Abstract
We recently delineated the importance of a type VI secretion system (T6SS) gene cluster in the virulence of diarrheal isolate SSU of Aeromonas hydrophila and showed that VasH, a σ54 activator and T6SS component, was involved in the production of its associated effectors, e.g., hemolysin-coregulated protein. To identify additional T6SS effectors and/or secreted proteins, we subjected culture supernatants from deletion mutants of A. hydrophila, namely, a Δact mutant (a T2SS-associated cytotoxic enterotoxin-encoding gene) and a Δact ΔvasH mutant, to 2-dimensional gel electrophoresis and mass spectrometric analysis. Based on these approaches, we identified a member of the VgrG protein family, VgrG1, that contained a vegetative insecticidal protein (VIP-2) domain at its carboxyl-terminal end. Consequently, the vgrG1 gene was cloned in pBI-EGFP and pET-30a vectors to be expressed in HeLa Tet-Off cells and Escherichia coli, respectively. We assessed the ADP-ribosyltransferase (ADPRT) activity of various domains of purified recombinant VgrG1 (rVgrG1) and provided evidence that only the full-length VgrG1, as well as its carboxyl-terminal domain encoding the VIP-2 domain, showed ADPRT activity. Importantly, bacterium-host cell interaction was needed for the T6SS to induce cytotoxicity in eukaryotic cells, and we demonstrated translocation of VgrG1. Furthermore, our data indicated that expression of the genes encoding the full-length VgrG1 and its carboxyl-terminal domain in HeLa Tet-Off cells disrupted the actin cytoskeleton, which was followed by a decrease in cell viability and an increase in apoptosis. Taken together, these findings demonstrated for the first time that VgrG1 of A. hydrophila possessed actin ADPRT activity associated with its VIP-2 domain and that this domain alone was able to induce a rounded phenotype in HeLa Tet-Off cells, followed by apoptosis mediated by caspase 9 activation.
The presence of Aeromonas species in water distribution systems and different foods indicates their potential as a food-borne pathogen, and therefore, this organism represents a public health concern (10). Consequently, our laboratory identified and characterized multiple virulence factors from diarrheal isolate SSU of Aeromonas hydrophila. One of the most potent virulence factors of this A. hydrophila strain is the cytotoxic enterotoxin, designated Act, which is secreted via a type II secretion system (T2SS) (14, 15, 37). The other recently identified virulence factor from this isolate is the type III secretion system (T3SS)-associated effector AexU (41-43).
Microbial toxins with ADP-ribosylating activity have been grouped into four different families based on their respective host targets (20, 45). Type I toxin proteins target heteromeric GTP-binding proteins (e.g., cholera and pertussis toxins) (16, 21), type II proteins modify elongation factor 2 (e.g., diphtheria toxin and Pseudomonas aeruginosa exotoxin A) (47, 51), type III proteins target small GTP-binding proteins (e.g., Clostridium botulinum C3 exoenzyme) (3), and type IV proteins are specific for ADP ribosylation of G-actin. The last family of proteins includes C. botulinum C2 toxin, Clostridium perfringens iota toxin, Clostridium spiroforme toxin, Clostridium difficile binary toxin A, and Bacillus cereus vegetative insecticidal protein (VIP) (1, 2, 5, 18, 31, 32, 46).
The type VI secretion system (T6SS) was recently characterized from several gram-negative bacteria (6, 8, 36). Most of the genes located in the T6SS cluster do not exhibit any identity with the genes of the other secretion systems; however, ortholog proteins, such as IcmF and DotU, have been reported as part of the T4SS in Legionella pneumophila (39). The T6SS system is able to export effector proteins into the extracellular milieu and/or is able to translocate them into eukaryotic host cell cytoplasm, as type 3 and type 4 secretion systems do by an as-yet-unknown mechanism which is currently under study (9, 24, 27, 34, 35, 44). Importantly, known proteins that are exported or translocated through this system do not have any known signal peptide, which indicates that they are secreted in a Sec- or Tat-independent manner (8, 9, 29, 50).
Two T6SS-associated effectors, namely, hemolysin-coregulated protein (Hcp) and valine-glycine repeat G (VgrG), have recently been molecularly and structurally characterized from Vibrio cholerae and P. aeruginosa (24, 27, 34, 35). The V. cholerae genome has three copies of VgrG which are located both within and outside of the T6SS gene cluster (35). Our recent analysis of the complete genome of A. hydrophila ATCC 7966T, an environmental isolate, also indicated the presence of three copies of the vgrG gene with a distribution in the genome similar to that in V. cholerae (44). In some bacteria, vgrG genes are linked to the hcp gene (12), and the expression of the vgrG (vgrG2 and vgrG3) genes is required for Hcp secretion. Likewise, expression of the hcp gene is required for secretion of VgrG proteins in V. cholerae (34, 35). In addition, structural analysis of Hcp and VgrG from P. aeruginosa and V. cholerae showed that these proteins independently formed channel-like structures which could be used to transport macromolecules through them (23, 27, 34). Thus, these data suggested that VgrG and Hcp proteins could be part of the secretion apparatus.
Sequence analyses of VgrG proteins from different bacteria showed that all of them were highly conserved in their NH2-terminal domains. The domain called VgrG, or COG3501, shared similarities to the gp5 and gp27 proteins of the bacteriophage T4 tail spike (12, 34). Some VgrG proteins had different COOH-terminal extensions, which contained domains having different activities. For example, VgrG from P. aeruginosa carried a zinc-metalloprotease domain, while VgrG1 and VgrG3 from V. cholerae contained repeats in the structural toxin A (RtxA) and peptidoglycan binding domains, respectively (34). Only recently was it reported that VgrG1 from V. cholerae is translocated into eukaryotic host cells with deleterious effects (24), a finding that suggests VgrG proteins with extended COOH termini could have roles as T6SS effectors.
Recently, we reported the importance of vasH (σ54 activator) and vasK of A. hydrophila SSU in the expression of the T6SS gene cluster and the secretion and translocation of T6SS-associated effector proteins and their crucial roles in evoking mouse lethality (44). In our previous study (44) and now, in the current article, we demonstrated that the vasH isogenic mutant was unable to express and produce known T6SS proteins, such as Hcp and VgrG2/3. On the other hand, the vasK isogenic mutant was able to express and translocate Hcp into the host eukaryotic cell but unable to secrete it into the extracellular milieu (44). Since VasH is a member of the RpoN family of σ54 transcriptional factor regulators, which are associated with modulation of virulence factors in several bacteria (33), it is not surprising that the ΔvasH mutant of A. hydrophila SSU did not express genes present in the T6SS gene cluster.
The A. hydrophila SSU genome has not been sequenced; however, the proteomics analysis of the data indicated the existence of VgrG1, with its gene localized outside the T6SS gene cluster. Furthermore, we found that this protein has a COOH-terminal extension containing a vegetative insecticidal protein-2 (VIP-2) domain, known for its actin ADP-ribosylating activity (18). We noted that the expression of the gene encoding either the full-length VgrG1 or its COOH-terminal domain that contained only the VIP-2 domain induced apoptosis in HeLa Tet-Off cells. Overall, we demonstrated that VgrG1 is an important virulence factor of A. hydrophila that is secreted and also translocated via the T6SS. Furthermore, this is the first characterization of VgrG from any bacterium with actin ADP-ribosylating activity.
MATERIALS AND METHODS
Bacterial strains.
Wild-type A. hydrophila SSU and its various mutant strains were grown in Luria-Bertani (LB) medium at 37°C with continuous shaking (180 rpm). Deletion mutants of A. hydrophila SSU, with Δact and Δact ΔvasH mutations, previously developed in our laboratory (44), were grown in LB medium supplemented with 100 μg/ml of kanamycin (Sigma, St. Louis, MO) and streptomycin plus spectinomycin (100 μg/ml each; Sigma), respectively. The bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and vectors used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or referencea |
|---|---|---|
| A. hydrophila strains | ||
| SSU | Diarrheal isolate of A. hydrophila | CDC |
| SSU-R | Rifampin-resistant (Rifr) strain | Laboratory stock |
| Δact strain | Isogenic act gene mutant of SSU-R; Rifr Kmr | 14 |
| Δact ΔvasH strain | Isogenic vasH gene mutant of SSU-R Δact; Rifr Kmr Smr/Spr | 44 |
| Δact ΔvasH vasH strain | Isogenic vasH gene mutant of SSU-R Δact strain complemented with vasH in pBR322; Rifr Kmr Smr/Spr Apr | This study |
| Δact vgrG1::bla strain | Isogenic act gene mutant of SSU-R expressing the vgrG1::bla gene in pGEN222; Rifr Kmr Apr | This study |
| Δact vgrG1-NH2::bla strain | Isogenic act gene mutant of SSU-R expressing the vgrG1-NH2::bla gene in pGEN222; Rifr Kmr Apr | This study |
| Δact ΔvasH vgrG1::bla strain | Isogenic vasH gene mutant of SSU-R Δact strain expressing the vgrG1::bla gene in pGEN222; Rifr Kmr Smr/Spr Apr | This study |
| Δact ΔvasH vgrG1-NH2::bla strain | Isogenic vasH gene mutant of SSU-R Δact expressing the vgrG1-NH2::bla gene in pGEN222; Rifr Kmr Smr/Spr Apr | This study |
| ATCC 7966 | Environmental isolate | ATCC |
| E. coli strains | ||
| DH5α | Production of recombinant plasmids; recA gyrA | Life Technologies |
| HMS174-DE3 | Production of recombinant proteins cloned in pET-30a, carries a T7 RNA polymerase copy under the control of lacUV5 promoter | Novagen |
| Plasmids | ||
| pET-30a | pBR322-derived expression vector with T7 lac promoter, expression and production of recombinant proteins with 6×His tag; Kmr | Novagen |
| pBI-EGFP | Expression and production of recombinant proteins in Tet-Off eukaryotic system; Apr | Clontech |
| pBR322 | Apr Tcr | GE Healthcare |
| pGEN222 | Apr | 11 |
CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection.
Vectors.
The genes encoding full-length VgrG1 or its NH2- or COOH-terminal domain from A. hydrophila ATCC 7966 were cloned into a pET-30a vector (Novagen, Madison, WI) for hyperexpression and purification purposes. The DNA fragments were cloned into the BglII and XhoI or SalI restriction enzyme sites of the vector. The recombinant proteins contained a histidine tag for nickel affinity chromatography at the NH2-terminal end. The pBI-EGFP (enhanced green fluorescent protein) vector (Clontech, Mountain View, CA) was used to express and produce recombinant proteins from a HeLa Tet-Off cell system (Clontech). The fragments encoding the full-length VgrG1 or the NH2- or COOH-terminal domain of VgrG1 were cloned into the MluI and NheI restriction enzyme sites.
For translocation studies, the gene segment from A. hydrophila ATCC 7966 encoding either full-length VgrG1 or its NH2-terminal domain without the stop codon was cloned in the pGEN222 vector (11), between the ClaI and MluI restriction enzyme sites. Downstream from the above-mentioned vgrG1 genes, the β-lactamase gene (blaM; without the region encoding the signal peptide) was cloned between the MluI and SalI restriction enzyme sites to produce in-frame fusions. These recombinant plasmids were electroporated into A. hydrophila SSU Δact and Δact ΔvasH mutant strains. The production of fusion proteins from these bacterial strains was tested by Western blot analysis using antibodies against Bla (Abcam, Cambridge, MA), the VIP-2 domain of VgrG1, and VgrG2 after 2 h of cultivation at 37°C in Dulbecco's modified Eagle's medium (DMEM) containing 75 mM NaCl to induce the expression of the target gene. Table 1 lists the plasmids used in this study. The primers, having different restriction enzyme sites and used to clone various DNA fragments, were synthesized by Integrated DNA Technologies (Cedar Rapids, IA) and are shown in Table 2, along with their sequences.
TABLE 2.
Primer sequences used in this study
| Primerb | Sequencea | Purpose |
|---|---|---|
| vgrG1-Full-MluI | 5′-TCATACGCGTATGGCAGACAGCACAGG-3′ | PCR amplification of full-length VgrG1-encoding region for cloning in pBI-EGFP |
| vgrG1-Full-NheI | 5′-GGTCGCTAGCTTATAATACGGAAACCTC-3′ | |
| vgrG1-NH2-MluI | 5′-TCATACGCGTATGGCAGACAGCACAGG-3′ | PCR amplification of VgrG1 NH2-terminal-domain-encoding region for cloning in pBI-EGFP |
| vgrG1-NH2-ter-NheI | 5′-GGTCGCTAGCTTATGGGCCTCCGGCAAGCTGG-3′ | |
| vgrG1-COOH-ter-MluI | 5′-TCATACGCGTATGATGCCACCGCCGCCGCC-3′ | PCR amplification of VgrG1 COOH-terminal-domain-encoding region for cloning in pBI-EGFP |
| vgrG1-COOH-ter-NheI | 5′-GGTCGCTAGCTTATAATACGGAAACCTC-3′ | |
| vgrG1-Full-BglII | 5′-TCATAGATCTAATGGCAGACAGCACAGG-3′ | PCR amplification of full-length VgrG1-encoding region for cloning in pET-30a |
| vgrG1-Full-SalI | 5′-GGTCGTCGACTTATAATACGGAAACCTC-3′ | |
| vgrG1-NH2-ter-BglII | 5′-TCATAGATCTAATGGCAGACAGCACAGG-3′ | PCR amplification of VgrG1 NH2-terminal-domain-encoding region for cloning in pET-30a |
| vgrG1-NH2-ter-SalI | 5′-GGTCGTCGACTTATGGGCCTCCGGCAAGCTGG-3′ | |
| vgrG1-COOH-ter-BglII | 5′-TCATAGATCTAATGATGCCACCGCCGCCGCC-3′ | PCR amplification of VgrG1 COOH-terminal-domain-encoding region for cloning in pET-30a |
| vgrG1-COOH-ter-XhoI | 5′-GGTCCTCGAGTCATAATACGGAAACCTCAATC-3′ | |
| vgrG2-MluI | 5′-TCATACGCGTAATGGCAGACAGCACAGG-3′ | PCR amplification of full-length VgrG2-encoding region for cloning in pBI-EGFP |
| vgrG2-NheI | 5′-GGTCGCTAGCTCAGCCACCACCCTCCTGTCTGG-3′ | |
| vgrG2-BglII | 5′-TCATAGATCTAATGGCAGACAGCACAGG-3′ | PCR amplification of full-length VgrG2-encoding region for cloning in pET-30a |
| vgrG2-XhoI | 5′-GGTCCTCGAGTCAGCCACCACCCTCCTGTCTGG-3′ | |
| vgrG1-Full-ClaI | 5′-TCATATCGATAATGGCAGACAGCACAGG-3′ | PCR amplification of full-length vgrG1 without the stop codon for cloning in pGEN222 |
| vgrG1-Full-MluI | 5′-GGTCACGCGTTAATACGGAAACCTC-3′ | |
| vgrG1-NH2-ClaI | 5′-TCATATCGATAATGGCAGACAGCACAGG-3′ | PCR amplification of VgrG1 NH2-terminal-domain-encoding region without the stop codon for cloning in pGEN222 |
| vgrG1-NH2-MluI | 5′-GGTCACGCGTTGGGCCTCCGGCAAGCTGG-3′ | |
| bla-MluI | 5′-GGTCACGCGTATGCACCCAGAAACGCTGGTG-3′ | PCR amplification of blaM without the signal sequence for cloning in pGEN222-vgrG1s |
| bla-SalI | 5′-GGTCGTCGACTTACCAATGCTTAATCAGTG-3′ |
Underlined sequences represent restriction enzyme sites.
The designations NH2-ter and COOH-ter refer to the NH2- and COOH-terminal domains of VgrG1.
Cell lines.
HeLa Tet-Off cells (Clontech), a human cervical epithelial cell line, were grown in DMEM with high glucose (Invitrogen/Gibco Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Tet system approved; Clontech) and 100 μg/ml of G418 (Cellgro, Herndon, VA) under standard tissue culture conditions at 37°C and 5% CO2 in a humid atmosphere. The HeLa Tet-Off cells were transfected by electroporation with the recombinant pBI-EGFP plasmids containing different vgrG1 fragments. Briefly, single-cell suspensions (∼5 × 106 cells/ml) were transfected with different plasmid constructs in 4-mm cuvettes (Bio-Rad, Hercules, CA) by using an exponential protocol (300 V, 950 μF, and ∞Ω) in a Gene Pulser Xcell (Bio-Rad). The cells were then recovered in complete medium, plated, and grown under standard tissue culture conditions.
Normal HeLa cells (without the Tet-Off system) were used for assays that did not require transfection. These cells were grown in high-glucose DMEM supplemented with 10% FBS.
Recombinant protein production.
Escherichia coli HMS174-DE3 cells containing pET-30a recombinant plasmids (i.e., coding for VgrG2 and the VIP-2 domain of VgrG1) were grown in LB medium and induced with 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) for 4 h at 37°C. Recombinant proteins were purified by using the ProBond purification system (Invitrogen) and following the nondenaturing protocol as described by the manufacturer. One-milliliter fractions were collected and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining. Fractions containing the protein of interest were mixed and dialyzed overnight against phosphate-buffered saline (PBS) at 4°C. The protein concentration was measured by using a Bradford assay (Bio-Rad).
Antibody production.
Female Swiss Webster mice (n = 5; Taconic Farms, Germantown, NY) were immunized via the intraperitoneal route with 10 μg of purified recombinant protein mixed with complete Freund's adjuvant (Sigma) and boosted on day 15 by using incomplete Freund's adjuvant (Sigma). Sera were obtained from mice at weeks 2 and 4 after immunization. The antibody specificity was determined by Western blot analysis by using whole E. coli lysates containing recombinant proteins, as well as the respective purified recombinant protein as the source of antigen. Antibodies against recombinant Hcp (rHcp) were previously developed in our laboratory (44).
Since we used recombinant VgrG2 (rVgrG2) for antibody production, the sera obtained did not differentiate between VgrG2 and VgrG3 proteins of A. hydrophila SSU due to the high homology (∼90%) between the proteins and their similar sizes (∼77 kDa) on Western blots. As a result, we refer to VgrG2 as VgrG2/3 in our Western blot analysis data.
Western blot analysis.
HeLa Tet-Off cells were lysed in Tris-glycine buffer after 24 h of transfection and subjected to SDS-PAGE. The proteins from the gels were then transferred to Hybond-ECL nitrocellulose membranes (GE Healthcare, Piscataway, NJ), which were blocked and then treated with specific primary and secondary antibodies, as we previously described (44). SuperSignal West pico or femto chemiluminescence substrate (Thermo Scientific, Rockford, IL) was used to develop the blots, followed by X-ray film exposure.
Two-dimensional (2-D) gel electrophoresis and mass spectrometry.
The LB cultures of the A. hydrophila SSU Δact and Δact ΔvasH mutants were grown for 2 h to a cell density of 5 × 106 cells/ml in 10 ml of DMEM with high glucose and without FBS at 37°C. The supernatants obtained after centrifugation (6,000 × g for 10 min) were filtered through 0.2-μm filters. Next, the proteins in the supernatants were precipitated with trichloroacetic acid (TCA) (10% [vol/vol] final concentration; Sigma), and the pellet was collected by centrifugation at 14,000 × g for 20 min at 4°C. The protein pellets were washed 3 times with cold acetone, dried, and resuspended in 200 μl of DeStreak rehydration solution (GE Healthcare).
For the first dimension, the proteins were separated in 13-cm, nonlinear pH 3 to 10 strips (GE Healthcare) by using the following protocol in an IPGphor apparatus (GE Healthcare): rehydration, 50 V for 10 h, 250 V for 1 h, 500 V for 1 h, 1,000 V for 2 h, and 8,000 V for 7 h at 75 μA/strip and 20°C. Subsequently, the strips were incubated in equilibrium buffer (GE Healthcare) for 10 min followed by SDS-4-to-20% PAGE. The gels were stained with Sypro ruby (Bio-Rad), and images for analysis were acquired in a Gel Doc system (Bio-Rad).
As an analysis strategy for the 2-D gels, the proteins present in supernatants of the Δact mutant of A. hydrophila SSU but not in the supernatants of the Δact ΔvasH mutant were considered prime candidates for future study. For this analysis, we used 3 gels per bacterial mutant strain, and the differential spots from the 2-D gels were identified by using Progenesis Samespot version 2.0.2733.19819 software (Nonlinear Dynamics, Durham, NC). Different spots were manually picked, trypsin digested, and analyzed by mass spectrometry (matrix-assisted laser desorption ionization-time of flight [MALDI-TOF]). The peptide sequences then were matched against the proteobacterial database, and results with high homologies (low expectation numbers) (Table 3) were examined.
TABLE 3.
Mass spectrometric alignment of differentially produced protein spots from supernatants of A. hydrophila SSU Δact and Δact ΔvasH strains, based on 2-D gel electrophoresisa
| Spot | Rank | Protein with highest homology | Protein expectation score |
|---|---|---|---|
| 5_4804 | 1 | VgrG3 protein of Aeromonas hydrophila | 9.97631E-70 |
| 5_5573 | 1 | VgrG3 protein of Aeromonas hydrophila | 2.50594E-65 |
| 5_5562 | 1 | VgrG2 protein of Aeromonas hydrophila | 9.97631E-53 |
| 5_4798 | 1 | VgrG3 protein of Aeromonas hydrophila | 6.29463E-46 |
| 5_6426 | 1 | VgrG3 protein of Aeromonas hydrophila | 3.15479E-43 |
| 5_4785 | 1 | VgrG3 protein of Aeromonas hydrophila | 6.29463E-39 |
| 3_3026 | 1 | Putative hemolysin-coregulated protein of Aeromonas hydrophila | 1.99054E-35 |
| 5_4809 | 1 | Rhselement Vgr family protein ofAeromonas hydrophilasubsp.hydrophilaATCC 7966 | 3.97164E-34 |
| 5_5996 | 1 | VgrG3 protein of Aeromonas hydrophila | 3.97164E-33 |
| 5_6503 | 1 | Putative VgrG protein of Aeromonas hydrophila | 9.97631E-26 |
| 5_5611 | 1 | Phosphatidylcholine-sterol acyltransferase of Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 9.97631E-20 |
| 5_6093 | 1 | Putative VgrG protein of Aeromonas hydrophila | 7.92447E-14 |
| 3_5936 | 1 | Rhselement Vgr family protein ofAeromonashydrophilasubsp.hydrophilaATCC 7966 | 1.25594E-13 |
| 5_5992 | 1 | VgrG3 protein of Aeromonas hydrophila | 9.97631E-10 |
| 3_4252 | 1 | HcpA homolog of Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 9.97631E-09 |
| 3_6571 | 1 | HcpA homolog of Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 1.99054E-08 |
| 5_5746 | 1 | Hypothetical protein AHA_3948 of Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 1.25594E-05 |
| 5_6471 | 1 | Inorganic pyrophosphatase of Aeromonas hydrophila subsp. hydrophila ATCC 7966 | 0.007924466 |
| 5_4403 | 1 | VgrG protein of Aeromonas salmonicida subsp. salmonicida A449 | 0.019905359 |
| 5_539 | 1 | Type III restriction protein Res subunit of Xanthobacter autotrophicus Py2 | 0.019905359 |
| 5_260 | 1 | Ribosomal large subunit pseudouridine synthase B of Aeromonas salmonicida subsp. salmonicida A449 | 0.039716412 |
| 5_4807 | 1 | Transcriptional regulator of GntR family of Mesorhizobium sp. BNC1 | 0.199053585 |
| 3_6711 | 1 | Hypothetical protein ECA4275 of Erwinia carotovora subsp. atroseptica SCRI1043 | 0.629462706 |
| 3_6570 | 1 | Hypothetical protein Acry_2197 of Acidiphilium cryptum JF-5 | 1.255943216 |
| 5_5609 | 1 | Flagellar hook-associated 2 domain protein of Shewanella baltica OS155 | 1.255943216 |
| 5_6215 | 1 | Hypothetical protein msr9585 of Mesorhizobium loti MAFF303099 | 1.255943216 |
| 5_4784 | 1 | Hypothetical protein RPC_0976 of Rhodopseudomonas palustris BisB18 | 2.505936168 |
| 5_5356 | 1 | Regulatory protein of Oceanicola granulosus HTCC2516 | 6.294627059 |
| 5_548 | 1 | Hypothetical protein HI0688 of Haemophilus influenzae Rd KW20 | 6.294627059 |
| 5_6707 | 1 | Peptidyl-prolyl cis-trans isomerase C of Yersinia pestis KIM | 6.294627059 |
| 5_6113 | 1 | Putative ABC-type multidrug transport system, ATPase component, of Photobacterium profundum 3TCK | 7.924465962 |
| 5_6708 | 1 | General secretion pathway protein L of Vibrio cholerae MZO-3 | 15.8113883 |
Data in bold font represent protein spots matching VgrG1 from A. hydrophila ATCC 7966.
ADP ribosylation assay.
HeLa cells in suspension were lysed by sonication in a buffer containing 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol (DTT), 5 mM MgCl2, and protease inhibitors. The whole lysate was centrifuged at 14,000 × g for 10 min at 4°C, and the supernatant was used for the ADP ribosylation assay (19). Briefly, 50 μg of normal HeLa cell lysate proteins or 1 μg of recombinant nonmuscle actin (Cytoskeleton, Denver, CO) as the target protein for ADP ribosylation was incubated with 1 μg of different purified rVgrG1 proteins and 10 μM NAD conjugated with biotin (R&D Systems, Minneapolis, MN) for 30 min at 37°C. The reaction was stopped by adding SDS-sample buffer and the proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes (GE Healthcare), and incubated with streptavidin-horseradish peroxidase (HRP) after nonspecific sites were blocked with 1% bovine serum albumin (BSA). SuperSignal West pico chemiluminescence substrate was used to develop the blot, followed by X-ray film exposure.
Translocation of VgrG1.
For studying translocation of VgrG1::Bla fusion proteins in HeLa cells, we used CCF4-AM as a substrate for Bla. CCF4-AM, a fluorescence resonance energy transfer (FRET)-based substrate for β-lactamase, contains a cephalosporin core linked to coumarin and fluorescein. CCF4 requires an excitation of 408 nm and produces emissions of 460 nm (blue from coumarin) and 530 nm (green from fluorescein). In the presence of β-lactamase activity, the substrate is cleaved, resulting in the breakage of FRET and loss of the 530-nm emission (green).
For translocation purposes, the full-length VgrG1::Bla or the NH2-terminal VgrG1::Bla from the pGEN222 vector was produced from A. hydrophila SSU Δact and Δact ΔvasH mutant strains. The mutant strains harboring only the vector were used as negative controls. HeLa cells in suspension were placed in flow cytometry tubes in DMEM without serum at a concentration of 1 × 106 cells/ml. These cells were infected at a multiplicity of infection (MOI) of 10, with A. hydrophila SSU Δact and Δact ΔvasH mutant strains producing different forms of VgrG1::Bla fusion proteins. After 15 min of infection, CCF4-AM working solution (6×) (Invitrogen) was added and the tubes incubated for 45 min. The cells were then acquired in a Becton Dickinson FACSAria flow cytometer in which a violet laser diode (408 nm) was used for excitation and which detected emissions at 450/40 (blue) and 530/30 (green). Ratios between percentages of blue HeLa cells and percentages of green HeLa cells were calculated for analysis.
Host cell morphology.
HeLa cells plated in 6-well plates were cocultured with different mutant strains of A. hydrophila SSU at an MOI of 5 in DMEM without FBS. Changes in morphology were evaluated by phase-contrast microscopy over a 90- to 120-min period. In parallel, to test the importance of bacterium-host cell contact in morphological changes of the HeLa cells, we performed cocultures of bacteria and host cells by using transwell inserts having a 0.4-μm pore size (Costar, Corning, NY).
HeLa Tet-Off cells were electroporated with various vgrG1 constructs and, after 24 h of incubation, they were stained with Alexa Fluor 568-conjugated phalloidin (Invitrogen-Gibco) following the manufacturer's instructions. Briefly, the cells were fixed and permeabilized in situ with Cytofix/Cytoperm (Becton Dickinson, San Jose, CA) for 20 min at 4°C. Then, the cells were scraped and stained with Alexa Fluor 568-conjugated phalloidin for 1 h at room temperature and washed with Perm/Wash solution (Becton Dickinson). The cells were acquired in a FACScan flow cytometer and analyzed with FACSDiva software (Becton Dickinson). Additionally, the stained cells were placed on glass slides by using a Shandon cytospin cytocentrifuge (Thermo Scientific, Waltham, MA) at 1,500 rpm for 5 min. The coverslips were mounted with medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector, Burlingame, CA), and the images were then acquired and analyzed by fluorescence microscopy (Olympus BX51, DPManager version 1.2.1.107, and DPController version 1.2.1.108; Olympus Optical Co., Ltd.).
Quantification of G- and F-actin.
A G-actin/F-actin in vivo assay kit (Cytoskeleton) was used to quantify the amounts of G- and F-actin present in HeLa Tet-Off cells transfected with the pBI-EGFP vector expressing and producing various domains of VgrG1. Likewise, G-actin/F-actin ratios were determined in HeLa cells cocultured with different mutant strains of A. hydrophila SSU following the manufacturer's instructions. Briefly, HeLa cells plated in 12-well plates were lysed with 300 μl of lysis buffer. The cells were then scraped and passed through a 25-gauge needle. The total cell lysates were centrifuged at 100,000 × g for 1 h at 37°C, and the supernatants collected. The pellets were resuspended in the same volume (300 μl) of the SDS-PAGE loading buffer. Supernatants containing G-actin and pellets containing F-actin were subjected to SDS-10% PAGE and Western blot analysis by using an antiactin antibody (Cytoskeleton). The density of each band was calculated by using AlphaEasyFC software (Alpha Innotech, San Leandro, CA). The results were reported as the percentages of F- and G-actin per sample. The sum total of G- and F-actin corresponded to 100%.
Host cell viability.
Incorporation of 7-amino actinomycin D (7-AAD) (Becton Dickinson) was used to determine the HeLa Tet-Off cell viability. HeLa Tet-Off cells expressing different vgrG1 fragments were detached, washed, and then incubated for 10 min with 7-AAD (5 μl per tube). Immediately after staining, the cells were analyzed by using flow cytometry.
Host cell apoptosis.
We evaluated the extent of apoptosis of HeLa Tet-Off cells expressing different vgrG1 fragments by detection of cytoplasmic nucleosomes and measurement of caspase 3 and 9 activation, as we previously described (43).
Statistical analysis.
A two-way analysis of variance (ANOVA) and Bonferroni's posttest were used for statistical analysis of the data, using GraphPad Prism version 4.02 for Windows (Software MacKiev, San Diego, CA).
RESULTS
A. hydrophila SSU secretes VgrG1 via the T6SS.
We analyzed supernatants from A. hydrophila SSU Δact and Δact ΔvasH mutant strains by 2-D gel electrophoresis and identified differentially expressed and produced protein spots by mass spectrometry. We chose to use the A. hydrophila SSU Δact mutant as the parental strain to avoid any interference of T6SS-associated cytotoxicity with that contributed by Act. It has been shown that deletion of the vasH gene prevents the transcription, translocation, and secretion of effector proteins via the T6SS (i.e., Hcp) (35, 44). The approach used in this analysis was to identify protein spots present in the Δact mutant strain but not in the Δact ΔvasH mutant, thus allowing us to specifically target T6SS-secreted proteins. Among the 32 differentially expressed and produced spots (Table 3), we detected homology to the known T6SS proteins, such as Hcp-1, Hcp-2, VgrG1, VgrG2, and VgrG3. The differentially produced proteins related to the T6SS gene cluster showed low expectation scores (high homology) in comparison to the scores of those proteins not related to the T6SS gene cluster. However, those proteins with high expectation scores (low homology) need to be analyzed carefully (Table 3).
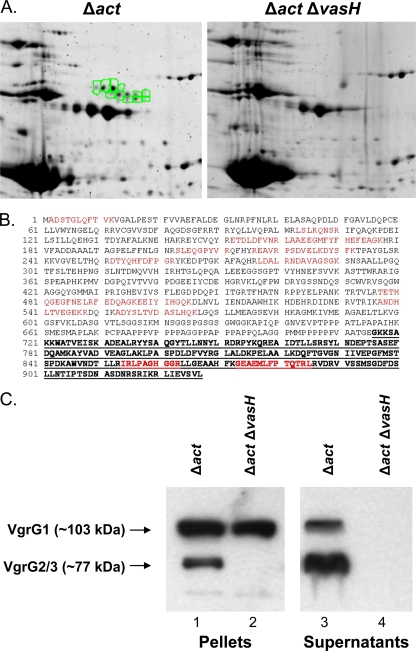
We identified by 2-D gel electrophoresis well-defined clusters in the culture supernatant of the Δact mutant strain that were absent from the Δact ΔvasH mutant strain. One of these clusters contained various isoforms of Hcp, while the other contained VgrG homologs. From the VgrG cluster (Fig. 1A, highlighted in green), we identified proteins (isoforms) with high homology (expectation score, 3.97E-34) to VgrG1 (gi|117619461) of the environmental isolate ATCC 7966 of A. hydrophila, whose genome sequence was recently annotated (38). Figure 1B depicts the alignment of sequences of 12 peptides (highlighted in red) obtained by mass spectrometric analysis of VgrG1 from A. hydrophila SSU with those from the sequence of A. hydrophila ATCC 7966 VgrG1; they were exactly identical (38). Since the genome of A. hydrophila SSU is not totally sequenced yet and we have sequenced only T6SS and T3SS gene clusters (41, 44), we were unable to detect, based on computer alignment programs, matches of VgrG1 of A. hydrophila SSU with VgrG1 of A. hydrophila ATCC 7966. This is because the vgrG1 gene is located on the bacterial chromosome outside the T6SS gene cluster, in contrast to the vgrG2 and vgrG3 genes, which reside within the T6SS gene cluster.
FIG. 1.
Identification of proteins secreted via T6SS in the supernatant of A. hydrophila SSU. (A) Comparison of 2-D gels containing proteins from the supernatants of the A. hydrophila SSU Δact (left) and Δact ΔvasH (right) mutant strains. Highlighted spots (in green) represent a cluster of proteins secreted via the T6SS which were identified by mass spectrometric analysis. (B) Alignment of VgrG1 from A. hydrophila ATCC 7966 (gi|117619461) with peptides identified via mass spectrometry (red) in one of the secreted proteins (VgrG1) from A. hydrophila SSU. The bold, underlined sequence represents the VIP-2 domain. (C) Western blot analysis of pellet and supernatant fractions of A. hydrophila SSU Δact (lanes 1 and 3) and Δact ΔvasH (lanes 2 and 4) mutant strains using antibodies specific to the VIP-2 domain of VgrG1 in combination with specific antibodies against VgrG2.
Importantly, we identified the presence of a VIP-2 domain in the COOH-terminal portion (amino acid residues 702 to 927) of VgrG1 of A. hydrophila (both the SSU and ATCC 7966 strain) (E-value, 5E-10) by using a specialized BLAST search of conserved domains (Fig. 1B, indicated by underlined bold font).
Figure SI in the supplemental material depicts amino acid residues that are conserved between the reference sequence (cd00233) and the VIP-2 domain of A. hydrophila ATCC 7966 (25) in red. To confirm the secretion of VgrG proteins by A. hydrophila SSU, we performed Western blot analysis on supernatants from both the Δact and Δact ΔvasH mutant strains of A. hydrophila SSU by using antibodies specific to the VIP-2 domain of VgrG1, as well as antibodies against VgrG2 (which recognize all members of the VgrG family) (Fig. 1C).
We were able to detect VgrG1 (∼103 kDa) and VgrG2/3 (exhibiting similar molecular masses of ∼77 kDa) in the supernatant and pellet fractions of the A. hydrophila SSU Δact mutant (Fig. 1C, lanes 1 and 3). However, the Δact ΔvasH mutant of A. hydrophila SSU was unable to secrete any member of the VgrG family of proteins (Fig. 1C, lane 4). Importantly, we could detect the presence of VgrG1 but not VgrG2/3 in the pellet fraction of the A. hydrophila SSU Δact ΔvasH mutant (Fig. 1C, lane 2), which signified that, since the vgrG1 gene is outside the T6SS gene cluster, its regulation is possibly independent of VasH. However, the inability of the Δact ΔvasH mutant to secrete VgrG1 (Fig. 1C, lane 4) indicated that a T6SS apparatus was needed for the secretion of VgrG1.
We noted that the NH2-terminal domain of VgrG1 (amino acid residues 1 to 701) harboring the VgrG domain is present in all members of the VgrG protein family (see Fig. SIIA). Interestingly, within the VgrG domain, there are the phage GPD superfamily region (c|01392), which is present in the bacteriophage T4 tail spike, and the DUF586 (pfam04524) region, which has no known function (see Fig. SIIA). As noted in Fig. SIIB, the COOH-terminal extension of V. cholerae VgrG1 has an RtxA domain, unlike the VIP-2 domain (cd00233/pfam01129) found in VgrG1 of A. hydrophila SSU and ATCC 7966 strains (see Fig. SIIA). Recent reports have shown structural similarities between the VgrG domain and gp27 and gp5, as well as with Hcp-hexameric rings and gp25 of bacteriophage T4 (23, 34; also see Fig. SIIB). These findings highlighted the similarity of the T6SS apparatus to the bacteriophage T4 needlelike structure and suggested that T6SS and bacteriophage have similar functions in the translocation of proteins through host cell membranes.
VgrG1 from A. hydrophila possesses ADPRT activity.
Since the COOH-terminal domains of VgrG1 were highly conserved between A. hydrophila ATCC 7966 and A. hydrophila SSU and because the NH2-terminal domains of all known VgrGs have significant homologies, we tested the actin ADP-ribosyltransferase (ADPRT) activity associated with the VgrG1 of A. hydrophila ATCC 7966. We cloned the full-length vgrG1 gene and its NH2-terminal-domain-encoding DNA fragment (bp 1 to 2106 [amino acid residues 1 to 701]) and the DNA fragment encoding the COOH-terminal end (bp 2107 to 2784 [amino acid residues 702 to 927]), which harbors only the VIP-2 domain, in the pET-30a vector and produced the recombinant proteins in E. coli. The purified rVgrG1 full-length and rVgrG1 COOH-terminal domain proteins were able to catalyze the incorporation of biotin-ADP (from 6-biotin-17-NAD) into an approximately 45-kDa target protein in HeLa cell lysate (Fig. 2A, lanes 5 and 6) or into recombinant nonmuscle actin (Fig. 2B, lanes 5 and 6). In contrast, the purified rVgrG1 NH2-terminal domain protein was not able to catalyze this reaction (Fig. 2A and B, lanes 4). The appropriate negative controls, which included HeLa cell lysate or recombinant actin alone (Fig. 2A and B, lanes 1) or in conjunction with NAD (lanes 2), as well as NAD alone (lanes 3), did not exhibit any ADPRT activity.
FIG. 2.
VgrG1 has ADP-ribosyltransferase (ADPRT) activity, which is associated with the presence of the VIP-2 domain. ADPRT assays were performed by using 6-biotin-17-NAD, purified rVgrG proteins, and HeLa cell lysates (A) or recombinant nonmuscle actin (B) as a source of target protein. The reaction mixtures were separated on SDS-12% PAGE gels and electroblotted to nitrocellulose membranes, and the incorporation of biotin-ADP by the target protein was detected by using streptavidin-HRP. The nature of the sample loaded in each lane of the gels is depicted in a table below panel B.
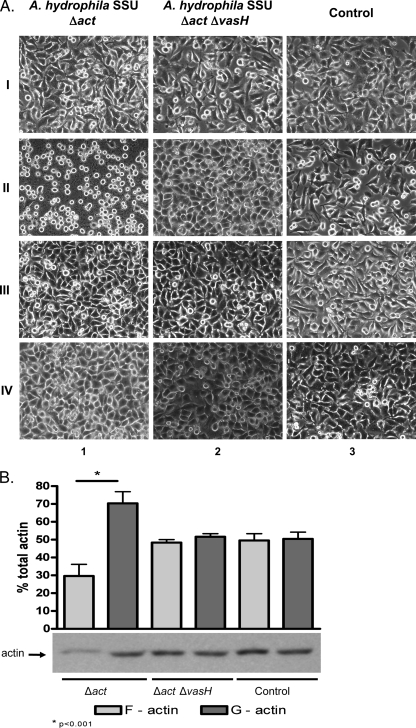
The A. hydrophila SSU Δact mutant strain induces a rounded phenotype in HeLa cells which is cell contact dependent.
To test the cytotoxic effect of the T6SS effector proteins of A. hydrophila on eukaryotic cells, changes in the morphology of the latter were evaluated in cocultures of normal HeLa cells infected with either the A. hydrophila SSU Δact mutant (with functional T6SS) or the Δact ΔvasH mutant strain (with nonfunctional T6SS). We noted that HeLa cells cocultured with the A. hydrophila SSU Δact mutant showed a host-bacterium contact-dependent rounded morphology after 90 min of coculture (Fig. 3A, row II, column 1). In contrast, this phenotype was observed to be minimal in cocultures of HeLa cells with the A. hydrophila SSU Δact ΔvasH mutant (Fig. 3A, row II, column 2). Importantly, we could demonstrate reversal of this rounding phenotype of HeLa cells when the A. hydrophila SSU Δact ΔvasH mutant was complemented with the vasH gene in trans by using the pBR322 vector (see Fig. SIIIA in the supplemental material).
FIG. 3.
(A) Induction of HeLa rounded-cell phenotype in cocultures with different strains of A. hydrophila SSU. HeLa cells were cocultured for 90 min with the A. hydrophila SSU Δact mutant (column 1) and Δact ΔvasH mutant (column 2) or cultured alone (noninfected HeLa cell control; column 3) in direct bacterium-host cell contact (II) or by using transwell inserts (III). Supernatants from cocultures in direct cell-to-cell contact were collected after 90 min and used as preconditioned media on fresh HeLa cell cultures (IV). The initial morphology of HeLa cells at 0 min is shown in row I, and that of noninfected HeLa cells (control) in column 3. Original magnification, ×400. (B) Quantification of G- and F-actin by Western blot and densitometric analyses. HeLa cells in direct contact with different strains of A. hydrophila SSU were harvested after 90 min of coculture. Cells were lysed and processed as indicated in Materials and Methods. The bar graph represents percentages (means ± standard deviations) of G- and F-actin in HeLa cells infected with different mutant strains from three independent experiments, and the Western blot image is representative of all of them.
In parallel, cocultures using transwell inserts were employed to test whether host-bacterium cell contact was indeed needed for the induction of the cell-rounding phenotype. Consequently, A. hydrophila SSU mutant strains (Δact or Δact ΔvasH) were placed in the upper chamber, while the HeLa cells were placed in the bottom chamber to avoid direct host-bacterium cell contact. Importantly, in contrast to direct-contact cocultures, HeLa cells cocultured by using transwell inserts with different A. hydrophila SSU mutant strains had a cell morphology similar to that of the control cells without the bacteria (Fig. 3A, row III, columns 1 and 2 versus 3).
Subsequently, supernatants of cocultures of HeLa cells with different strains of A. hydrophila SSU were removed after 90 min, filtered through a 0.2-μm membrane, and used as preconditioned media on fresh HeLa cells. Host cells exposed to these preconditioned media did not show any changes in their morphology even after 2 h of exposure (Fig. 3A, row IV, columns 1 and 2 versus control HeLa cells depicted in column 3). These results indicated that induction of the cell-rounding phenotype of HeLa cells required direct bacterium-host cell contact and that the protein effector(s) was translocated directly into the eukaryotic cell cytoplasm via the T6SS. Figure SIIIB in the supplemental material shows the presence of Hcp and VgrG2/3 in conditioned medium before (Pre) and after (Post) incubation (for 90 min) with fresh HeLa cells, based on Western blot analysis. It is unclear why we detected lesser amounts of Hcp and VgrG2/3 in the postconditioned media. The decrease in Hcp was more prominent than that of VgrG2/3, and it could possibly be related to either degradation of the proteins or their binding to the host cells (44).
To evaluate a relationship between the rounded phenotype and actin ADP ribosylation, we quantified the ratios of G- and F-actin by Western blot analysis in HeLa cells cocultured with different strains of A. hydrophila SSU. As seen by the data in Fig. 3B, HeLa cells cocultured with the A. hydrophila SSU Δact mutant showed a significant change in the ratio of G-actin/F-actin (70%/30%) compared to the ratios in HeLa cells cocultured with the A. hydrophila Δact ΔvasH mutant (52%/48%) and control HeLa cells (51%/49%). These results correlated with findings regarding the rounded phenotype of HeLa cells cocultured with the parental (Δact) A. hydrophila SSU strain (Fig. 3A, row II, column 1).
A. hydrophila SSU is able to translocate VgrG1 into the eukaryotic host cell cytoplasm via the T6SS.
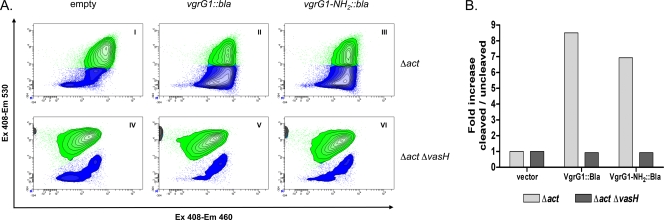
We examined the importance of cell-to-cell contact for the induction of the rounded phenotype of HeLa cells by evaluating the translocation of VgrG1 into the host cell cytoplasm. For these studies, we first showed, by Western blot analysis using antibodies to β-lactamase, the presence of fusion proteins (full-length VgrG1::Bla and VgrG1-NH2::Bla) in the bacterial pellet and the supernatant of the parental A. hydrophila SSU Δact strain (see Fig. SIV, lanes 2 and 3, in the supplemental material). In contrast, by using the Δact ΔvasH mutant strain, we could detect the fusion proteins in the bacterial pellet but not in the supernatant (see Fig. SIV, lanes 5 and 6). Appropriate negative controls did not show the presence of fusion proteins in either the pellet or the supernatant fraction of the above-mentioned mutant bacteria (see Fig. SIV, lanes 1 and 4). Similarly sized fusion proteins were detected on Western blots when antibodies to VgrG2 and the VIP-2 domain of VgrG1 were used (data not shown).
These bacterial strains were then used to infect HeLa cells, and we tracked the β-lactamase activities associated with VgrG1 fusion proteins in the host cell cytoplasm by using the CCF4 FRET-based substrate. With flow cytometry, we showed that the A. hydrophila SSU Δact mutant strain expressing and producing either the full-length VgrG1::Bla or the VgrG1-NH2::Bla was able to cleave the substrate, turning HeLa cells from green to blue (Fig. 4A, panel I [15% blue/85% green] versus panel II [60% blue/40% green] and panel III [55% blue/45% green]). Figure 4A, panel I, shows HeLa cells transfected with the A. hydrophila SSU Δact mutant strain with vector alone, which served as a negative control. In contrast, when we compared them to the controls, A. hydrophila SSU Δact ΔvasH mutant strains expressing and producing the same fusion proteins were found not to have induced any significant changes in the proportion of blue/green HeLa cells (Fig. 4A, panels IV [16% blue/84% green] versus V [17% blue/83% green] and VI [15% blue/85% green]). Figure 4B shows the fold increase in the proportion of blue cells (with cleaved substrate) to green cells (with uncleaved substrate) in an experiment that is representative of 5 independent experiments.
FIG. 4.
Translocation of VgrG1 into HeLa cell cytoplasm. HeLa cells were infected with the A. hydrophila SSU Δact or Δact ΔvasH mutant strain expressing and producing full-length VgrG1::Bla (referred to as VgrG1::Bla) or VgrG1-NH2::Bla. As a control, HeLa cells were infected with bacteria containing the empty vector. (A) Flow cytometric density plots showing disruption of CCF4 FRET (from green to blue) due to translocation of Bla into cytoplasm of HeLa cells infected with the A. hydrophila Δact parental strain (II and III) compared to its disruption in host cells infected with A. hydrophila SSU containing the empty vector (I and IV). HeLa cells infected with the A. hydrophila Δact ΔvasH mutant expressing and producing the fusion proteins were not able to translocate Bla and did not disrupt the CCF4 FRET (V and VI). For analysis, 2 × 105 HeLa cells were acquired and gated in side forward/side scatter patterns to avoid aggregates. Ex, excitation wavelength; Em, emission wavelength. (B) Fold increases in percentages of blue HeLa cells with cleaved substrate versus green HeLa cells with uncleaved substrate (after infection with A. hydrophila SSU Δact and Δact ΔvasH mutant strains) compared to the results for HeLa cells infected with bacteria carrying the empty vector. The graph shows data from a representative experiment.
Expression of the vgrG1 gene in HeLa Tet-Off cells induces a rounded phenotype.
To determine the functionality of the COOH-terminal VIP-2 domain present in VgrG1, the full-length gene and its NH2- and COOH-terminal-domain-encoding gene segments from A. hydrophila ATCC 7966 were cloned into a pBI-EGFP vector and then expressed in HeLa Tet-Off cells. The pBI-EGFP vector allowed coexpression of the gene of interest together with the expression of the gene encoding EGFP to differentiate the transfected from untransfected host cells. The transfected HeLa Tet-Off cells were then evaluated by Western blot analysis by using antibodies specific for VgrG2 and the VIP-2 domain of VgrG1 (Fig. 5A).
FIG. 5.
(A) Western blot analysis of HeLa Tet-Off cell lysates expressing and producing different fragments of VgrG1. The various forms of VgrG1 were detected by using two types of sera. We used sera from mice immunized with rVgrG2 of A. hydrophila SSU which cross-reacted with the NH2-terminal portion of VgrG1 (left panel) and from mice immunized with the recombinant VIP-2 domain of VgrG1 of A. hydrophila ATCC 7966 which recognized only VgrG1 and its COOH-terminal domain. The samples loaded in the lanes are depicted on the top. α, anti. (B) Morphological changes of HeLa Tet-Off cells induced by the expression of different VgrG1 fragments of A. hydrophila ATCC 7966. Host cells were stained for actin cytoskeleton by using Alexa Fluor 568-phalloidin (red), and the expression of the enhanced fluorescent green protein (EGFP) gene was detected in cells successfully transfected with the pBI-EGFP vector alone (I) or with the vector containing genes encoding NH2-terminal (II), full-length (III), or COOH-terminal (IV) fragments of VgrG1. The blue strain (DAPI) shows nuclei. Original magnification, ×400. (C) Quantification of actin cytoskeleton (F-actin) as measured by fluorescent phalloidin staining of HeLa Tet-Off cells expressing and producing different VgrG1 fragments. Flow cytometry dot plots show results for HeLa Tet-Off cells stained with Alexa Fluor 568-phalloidin and expressing different fragments of VgrG1. The analysis was performed on EGFP-positive cells. The percentages of positive cells from representative experiments (72 h) are shown in the plotted quadrants (left), and mean fluorescent intensity values (MFI) from three different assays at 24 h and 72 h were determined (right). Statistical differences at 24 h (P < 0.01) and 72 h (P < 0.001) were noted between results for cells expressing vector alone (pBI-EGFP) and cells expressing and producing full-length VgrG1 and the COOH-terminal fragment and between results for cells expressing and producing the NH2-terminal fragment and cells expressing full-length VgrG1 and the COOH-terminal fragment. (D) Quantification of F-actin and G-actin present in HeLa Tet-Off cells expressing and producing different VgrG1 fragments. The percentages of F- and G-actin per sample (30 μl) were analyzed by Western blot analysis and by using antibodies to actin followed by densitometric scanning of the blots. Densitometric quantification of the results from three different assays and a representative Western blot image are shown. Statistical differences at 24 h (P < 0.001) were noted between fractions containing F- and G-actin in HeLa cells expressing full-length VgrG1 and the COOH-terminal fragments. Asterisks indicate statistically significant differences. The designation ter refers to NH2- or COOH-terminal domains.
Since VgrG2 is highly homologous to its family members, antibodies to VgrG2 recognized the full length (∼103 kDa) and the NH2-terminal domain (∼77 kDa) (Fig. 5A, lanes 2 and 3) but not the COOH-terminal domain (∼25 kDa) of VgrG1 in HeLa Tet-Off cell lysates, as VgrG2 does not possess the VIP-2 domain (Fig. 5A, lane 1). As expected, antibodies to the VIP-2 domain reacted only with the full-length VgrG1 and the COOH-terminal domain of VgrG1 (Fig. 5A, lanes 5 and 7) and not with the NH2-terminal domain of VgrG1 (Fig. 5A, lane 6). Lanes 4 and 8 in Fig. 5A represent the results for the cell lysates from HeLa Tet-Off cells expressing only the pBI-EGFP vector, which served as a negative control.
After 24 h of transfection, the HeLa-Tet-Off cells expressing either the full-length vgrG1 (Fig. 5B, panel III) or its COOH-terminal domain (Fig. 5B, panel IV) showed a rounded morphology, in contrast to the morphology of the HeLa Tet-Off cells expressing only the NH2-terminal domain of vgrG1 (Fig. 5B, panel II), the vector alone (pBI-EGFP) (Fig. 5B, panel I), or the full-length vgrG2 (see Fig. SV in the supplemental material), which maintained normal morphology. In order to examine alterations in the actin cytoskeleton of the HeLa Tet-Off cells expressing and producing different forms of VgrG1, we evaluated the ratios of G-actin/F-actin by Western blot analysis. In addition, the host cells were also stained with Alexa Fluor 568-phalloidin (red stain) and analyzed by fluorescent microscopy and flow cytometry.
As shown in Fig. 5B, panels III and IV, HeLa Tet-Off cells expressing and producing full-length VgrG1 and the COOH-terminal domain (EGFP-positive cells) showed a rounded phenotype, and the actin cytoskeleton was severely disrupted, as evidenced by little or no phalloidin staining. On the other hand, HeLa Tet-Off cells expressing/producing only the vector (Fig. 5B, panel I) or the VgrG1 NH2-terminal domain (Fig. 5B, panel II) exhibited a normal phenotype with an intact actin cytoskeleton (phalloidin staining) having a morphology similar to that of adjacent nontransfected cells (no EGFP, but phalloidin-positive cells).
Changes in the actin cytoskeleton were quantified by flow cytometry after phalloidin staining of the transfected cells expressing and producing EGFP. We found a lower percentage of phalloidin-positive HeLa Tet-Off cells, producing full-length VgrG1 (29%) and its COOH-terminal domain (25%), than of HeLa Tet-Off cells that produced the VgrG1 NH2-terminal domain (86%) or expressed the vector alone (82%) (Fig. 5C, left panel). In addition, we analyzed the mean fluorescence intensity (MFI) of the actin cytoskeleton staining in the transfected HeLa Tet-Off cells. As shown in Fig. 5C, right, after 24 and 72 h of transfection, the MFIs of HeLa Tet-Off cells expressing and producing the full-length VgrG1 and the VgrG1 COOH-terminal domain were significantly lower than those of HeLa Tet-Off cells producing the VgrG1 NH2-terminal domain or expressing the vector alone (P < 0.001).
To confirm the phalloidin staining data, we quantified the amounts of G- and F-actin present in these samples by Western blot analysis and densitometric scanning of the blots (Fig. 5D). HeLa cells expressing genes encoding either the full-length VgrG1 or its COOH-terminal domain showed increased amounts of G-actin (63% and 67%, respectively), in contrast to the amounts in cells expressing the gene encoding the NH2-terminal domain or HeLa cells transfected with the vector alone (49% and 48%, respectively). Together, these results showed the importance of the VIP-2 domain of VgrG1 in the induction of the rounded phenotype of host cells via ADP ribosylation of actin.
Expression of the vgrG1 gene in HeLa Tet-Off cells induces apoptosis.
The viability of the HeLa cells expressing and producing VgrG1 was evaluated by the incorporation of 7-AAD. At 72 h, the percentage of 7-AAD-positive cells was significantly higher in HeLa Tet-Off cells producing the full-length VgrG1 (44.7% ± 7.7% [mean ± standard deviation]) and the COOH-terminal domain (46.3% ± 5.5%) of VgrG1 than in HeLa Tet-Off cells expressing and producing the NH2-terminal domain of VgrG1 (14% ± 4.2%) (P < 0.01) or the vector alone (13.6% ± 5.7%) (P < 0.001) (Fig. 6).
FIG. 6.
Viability of HeLa Tet-Off cells expressing genes encoding different fragments of VgrG1 from A. hydrophila ATCC 7966. Percentages of dead and/or dying cells were quantified by incorporation of 7-AAD and flow cytometry of HeLa Tet-Off cells expressing vector alone (pBI-EGFP) or producing different fragments of VgrG1 (NH2- and COOH-terminal domains and full-length) after 24 h and 72 h of transfection. Means ± standard deviations of the results from three different assays are shown, and statistical significance is indicated. The designation ter refers to NH2- or COOH-terminal domains.
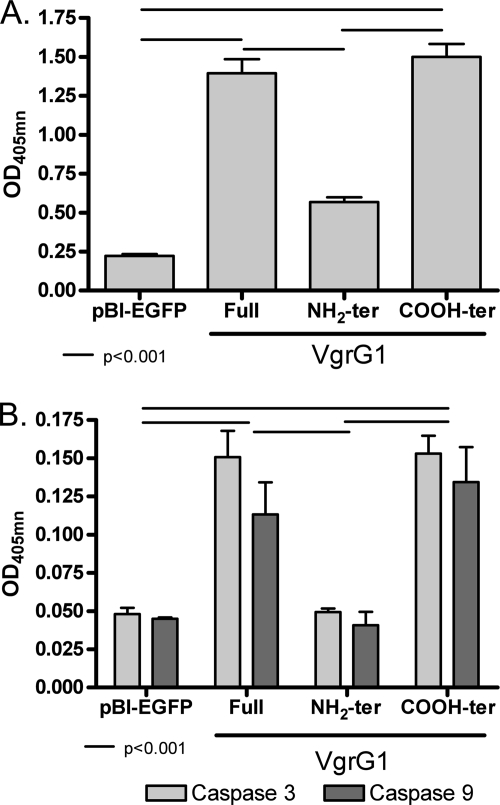
We then evaluated the rate of apoptosis induced by vgrG1 expression in HeLa Tet-Off cells. We found significant increases (P < 0.001) in the amounts of cytoplasmic nucleosomes of HeLa Tet-Off cells expressing and producing full-length VgrG1 and the COOH-terminal domain of VgrG1 in comparison to the amounts for HeLa Tet-Off cells producing the NH2-terminal domain of VgrG1 or expressing the vector alone after 24 h of transfection (Fig. 7A). In accord with these results, the activation of caspase 3 and 9 was significantly greater (P < 0.001) in the HeLa Tet-Off cells producing the full-length VgrG1 and its COOH-terminal domain than in cells producing the NH2-terminal domain of VgrG1 or expressing the vector alone (Fig. 7B).
FIG. 7.
Apoptosis of HeLa Tet-Off cells expressing genes encoding different fragments of VgrG1. Apoptosis rates were measured by quantification of cytoplasmic nucleosomes (A) and levels of caspase 3 and caspase 9 activity (B) in lysates of HeLa Tet-Off cells expressing vector alone (pBI-EGFP) or genes encoding NH2- and COOH-terminal fragments and full-length VgrG1 from A. hydrophila ATCC 7966. Means ± standard deviations of the results from three different assays are shown, and statistical significance (P < 0.001) is indicated. The designation ter refers to NH2- or COOH-terminal domains. OD405nm, optical density at 405 nm.
DISCUSSION
In this study, we report that the VgrG1 protein from A. hydrophila carries a VIP-2 domain, which has ADPRT activity with deleterious effects on the host cells. Thus far, this is the only member of the VgrG family known to have the VIP-2 domain, and it is present in tested A. hydrophila strains, namely, SSU and ATCC 7966. Since we do not have the full genome sequence of A. hydrophila SSU, the precise location of the vgrG1 gene on its chromosome is currently unknown. Consequently, we preferred to conduct studies with rVgrG1 of A. hydrophila ATCC 7966. The VgrG protein family was initially associated with the Rhs (recombination hot spot) family, and although the VgrG and RhsG proteins are not homologous, they do share some common characteristics, such as their hydrophilic nature, large size, and regularly repeated peptide motifs (12, 48). Based on the similarities between the RhsG and VgrG family proteins, it was hypothesized that VgrG could have acquired the multiple COOH-terminal extensions by horizontal transfer (6, 12).
It is important to note that the presence of VgrG1 was detected in the bacterial culture supernatants by Western blot analysis only when antibodies against VgrG2 were combined with antibodies against the VIP-2 domain of VgrG1 and by using a highly sensitive West femto chemiluminescence substrate (Fig. 1C). These data might suggest the availability of a limited number of immunoreactive epitopes in the VIP-2 domain to which antibodies could be generated. This, in combination with the low concentration of VgrG1 present in the bacterial culture supernatant, might cause difficulties in its detection.
Interestingly, VgrG1 could be detected and identified in the concentrated supernatants of bacterial cultures through highly sensitive fluorescent Sypro ruby staining used for 2-D gels. On the other hand, VgrG2/3 could be easily detected in bacterial culture supernatants (Fig. 1C, lane 3) and in tissue culture medium of bacterium-host cell cocultures by Western blot analysis by using only antibodies against VgrG2 (see Fig. SIIIB in the supplemental material). We believe that this difference in the detection of VgrG2/3 and VgrG1 by Western blot analysis could be due to varying amounts of VgrGs present in the culture supernatants. We could easily detect VgrG2/3 because of possible higher levels of expression of their genes and more secretion of the corresponding proteins and/or their possible lower rates of degradation. Further, the accumulation of both of these proteins in a single band due to their similar molecular sizes could result in a band which is much stronger in intensity. In HeLa Tet-Off cell lysates, we could also easily detect the presence of VgrG1 on Western blots by employing either VgrG2 or VIP-2-specific antibodies (Fig. 5A), because highly concentrated protein preparations were used for the assay and the gene was hyperexpressed using the pBI vector.
By bioinformatic and structural analyses, VgrG proteins were shown to share structural features of a bacteriophage T4 tail spike (7, 34). During phage infection, the tail spike is inserted into the bacterial outer membrane, and due to the shared structural features, VgrG could act similarly, i.e., as a needle tip that, in conjunction with the tubelike structure formed by Hcp, could puncture the host cellular membrane and translocate effector protein(s) (7, 12, 23, 24, 30). In these models, the COOH-terminal extensions from some VgrG proteins, such as VgrG1 from V. cholerae, with actin cross-linking activity (see Fig. SIIB in the supplemental material), and VgrG1 from A. hydrophila, with actin ADPRT activity which impedes actin polymerization (see Fig. SIIA), might be introduced directly into the host cellular cytoplasm where they exhibit their action. Likewise, V. cholerae VgrG3, with a peptidoglycan binding domain; P. aeruginosa VgrG, with a zinc metalloprotease domain; and Y. pestis VgrGs, with tropomyosinlike, YadA-like, and pertactinlike domains (34), could induce alteration in host cells.
Importantly, although VgrG1s from V. cholerae and A. hydrophila were able to induce a rounded-cell phenotype, its induction was by different mechanisms. Finally, sequence alignment of the VgrG1s from V. cholerae and A. hydrophila ATCC 7966 showed a high homology in the VgrG NH2-terminal domains (55%) but a low homology (8%) in the COOH-terminal regions where the RtxA and the VIP-2 domain, respectively, are located (see Fig. SIIA and B in the supplemental material).
In our earlier study, we characterized a T3SS effector protein, AexU, which also has ADPRT activity. This effector induced a rounded phenotype when its corresponding gene was expressed in HeLa Tet-Off cells (43). In addition, the results of our previous study indicated that the production and translocation of AexU was not altered when the vasH gene was deleted from the parental A. hydrophila strain; however, this vasH mutant showed delayed cytotoxicity in HeLa cells, as well as in RAW 264.7 murine macrophages (44). Together, these data suggested that even though AexU and VgrG1 could induce a rounded phenotype in the host cell via the ADPRT activity, their mechanisms of regulation could be different and their corresponding genes might be activated or repressed under different stimuli. Furthermore, unidentified other T3SS effectors and proteins secreted by other mechanisms might also lead to host cell toxicity.
Recently published data showed the translocation of VgrG1 from V. cholerae into the eukaryotic cell cytoplasm (24). Similarly, we showed translocation of VgrG1 from the A. hydrophila SSU Δact mutant strain but not from the Δact ΔvasH mutant in HeLa cells. These data indicated an absolute need for the T6SS to translocate this effector.
Yu et al. (53) reported the presence of a sequence containing vgrG, vsdC, and a T3SS-related protein gene (AY376445) in A. hydrophila strain PPD134/91. BLAST conserved-domain analysis of this sequence showed that VgrG (gi|60328264) corresponded to a protein without the COOH-terminal extension and that VsdC (gi|60328266) has a VIP-2 motif. Alignment of a similar sequence found in A. hydrophila ATCC 7966 with that of A. hydrophila PPD134/91 showed high identity/homology with two open reading frames corresponding to VgrG1 (gi|117619461) and a T3SS-related protein (gi|117621298). Although vgrG and vsdC represented two independent open reading frames in A. hydrophila strain PPD134/91, a similar sequence in A. hydrophila ATCC 7966 was represented by only one vgrG1 gene (gi|117619461). Likewise, in A. hydrophila SSU, based on our proteomics and Western blot analysis data, it is apparent that the VIP-2 domain is linked to the VgrG core in a single open reading frame. In contrast, the T3SS-related protein-encoding open reading frame was present in both A. hydrophila ATCC 7966 and A. hydrophila PPD134/91.
It is known that surface epithelial cells become apoptotic after detaching from their underlying basement membrane by a process called anoikis (13), which is characterized by the loss of mitochondrial membrane potential through the action of Bax/Baf that results in caspase activation and DNA fragmentation (17, 22, 28). After depolymerization of actin filaments induced by latrunculin A or cytochalasin D, Bax protein is localized in the outer mitochondrial membrane, where it interacts with and keeps open the voltage-dependent anion channels and/or forms oligomeric pores in the outer membrane which allow the loss of membrane potential and the release of both cytochrome c and other proapoptotic factors (26, 49, 52). Apoptosis induced by VgrG1 of A. hydrophila could be mediated by perturbation of the mitochondrial function. Thus, after the alteration of the actin cytoskeleton by VgrG1-ADPRT activity, the release of cytochrome c could activate caspase 9 and, subsequently, caspase 3, as our results indicated.
Gastrointestinal infections with A. hydrophila induce severe diarrhea, and we previously reported the participation of three different enterotoxins produced by A. hydrophila SSU as players in inducing bloody and nonbloody diarrhea (4, 40). The studies described in this paper have introduced another possible player, namely, VgrG1, which, by actin ADPRT activity, could trigger actin depolymerization with fatal effects on the intestinal epithelial barrier, thus allowing the entry of other virulence factors associated with A. hydrophila SSU pathogenesis. Our future studies will be targeted at developing vgrG1 or vgrG2/3 null mutants, as well as at deleting the ascV gene from the Δact ΔvasH mutant of A. hydrophila SSU, so that the contributions of T3- and T6SS effectors on host cell toxicity can be fully investigated.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH/NIAID (AI041611) and the Environmental Protection Agency. Johanna C. Sierra and Giovanni Suarez were supported by J. W. McLaughlin Endowment predoctoral fellowships, University of Texas Medical Branch at Galveston.
We acknowledge Biomolecular Resource Facility Cores at UTMB for providing their expertise in performing 2-D gel electrophoresis image analysis and mass spectrometry analysis. We thank Mark Griffin for providing his proficiency in the flow cytometry analysis. We thank Mardelle Susman for editorial assistance.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aktories, K., M. Barmann, I. Ohishi, S. Tsuyama, K. H. Jakobs, and E. Habermann. 1986. Botulinum C2 toxin ADP-ribosylates actin. Nature 322:390-392. [DOI] [PubMed] [Google Scholar]
- 2.Aktories, K., and A. Wegner. 1989. ADP-ribosylation of actin by clostridial toxins. J. Cell Biol. 109:1385-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aktories, K., U. Weller, and G. S. Chhatwal. 1987. Clostridium botulinum type C produces a novel ADP-ribosyltransferase distinct from botulinum C2 toxin. FEBS Lett. 212:109-113. [DOI] [PubMed] [Google Scholar]
- 4.Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth, H., K. Aktories, M. R. Popoff, and B. G. Stiles. 2004. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev. 68:373-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingle, L. E., C. M. Bailey, and M. J. Pallen. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3-8. [DOI] [PubMed] [Google Scholar]
- 7.Cascales, E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3:287-300. [PubMed] [Google Scholar]
- 9.Dudley, E. G., N. R. Thomson, J. Parkhill, N. P. Morin, and J. P. Nataro. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61:1267-1282. [DOI] [PubMed] [Google Scholar]
- 10.Edberg, S. C., F. A. Browne, and M. J. Allen. 2007. Issues for microbial regulation: Aeromonas as a model. Crit. Rev. Microbiol. 33:89-100. [DOI] [PubMed] [Google Scholar]
- 11.Fang, C. M., J. Y. Wang, M. Chinchilla, M. M. Levine, W. C. Blackwelder, and J. E. Galen. 2008. Use of mchI encoding immunity to the antimicrobial peptide microcin H47 as a plasmid selection marker in attenuated bacterial live vectors. Infect. Immun. 76:4422-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filloux, A., A. Hachani, and S. Bleves. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570-1583. [DOI] [PubMed] [Google Scholar]
- 13.Frisch, S. M., and H. Francis. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galindo, C. L., A. A. Fadl, J. Sha, C. Gutierrez, Jr., V. L. Popov, I. Boldogh, B. B. Aggarwal, and A. K. Chopra. 2004. Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J. Biol. Chem. 279:37597-37612. [DOI] [PubMed] [Google Scholar]
- 15.Galindo, C. L., J. Sha, A. A. Fadl, L. Pillai, and A. K. Chopra. 2006. Host immune responses to Aeromonas virulence factors. Curr. Immunol. Rev. 2:13-26. [Google Scholar]
- 16.Gill, D. M., and R. Meren. 1978. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 75:3050-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore, A. P., A. D. Metcalfe, L. H. Romer, and C. H. Streuli. 2000. Integrin-mediated survival signals regulate the apoptotic function of Bax through its conformation and subcellular localization. J. Cell Biol. 149:431-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, S., J. A. Craig, C. D. Putnam, N. B. Carozzi, and J. A. Tainer. 1999. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 6:932-936. [DOI] [PubMed] [Google Scholar]
- 19.Heine, K., S. Pust, S. Enzenmuller, and H. Barth. 2008. ADP-ribosylation of actin by the Clostridium botulinum C2 toxin in mammalian cells results in delayed caspase-dependent apoptotic cell death. Infect. Immun. 76:4600-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holbourn, K. P., C. C. Shone, and K. R. Acharya. 2006. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273:4579-4593. [DOI] [PubMed] [Google Scholar]
- 21.Katada, T., and M. Ui. 1982. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc. Natl. Acad. Sci. U. S. A. 79:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koya, R. C., H. Fujita, S. Shimizu, M. Ohtsu, M. Takimoto, Y. Tsujimoto, and N. Kuzumaki. 2000. Gelsolin inhibits apoptosis by blocking mitochondrial membrane potential loss and cytochrome c release. J. Biol. Chem. 275:15343-15349. [DOI] [PubMed] [Google Scholar]
- 23.Leiman, P. G., M. Basler, U. A. Ramagopal, J. B. Bonanno, J. M. Sauder, S. Pukatzki, S. K. Burley, S. C. Almo, and J. J. Mekalanos. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, A. T., S. McAuley, S. Pukatzki, and J. J. Mekalanos. 2009. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35:D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, S. S., and P. Leder. 2001. Human MCF10A mammary epithelial cells undergo apoptosis following actin depolymerization that is independent of attachment and rescued by Bcl-2. Mol. Cell. Biol. 21:6529-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsu, M., N. Sakai, H. Fujita, M. Kashiwagi, S. Gasa, S. Shimizu, Y. Eguchi, Y. Tsujimoto, Y. Sakiyama, K. Kobayashi, and N. Kuzumaki. 1997. Inhibition of apoptosis by the actin-regulatory protein gelsolin. EMBO J. 16:4650-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 30.Pell, L. G., V. Kanelis, L. W. Donaldson, P. L. Howell, and A. R. Davidson. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popoff, M. R., and P. Boquet. 1988. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem. Biophys. Res. Commun. 152:1361-1368. [DOI] [PubMed] [Google Scholar]
- 32.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potvin, E., F. Sanschagrin, and R. C. Levesque. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32:38-55. [DOI] [PubMed] [Google Scholar]
- 34.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53:573-586. [DOI] [PubMed] [Google Scholar]
- 37.Ribardo, D. A., K. R. Kuhl, I. Boldogh, J. W. Peterson, C. W. Houston, and A. K. Chopra. 2002. Early cell signaling by the cytotoxic enterotoxin of Aeromonas hydrophila in macrophages. Microb. Pathog. 32:149-163. [DOI] [PubMed] [Google Scholar]
- 38.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72:5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sha, J., S. F. Wang, G. Suarez, J. C. Sierra, A. A. Fadl, T. E. Erova, S. M. Foltz, B. K. Khajanchi, A. Silver, J. Graf, C. H. Schein, and A. K. Chopra. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—Part I. Microb. Pathog. 43:127-146. [DOI] [PubMed] [Google Scholar]
- 43.Sierra, J. C., G. Suarez, J. Sha, S. M. Foltz, V. L. Popov, C. L. Galindo, H. R. Garner, and A. K. Chopra. 2007. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—Part II. Microb. Pathog. 43:147-160. [DOI] [PubMed] [Google Scholar]
- 44.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuge, H., M. Nagahama, M. Oda, S. Iwamoto, H. Utsunomiya, V. E. Marquez, N. Katunuma, M. Nishizawa, and J. Sakurai. 2008. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc. Natl. Acad. Sci. U. S. A. 105:7399-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandekerckhove, J., B. Schering, M. Barmann, and K. Aktories. 1987. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 225:48-52. [DOI] [PubMed] [Google Scholar]
- 47.Van Ness, B. G., J. B. Howard, and J. W. Bodley. 1980. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J. Biol. Chem. 255:10717-10720. [PubMed] [Google Scholar]
- 48.Wang, Y. D., S. Zhao, and C. W. Hill. 1998. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 180:4102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White, S. R., P. Williams, K. R. Wojcik, S. Sun, P. S. Hiemstra, K. F. Rabe, and D. R. Dorscheid. 2001. Initiation of apoptosis by actin cytoskeletal derangement in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 24:282-294. [DOI] [PubMed] [Google Scholar]
- 50.Williams, S. G., L. T. Varcoe, S. R. Attridge, and P. A. Manning. 1996. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64:283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, B. A., and R. J. Collier. 1992. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active-site structure and enzymic mechanism. Curr. Top. Microbiol. Immunol. 175:27-41. [DOI] [PubMed] [Google Scholar]
- 52.Wolter, K. G., Y. T. Hsu, C. L. Smith, A. Nechushtan, X. G. Xi, and R. J. Youle. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol. 139:1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, H. B., Y. L. Zhang, Y. L. Lau, F. Yao, S. Vilches, S. Merino, J. M. Tomas, S. P. Howard, and K. Y. Leung. 2005. Identification and characterization of putative virulence genes and gene clusters in Aeromonas hydrophila PPD134/91. Appl. Environ. Microbiol. 71:4469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.