
The conditions for the NaMN:DMB phosphoribosyltransferase activity assay were as described previously (30). CobT proteins were diluted with glycine-NaOH buffer (50 mM, pH 10.0) containing bovine serum albumin (0.05%, wt/vol) and sodium azide (4.6 mM). All reaction mixtures contained glycine-NaOH buffer (2.5 mmol), NaMN (100 nmol), DMB (1.5 nmol), and [14C,C-2]DMB (0.5 nmol) in a final volume of 20 μl. CobTWT protein (2.5 ng) was used as a positive control. The activities of the CobT(C160A) and CobT(G257A) proteins were assayed using 2.5 ng of total protein, while the activities of the CobT(G320D), CobT(G171D), and CobT(G257D) proteins were assayed using 0.5, 0.2, and 0.05 μg of protein, respectively. All reaction mixtures were incubated at 37°C, and samples were removed at 0-, 3-, 5-, 10-, 15-, 20-, and 30-min intervals. Reaction mixtures were heated to 80°C for 10 min to inactivate the enzyme.
(vi) Kinetic analysis of variant CobT enzymes.
A unit of CobT activity was defined as the amount of protein needed to make 1 μmol of product (α-ribazole-5′-phosphate) per min. The Km of class M CobT proteins for NaMN was determined as described above, under DMB saturating conditions and at various concentrations of NaMN (0.1 to 6 mM). The assay times for the CobTWT, CobT(C160A), CobT(G257A), CobT(G320D), CobT(G171D), and CobT(G257D) proteins were 5, 5, 12, 10, 15, and 4 min, respectively. KmNaMN values for the CobT variant proteins were obtained from Michaelis-Menten plots by using the Prism v4 graphic and statistical software package (GraphPad). Each point was performed in triplicate.
RESULTS
Identities of the mutations in alleles cobT296, cobT298, cobT304, cobT313, cobT1087, and cobT309.
We determined the DNA sequences of chromosomal cobT alleles encoding class M CobT variants (12). Alleles of interest were PCR amplified directly from the chromosome and sequenced. All sequencing reactions were performed in duplicate, and both strands were sequenced. All of the mutations detected were either a guanine-to-adenine change or a cytosine-to-thymine change (i.e., transitions), consistent with the use of hydroxylamine as the mutagen (21).
Mutations in strains JE766 (cobT296), JE768 (cobT298), JE774 (cobT304), JE783 (cobT313), JE799 (cobT1087), and JE779 (cobT309) resulted in substitutions G171D, G257D, G320D, G231D, G257D, and S269L A270V, respectively. All of the alleles, except cobT309, had a glycyl residue replaced with an aspartyl residue. Three of these alleles (cobT296, cobT298, and cobT304) were used in subsequent studies. Variant CobT(S269L A270V) was not studied because it had two substitutions.
Chromosomal levels of class M CobT proteins are low but detectable.
We performed Western blot analysis to determine whether the cell made detectable levels of class M mutant proteins from genome-encoded alleles. To generate CFE containing maximal levels of CobT, transcription of the cobT gene was induced by growing cells in the presence of 1,2-propanediol (23). Western blot analysis was performed using rabbit polyclonal antibodies against CobT. To avoid variations in the total amount of protein loaded onto SDS-PAGE gels, we added known amounts of CobU protein to the mixture as an internal control; CobU protein was detected using rabbit polyclonal antibodies against CobU. CFE of strains JE4124 (cobT296), JE4125 (cobT304), and JE4126 (cobT298) resulted in decreased but detectable levels of CobT protein relative to the extract of the cobT+ strain (see Fig. S6 in the supplemental material).
In vivo assessment of the enzymatic activity associated with class M CobT proteins.
We tested the ability of cobT mutant strains to grow under conditions that demanded different levels of Cbl. Cbl-dependent synthesis of l-Met requires low intracellular levels of Cbl (∼100 molecules per cell), whereas optimal growth on ethanolamine demands several thousand Cbl molecules per cell (1). When growth depended on MetH (Cbl-dependent methionine synthase) function, plasmid-encoded CobT activity was assessed by using strain JE2607 (cobB cobT recA) (Table 2). Performance of these studies with recombination-deficient (recA) strains provided confidence that growth of the strains was due to the enzymatic activity of plasmid-encoded CobT proteins. Inactivation of the cobB gene was needed to eliminate CobT-like activity associated with CobB (31). Growth on ethanolamine as the sole source of carbon and energy was assessed by using strain JE2423 (cobT recA). In this case, the cobB gene was not inactivated because its CobT-like activity is not sufficient to support the high demand for AdoCbl and because the absence of CobB prevents growth on ethanolamine for reasons unrelated to AdoCbl biosynthesis (27).
TABLE 2.
In vivo assessment of function of class M CobT variants
| Plasmid, encoded CobT protein, and supplement(s) | Generation time (h) on: |
|
|---|---|---|
| Ethanolaminea | Glycerolb | |
| pT7-5, none (cloning vector)c | ||
| Cbi | NGf | NG |
| Cbi + DMBd | NG | NG |
| Cbl | 4 ± 0.3 | 1 ± 0.1 |
| pJO27, CobTWT | ||
| Cbi | 5 ± 1 | 1 ± 0.1 |
| Cbi + DMBe | 4 ± 2 | 1 ± 0.1 |
| Cbl | 4 ± 0.2 | 1 ± 0.1 |
| pCOBT20, CobT(G320D) | ||
| Cbi | NG | NG |
| Cbi + DMBe | NG | 2 ± 0.1 |
| Cbl | 4 ± 0.2 | 1 ± 0.1 |
| pCOBT70, CobT(G320D H289A) | ||
| Cbi | NG | NG |
| Cbi + DMBe | 4 ± 0.1 | 1 ± 0.1 |
| Cbl | 3 ± 0.1 | 1 ± 0.1 |
| pCOBT72, CobT(H289A) | ||
| Cbi | NG | NG |
| Cbi + DMBe | 6 ± 2 | 1 ± 0.1 |
| Cbl | 4 ± 0.2 | 1 ± 0 |
| pCOBT30, CobT(G171D) | ||
| Cbi | NG | NG |
| Cbi + DMBd | NG | 3 ± 0.4 |
| Cbl | 4 ± 0.5 | 1 ± 0.1 |
| pCOBT76, CobT(G171D M325A) | ||
| Cbi | NG | NG |
| Cbi + DMBd | NG | NG |
| Cbl | 4 ± 0.1 | 1 ± 0.1 |
| pCOBT73, CobT(M325A) | ||
| Cbi | 11 ± 0.1 | 1 ± 0.1 |
| Cbi + DMBd | 4 ± 0.1 | 1 ± 0.1 |
| Cbl | 4 ± 0.1 | 1 ± 0.1 |
| pCOBT79, CobT(D171N) | ||
| Cbi | NG | 5 ± 2 |
| Cbi + DMBe | 5 ± 0.1 | 1 ± 0.1 |
| Cbl | 5 ± 0.1 | 1 ± 0.1 |
| pCOBT33, CobT(G257D) | ||
| Cbi | NG | NG |
| Cbi + DMBe | NG | NG |
| Cbl | 3 | 1 |
| pCOBT69, CobT(G257D P12G) | ||
| Cbi | NG | NG |
| Cbi + DMBd | NG | NG |
| Cbl | 4 ± 0.1 | 1 ± 0 |
| pCOBT71, CobT(P12G) | ||
| Cbi | 6 ± 1 | 1 ± 0 |
| Cbi + DMBd | 4 ± 0.3 | 1 ± 0.1 |
| Cbl | 5 ± 0.1 | 1 ± 0.1 |
| pCOBT39, CobT(G257A) | ||
| Cbi | 3 ± 1 | 1 ± 0 |
| Cbi + DMBe | 3 ± 1 | 1 ± 0 |
| Cbl | 3 ± 1 | 1 ± 0 |
| pCOBT75, CobT(G257N) | ||
| Cbi | 3 ± 0.8 | 11 ± 1 |
| Cbi + DMBe | 3 ± 0.5 | 2 ± 0.1 |
| Cbl | 3 ± 0.9 | 1 ± 0 |
NCE minimal medium containing ethanolamine (30 mM) as a carbon and energy source, MgSO4 (1 mM), and (CN)2Cbi (150 nM) plus DMB (as indicated below) or CNCbl (150 nM). Strain JE2423 was the recipient of each of the plasmids tested. The cobB+ allele was retained in JE2423 because the lack of CobB causes a phenotype unrelated to AdoCbl biosynthesis (26).
NCE minimal medium containing glycerol (30 mM) as a carbon and energy source, MgSO4 (1 mM), and (CN)2Cbi (15 nM) plus DMB (as indicated below) or CNCbl (15 nM). Strain JE2607 was the recipient of each of the plasmids tested.
Plasmid pT7-5 (29).
DMB concentration on glycerol and ethanolamine (600 μM).
DMB concentration on glycerol (150 μM) or ethanolamine (600 μM).
NG, no growth.
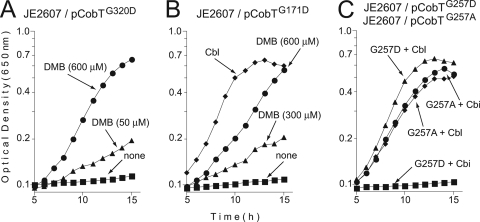
In some cases, when class M cobT alleles were placed in trans on a high-copy-number plasmid, exogenous DMB supported Cbl-dependent l-Met synthesis in strain JE2607. Variant proteins CobT(G320D) and CobT(G171D) retained enough activity to support growth, but only when DMB was present in the medium (Fig. 2A and B). In contrast, a strain that synthesized CobT(G257D) did not respond to DMB and required Cbl to grow (Table 2). The CobT(G320D), CobT(G171D), and CobT(G257D) proteins did not support the growth of strain JE2423 on ethanolamine in medium that contained (CN)2Cbi but lacked DMB (Table 2).
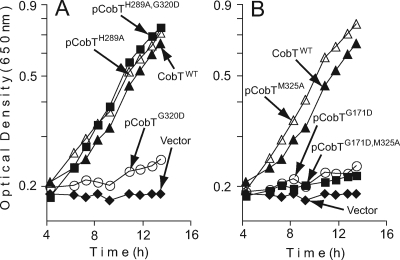
FIG. 2.
In vivo functional analysis of class M CobT variant proteins. Strains were grown in NCE minimal medium supplemented with glycerol and Cbl (positive control) or Cbi with or without DMB (for details, see Materials and Methods). The chromosomal copies of the cobB and recA genes in strain JE2607 (cobT cobB recA) were inactivated to eliminate nonspecific catalysis of the CobT reaction by the CobB sirtuin and to prevent homologous recombination between the plasmid and chromosomal alleles of cobT.
We pursued two possible explanations for the effects of the aspartate substitutions in residues G171, G257, and G320. These residues are important for CobT structural stability, the negative charge of the aspartyl side chain changes CobT into a nonfunctional conformation, or both. We focused on residue G257. For this purpose, we constructed two mutant cobT alleles, namely, cobT1278 and cobT1276, which directed the synthesis of variants CobT(G257A) and CobT(G257N), respectively. The G257A substitution would reduce the steric bulk of the side chain and eliminate the negative charge, while G257N would change the charge from negative to positive and retain the hydrogen-bonding capacity of the side chain. A strain that synthesized CobT(G257A) grew as well as the strain that synthesized CobTWT and did not require exogenous DMB (Table 2). Unlike a strain that synthesized CobT(G257D), the strain that made CobT(G257N) responded to exogenous DMB under conditions that demanded low levels of Cbl synthesis and grew as well as the wild-type strain on ethanolamine, even in the absence of DMB (Table 2).
The disulfide bond between C160 and C256 is not required for CobT function.
The crystal structure of the CobT protein contains a disulfide bond formed between residues C160 and C256 (9). We tested the relevance of this bond by constructing allele cobT1275 [encodes CobT(C160A)]. A strain that made CobT(C160A) displayed no discernible growth defect in NCE medium containing (CN)2Cbi and glycerol or ethanolamine relative to that of a strain that synthesized CobTWT (data not shown). The kinetic analysis of the CobT(C160A) variant did show some differences from CobTWT (see below), but these differences were not significant enough to impact growth behavior under the conditions tested.
Modeling.
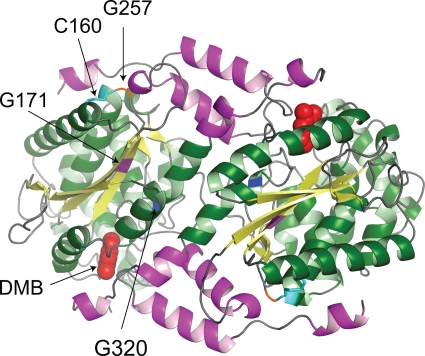
We used the structural information about CobT (7-9) in an attempt to understand the effect of class M mutations on CobT function. All modeling was done by using the MacPymol Molecular Graphic System (0.99rev8, 2007; DeLano Scientific LLC, Palo Alto, CA). The analysis of these mutations took into consideration that CobT is a dimer in its biologically active form, with the two active sites located at the subunit interface and part of the active site contributed by the small domain of the neighboring subunit (9). As shown in Fig. 3, residues G320 and G171 are located within or near the active site but residue G257 is not. Given the location of residue G257, it was of interest that the G257D substitution had the most severe growth defect of all of the single-amino-acid changes analyzed.
FIG. 3.
Crystal structure model of wild-type CobT protein from S. enterica. Shown is the biologically active dimeric structure of the S. enterica CobT protein in complex with DMB (Research Collaboratory for Structural Bioinformatics Protein Data Bank 1d0s). DMB is shown as space filled in the active sites located at the subunit interface. The arrows indicate the locations of residues affected by class M mutations (G171, G257, and G320). Also shown is the location of residue C160, which forms a disulfide bond with residue C256.
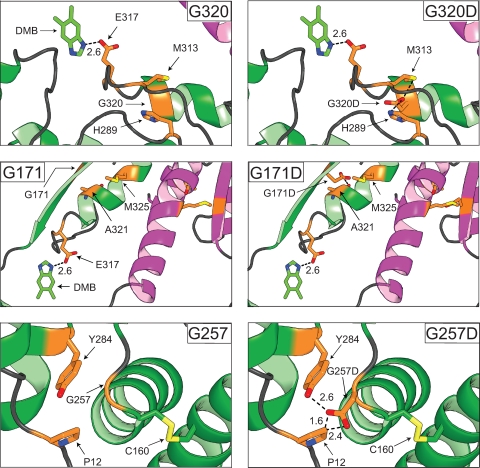
Putative effects induced by aspartate substitutions at positions G171, G320, and G257.
Model structures shown in Fig. 4 helped us visualize possible effects. The following observations were made by assuming that any given substitution did not significantly change the conformation of the wild-type protein. (i) In the CobT(G320D) protein, both oxygen atoms of the carboxylate function of the aspartyl side chain would be <3 Å away from an N atom of the imidazole ring of H289. (ii) In the CobT(G171D) protein, the methyl group of M325 would be 1 Å away from an O atom of the carboxylate group of D171. (iii) In the CobT(G257D) protein, the carboxylic group of D257 could introduce a new interaction with the hydroxyl group of residue Y284. (iv) In the CobT(G257A) protein, the alanyl residue would not create any hindrance or ionic interaction. (v) In the CobT(C160A) protein, the alanyl residue would not generate hindrances with the side chains of neighboring amino acids, but the loss of the side chain of the cysteinyl residue at position 160 would prevent disulfide bond formation with residue C256.
FIG. 4.
Modeling of class M CobT variant proteins. The MacPymol 0.99rev8 software package was used to model in the mutations of interest. As a comparison, the CobTWT protein is located in the left panels while the variant proteins are located in the right panels. One subunit of the proteins is magenta, and the neighboring subunit is green. The variant residues are yellow, and any residues they interact with are green. Interactions between the proposed catalytic residue (E317) and DMB are shown to illustrate how class M substitutions may affect CobT activity. The disulfide bond formed between residues C160 and C256 is yellow. Oxygen and nitrogen atoms in side chains are orange and blue, respectively. Distances are reported in angstroms.
Effects of putative compensatory changes.
We used the available crystal structures of CobT to determine whether the negative effects of aspartate substitutions at positions G257, G320, and G171 would be relieved by an Ala or Gly substitution at residue Pro12, His289, or Met325. We performed phenotypic analyses of strains synthesizing CobT variants with one or two substitutions. The sensitivity of the bioassays would detect CobT variants with very low residual activity (growth on glycerol; Table 2) or CobT enzymes with substantial activity (growth on ethanolamine as a carbon and energy source; Fig. 5).
FIG. 5.
Phenotypic analyses of strains that synthesize class M mutant CobT proteins with putative compensatory substitutions. We tested in vivo the ability of class M variant CobT proteins with putative compensatory substitutions to restore Cbl synthesis in strain JE2607 (cobT cobB recA1). Conditions for the assessment of growth on ethanolamine as a carbon and energy source were as described in Materials and Methods. All of the media tested contained (CN)2Cbi and DMB (600 μM); CNCbl was used as a positive control.
An H289A substitution counteracts the effect of the G320D substitution.
Strains that synthesized CobT(H289A) failed to grow on glycerol in the absence of DMB, but the addition of the latter fully corrected the growth defect. On ethanolamine, the same strain failed to grow in the absence of DMB but had only a slight growth defect when DMB was present in the medium. Strains that synthesized CobT(G320D) or CobT(G320D H289A) did not grow on ethanolamine in the absence of DMB, but unlike variant CobT(G320D), variant CobT(G320D H289A) supported growth on ethanolamine as well as CobTWT when DMB was present in the medium (Table 2; Fig. 5A).
An M325A substitution does not counteract the effect of the G171D mutation.
When the DMB concentration in the medium was high (600 μM), a strain synthesizing the CobT(G171D) protein had a subtle growth defect (threefold slower generation time) relative to a strain making CobTWT (Table 2), and the same strain failed to grow on ethanolamine (Fig. 5B; Table 2). A strain making CobT(G171D M325A) required Cbl for growth regardless of the carbon source used. In contrast, the control strain synthesizing CobT(M325A) grew in the absence of exogenous DMB, albeit at a rate slower than that of a strain making CobTWT. Addition of DMB to the medium allowed the strain making CobT(M325A) to grow as fast as a strain making CobTWT (Fig. 5B; Table 2).
Isolation of a mutation that compensates the effect of the G171D substitution.
We took a nonbiased, random mutagenic approach to isolate mutations in cobT296 [CobT(G171D)] that would reverse the negative effect of the G171D substitution. We screened the population of mutagenized plasmid pCOBT30 (cobT296), looking for growth on Cbi in the absence of exogenous DMB. Six of ∼23,000 colonies screened grew in the absence of DMB. Sequencing data showed that residue D171 was changed from the starting aspartate to asparagine [cobT1342, CobT(D171N)]; all six clones had the D171N substitution. Strain JE2607, carrying allele cobT1342 [CobT(D171N)], did not require exogenous DMB during growth on glycerol, although it still had a measurable growth defect compared to a strain synthesizing the CobTWT protein (generation times = 5 and 1 h, respectively) (Table 2).
A P12G substitution does not alter the effect of the G257D mutation.
CobT(G257D) and CobT(G257D P12G) were Cbl auxotrophs on glycerol (Table 2) or ethanolamine. In contrast, the control strain synthesizing the CobT(P12G) protein did not require exogenous DMB for growth on glycerol or ethanolamine (Table 2).
Purification of variant CobT proteins.
Under the conditions used, the biomass yield of strains overexpressing mutant cobT alleles was approximately 2.5 g of cell paste per liter of culture. The total amounts of the CobTWT, CobT(C160A), and CobT(G257A) proteins were 5, 10, and 1 mg/g of cell paste, respectively. These proteins were stored at −80°C until used. In contrast, only small amounts of the CobT(G320D), CobT(G171D), and CobT(G257D) proteins were isolated (0.1, 0.2, and 0.03 mg/g of cells paste, respectively), and because they were unstable, they were not stored at −80°C; instead, they were stored at 4°C and their activity was assessed immediately after isolation.
Kinetic parameters of class M CobT enzymes.
The phosphoribosyltransferase activities of the CobT(C160A), CobT(G320D), CobT(G171D), CobT(G275A), and CobT(G257D) proteins were quantified. The results of these activity measurements are shown in Table 3.
TABLE 3.
Kinetic parameters of wild-type and class M CobT proteinsa
| Protein | Sp act (μmol/min/mg) | KmNaMN (mM) | kcat (s−1) | kcat/Km(s−1 mM−1) | % Loss of catalytic efficiency |
|---|---|---|---|---|---|
| CobTWT | 30 ± 2 | 0.3 ± 0.03 | 38 ± 2 | 138 ± 22 | 0 |
| CobT(C160A) | 25 ± 2 | 1.2 ± 0.5 | 31 ± 3 | 29 ± 12 | 79 |
| CobT(G257A) | 11 ± 3 | 0.3 ± 0.05 | 14 ± 3 | 50 ± 14 | 64 |
| CobT(G257D) | 0.1 ± 0.01 | 2.2 ± 1.0 | 0.2 ± 0.1 | 0.1 ± 0.1 | >99 |
| CobT(G171D) | 0.5 ± 0.05 | 0.9 ± 0.15 | 0.6 ± 0.06 | 0.7 ± 0.04 | >99 |
| CobT(G320D) | 0.2 ± 0.1 | 1.0 ± 0.1 | 0.2 ± 0.01 | 0.2 ± 0.02 | >99 |
The amount of total protein used in the assays was adjusted to a concentration that resulted in less than 10% of product formed. The activities of the CobTWT, CobT(C160A), and CobT(G257A) proteins were assayed by using 2.5 ng of total protein, while the activities of the CobT(G320D), CobT(G171D), and CobT(G257D) proteins were assayed by using 500, 50, and 200 ng of protein, respectively. The activities of the CobTWT, CobT(C160A), CobT(G257A), CobT(G320D), CobT(G171D), and CobT(G257D) proteins were assayed at 5, 5, 12, 10, 15, and 4 min, respectively.
A change at residue G171 or G257 results in a less efficient enzyme.
The CobTWT and CobT(G257A) enzymes had similar KmNaMN values, but the CobT(G257A) variant was about threefold slower (kcat = 14 s−1) than the CobTWT enzyme (kcat = 38 s−1). However, the G257D substitution had a negative effect on the KmNaMN of the enzyme; that is, the KmNaMN (2.2 mM) was sevenfold greater than that of the CobTWT enzyme (0.3 mM). More drastic was the effect of the G257D substitution on the kcat, which was reduced 190-fold. The net effect of the G257D substitution was a 3-order-of-magnitude decrease in the catalytic efficiency of the enzyme. The effect of the G171D substitution was equally strong, increasing the KmNaMN by a factor of 3, slowing down the reaction by a factor of 63, resulting in an enzyme ∼200-fold less efficient than CobTWT.
Effect of the G320D substitution.
The G320D change increased the KmNaMN by a factor of 3 and reduced the kcat 190 fold, resulting in a much less efficient enzyme (690-fold) than CobTWT.
Effect of the C160A substitution.
The effect of the C160A change on the kcat was minimal (20% reduction), but we measured a fourfold increase in the Km for NaMN. The net effect was an enzyme fivefold less efficient than CobTWT.
DISCUSSION
Insights into the mechanism of CobT function were obtained through in vivo and in vitro analyses of the effect of class M CobT variants on coenzyme B12 synthesis in Salmonella enterica.
In vivo assessments of CobT variant functionality reveal degrees of severity.
The in vivo quantification of enzymatic activity associated with class M CobT proteins was based on our ability to discern degrees of function by tuning the demand for Cbl up or down. We identified CobT variants with low but detectable activities by demanding methionine synthesis via the Cbl-dependent methionine synthase (MetH) enzyme. CobT variants with higher levels of activity were revealed during growth on ethanolamine as a carbon and energy source.
The CobT(G257D) variant is unstable.
The CobT(G257D) variant lost >99% of its catalytic efficiency (Table 3) and did not support cell growth, even under conditions where the demand for Cbl was low (i.e., methionine synthesis). The fact that we could only purify minute amounts of the CobT(G257D) protein suggests rapid turnover of the protein. Modeling (Fig. 4) suggests that the above-mentioned effects may be due to ionic interactions introduced by the carboxylate function of the side chain of Asp. Residue G257 itself is not critical to CobT function, as shown by in vivo (Fig. 2C) and in vitro (Table 3) data obtained with a CobT(G257A) variant.
The CobT(G171D) protein may have low affinity for DMB.
Although the G171D substitution severely impairs CobT function (Fig. 2A and B), the enzyme is stable (see Fig. S6 in the supplemental material) and its activity is recovered with high concentrations of DMB (Fig. 2B). Based on this result, we suggest that the CobT(G171D) protein may have lower affinity for DMB, or alternatively, the effect might be due to restricted access of DMB to the active site. Unfortunately, for technical reasons, we have not been able to accurately measure the Km of the enzyme for DMB, but we suspect it to be in the low nanomolar range.
The G320D substitution may block access to the active site.
Structural modeling provides additional insight into how the G320D substitution may affect enzyme activity. Residue G320 is found in an α helix along the dimer interface (Fig. 3). The helixes in both subunits lie beside each other along the dimer interface in opposite orientations. A G320D change in both subunits may restrict the access of DMB to the active site. This effect may arise as a result of hydrogen bond interactions between D320 and H289 (Fig. 4), an idea that is supported by results obtained with the CobT(G320D H289A) enzyme, which was as active as CobTWT when DMB (600 μM) was present in the medium (Fig. 5A).
The disulfide bond in CobT is not critical to enzyme function.
Results from in vivo and in vitro analyses of CobT(C160A) function indicate that this variant is fivefold less efficient than CobTWT, mostly due to a fourfold increase in the Km for NaMN (Table 3). On the basis of this information, we suggest that the disulfide bond formed between C256 and C160 plays a fine-tuning role in the activity of the enzyme as a function of the NaMN concentration, and under some stressful conditions, the role of the disulfide bond may become more evident. Under the conditions tested, the disulfide bond was not critical for CobT function.
Why is intragenic complementation between mutant cobT alleles not observed?
We propose that the lack of intragenic complementation reported among class M cobT alleles is caused by the formation of dimers between two mutant CobT proteins with residual activity that is so low that the resulting CobT dimer cannot synthesize sufficient α-RP to keep up with even the lowest demand for AdoCbl.
Supplementary Material
Acknowledgments
This work was supported by PHS grant R01-GM40313 to J.C.E.-S.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersson, D. I., and J. R. Roth. 1989. Mutations affecting regulation of cobinamide biosynthesis in Salmonella typhimurium. J. Bacteriol. 171:6726-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 2002. Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica. J. Biol. Chem. 277:41120-41127. [DOI] [PubMed] [Google Scholar]
- 8.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 2001. Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica. J. Biol. Chem. 276:37612-37620. [DOI] [PubMed] [Google Scholar]
- 9.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 1999. The three-dimensional structures of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella typhimurium complexed with 5,6-dimethybenzimidazole and its reaction products determined to 1.9Å resolution. Biochemistry 38:16125-16135. [DOI] [PubMed] [Google Scholar]
- 10.Cormack, B. 1997. Directed mutagenesis using the polymerase chain reaction. Greene Publishing Associates & Wiley Interscience, New York, NY.
- 11.Escalante-Semerena, J. C. 2007. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J. Bacteriol. 189:4555-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escalante-Semerena, J. C., M. G. Johnson, and J. R. Roth. 1992. The CobII and CobIII regions of the cobalamin (vitamin B12) biosynthetic operon of Salmonella typhimurium. J. Bacteriol. 174:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escalante-Semerena, J. C., and J. R. Roth. 1987. Regulation of cobalamin biosynthetic operons in Salmonella typhimurium. J. Bacteriol. 169:2251-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escalante-Semerena, J. C., S. J. Suh, and J. R. Roth. 1990. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeter, R. M., B. M. Olivera, and J. R. Roth. 1984. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159:206-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Maggio-Hall, L. A., P. C. Dorrestein, J. C. Escalante-Semerena, and T. P. Begley. 2003. Formation of the dimethylbenzimidazole ligand of coenzyme B12 under physiological conditions by a facile oxidative cascade. Org. Lett. 5:2211-2213. [DOI] [PubMed] [Google Scholar]
- 18.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 2003. Alpha-5,6-dimethylbenzimidazole adenine dinucleotide (alpha-DAD), a putative new intermediate of coenzyme B12 biosynthesis in Salmonella typhimurium. Microbiology 149:983-990. [DOI] [PubMed] [Google Scholar]
- 19.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 1999. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. USA 96:11798-11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez, E., B. Bartolomé, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Moore, D. D. 2001. Commonly used reagents and equipment, p. A.2.1-A.2.8. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Appendix 2. http://mrw.interscience.wiley.com/emrw/9780471142720/cp/cpmb/article/mba02/current/pdf. [DOI] [PubMed]
- 23.Rondon, M. R. 1995. Regulation of the cobalamin biosynthetic (cob) and 1,2-propanediol utilization (pdu) genes of Salmonella typhimurium by the PocR protein and other cellular factors. Ph.D. dissertation. University of Wisconsin—Madison, Madison, WI.
- 24.Ryu, J., and R. J. Hartin. 1990. Quick transformation in Salmonella typhimurium LT2. BioTechniques 8:43-45. [PubMed] [Google Scholar]
- 25.Sasse, J. 1991. Detection of proteins, p.10.6.1-10.6.8. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Wiley Interscience, New York, NY. [Google Scholar]
- 26.Starai, V. J., J. Garrity, and J. C. Escalante-Semerena. 2005. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology 151:3793-3801. [DOI] [PubMed] [Google Scholar]
- 27.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 29.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1.-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, NY. [Google Scholar]
- 30.Trzebiatowski, J. R., and J. C. Escalante-Semerena. 1997. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J. Biol. Chem. 272:17662-17667. [DOI] [PubMed] [Google Scholar]
- 31.Trzebiatowski, J. R., G. A. O'Toole, and J. C. Escalante-Semerena. 1994. The cobT gene of Salmonella typhimurium encodes the NaMN:5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-α-d-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 176:3568-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren, M. J., E. Raux, H. L. Schubert, and J. C. Escalante-Semerena. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19:390-412. [DOI] [PubMed] [Google Scholar]
- 33.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 34.Zayas, C. L., and J. C. Escalante-Semerena. 2007. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosylcobalamin-5′-phosphate by the CobC phosphatase is the last step of the pathway. J. Bacteriol. 189:2210-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.