Abstract
The virulence of Bacillus cereus requires that bacteria have the capacity to colonize their host, degrade specific tissues, and circumvent the host immune system. To study this aspect of pathogenesis, we focused on three metalloproteases, InhA1, InhA2, and InhA3, which share more than 66% identity. The expression of these metalloprotease genes was assessed by transcriptional fusions with a lacZ reporter gene. The expression profiles suggest a complementary time course of InhA production. Indeed, the genes are simultaneously expressed but are oppositely controlled during stationary phase. We constructed single and multiple inhA mutants and assessed the bacterial locations of the proteins as well as their individual or additive roles in macrophage escape and toxicity, antibacterial-peptide cleavage, and virulence. InhA1, a major component of the spore exosporium, is the only InhA metalloprotease involved in bacterial escape from macrophages. A mutant lacking inhA1, inhA2, and inhA3 shows a strong decrease in the level of virulence for insects. Taken together, these results show that the InhA metalloproteases of B. cereus are important virulence factors that may allow the bacteria to counteract the host immune system.
Bacillus thuringiensis, Bacillus cereus, and Bacillus anthracis are gram-positive sporulating bacteria belonging to the B. cereus group (30). B. anthracis is highly virulent to mammals and is the causative agent of anthrax (47). B. thuringiensis is well known for its entomopathogenic properties (54). B. thuringiensis and B. cereus are responsible for food poisoning characterized by gastroenteritis (28, 56). Several strains are also opportunistic human pathogens associated with local and systemic infections (12, 13, 21, 32). Some B. cereus isolates have been implicated in lethal infections, similar in clinical presentation to B. anthracis infections, and several cases of bacteremia in preterm neonates, posing a potential public health problem, have been described (33, 34, 45).
B. cereus produces several secreted proteins whose expression is controlled by the pleiotropic transcriptional activator PlcR (25). These include enterotoxins (Hbl, Nhe), cytolysins (CytK), and phospholipases (phosphatidylcholine-specific phospholipase C), which may contribute to diarrhea by disrupting the epithelial layer (27, 49). When the bacteria enter the sporulation process, plcR transcription, and consequently PlcR-regulated gene expression, is repressed by the key sporulation regulator Spo0A (40). Deletion of plcR reduces, but does not abolish, the virulence of the bacteria in insects and mice, and during endophthalmitis in rabbits (6, 52), suggesting that other factors may be involved. Moreover, B. anthracis does not express plcR, and introduction of a functional copy does not increase its virulence in a murine model (44).
B. thuringiensis and B. cereus establish persistent infections with infiltration of phagocytes, implying that the bacteria circumvent the host immune system (2, 14, 31). However, interactions between the host immune cells and these bacteria are still poorly characterized, and how they resist the immune defense system remains unknown. The metalloprotease InhA1 may be involved in this process. InhA1 contains the zinc-binding and catalytic active-site residues common to metalloproteases (17, 43, 48). InhA1 has a lethal effect following injection into the insect hemocoel, presumably due to interactions with host components (55). InhA1 hydrolyzes cecropin and attacin, two antibacterial proteins found in the hemolymph of insects (11). B. anthracis InhA1 shares 91% identity with InhA1 of B. cereus and is one of the major proteases isolated in the culture supernatant (9). It digests various substrates, including extracellular matrix proteins, and cleaves tissue components such as fibronectin, laminin, and type I and IV collagens (10). The massive tissue degeneration caused by metalloproteases with collagenase function may help bacteria to cross the host barrier and gain access to deeper tissues (46). However, the virulence of a B. thuringiensis inhA1-deficient mutant is not significantly affected in various insect models (17). Another putative role of InhA1 is suggested by the fact that InhA1 is both secreted throughout the growth cycle of B. cereus and associated with the spore exosporium (8). The spores are internalized by macrophages but are able to survive and escape from this hostile environment; InhA1 is a key effector involved in this process (50).
Previous studies have characterized a gene encoding another putative metalloprotease that has 66% identity to InhA1 and also harbors a zinc-binding domain. This protein, named InhA2, has a role in virulence after oral inoculation of spores into Galleria mellonella (17). However, InhA2 alone is not sufficient to provide virulence in the absence of the other PlcR-regulated genes (16). The transcription of inhA2 depends on PlcR and, in contrast to that of inhA1, is repressed by Spo0A (16, 25). The Spo0A-dependent regulation of inhA1 expression depends on AbrB, which is known to regulate the expression of transition state and sporulation genes in Bacillus subtilis (26).
A third protein, InhA3, 72% identical to InhA1, is also found in the culture supernatant of B. cereus, but only in trace amounts (24). Its function and regulation have not yet been described.
In this study, we constructed single and multiple mutants of the InhA1, InhA2, and InhA3 proteases and assessed their individual and relative roles in macrophage escape and toxicity, antibacterial peptide cleavage, and virulence. We explored the need for this apparent duplication of genes by studying their respective locations and time courses of expression during bacterial growth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The acrystalliferous B. thuringiensis strain 407 (Cry−) was used as the wild-type background and was transformed as described previously (42). The mutant strains B. thuringiensis 407 ΔplcR, 407 ΔinhA1, 407 ΔinhA2, and 407 ΔinhA1 ΔinhA2 have been described previously (17, 52). The transcriptional fusions 407 (pHT304-inhA1′Z) and 407 (pHT304-inhA2′Z) are described elsewhere (17, 26). Escherichia coli TG1 was used in the antibacterial peptide cleavage assay.
All strains were grown in LB medium at 37°C or in HCT medium at 30°C with shaking (38). Bacteria were harvested by centrifugation for 10 min at 3,500 rpm. Pellets were washed twice with phosphate-buffered saline and were resuspended in phosphate-buffered saline at 108 bacteria/ml. The culture supernatant was filtered through a 0.22-μm-pore-size filter. To prepare spores, B. thuringiensis was grown at 30°C in the sporulation-specific medium HCT plus 0.3% glucose as described in reference 17.
Plasmid and mutant strain constructions.
The inhA3 gene was disrupted as follows. BamHI-XbaI (1,148-bp) and SphI-HindIII (1,025-bp) DNA fragments corresponding to upstream and downstream regions of the inhA3 gene were generated from the B. thuringiensis 407 chromosome by PCR using primer pairs inhA3-1 (5′-CGCGGATCCGGCGAACTCAACCGTTTGGGG-3′)-inhA3-2 (5′-GCTCTAGACGAACAGCTCCATTGTATTC-3′) and inhA3-3 (5′-ACATGCATGCGGCGTTCATCCAGGTGAAGG-3′)-inhA3-4 (5′-CCCAAGCTTCCCCTGTACCCCTCTTATCC-3′). A Tetr cassette carrying a tet gene was purified from pHTS1 (53) as a 1.6-kb XbaI-SphI fragment. The amplified DNA fragments were digested with the appropriate enzymes and inserted between the HindIII and BamHI sites of pRN5101 (58). The resulting plasmid was introduced into strain 407, strain 407 ΔinhA1, and strain 407 ΔinhA1 ΔinhA2 by electroporation (4), and the inhA3 gene was deleted by a double-crossover event as previously described (41). Chromosomal allele exchange was confirmed by PCR with oligonucleotide primers located upstream of InhA3-1 (InhA3-5 [5′-CGCGGATCCGGTATTGGAGGAGGTCTTTG-3′]), downstream of InhA3-4 (InhA3-6 [5′-CCGGAATTCGCTAAACGAAGTCTAAATGG-3′]), and in the Tetr cassette (5′-CGGGTCGGTAATTGGGTTTG-3′ and 5′-GCAGCTGCACCAGCCCCTTG-3′). The insertion mutant strains were designated 407 ΔinhA3, 407 ΔinhA1 ΔinhA3, and 407 ΔinhA1 ΔinhA2 ΔinhA3.
The inhA3 gene, including the coding sequence and the presumed promoter region, was cloned using the primer pair inhA3-7 (5′-CGCGGATCCGGTATTGGAGGAGGTCTTTG-3′)-inhA3-8 (5′-CCGGAATTCGCTAAACGAAGTCTAAATGG-3′). The fragment was inserted between the BamHI and EcoRI sites of the pHT304 vector, which was then used to transform strains 407 ΔinhA3 and 407 ΔinhA1 ΔinhA2 ΔinhA3 by electroporation. Transformants were selected for resistance to erythromycin. The resulting new complemented strains were designated 407 ΔinhA3/inhA3 and 407 ΔinhA1 ΔinhA2 ΔinhA3/inhA3.
Strain 407 ΔplcRΔinhA1 was constructed by disrupting the inhA1 gene in strain 407 ΔplcR. For this purpose, the pRN5101 plasmid containing the inhA1-lacZ fusion (26) was introduced by electroporation into strain 407 ΔplcR. Chromosomal allele exchange was confirmed by PCR using the sin14-sin15 primer pair (26).
A transcriptional inhA3′-lacZ fusion was constructed by using an XbaI-PstI DNA fragment corresponding to the promoter region of inhA3, generated by PCR using primer pair pinhA3-1 (5′-CCCAAGCTTGTAACAGAGAATCACGCTATA-3′)-pinhA3-2 (5′-CGGGATCCAGCATGCGCTGCGTGACCTCC-3′). The PCR fragment was digested with the appropriate enzymes and inserted between the XbaI and PstI sites of pHT304-18Z (1). The recombinant plasmid, designated pHT304-inhA3′Z, was introduced into strain 407 and into mutant strains 407 ΔplcR and 407 Δspo0A by electroporation. Transformants were named 407 (pHT304-inhA3′Z), 407 ΔplcR (pHT304-inhA3′Z), and 407 Δspo0A (pHT304-inhA3′Z).
InhA protein sequence analysis.
Twenty-four protein sequences from international databases were compared using ClustalW (57), and they were curated using GBlocks (7). There were a total of 690 positions in the final data set. The evolutionary distances were computed using the JTT matrix-based method (35). A phylogenetic tree was constructed using the BioNJ method (23), and phylogenetic analyses were conducted in PHYLIP (19). NJplot software was used to generate a graphic representation of the resulting tree. Bootstrap estimates (18) were obtained from 1,000 replicates.
The SignalP, version 1.1, prediction server was used to identify potential cleavage sites. The strain 407 inhA3 sequence was determined by sequencing a PCR fragment amplified from the strain 407 chromosome using primers InhA3-5 and InhA3-6 (Sigma-Aldrich).
Purification of the InhA2 and InhA3 proteins.
Plasmids pGEX6P1-GST-InhA2 and pGEX6P1-GST-InhA3 were constructed as follows. The inhA2 and inhA3 genes were amplified from the strain 407 chromosome by PCR using primer pairs InhA2-GST-1 (5′-TCCCCCGGGAGAAACGGTAGCAAAAGAG-3′)-InhA2-GST-2 (5′-CCGCTCGAGTTAACGTTTAATCCAAACAGC-3′) and InhA3-GST-1 (5′-TCCCCCGGGTGAGACACCAACATCATCTCT-3′)-InhA3-GST-2 (5′-CCGCTCGAGTTATCGATGAAGCCACACTGC-3′), respectively. The corresponding DNA fragment was inserted between the SmaI and XhoI sites of plasmid pGEX6P1 (GE Healthcare), and the resulting plasmid was introduced into E. coli M15(pREP4) (Qiagen). E. coli M15(pREP4) harboring plasmid pGEX6P1-GST-InhA2 or pGEX6P1-GST-InhA3 was grown at 30°C until an optical density at 600 nm of 0.8 was reached, and protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was continued for 2 h after IPTG induction. Bacteria were then collected by centrifugation at 7,700 × g for 10 min. Pellets were lysed using 1% Triton X-100 and sonication in 50 mM Tris-8 M urea. After centrifugation, the tagged InhA2 or InhA3 contained in the supernatant was dialyzed against 50 mM Tris, pH 8, and was purified using the Bulk GST purification module (GE Healthcare) according to the manufacturer's instructions. The protein concentration was quantified on a sodium dodecyl sulfate-polyacrylamide gel with the Image Master 1D program (Amersham Pharmacia Biotech).
β-Galactosidase assays.
Strains harboring plasmid transcriptional lacZ fusions were grown in LB or HCT medium at 37°C or 30°C, respectively. β-Galactosidase activity was determined as previously described (5). β-Galactosidase assays of bacteria from insect larvae infected by intrahemocoelic injection were performed as described previously (15). Specific activities are expressed in units of β-galactosidase per milligram of protein (Miller units). Results are shown as means and standard deviations for at least three independent tests.
Protein localization.
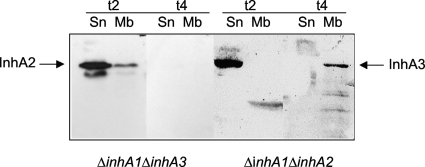
Bacteria were grown on LB medium and were harvested 2 and 4 h after the onset of stationary phase (t2 and t4, respectively). The supernatant was filtered through a 0.22-μm-pore-size filter. The bacterial pellet was disrupted with glass beads (diameter, 212 to 300 μm; Sigma) in a Fast-Prep machine (Savant), and a bacterial extract was obtained after centrifugation: the pellet corresponded to the membrane fraction and the supernatant to the cytoplasmic fraction. The different fractions were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel. The gel was then transferred to a nitrocellulose membrane (Amersham), which was blotted with a rabbit polyclonal antibody recognizing the different InhA proteins, and the proteins were revealed using the ECL system (Pierce). The image shown in Fig. 4 represents one out of four different gels with similar results.
FIG. 4.
Bacterial localization of the InhA2 and InhA3 proteins. Strains 407 ΔinhA1 ΔinhA3 and 407 ΔinhA1 ΔinhA2 were grown on LB medium and were harvested at t2 and t4. Supernatant (Sn) and membrane (Mb) fractions were blotted with a rabbit polyclonal antibody recognizing the different InhA proteins, and the specific locations of InhA2 and InhA3 were determined.
Inhibition zone assay.
Inhibition zone assays consisted of plating E. coli onto LB medium and adding synthetic cecropin A (Sigma) on a small paper disk. Cecropin A prevents the growth of E. coli, and an inhibition zone is seen around the disk. Purified proteins or culture filtrates of B. cereus mutant strains grown in LB medium and collected at t2 are added to the disk, and degradation of cecropin A is observed by the disappearance of the inhibition zone.
Protease assay.
The protease cleavage activities of the InhA proteins were tested by measuring the lysis zones on milk-casein agar plates. Purified proteins (1 μg) were inoculated into holes on agar plates containing 4 g/liter peptone, 2 g/liter casein, and 10 g/liter skim milk powder. Plates were incubated for 24 h at 37°C, and protease activity was evaluated by measuring the lysis zones around the protein.
Cell culture and macrophage infection.
Spore release infection experiments and cytotoxicity assays on J774 murine macrophage-like cells were performed as previously described (49, 50). For cytotoxicity, cells were infected with various dilutions of the culture supernatant taken at t2. After 2 h of incubation, trypan blue dye was added to the cells. Nonpermeabilized (viable) cells remained unstained, whereas permeabilized (killed) cells allowed the dye to enter the cytoplasm and were therefore stained blue. Cytotoxicity is expressed as the percentage of cells that were stained blue. Results are means for three independent experiments.
Insects and in vivo experiments.
Infection by injection into the hemolymph of Galleria mellonella or Bombyx mori larvae was performed as described previously (17, 52). For groups of 15 last-instar larvae, the base of the last proleg was injected with a 10-μl suspension containing 7.5 × 103 vegetative bacteria or spores for G. mellonella, or 50 vegetative bacteria for B. mori. Insect mortality was recorded after 24 h. To estimate the number of bacteria in living or dead larvae, insects were crushed after infection and homogenized in sterile water; dilutions were plated onto LB agar plates. All tests were run at least three times.
Nucleotide sequence accession number.
The sequence of the B. thuringiensis strain 407 inhA3 gene has been submitted to GenBank under accession number FJ717416.
RESULTS
Presence and comparison of the inhA1, inhA2, and inhA3 genes in sequenced strains of the B. cereus group.
The amino acid sequence of InhA1 from B. cereus ATCC 14579 was used to search the entire genome of B. cereus ATCC 14579. This revealed the presence of three highly similar peptide sequences: InhA1 (Bc1284), InhA2 (Bc0666), and InhA3 (Bc2984). The inhA1, inhA2, and inhA3 genes were also present in the unsequenced B. thuringiensis strain 407 used in this study, since they were all amplified by PCR from this strain. Since inhA1 and inhA2 from this strain were already sequenced and aligned (17, 26), the strain 407 inhA3 gene was sequenced, and the three gene sequences were compared (Fig. 1). InhA1 and InhA2 shared 66% identity. InhA1 and InhA3 shared 74% identity, and InhA2 and InhA3 shared 71% identity.
FIG. 1.
Alignment of the B. thuringiensis 407 InhA1, InhA2, and InhA3 amino acid sequences. Numbers indicate positions in the amino acid sequence. Identical residues are shaded. The conserved zinc-binding domain (HEXXH) is boxed in gray. The putative cleavage site (AYAET) is boxed, and the lipobox-like (IIGC) signal in InhA2 is at positions 18 to 21.
InhA1, InhA2, and InhA3 have calculated molecular masses of 86.6 kDa, 87.9 kDa, and 87.2 kDa, respectively. The three amino acid sequences each contain the zinc-binding motif (HEXXH), which is characteristic of the zinc metalloprotease family (36). The amino acid sequences were submitted to the SignalP, version 1.1, prediction server, Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/SignalP). Potential cleavage sites (AYA-ET) were found between positions 31 and 32 for InhA1 and between positions 32 and 33 for InhA2 and InhA3, suggesting that the proteases are exported. However, the InhA2 sequence contains a lipobox-like (IIGC) signal of peptidase 2 (LXXC) (prokaryotic membrane lipoprotein lipid attachment site profile). The sequence upstream of this lipobox-like signal contains several charged amino acids (MRRK) followed by a long hydrophobic zone (APFKVLSSLAIAA), which may act as a cleavage site, implying that the lipobox might be functional. InhA2 might therefore be a transmembrane protein.
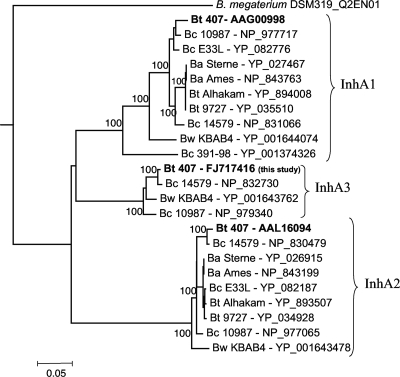
A phylogenetic tree was constructed based on the sequence similarities of 24 protein sequences from international databases using Clustal W (57) (Fig. 2). This revealed the presence of one to three highly similar peptide sequences depending on the strains. inhA1 is present in all the strains (Fig. 2), and the gene environment is highly conserved (not shown). inhA2 is present in 9 of the 10 strains. The inhA3 gene was found only in the B. cereus strains ATCC 14579 and ATCC 10987, in the Bacillus weihenstephanensis strain KBAB4, and in the B. thuringiensis strain 407. It is interesting that this gene is absent in B. anthracis strains.
FIG. 2.
Phylogenetic tree based on InhA protein sequence analysis. Protein sequences were from international databases and are designated as the strain name followed by the accession number for the NCBI or GenBank reference sequence. The evolutionary history was inferred using an improved neighbor-joining method. Numbers on branches are bootstrap values above 80%. The bar (0.05 unit) indicates the scale of evolutionary distance; the unit is the number of amino acid substitutions per site. Bt, B. thuringiensis; Bc, B. cereus; Ba, B. anthracis; Bw, B. weihenstephanensis.
Expression and regulation of the inhA genes during bacterial growth.
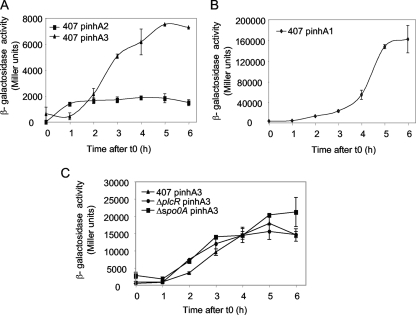
The transcriptional activities of the inhA1, inhA2, and inhA3 promoters were assayed using transcriptional fusions between the promoter regions and the promoterless lacZ gene. B. thuringiensis 407 strains carrying the corresponding plasmids were grown in rich LB medium at 37°C or in sporulation-specific HCT medium at 30°C. The β-galactosidase activity was measured at various stages of growth between t1 and t6. In LB medium (Fig. 3A and B), the expression of inhA1, inhA2, and inhA3 was low during the exponential phase of growth and increased during the first stages of stationary phase. Expression of inhA1 started at t0 (4,000 Miller units) and increased up to t6 (160,000 Miller units). inhA2 was expressed from t1 (1,400 Miller units) to t4 (1,900 Miller units). Expression of inhA3 started at t1 (600 Miller units) and increased up to t5 (7,500 Miller units). In HCT medium, no β-galactosidase activity was detected for inhA2, as previously reported (17; also data not shown). In contrast, inhA3 expression was high in HCT medium, reaching 18,000 Miller units at t5 (Fig. 3C). The inhA3 expression profiles in LB and HCT media were similar, with expression from t0 to t5.
FIG. 3.
Expression of the inhA1, inhA2, and inhA3 genes under various growth conditions. The specific β-galactosidase activities of strains 407 (A, B, and C), 407 ΔplcR (C), and 407 Δspo0A (C) harboring the transcriptional inhA1′-lacZ (pinhA1) (B), inhA2′-lacZ (pinhA2) (A), or inhA3′-lacZ (pinhA3) (A and C) fusion were measured when bacteria were grown in LB medium at 37°C (A and B) or in HCT medium at 30°C (C).
It has been shown previously that inhA1 expression is repressed by abrB and is therefore dependent on Spo0A, which represses abrB expression (26). In sharp contrast, inhA2 expression is activated by PlcR (16) and is thus negatively regulated by Spo0A, which represses plcR expression (40). In comparison with inhA1 and inhA2, inhA3 expression is independent of PlcR and Spo0A; no statistical difference in β-galactosidase activities between the wild-type strain and the Δspo0A or the ΔplcR mutant strain was observed (Fig. 3C). Therefore, the three genes are coexpressed during bacterial growth, although differentially regulated.
Protein localization.
All three InhA proteins are secreted (24). InhA1 is also associated with the spore surface (8). We therefore questioned whether InhA2 and InhA3 could have specific bacterial locations. The localization of the proteins was assessed by Western blotting of the different bacterial fractions of strain 407 ΔinhA1 ΔinhA2 (for the localization of InhA3) and strain 407 ΔinhA1 ΔinhA3 (for the localization of InhA2). InhA2 was found in the supernatant and at the bacterial membrane at t2 (Fig. 4). No detectable amount of InhA2 was found at t4. InhA3 was found secreted at t2 and at the bacterial surface at t4 (Fig. 4). This implies that InhA3 is first secreted and that the extracellular protein might be then readsorbed to the bacterial surface.
Anticecropin activity.
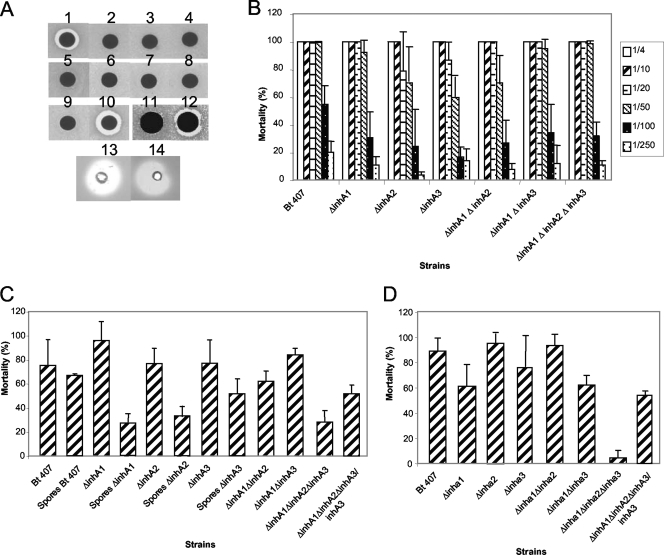
Culture filtrates of mutant strains collected at t2 were used to study their protective effects on E. coli against the antibacterial peptide cecropin. As shown in Fig. 5A, supernatants of strains 407, 407 ΔinhA1, 407 ΔinhA2, 407 ΔinhA3, 407 ΔinhA1 ΔinhA2, 407 ΔinhA1 ΔinhA3, and 407 ΔinhA1 ΔinhA2 ΔinhA3 protected E. coli from cecropin activity. This indicates that degradation of cecropin is not achieved exclusively by an InhA-specific proteolytic activity. Consistently, although strain 407 ΔplcR had anticecropin activity, the mutant 407 ΔplcR ΔinhA1 did not (Fig. 5A). This implies that anticecropin activity is due to a combination of InhA1 and one or several PlcR-dependent components. Since inhA3 expression is independent of PlcR, this result suggests that InhA3 has no anticecropin activity. To confirm this, purified InhA2 and InhA3 were tested for their capacities to degrade cecropin. Figure 5A shows that InhA2 has anticecropin activity, whereas InhA3 does not. Thereafter, the capacity to degrade the antimicrobial peptide cecropin is specific to InhA1 and InhA2, and InhA3 does not seem to have this activity. To test whether InhA3 shared common activities with the other InhA proteins, the purified proteins were tested for their abilities to cleave casein. Both InhA2 and InhA3 induced lysis plaques on casein-agar plates (Fig. 5A), indicating that the two purified proteins have protease activity.
FIG. 5.
inhA mutant phenotypes. Strains 407, 407 ΔinhA1, 407 ΔinhA2, 407 ΔinhA3, 407 ΔinhA1 ΔinhA2, 407 ΔinhA1 ΔinhA3, 407 ΔinhA1 ΔinhA2 ΔinhA3, 407 ΔinhA1 ΔinhA2 ΔinhA3/inhA3, 407 ΔplcR, and 407 Δplcr ΔinhA1 were tested for their capacities to cleave the antibacterial peptide cecropin (A), to induce macrophage membrane permeability at various dilutions (B), and to kill Galleria mellonella (C) or Bombyx mori (D) larvae. In panel A, the following strains were tested: 2, strain 407; 3, strain 407 ΔinhA1; 4, strain 407 ΔinhA2; 5, strain 407 ΔinhA3; 6, strain 407 ΔinhA1 ΔinhA2; 7, strain 407 ΔinhA1 ΔinhA3; 8, strain 407 ΔinhA1 ΔinhA2 ΔinhA3; 9, strain 407 ΔplcR; 10, strain 407 ΔplcR ΔinhA1. Disk 1 shows the result for cecropin alone. Additionally, purified InhA2 and InhA3 proteins were tested for their capacities to cleave cecropin (disks 11 and 12, respectively) and casein (disks 13 and 14, respectively).
Role of InhA in cytotoxicity for macrophages and in spore escape.
We tested the abilities of single and multiple mutants to induce toxicity on macrophages (Fig. 5B). A trypan blue test was used to assess the effects of supernatants on membrane alterations and to quantify the effects. As expected, the supernatant of the wild-type strain B. thuringiensis 407 was highly cytotoxic, with 100% cytotoxicity from the 1/4 to the 1/50 dilution, 55% cytotoxicity for the 1/100 dilution, and 20% cytotoxicity for the 1/250 dilution. Supernatants of all the strains tested had the same toxic effect on macrophages as the supernatant of the wild-type strain. Therefore, deletion of the three metalloproteases together does not prevent cytotoxicity for eukaryotic macrophages.
We have previously shown that spores of the ΔinhA1 mutant were retained inside macrophages, whereas spores of the wild-type B. thuringiensis strain 407 were capable of escaping from the cells (50). However, spores of the 407 ΔinhA2 and 407 ΔinhA3 mutants are still capable of escaping from macrophages (data not shown), implying that only InhA1 is involved in this process. This is in agreement with the location of InhA1 in the exosporium (8).
Role in virulence.
To establish the contribution of the InhA proteases to virulence, we tested the pathogenicities of single and multiple mutants in their insect hosts. Vegetative bacteria or spores were injected into the hemocoel of G. mellonella (Fig. 5C) or B. mori (Fig. 5D), and mortality was recorded 24 h postinfection.
When vegetative bacteria were injected at a dose of 7,500 bacteria/insect into the hemocoel of G. mellonella, only the virulence of the triple mutant strain 407 ΔinhA1 ΔinhA2 ΔinhA3 was strongly affected (28% insect mortality for the mutant and 75% for the wild type; P < 0.002 by the Student t test). The 50% lethal dose, established with the Log-Probit program (20), was 2.8 × 103 bacteria for the wild type and 24 × 103 for the mutant. Complementation of this mutant with the inhA3 gene resulted in a level of virulence comparable to that of strain 407 ΔinhA1 ΔinhA2 (56% and 62% mortality, respectively; P > 0.07 [not statistically different]) (Fig. 5C).
To estimate the number of bacteria in infected larvae, insects were crushed 24 h after infection and homogenized in sterile water; dilutions were plated onto LB agar plates. The level of survival after infection was (2.2 ± 2) × 109 CFU/larva after infection with the wild-type strain and (2.7 ± 2.5) × 104 CFU/larva after infection with the 407 ΔinhA1 ΔinhA2 ΔinhA3 mutant (data not shown). These results imply that the mutant proliferates less than the wild type inside the insect host.
Since InhA1 is located at the spore surface, we assessed the virulence of the spores of an inhA1-deficient mutant. The virulence of the ΔinhA1 mutant spores after injection was strongly reduced from that of the wild type (26% and 70% mortality, respectively; P < 0.04). The virulence of the ΔinhA2 spores was also strongly reduced (33% mortality; P < 0.01) from that of the wild type, but the virulence of the ΔinhA3 spores was comparable to that of the wild type (63%) (Fig. 5C).
The results of infection of B. mori larvae (Fig. 5D) were comparable to those obtained with G. mellonella. Indeed, the virulence of the multiple mutant strain 407 ΔinhA1 ΔinhA2 ΔinhA3 was greatly affected; it induced only 4% mortality, compared to 89% mortality with the wild type (P < 0.002) and 52% mortality with the inhA3-complemented strain (not significantly different from the wild type; P > 0.08).
Taken together, these data show that in the two insects G. mellonella and B. mori, deletion of the three metalloprotease genes inhA1, inhA2, and inhA3 induces a strong reduction of virulence, demonstrating that these proteins play an important role during the infection process.
InhA expression during insect infection.
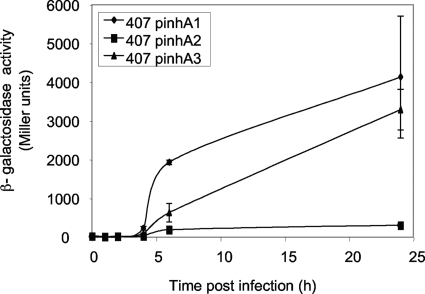
To assess the expression of the three genes in vivo, G. mellonella larvae were crushed after infection with strains carrying the inhA1, inhA2, and inhA3 promoter transcriptional fusions, and β-galactosidase production was measured (Fig. 6). Expression of the three genes began 4 h postinfection and continued until 24 h postinfection, when it reached 4,000 Miller units for inhA1 and 3,300 Miller units for inhA3, although inhA2 was only poorly expressed in vivo, with a maximum of 300 Miller units at t24. The three genes were expressed in vivo long before insect death (usually occurring 18 h after infection), showing the importance of these genes during virulence.
FIG. 6.
Expression of the inhA1, inhA2, and inhA3 genes in vivo. The specific β-galactosidase activity of strain 407 harboring the transcriptional inhA1′-lacZ (pinhA1), inhA2′-lacZ (pinhA2), or inhA3′-lacZ (pinhA3) fusion was measured after injection into Galleria mellonella larvae.
DISCUSSION
All bacterial zinc metalloproteases belong to the zincin superfamily, which contains the conserved zinc-binding consensus motif HEXXH. The two histidines are zinc ligands, and the glutamic acid is involved in enzymatic activity (46). In silico analysis reveals the presence of one to three highly similar zinc metalloproteases, InhA proteins, in the genomes of several members of the B. cereus group. In the reference B. cereus strain ATCC 14579 and in B. thuringiensis 407, the three inhA genes coexist and share more than 66% identity. inhA3 appeared to be present in only a few members of the B. cereus group.
Transcriptional analysis of the inhA genes shows that the expression of the genes is subject to different regulations. While inhA1 expression is activated from t1 to t6 in LB medium, inhA2 expression is activated from t1 to t4, and inhA3 is expressed from t1 to t5. Moreover, inhA1 expression is almost 10-fold higher than inhA2 expression in LB medium. However, the use of plasmid constructs rather than chromosomal transcriptional fusions to analyze expression might result in false quantitative comparisons. In the case of inhA1, the plasmid transcriptional fusion probably causes a dosage effect, reducing the availability of the repressor AbrR and thus resulting in overexpression of the inhA1′-lacZ transcriptional fusion. inhA2 transcription is repressed by Spo0A, and it is not expressed in the HCT sporulation medium (16), whereas inhA3 is expressed in HCT medium at a level threefold higher than that in the richer LB medium. Therefore, expression of inhA2 requires a rich medium, while inhA3 is preferentially expressed when the bacteria grow in a relatively poor sporulation-specific medium (HCT). This might explain why, in infected moths, likely a relatively poor medium, inhA2 is poorly expressed and why inhA1 and inhA3 have similar expression patterns. The expression of inhA2 is dependent on the pleiotropic regulator PlcR (16). The expression of inhA1 and inhA3 does not depend on plcR. PlcR is not functional in B. anthracis (44), and the inhA3 gene is absent in this species (51). Therefore, it is likely that only inhA1 is expressed in B. anthracis. In contrast to inhA1 and inhA2 expression (16, 26), Spo0A does not affect the expression of inhA3. PlcR, Spo0A, and AbrB are the main regulators described for B. cereus. Thereafter, inhA3 is likely regulated by another, as yet undescribed regulatory system.
Major bacterial processes, such as the production of antibiotics, induction of competence, biofilm formation, sporulation, and expression of virulence factors, are controlled in a multicellular manner. The functional significance of the duplication of the inhA genes and their complementary regulation systems might reflect a physiological regulatory mechanism, which enables bacteria to cope with adverse environmental conditions. This prompted us to study whether these three proteases might have redundant functions. We first examined their bacterial localizations. The putative signal peptide cleavage site, located in almost the same positions for the three InhA proteins, suggests that they are exported. InhA1, InhA2, and InhA3 are accordingly found in the supernatant of B. cereus (24) (Fig. 4). According to proteomics results, the concentration of InhA1 in the supernatant was almost 25-fold lower than that of InhA2 and increased only after t1, whereas the concentration of InhA2 increased from t0. The two concentrations peaked at t2 and were barely detectable at t3 for InhA1 and t3.5 for InhA2 (24). Thus, these two proteases were found in the culture supernatant only during a window of a few hours in early-stationary phase. InhA3 was found, although at trace levels (not quantifiable), in LB medium from t2 to t5 (N. Gilois and M. Gohar, personal communication). However, lacZ fusions indicate that both inhA1 and inhA3 are activated from t1 to t5. inhA2 is expressed but stops being activated from t1 on; the curve plateaus from t1 to t5. The apparent discrepancy between the proteomic and expression data suggests that the proteins might be located elsewhere after being secreted. Our results show that the destinations of the three InhA proteins differ. InhA1 is likely adsorbed to the spore exosporium (8). InhA2 and InhA3 were found at the bacterial surface, at t2 for InhA2 and t4 for InhA3. At t4, no detectable amount of InhA2 was found, either in the supernatant or in the membrane. This suggests that InhA2, whose expression is weak and whose promoter stops being activated at t1 (plateauing from t1 on), is rapidly degraded. At t4, InhA3 was found only in the membrane fraction, suggesting that all InhA3 protein produced at this time was located solely in the membrane and did not remain extracellularly or was degraded, or both.
Interestingly, a new stress-related metalloprotease highly similar to InhA3 of B. cereus 14579 was identified in Bacillus megaterium (59). It is especially expressed under nutrient-limiting conditions, is strongly retained in the bacteria. and is found both cell associated and secreted in the culture medium.
The virulence of the mutant lacking inhA1, inhA2, and inhA3 is strongly affected after injection into the hemocoels of G. mellonella and B. mori larvae. This is striking, since the B. thuringiensis 407 ΔinhA1 ΔinhA2 ΔinhA3 mutant is the first B. cereus group strain whose growth is not affected in LB medium at 30°C (culture temperature for B. mori [data not shown]) that has reduced virulence for B. mori. Single or double mutants are less affected than the triple mutant, implying that the three metalloproteases act concomitantly to induce insect death. The inhA2 mutant has no effect on insect mortality after injection of vegetative bacteria into the hemocoel, as previously described (17). However, the virulence of spores of the inhA2-deficient mutant is affected after injection into the G. mellonella hemocoel. Moreover, InhA2 is required, although not sufficient, for pathogenicity in insects infected orally, and InhA1 seems less important at this stage (16, 17). InhA2 might therefore have a role during the early steps of infection, to help the bacteria cross the intestinal barrier (3). The virulence of spores of the inhA1-deficient mutant is affected.
The innate immune systems of insects, which lack adaptive immunity, are based on two major responses: the synthesis of microbial peptides and the cellular immune system (plasmatocytes, lamellocytes, and granulocytes). Antimicrobial peptides act preferentially on membranes rich in anionic phospholipids in prokaryotes, and eukaryotic cell membranes are composed mainly of neutral phospholipids and cholesterol, which seem to inhibit peptide incorporation (22). In the hemolymph, the concentration of antibacterial peptides may reach 300 to 500 μM upon infection (39). Two antibacterial peptides isolated from B. mori show activity against B. thuringiensis and B. cereus (29, 37): enbocin (cecropin family) and moricin. Purified InhA1 of B. thuringiensis has been shown to cleave cecropin (11), but we show that InhA1 is not necessary to cleave cecropin, since the inhA1 mutant still has anticecropin activity. Moreover, like the plcR mutant, the single and double inhA mutants and the triple mutant lacking inhA1, inh2, and inh3 also have anticecropin activity. However, the mutant lacking plcR and inhA1 was deficient in anticecropin activity, showing that this activity can also be performed by a PlcR-dependent protein other than InhA2. This suggests that although purified InhA1 (11; also data not shown) and InhA2 (Fig. 5) have anticecropin activity, they are not essential for cecropin resistance in B. thuringiensis.
Macrophages are major effectors of the host immune system against bacterial infections. For successful infection, pathogens must defeat or avoid cells of the host immune system. We have previously demonstrated that Bacillus spores are able to survive the macrophage intracellular environment and to escape from phagocytes (50) and that this ability depends on the metalloprotease InhA1. Here we show that a mutant lacking the three InhA proteases is still cytotoxic toward macrophages, although it loses its virulence capacity. Thus, the bacterial mechanism inducing insect death is not limited to the bacterial capacity to kill professional phagocytes.
We have identified new factors that act concomitantly to allow bacteria to infect a susceptible host. The three genes are coexpressed in vitro and in vivo. They are, however, regulated by different mechanisms, and their final destinations differ. Precise characterization of these genetic determinants will allow the development of antimicrobial strategies.
Acknowledgments
We thank Myriam Hajaij-Ellouze and Christina Nielsen-Leroux for the gift of the anti-InhA antibody. We thank all members of the GME laboratory for helpful discussions and Maria Mavris for proofreading the manuscript.
This work was supported by grant ANR-05-PNRA-013.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1994. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J. Bacteriol. 176:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfaro, D. V., III, S. J. Hudson, J. J. Offele, A. A. Bevin, M. Mines, R. M. Laughlin, and R. J. Schoderbek. 1996. Experimental posttraumatic Bacillus cereus endophthalmitis in a swine model. Efficacy of intravitreal ciprofloxacin, vancomycin, and imipenem. Retina 16:317-323. [DOI] [PubMed] [Google Scholar]
- 3.Altincicek, B., M. Linder, D. Linder, K. T. Preissner, and A. Vilcinskas. 2007. Microbial metalloproteinases mediate sensing of invading pathogens and activate innate immune responses in the lepidopteran model host Galleria mellonella. Infect. Immun. 75:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 5.Bouillaut, L., N. Ramarao, C. Buisson, N. Gilois, M. Gohar, D. Lereclus, and C. Nilesen-LeRoux. 2005. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 71:8903-8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callegan, M. C., S. T. Kane, D. C. Cochran, M. S. Gilmore, M. Gominet, and D. Lereclus. 2003. Relationship of PlcR-regulated factors to Bacillus endophthalmitis virulence. Infect. Immun. 71:3116-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 8.Charlton, S., A. J. Baillie, and A. Moir. 1999. Characterisation of exosporium of Bacillus cereus. J. Appl. Microbiol. 87:241-245. [DOI] [PubMed] [Google Scholar]
- 9.Chitlaru, T., O. Gat, Y. Gozlan, N. Ariel, and A. Shafferman. 2006. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 188:3551-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, M. C., T. G. Popova, B. A. Millis, D. V. Mukherjee, W. Zhou, L. A. Liotta, E. F. Petricoin, V. Chandhoke, C. Bailey, and S. G. Popov. 2006. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J. Biol. Chem. 281:31408-31418. [DOI] [PubMed] [Google Scholar]
- 11.Dalhammar, G., and H. Steiner. 1984. Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. Eur. J. Biochem. 139:247-252. [DOI] [PubMed] [Google Scholar]
- 12.Damgaard, P. H., P. E. Granum, J. Bresciani, M. V. Torregrossa, J. Eilenberg, and L. Valentino. 1997. Characterization of Bacillus thuringiensis isolated from infections in burn wounds. FEMS Immunol. Med. Microbiol. 18:47-53. [DOI] [PubMed] [Google Scholar]
- 13.Dancer, S. J., D. McNair, P. Finn, and A. B. Kolsto. 2002. Bacillus cereus cellulitis from contaminated heroin. J. Med. Microbiol. 51:278-281. [DOI] [PubMed] [Google Scholar]
- 14.Edlund, T., I. Sidén, and H. G. Boman. 1976. Evidence for two immune inhibitors from Bacillus thuringiensis interfering with the humoral defense system of saturniid pupae. Infect. Immun. 14:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedhila, S., N. Daou, D. Lereclus, and C. Nielsen-LeRoux. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62:339-355. [DOI] [PubMed] [Google Scholar]
- 16.Fedhila, S., M. Gohar, L. Slamti, P. Nel, and D. Lereclus. 2003. The Bacillus thuringiensis PlcR-regulated gene inhA2 is necessary, but not sufficient, for virulence. J. Bacteriol. 185:2820-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedhila, S., P. Nel, and D. Lereclus. 2002. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J. Bacteriol. 184:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 20.Finney, D. J. 1971. Probit analysis. Cambridge University Press, London, United Kingdom.
- 21.Frankard, J., R. Li, F. Taccone, M. J. Struelens, F. Jacobs, and A. Kentos. 2004. Bacillus cereus pneumonia in a patient with acute lymphoblastic leukemia. Eur. J. Clin. Microbiol. Infect. Dis. 23:725-728. [DOI] [PubMed] [Google Scholar]
- 22.Ganz, T., and R. I. Lehrer. 1999. Antibiotic peptides from higher eukaryotes: biology and applications. Mol. Med. Today 5:292-297. [DOI] [PubMed] [Google Scholar]
- 23.Gascuel, O. 1997. An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 24.Gilois, N., N. Ramarao, L. Bouillaut, S. Perchat, S. Aymerich, C. Nielsen-Leroux, D. Lereclus, and M. Gohar. 2007. Growth-related variations in the Bacillus cereus secretome. Proteomics 7:1719-1728. [DOI] [PubMed] [Google Scholar]
- 25.Gohar, M., K. Faegri, S. Perchat, S. Ravnum, O. A. Økstad, M. Gominet, A. B. Kolstø, and D. Lereclus. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandvalet, C., M. Gominet, and D. Lereclus. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805-1813. [DOI] [PubMed] [Google Scholar]
- 27.Guinebretière, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen, B. M., and S. Salamitou. 2000. Virulence of Bacillus thuringiensis, p. 41-64. In J.-F. Charles, A. Delécluse, and C. Nielsen-Le Roux (ed.), Entomopathogenic bacteria: from laboratory to field application. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 29.Hara, S., and M. Yamakawa. 1995. A novel antibacterial peptide family isolated from silkworm, Bombyx mori. Biochem. J. 310:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez, E., F. Ramisse, T. Cruel, R. le Vagueresse, and J. D. Cavallo. 1999. Bacillus thuringiensis serotype H34 isolated from human and insecticidal strains serotypes 3a3b and H14 can lead to death of immunocompetent mice after pulmonary infection. FEMS Immunol. Med. Microbiol. 24:43-47. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez, E., F. Ramisse, J. P. Ducoureau, T. Cruel, and J. D. Cavallo. 1998. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. J. Clin. Microbiol. 36:2138-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilliard, N. J., R. L. Schelonka, and K. B. Waites. 2003. Bacillus cereus bacteremia in a preterm neonate. J. Clin. Microbiol. 41:3441-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. U. S. A. 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 36.Jongeneel, C. V., J. Bouvier, and A. Bairoch. 1989. A unique signature identifies a family of zinc-dependent metallopeptidase. FEBS Lett. 242:211-214. [DOI] [PubMed] [Google Scholar]
- 37.Kim, S. H., B. S. Park, E. Y. Yun, Y. H. Je, S. D. Woo, S. W. Kang, K. Y. Kim, and S. Kang. 1998. Cloning and expression of a novel gene encoding a new antibacterial peptide from silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 246:388-392. [DOI] [PubMed] [Google Scholar]
- 38.Lecadet, M. M., M. O. Blondel, and J. Ribier. 1980. Generalized transduction in Bacillus thuringiensis var. berliner 1715, using bacteriophage CP54 Ber. J. Gen. Microbiol. 121:203-212. [DOI] [PubMed] [Google Scholar]
- 39.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U. S. A. 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lereclus, D., H. Agaisse, C. Grandvalet, S. Salamitou, and M. Gominet. 2000. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int. J. Med. Microbiol. 290:295-299. [DOI] [PubMed] [Google Scholar]
- 41.Lereclus, D., and O. Arantes. 1992. spbA locus ensures the segregational stability of pHT1030, a novel type of gram-positive replicon. Mol. Microbiol. 6:35-46. [DOI] [PubMed] [Google Scholar]
- 42.Lereclus, D., O. Arantes, J. Chaufaux, and M.-M. Lecadet. 1989. Transformation and expression of a cloned ∂-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 51:211-217. [DOI] [PubMed] [Google Scholar]
- 43.Lövgren, A., M. Zhang, A. Engström, G. Dalhammar, and R. Landén. 1990. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol. Microbiol. 4:2137-2146. [DOI] [PubMed] [Google Scholar]
- 44.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 45.Miller, J. M., J. G. Hair, M. Hebert, L. Hebert, F. J. Roberts, Jr., and R. S. Weyant. 1997. Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 35:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyoshi, S. I., and S. Shinoda. 2000. Microbial metalloprotease and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 47.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 48.Ogierman, M. A., A. Fallarino, T. Riess, S. G. Williams, S. R. Attridge, and P. A. Manning. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J. Bacteriol. 179:7072-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramarao, N., and D. Lereclus. 2006. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 8:1483-1491. [DOI] [PubMed] [Google Scholar]
- 50.Ramarao, N., and D. Lereclus. 2005. The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell. Microbiol. 7:1357-1364. [DOI] [PubMed] [Google Scholar]
- 51.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 52.Salamitou, S., F. Ramisse, M. Brehélin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 53.Sanchis, V., H. Agaisse, J. Chaufaux, and D. Lereclus. 1996. Construction of new insecticidal Bacillus thuringiensis recombinant strains by using the sporulation non-dependent expression system of cryIIIA and a site specific recombination vector. J. Biotechnol. 48:81-96. [DOI] [PubMed] [Google Scholar]
- 54.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidén, I., G. Dalhammar, B. Telander, H. G. Boman, and H. Somerville. 1979. Virulence factors in Bacillus thuringiensis: purification and properties of a protein inhibitor of immunity in insects. J. Gen. Microbiol. 114:45-52. [DOI] [PubMed] [Google Scholar]
- 56.Stenfors Arnesen, L. P., A. Fagerlund, and P. E. Granum. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579-606. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, W., J. Sun, R. Hollmann, A. P. Zeng, and W. D. Deckwer. 2006. Proteomic characterization of transient expression and secretion of a stress-related metalloprotease in high cell density culture of Bacillus megaterium. J. Biotechnol. 126:313-324. [DOI] [PubMed] [Google Scholar]