Abstract
Proteins of 50 or fewer amino acids are poorly characterized in all organisms. The corresponding genes are challenging to reliably annotate, and it is difficult to purify and characterize the small protein products. Due to these technical limitations, little is known about the abundance of small proteins, not to mention their biological functions. To begin to characterize these small proteins in Escherichia coli, we assayed their accumulation under a variety of growth conditions and after exposure to stress. We found that many small proteins accumulate under specific growth conditions or are stress induced. For some genes, the observed changes in protein levels were consistent with known transcriptional regulation, such as ArcA activation of the operons encoding yccB and ybgT. However, we also identified novel regulation, such as Zur repression of ykgMO, cyclic AMP response protein (CRP) repression of azuC, and CRP activation of ykgR. The levels of 11 small proteins increase after heat shock, and induction of at least 1 of these, YobF, occurs at a posttranscriptional level. These results show that small proteins are an overlooked subset of stress response proteins in E. coli and provide information that will be valuable for determining the functions of these proteins.
A challenge of whole-proteome studies of any biological system is the identification and characterization of small proteins, herein defined as those having 50 or fewer amino acids. These small proteins are difficult to detect using standard biochemical techniques. For the analysis of proteins in complex lysates, many studies have employed two-dimensional gel electrophoresis, a technique that is biased toward abundant proteins of standard size (30 to 200 kDa) (21, 29). Proteins that are present at low levels or have extremely large or small molecular masses are usually missed in these experiments (11, 21, 29, 38). Recently, methods for using mass spectroscopy to analyze proteins from crude mixtures have been developed, and they can provide higher sensitivity and resolution (21). However, small proteins are still difficult to identify using these methods. This is in part due to the fact that fewer peptides are produced from peptidic degradation of small proteins, making it more difficult to identify these proteins with confidence. The result of these experimental constraints is a paucity of information about the number and identity of small proteins that are present even under standard growth conditions in any biological system. Even less is known about stress-induced accumulation of these proteins.
Those small proteins that have been identified and characterized, however, indicate that they can have important roles in both bacteria and eukaryotes. In Salmonella, the 30-amino-acid MgtR protein negatively regulates the MgtC virulence factor by binding to and facilitating its degradation (2). The 46-amino-acid Bacillus subtilis Sda protein represses sporulation by inhibiting the activity of the KinA kinase (4, 37). A group of 20- to 22-amino-acid proteins that are excreted by Staphylococcus aureus during infection act to disrupt neutrophil membranes and cause cell lysis (47). In Drosophila melanogaster, a family of 11-amino-acid peptides encoded on a polycistronic mRNA have been implicated in leg development (14), and the 31-amino-acid amphipathic helical sarcolipin protein from rabbits regulates the activity of the sarcoplasmic reticulum Ca2+-ATPase (48). These examples illustrate the diverse role of small proteins in cell physiology, as well as their universal distribution.
In a previous study, we confirmed the synthesis of 20 previously annotated small proteins in Escherichia coli by integrating the sequential peptide affinity (SPA) tag upstream of the stop codon on the chromosome (22). Eighteen of the 20 proteins were expressed in rich medium, while 2 were only detected under specific conditions. We also predicted numerous new small protein genes by using sequence conservation and ribosome binding site models and confirmed the synthesis of 18 of these proteins.
To further characterize 51 previously detected or predicted proteins, we assayed the levels of the SPA-tagged proteins under different growth conditions and after exposure to stress and found that a number of the small proteins are synthesized under specific growth conditions. Four of these, YkgO, AzuC, YkgR, and YobF, were selected for further investigation of the mechanisms of their regulation. We found that transcription of the ykgMO operon is repressed by Zur and that azuC and ykgR transcription is repressed and activated by CRP, respectively. We also found that the increased accumulation of YobF in response to heat shock occurs at a posttranscriptional level.
MATERIALS AND METHODS
Strain constructions.
All strains and oligonucleotides used in the study are listed in Tables S1 and S2, respectively, of the supplemental material. The SPA-tagged strains constructed for this study were generated as previously described (22). The SPA fusion to the small open reading frame (ORF) predicted to overlap pyrG (pyrG-mazG) was generated in NM400 as for the other strains but was not moved into MG1655. In this case, the NM400 strain was analyzed. The strains used for the dot blot assays retained the kanamycin cassette downstream of the SPA tag sequence to facilitate handling of the large number of cultures. Prior to the confirmatory Western blot assays, the kanamycin cassettes were removed by transformation with a plasmid expressing the FLP recombinase (pCP20) (7). Excision of the kanamycin cassette was confirmed by PCR. The Δzur::kan and ΔcadC::kan mutants were generated by homologous recombination with a PCR fragment obtained by amplifying the kanamycin cassette on the pKD4 plasmid. The mutant alleles were sequenced, and the Δzur::kan allele was moved into the ykgO-SPA (Kans) strain, while the ΔcadC::kan allele was moved into the azuC-SPA (Kans) strain by P1 transduction. The Δcrp::cat allele from NRD352 (10) and the rpoS::Tn10 allele described in reference 52 were moved into the azuC-SPA (Kans) and ykgR-SPA (Kans) strains by P1 transduction. The ΔgadXYW::kan allele from PM1293 (31) and the ΔgadE::kan allele from EK551 (30) were moved into the azuC-SPA (Kans) strain, and the ΔoxyS::cat allele from GSO113 (43) and the Δlon::tet allele from ML30008 (28) were moved into the yobF-SPA (Kans) strain by P1 transduction.
Chromosomal transcriptional fusions between the azuC and ykgR promoters and the SPA tag were constructed by replacing the 5′-untranslated regions (UTRs) of azuC and ykgR with the 5′-UTR from multiple cloning site 3 (MCS3) in the pBAD24 plasmid (5′-ACCCGTTTTTTGGGCTAACAGGAGGAATTAACC-3′) (20) and the small protein ORF with the SPA tag sequence. Chromosomal translational fusions to the SPA tag were constructed by replacing the azuC and ykgR ORFs with the SPA tag sequence. The second codon in the SPA tag sequence encodes Met, and this codon was used as the start codon for the SPA reporters. For both sets of constructs, the SPA tag sequence and kanamycin cassette from the pJL148 plasmid were amplified by PCR, transformed into NM400, sequenced, and transduced by P1 into MG1655. For the transcriptional and translational fusion strains, the kanamycin cassette was retained in the strain after transduction.
The ykgR-SPA (Kans) strain was transformed with pSAKTtrc from CAG62093 (B.-M. Koo, unpublished data) to overexpress σH under the control of the trc promoter and with pTrc99A-rpoE from KMT249 (K. M. Thompson, unpublished data) to overexpress σE under the control of the trc promoter.
Growth conditions.
All strains except those carrying crp::cat were grown in Luria broth (LB) rich medium or M63 minimal medium containing 0.0005% vitamin B1 and either 0.2% glucose or 0.4% glycerol. For the crp::cat mutant strains, the minimal medium also contained 0.2% Casamino Acids.
For the dot blot assays, unless stated otherwise, all cultures were grown at 37°C as 5-ml cultures in 50-ml Falcon tubes with shaking at 250 rpm. LB cultures were inoculated with a 1:1,000 dilution of overnight cultures. Minimal glucose cultures were inoculated with a 1:500 dilution of overnight cultures, while minimal glycerol cultures were inoculated with a 1:2,000 dilution (to allow cultures to grow overnight). Oxygen-limited cultures were inoculated from LB-grown overnight cultures into 2 ml LB plus 0.2% glucose (deoxygenated) in 2-ml Eppendorf tubes and were grown without shaking. The corresponding aerobic control cultures were inoculated from the same overnight cultures into 5 ml LB plus 0.2% glucose in 50-ml Falcon tubes and were grown with shaking. Cells from both sets of samples were harvested at an optical density at 600 nm (OD600) of 0.4 to 0.5. To induce cell envelope stress, cultures grown to an OD600 of 0.2 to 0.3 were exposed to 0.025% SDS and 1 mM EDTA for 1 h. To induce acid stress, cultures grown overnight in LB were inoculated into LB-morpholineethanesulfonic acid (MES; 100 mM; pH 5.5 to 5.6) or LB-morpholinopropanesulfonic acid (MOPS; 100 mM; pH 7.5 to 7.6) medium and grown to an OD600 of 0.4 to 0.5. To induce heat shock, cultures grown at 30°C to an OD600 of 0.4 to 0.5 were transferred to a 45°C water bath for 20 min. To induce cold stress, cultures grown at 37°C to an OD600 of 0.2 to 0.3 were transferred to a 10°C water bath for 1 h. To induce oxidative stress or thiol stress, cultures grown to an OD600 of 0.2 to 0.3 were exposed to either 1 mM hydrogen peroxide or 1 mM diamide, respectively, for 30 min. For iron depletion stress, cultures grown to an OD600 of 0.1 to 0.2 were treated with 200 μM dipyridyl for 1 h. To induce DNA damage, cultures grown to an OD600 of 0.3 to 0.4 were treated with 2 μg/ml mitomycin C for 30 min. The OD600 was measured at time of harvest, and the control sample for each of the stress conditions other than oxygen limitation and acid stress were samples grown in LB medium to a similar OD600. Cultures were transferred to ice water baths in order to stop cell growth, and cells were collected from 1-ml aliquots of culture. The cell pellets were frozen on dry ice and stored at −80°C.
Larger samples, grown in 250-ml flasks, were used for the confirmation Western blot assay results. For the cell envelope stress Western blot assays, 30-ml LB cultures started with a 1:1,000 dilution from an overnight culture grown at 37°C were grown to an OD600 of 0.3 to 0.4 at 37°C. The cultures were then divided into four 5-ml aliquots, and water, SDS (0.025%), and/or 1 mM EDTA was added to each culture. Cultures were allowed to grow to an OD600 of 1.0 to 1.2 (1 h of growth for all cultures except those exposed to EDTA and SDS, which grew more slowly) and then harvested. For the acid stress Western blot assays, 30-ml LB-MOPS (pH 7.5) and LB-MES (pH 5.5) cultures were inoculated 1:1,000 from overnight LB cultures and grown until the OD600 was 0.4 to 0.5 at 37°C and harvested. For the heat shock confirmation Western blot assays, 30-ml LB cultures were inoculated 1:1,000 from overnight cultures grown at 30°C and were grown to an OD600 of 0.2 to 0.3 at 30°C. Then 10 ml of the culture was transferred to a 125-ml flask and incubated at 30°C for 20 min, while another 10 ml of culture was incubated at 45°C for 20 min.
Immunoblot assays.
For the dot blot assays, whole cells were resuspended in 1× sample buffer (0.5% SDS, 0.006 mg bromophenol blue, 13% glycerol, 50 mM sodium phosphate buffer [pH 8]) and heated at 95°C for 10 min. A 1-μl aliquot of each sample (equivalent to the cells creating an OD600 of 0.0028) was spotted on a nitrocellulose membrane (Invitrogen) followed by 1-μl aliquots of a half-dilution series. For cells expressing the YbgT-SPA, YnhF-SPA, and YbhT-SPA fusions, the levels of the proteins were significantly higher than the other tagged proteins, so the boiled extract was diluted 1:10 in 1× sample buffer prior to spotting on the membrane. After all spots were applied, the membrane was dried at room temperature for 10 min and then incubated in phosphate-buffered saline with Tween (PBS-T; KD Medical) for 30 min. The membranes were blocked with 3% milk and probed with anti-FLAG M2-AP monoclonal antibody (Sigma-Aldrich) in 2% milk. Signals were visualized using Lumi-Phos WB (Pierce) following standard methodologies. For the confirmation Western blot assays, samples were processed as previously described (22). In brief, a fraction equivalent to the cells in a culture with an OD600 of 0.057 was separated on a Novex 16% Tricine gel (Invitrogen) and transferred to a nitrocellulose membrane (Invitrogen). The membrane was blocked with 3% milk and then probed with anti-FLAG M2-AP antibody (1:1,000 dilution). Membranes were then washed with PBS-T and incubated with Lumi-Phos (Pierce) prior to exposure to film.
Quantification of protein levels.
For quantification of Western blot samples, dot blot dilution series were used to determine relative protein levels. The dilution series were performed similar to the dilution series for the survey dot blots; 1-μl aliquots of a half-dilution series were spotted on nitrocellulose membranes adjacent to the dilution series of other samples in the original Western blot assay. Comparison of spot intensities of the different dilution series allowed us to quantify the relative protein levels for each sample.
Northern analysis.
Northern analysis was conducted essentially as described previously (12). Total RNA was collected from 5 ml of culture by acid-phenol extraction. RNA (5 μg) was separated on 6% acrylamide gels, transferred to a Zeta-Probe membrane (Bio-Rad), and probed with oligonucleotide probes (see Table S2 in the supplemental material) end labeled with [32P]ATP using T4 polynucleotide kinase. Hybridization and wash steps were as described previously.
Primer extension analysis.
Primer extension analysis was conducted as described previously (51). Total RNA was collected from 5 ml of culture by TRIzol (Invitrogen) extraction. Oligonucleotide primers (see Table S2 in the supplemental material) end labeled with [32P]ATP using T4 polynucleotide kinase were incubated with 5 μg of total RNA, allowed to anneal, and then extended with reverse transcriptase (Life Sciences). To generate a sequence ladder, a PCR fragment encompassing the corresponding region was generated, and DNA sequencing reactions were carried out using the SequiTherm EXCEL II DNA sequencing kit (Epicentre) and the primer used in the primer extension reaction.
RESULTS
Small protein levels can be assayed in a high-throughput manner.
We previously confirmed the synthesis of 38 proteins of less than 50 amino acids by integrating the SPA tag upstream of the stop codon on the chromosome and assaying for accumulation by Western blot analysis (22). In our initial work we noted that some of the tagged proteins were expressed under very specific conditions. For example we observed the expected α-methylglucoside induction of SgrT (44). These observations suggested that the SPA tag did not interfere with the regulated synthesis of the proteins. We thus decided to examine the levels of the tagged proteins under a wide range of conditions. We were especially interested in determining if, under specific growth conditions, we could detect synthesis of full-length proteins that we had not observed in cells grown in rich media in our previous study. These predicted proteins included five previously annotated ORFs (Tpr, YlcH, DinQ, YoaI, and YjjY) and six ORFs predicted by our bioinformatic assays in the ymjC′-ycjY, ycgI′-minE, ykgD-ykgE, gmr-rnb, ydjA-sppA, and fabG-acpP intergenic regions (see Table S3 in the supplemental material). We also integrated the SPA tag upstream of the stop codons of two additional putative short ORFs: ymjB, which was originally annotated as a short ORF but is likely a pseudogene remnant, and a short ORF predicted to overlap the 5′ end of the pyrG ORF (see Table S3).
Due to the high specificity and sensitivity of the anti-FLAG antibody, it was possible to screen accumulation of the 51 proteins mentioned above in a high-throughput manner using dot blot assays (Fig. 1, Table 1; see also Fig. S1 in the supplemental material). Based on dilution series of control samples, the dot blot assays provided a detection range of greater than 1,000-fold, depending on the exposure of the blots. The large dynamic range allowed us to detect changes in protein accumulation of both the highly expressed small proteins as well as small proteins expressed at much lower levels. To reduce the number of false positives due to variations in the assay, only differences of fourfold or greater were considered significant. We did not detect the tagged proteins under any condition for the 11 short ORFs for which we had not observed synthesis in our previous study, or for the two putative short ORFs tagged specifically for this study. However, we did identify a number of proteins that were highly induced under specific growth conditions; many of these proteins were present at barely detectable levels under the previous conditions tested (22).
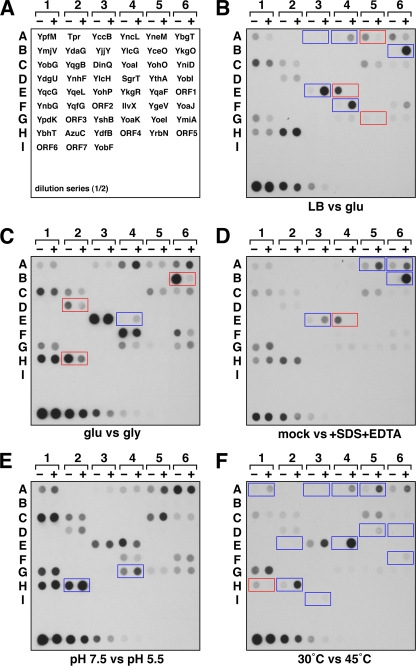
FIG. 1.
Dot blot analysis of small protein levels under different growth conditions. (A) Diagram of spotting patterns. Each tagged protein was assayed under two conditions, designated + or −. A twofold dilution series for YbgT-SPA from cells grown in LB is at the bottom of each blot, with a final dilution of 1:2,048. Predicted, unannotated ORFs for which we had not previously seen full-length protein expression are designated ORF1 to ORF7: ORF1, ymjC′-ycjY; ORF2, ycgI′-minE; ORF3, ykgD-ykgE; ORF4, gmr-rnb; ORF5, ydjA-sppA; ORF6, fabG-acpP; ORF7, pyrG-mazG (see Table S3 in the supplemental material). (B) Dot blot of protein levels in LB (−) versus minimal glucose medium (+). (C) Dot blot of protein levels in minimal glucose (−) versus minimal glycerol medium (+). (D) Dot blot of protein levels in LB (−) or LB plus 0.025% SDS and 1 mM EDTA (+). (E) Dot blot of protein levels in LB-MOPS (pH 7.5) (−) or LB-MES (pH 5.5) (+). (F) Dot blot of protein levels in LB at 30°C (−) or after transfer to 45°C (+). A longer exposure of this dot blot (presented in Fig. S2 of the supplemental material) shows the changes in the less abundant small proteins after heat shock. In all cases, strains were grown as 5-ml cultures in 50-ml Falcon tubes. With the exception of the cultures exposed to heat shock, all cells were grown at 37°C. Cells were collected and samples were analyzed by dot blotting as described in Materials and Methods. Proteins whose expression was induced under the + condition are boxed in blue, while proteins whose expression was reduced under the + condition are boxed in red.
TABLE 1.
Regulated expression of 21 small proteins
| Proteina | Comparison of growth under the two indicated conditions |
Mapb | RegulonDBc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LB vs minimal glucose | Minimal glucose vs minimal glycerol | Aerobic vs low oxygen | Mock treatment vs SDS + EDTA | pH 7.5 vs pH 5.5 | 30°C vs 45°C | 37°C vs 10°C | Mock treatment vs H2O2 | Mock treatment vs diamide | |||
| YpfM | ++ | Monocistronic | None | ||||||||
| YccB | ++d | ++++++ | ++ | appAB-yccB-appC | σ38, σ70, AppY, ArcA | ||||||
| YncL | ++ | +++ | Monocistronic | None | |||||||
| YneM | - -e | ++ | ++ | - - | Monocistronic | None | |||||
| YbgT | +++ | ++ | cydAB-ybgTE | σ70, ArcA, Fnr, FruR, HNS | |||||||
| YkgO | ++++++ | - - - - - - | ++++++ | ykgMO | σ70, Zur | ||||||
| YqgB | - - | yqgB-speAB | None | ||||||||
| YnhF | - - | ++ | Monocistronic | None | |||||||
| YthA | ++ | yjhBC-ythA | None | ||||||||
| YobI | ++ | Monocistronic | NL | ||||||||
| YqeL | ++ | yqeKL | None | ||||||||
| YohP | ++++ | ++++ | ++ | ++ | Monocistronic | NL | |||||
| YkgR | - - - - | ++ | - - - | ++++++ | - - - - | - - | ++ | Monocistronic | NL | ||
| IlvX | +++ | ilvXGMEDA | σ70, Lrp, IHF | ||||||||
| YoaJ | ++ | ++ | yeaP-yoaKJ | None | |||||||
| YpdK | - - - | Monocistronic | NL | ||||||||
| YoaK | ++ | - - | yeaP-yoaKJ | None | |||||||
| YoeI | - | - - | yoeI-yeeF | None | |||||||
| YbhT | - - | Monocistronic | None | ||||||||
| AzuC | - - - | - - | ++ | +++ | ++ | ++ | In IsrB sRNA | NL | |||
| YobF | ++ | yobF-cspC | None | ||||||||
All small proteins that had at least a 4-fold change in protein levels in at least one experiment are listed.
Predicted operonic organization of short ORFs.
Transcriptional regulators listed in RegulonDB that are upstream of either the short ORF or the first gene in the operon. NL, not listed.
Each + indicates a 2-fold increase in protein expression in cells grown under the second condition compared to the first condition.
Each - indicates a 2-fold decrease in protein expression in cells grown under the second condition compared to the first condition.
Small protein levels vary with growth conditions.
To determine if any of the small proteins were expressed differently in minimal medium compared to rich medium, protein levels were assayed during exponential-phase growth in LB or M63 medium supplemented with 0.2% glucose or 0.4% glycerol. Five proteins (YccB-SPA, YncL-SPA, YkgO-SPA, YohP-SPA, and IlvX-SPA) were present at 4-fold or higher levels in minimal glucose medium compared to LB medium, while the levels of three proteins (YneM-SPA, YkgR-SPA, and YoeI-SPA) were lower (Fig. 1B). Similar changes in accumulation were observed for the comparison between growth in glycerol minimal medium and LB, with some differences in the fold changes (see Fig. S1B in the supplemental material). Consistent with the observed regulation, the IlvX protein is encoded in the ilvXGEDA operon, which is induced in minimal medium to synthesize isoleucine (42).
To determine if any of the small proteins were expressed differently depending on the available carbon source, we examined protein levels in cells grown in minimal glucose versus minimal glycerol medium (Fig. 1C). Four differences were detected. The YkgO-SPA, YnhF-SPA, and AzuC-SPA proteins were all present at lower levels in minimal glycerol-grown cells compared to minimal glucose-grown cells, while YkgR-SPA levels were higher in minimal glycerol. The observed changes in protein levels could reflect regulation by cyclic AMP (cAMP) receptor protein (CRP), a transcription factor that modulates the expression of hundreds of genes, depending on glucose availability (18). CRP is able to bind DNA to positively or negatively regulate transcription after it complexes with cAMP, the levels of which increase in response to low glucose. Therefore, ykgO, ynhF, and azuC, for which we observed higher protein levels in glucose-grown cells, could be repressed by CRP-cAMP. In contrast, CRP could positively regulate ykgR, for which lower protein levels were detected in glucose-grown cells. Potential CRP binding sites can be found upstream of azuC and ykgR (see below).
Four proteins (YccB-SPA, YbgT-SPA, YnhF-SPA, and YohP-SPA) showed more-than-4-fold-higher levels during oxygen-limited growth compared to aerobic growth in rich medium (see Fig. S1C in the supplemental material). The most dramatic change was for the small protein YccB-SPA, with a greater-than-30-fold increase in protein levels. The yccB gene is in the appCBA operon, which encodes the subunits of the cytochrome bd II oxidase. The ybgT gene, a paralog of yccB, is in the cydAB operon, which encodes cytochrome bd I oxidase. Transcription of both of these operons has been shown to be induced under low-oxygen conditions (8, 9) and to be activated by the ArcA transcription factor (3, 8) (Table 1). Two proteins (YoeI-SPA and AzuC-SPA) showed reduced levels under low oxygen conditions, suggesting that synthesis of these proteins is repressed during oxygen-limited growth.
Levels of small proteins increase upon stress.
The synthesis of a number of low-molecular-mass proteins in E. coli is regulated upon exposure to stress. The IbpA (15.7-kDa) and IbpB (16.1-kDa) proteins accumulate upon heat shock (25, 32), and Csp family proteins (∼7 kDa) accumulate in response to a variety of stresses, including cold shock (49, 50). To test for stress-induced accumulation of our set of small proteins, we assayed the levels of the tagged proteins after exposure to cell envelope stress, acidic pH, heat shock, cold shock, oxidative stress, thiol stress, iron starvation, and the DNA-damaging agent mitomycin C. These stress conditions were chosen to sample a broad range of potential stress response pathways and hence possible small protein functions.
To examine small protein levels in cells undergoing cell envelope stress, cultures were exposed to SDS and EDTA for 1 h, and protein levels were compared to those in unstressed cells grown to similar optical densities (Fig. 1D). Four proteins (YkgO-SPA, YneM-SPA, YohP-SPA, and YbgT-SPA) were induced at least 4-fold. Western blot assays confirmed that the YkgO-SPA and YneM-SPA proteins accumulated after SDS-EDTA exposure (Fig. 2A). However, the levels of YkgO-SPA and YneM-SPA were similarly increased when cells were exposed to EDTA alone, suggesting that both proteins are likely induced in response to a decrease in cations due to chelation by EDTA. Consistent with our results, yneM transcription was recently found to be regulated by the two-component regulator PhoP in response to changes in Mg2+ levels (32a). Increased levels of YohP-SPA appear to be specific to cells exposed to both SDS and EDTA, suggesting that YohP is responding to cell envelope stress. The dot blot results indicated that the YbgT-SPA protein is present at higher levels upon cell envelope stress, but the levels of the protein were unchanged in the Western blot assays. Similarly according to the dot blot results, YkgR-SPA levels were slightly decreased by SDS and EDTA, but Western blot analysis showed that while YkgR-SPA was present at lower levels after SDS and/or EDTA exposure, the effect was slight. The differences between the dot blot and Western blot assays could be due to the absence of the kanamycin cassette in the tagged strains used for the Western blot assays, slight variations in growth, or differences between the two assays.
FIG. 2.
Western blot analysis of small protein levels. (A) Proteins induced by SDS and/or EDTA exposure. Cells grown overnight in LB were diluted into 30 ml of LB and grown to an OD600 of 0.2 to 0.3. Cultures were then split into five 5-ml aliquots and exposed to water, 0.025% SDS, 1 mM EDTA, or 0.025% SDS plus 1 mM EDTA. Cells were harvested before stress (T0) and at an OD600 of 1.2 to 1.7. (B) Proteins induced under acidic conditions. Cells grown overnight in LB were diluted into 30 ml of either LB-MOPS (pH 7.5) or LB-MES (pH 5.5) and harvested at an OD600 of 0.3 to 0.4. (C) Proteins induced by heat shock. Cells grown overnight in LB at 30°C were diluted into 30 ml of LB and grown at 30°C to an OD600 of 0.4. Cultures were then split into three 10-ml aliquots. One set of samples was transferred to 45°C, while the other half was kept at 30°C. Cells were harvested before transfer (T0) as well as 5 and 20 min after transfer. Western blot analysis using anti-FLAG, alkaline phosphatase-conjugated antibodies was carried out with whole-cell extracts harvested from the cultures above. Asterisks denote the band corresponding to the fusion protein. Exposure times were optimized for each panel for visualization here; therefore, the signal intensity shown does not indicate relative abundance between proteins.
Two proteins (AzuC-SPA and YoaK-SPA) were present at higher levels under acidic (pH 5.5) than neutral (pH 7.5) conditions (Fig. 1E); AzuC-SPA was strongly induced, while induction of YoaK-SPA was more modest (Fig. 2B). The 5′ end of the transcript encoding yoaK has not been mapped, but the gene may be encoded in a polycistronic message with the small gene yoaJ as well as yeaP, a gene encoding a GGDEF domain protein with diguanylate cyclase activity (39). Confirmation Western blot assays showed that YoaJ, but not YoaK, seems to accumulate to slightly higher levels in acidic medium (Fig. 2B). Again, the differences in the levels of the YoaJ and YoaK proteins in the two assays could be due to the removal of the kanamycin cassettes prior to the confirmation Western blot assays, which could alter transcription or translation of other genes encoded on the same mRNA. Further work is needed to determine if these two small proteins are cotranscribed and coordinately regulated.
Eleven small proteins (YkgR-SPA, YobF-SPA, YqeL-SPA, YoaJ-SPA, YncL-SPA, YpfM-Spa, YneM-SPA, YobI-SPA, AzuC-SPA, YthA-SPA, and YccB-SPA) were found to accumulate to at least 4-fold-higher levels after heat shock when cells were shifted from 30°C to 45°C for 20 min (Fig. 1F). Western blot experiments confirmed the heat shock induction of all of these proteins (Fig. 2C). However, the one protein that showed reduced levels after heat shock, YbhT-SPA, did not show the same response in the confirmation Western blot assays. Nine proteins (YkgR-SPA, YobF-SPA, YqeL-SPA, YoaJ-SPA, YncL-SPA, YpfM-Spa, YneM-SPA, YobI-SPA, and AzuC-SPA) were induced within 5 min after transfer from 30°C to 45°C, while two proteins (YthA-SPA and YccB-SPA) only showed induction after extended heat shock. No σ32 binding sites have been predicted for any of these genes by RegulonDB (40) (Table 1), although weak sites can be found upstream of ypfM, yneM, and yobI (V. Rhodius, unpublished data). Four genes, yobF, yqeL, ythA, and yccB, are predicted to be encoded in four different operons, but the other genes in the operons have no clear relation to heat shock. Two of the most strongly induced proteins, YkgR and YobF, were selected for further study (see below). In contrast to heat shock, no proteins were induced more than 2-fold by cold shock (see Fig. S1D in the supplemental material), suggesting that none of the small proteins tested is involved in the cold shock stress response. In fact, one heat shock-induced protein, YkgR-SPA, was strongly repressed after cold shock, suggesting that it may be detrimental under cold stress conditions.
To identify small proteins induced by oxidative stress, exponential-phase cells were exposed to hydrogen peroxide for 30 min and protein levels were compared with those in nonstressed cells. Two proteins (YohP-SPA and AzuC-SPA) were induced ∼4-fold by hydrogen peroxide treatment, and YkgR-SPA levels were reduced ∼4-fold (see Fig. S1E in the supplemental material). In a related experiment, cells were exposed to the thiol oxidant diamide for 30 min (see Fig. S1F in the supplemental material). Diamide-stressed cells showed increased levels of three proteins (AzuC-SPA, YkgR-SPA, and YoaK-SPA) as well as decreased levels of three proteins (YneM-SPA, YpdK-SPA, and YoaJ-SPA). Interestingly, there was little overlap in small protein accumulation when comparing oxidative stress caused by hydrogen peroxide and thiol stress caused by diamide; only AzuC-SPA was induced by both stresses. In contrast, the treatments had opposite effects on the levels of YkgR-SPA.
The levels of the tagged proteins were essentially unchanged after 1 h of dipyridyl treatment to induce iron starvation (data not shown). Similarly, exposure to the DNA damaging agent mitomycin C for 30 min did not significantly change the accumulation of any of the small proteins (data not shown).
ykgM-ykgO is repressed by Zur.
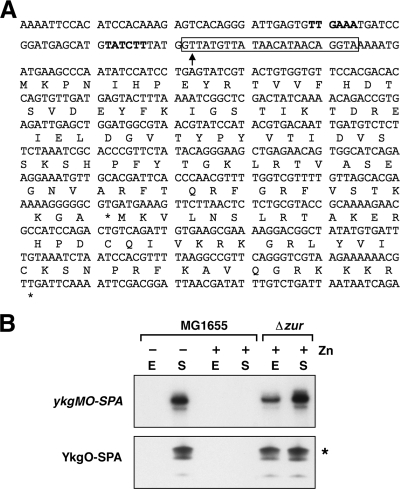
Five small proteins showed increased levels in cells grown in minimal glucose medium compared to cells grown in rich medium. One of the most highly induced proteins was YkgO, a paralog of the ribosomal protein RpmJ. We had observed induction of YkgO-SPA in minimal medium in our previous study and hypothesized that this effect was due to the regulation of the ykgM-ykgO operon by the Zur repressor (22). Zur, a transcription factor found in many species of bacteria, regulates gene expression in response to zinc levels (34). DNA binding by Zur requires zinc; under zinc-depleted conditions, Zur is released from the DNA, allowing transcription of repressed genes. The minimal medium used in our experiments does not contain added zinc, and we also observed strong induction of YkgO-SPA when cells were treated with EDTA, consistent with our prediction that the high levels of YkgO-SPA in cells grown in minimal medium might be due to Zur derepression. A previous bioinformatic search for Zur binding sites in E. coli identified a potential binding site overlapping the transcription start of the ykgM-ykgO operon of E. coli (34) (Fig. 3A). To test whether the induction was in fact zinc dependent, we added zinc to the medium and found that ykgM-ykgO-SPA mRNA levels and YkgO-SPA protein levels were strongly repressed by zinc (Fig. 3B). In contrast, the levels of both the mRNA and protein were high in the presence of zinc in a Δzur background. These data confirm the predicted Zur repression of the ykgM-ykgO operon. We noted that YkgO-SPA levels were high in both exponential and stationary phase with the Δzur mutant, but we only detected high levels of the protein in stationary-phase zur+ cells grown in minimal medium. A likely explanation for these observations is the presence of trace amounts of zinc in the minimal media used for these experiments; this level could be sufficient to repress ykgM-ykgO at low cell density but becomes limiting by stationary phase, allowing YkgO to be expressed in a majority of the cells. Consistent with this possibility, there was some variation in YkgO-SPA levels in exponential-phase cells grown in minimal medium, while YkgO-SPA levels were consistently high in stationary-phase cells (data not shown).
FIG. 3.
Zur repression of ykgO. (A) Sequence of the ykgM-ykgO promoter and coding region. The +1 site of transcription (24) is denoted with an arrow. Potential σ70 −10 and −35 sequences are indicated in bold, and the predicted Zur binding site is boxed. (B) ykgO-SPA mRNA (top) and YkgO-SPA protein (bottom) levels in MG1655 and Δzur cells grown in minimal glucose medium with (+) or without (−) zinc. Overnight cultures were grown in M63 containing 0.2% glucose and 100 μM zinc acetate. Cells were washed twice in M63 containing 0.2% glucose and diluted into M63 glucose medium lacking or containing 100 μM zinc acetate. Cells were harvested at exponential (E) and stationary (S) phase and tested for ykgO-SPA expression and YkgO-SPA synthesis. For the Northern analysis, total RNA (5 μg of each sample) was separated on a 6% acrylamide gel. RNA was transferred to nitrocellulose and probed with an end-labeled oligonucleotide complementary to the ykgO ORF. The band runs at ∼600 nucleotides, consistent with the expected size of the ykgM-ykgO-SPA transcript. Western blot analysis was performed as stated for Fig. 2. The asterisk denotes a band corresponding to the full-length SPA-tagged YkgO protein.
azuC is repressed by CRP.
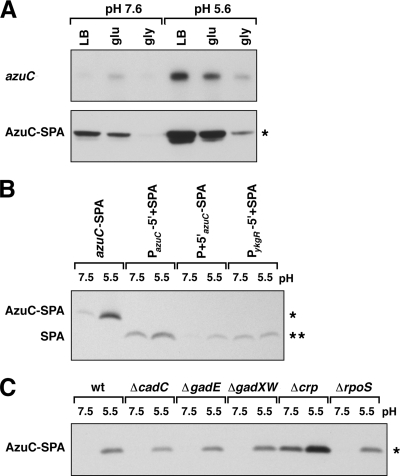
Three proteins were present at higher levels in minimal glucose medium than in minimal glycerol medium, suggesting that their synthesis may be repressed by the CRP transcription factor. One of these, AzuC, a basic (pKa 10.3), 28-amino-acid protein predicted to form an amphipathic α-helix by HeliQuest (15), is encoded by a transcript that was first identified as the ISO92 small RNA (6). The start of the azuC transcript was mapped to 42 nucleotides (nt) upstream of the azuC AUG by primer extension (see Fig. S2 in the supplemental material). Two locations within the promoter region contain DNA sequences similar to CRP DNA binding sites (26), one immediately upstream of the −35 hexamer and the other overlapping with the −10 hexamer (Fig. 4A). Binding of CRP to the putative downstream site would be expected to repress transcription initiation. Consistent with this prediction, deletion of the crp gene eliminated the carbon source regulation, resulting in similar levels of AzuC synthesis when cells were grown in media containing glucose or glycerol (Fig. 4B).
FIG. 4.
CRP repression of azuC. (A) Sequence of the azuC promoter and coding region. The +1 site of transcription (position 1986025 of the E. coli K-12 genome) is denoted with an arrow. Potential σ70 −10 and −35 sequences are indicated in bold, and the predicted CRP binding sites are boxed. (B) azuC-SPA mRNA (top) and AzuC-SPA protein (bottom) levels in MG1655 and Δcrp cells grown in minimal glucose (glu) and minimal glycerol (gly) media supplemented with 0.2% Casamino Acids and 0.0005% vitamin B1. M63 glucose and M63 glycerol cultures (30 ml inoculated with overnight cultures grown in the respective medium) were grown at 37°C to an OD600 of 0.3 to 0.4. Northern analysis was performed as described for Fig. 3. The RNA band runs at ∼400 nucleotides, consistent with the expected size of the azuC-SPA transcript. Western blot analysis was performed as stated for Fig. 2. The asterisk denotes a band corresponding to the full-length SPA-tagged AzuC protein.
In addition to being repressed by CRP, AzuC-SPA levels were repressed under low-oxygen conditions, moderately induced during heat shock, oxidative stress, and thiol stress, and substantially elevated during growth in acidic medium. The strong acid induction was also reflected in increased azuC-SPA mRNA levels (Fig. 5A). These observations suggested that CRP repression is alleviated upon acid stress or that other DNA binding proteins regulate azuC transcription. Alternatively, azuC mRNA levels might be subject to posttranscriptional regulation. To test if azuC induction by low pH occurs at a transcriptional or posttranscriptional level, we generated transcriptional and translational SPA fusions on the chromosome in which the azuC ORF was replaced with the SPA tag sequence, beginning with an ATG at the second codon of the tag sequence. For the transcriptional fusion, the azuC 5′-UTR was also replaced with the multiple-cloning site (MCS) 5′-UTR from the pBAD24 plasmid. Although the relative levels of the SPA peptide were different when expressed from different constructs, the levels from both azuC constructs were elevated during growth in acidic media but not from a control fusion to the ykgR promoter (Fig. 5B). Quantification of relative protein levels of the fusions showed that the fold difference between neutral- and acidic-grown cells was ∼8-fold for the full-length fusion, ∼4-fold for the transcriptional fusion, and ∼8-fold for the translational fusion (see Fig. S4A in the supplemental material). These results indicate that the acid induction occurs at both the transcriptional and posttranscriptional levels.
FIG. 5.
Acid induction of azuC. (A) azuC mRNA (top) and AzuC-SPA protein (bottom) levels in MG1655 in LB, minimal glucose (glu), and minimal glycerol (gly) medium buffered at pH 7.6 or 5.6. The RNA band runs at ∼400 nucleotides, consistent with the expected size of the azuC-SPA transcript. Cells were diluted into the respective medium from overnight cultures grown in LB. (B) Acid induction of azuC transcriptional and translational fusions. Extracts from strains containing the SPA-tagged azuC allele (azuC-SPA), a transcriptional fusion to the azuC promoter (PazuC-5′+SPA), a translational fusion to the azuC promoter and 5′ UTR (P + 5′azuC-SPA), or a control transcriptional fusion to the ykgR promoter (PykgR-5′+SPA) were probed for SPA expression in LB-MOPS (pH 7.5) and LB-MES (pH 5.5). In the transcriptional fusions, the azuC and ykgR 5′-UTRs were replaced by the MCS 5′-UTR from pBAD24, and the ORFs were replaced by the SPA tag. In the translational fusion, the azuC ORF was replaced just by the SPA tag (see Materials and Methods). Cultures grown overnight in LB were diluted into 10 ml of LB-MOPS (pH 7.6) or LB-MES (pH 5.6), and cells were harvested at an OD600 of 0.45 to 0.6. (C) AzuC-SPA expression in wild-type and mutant cells grown in neutral and acidic media. Cultures grown overnight in LB were diluted into 5 ml LB-MOPS (pH 7.6) or LB-MES (pH 5.6), and cells were harvested at an OD600 of 0.45 to 0.6. Northern analysis was performed as described for Fig. 3. Western blot analysis was performed as stated for Fig. 2. A single asterisk denotes a band corresponding to the full-length SPA-tagged AzuC protein, and a double asterisk denotes a band corresponding to the SPA peptide.
To examine the contribution of CRP as well as known transcriptional regulators of the E. coli responses to acidic conditions, we assayed AzuC-SPA levels in wild-type, Δcrp, ΔrpoS, ΔgadXW, ΔgadE, and ΔcadC cells exposed to pH 5.5 (Fig. 5C). No difference in the increase of AzuC-SPA accumulation was detected between the wild-type, ΔrpoS, ΔgadXW, ΔgadE, and ΔcadC cells (∼30-fold). In contrast, the degree of AzuC-SPA induction in acidic medium compared to neutral medium was reduced in a Δcrp background (∼4-fold) (see Fig. S4B in the supplemental material). These results suggest that CRP regulation contributes to the pH-dependent regulation of AzuC levels, most likely by repressing AzuC transcription in neutral media, but that there is also an as-yet-unidentified transcriptional regulator of azuC induction in acidic medium. In contrast to the strains carrying the full-length, transcriptional, and translational fusions for which results are shown in Fig. 5B, the kanamycin cassette associated with the azuC-SPA allele was flipped out in the strains assayed in Fig. 5C (see also Fig. S4B in the supplemental material), and we consistently observed greater changes for the AzuC-SPA fusion in strains lacking the kanamycin cassette.
ykgR is activated by CRP.
The dot blot assays showed that YkgR-SPA levels are higher in medium containing glycerol rather than glucose, suggesting that CRP activates ykgR transcription. Again, primer extension analysis was used to map the start of the ykgR mRNA to a single transcriptional start site 43 nt upstream of the ykgR AUG (see Fig. S2 in the supplemental material). While a potential −10 sequence was noted, the promoter lacked a potential −35 sequence (Fig. 6A). However, a potential CRP binding site could be found centered ∼41 nucleotides upstream of the transcriptional start, suggesting that the ykgR promoter may be a class II CRP-dependent promoter (27). At class II promoters, where the CRP binding site overlaps the −35 region and RNA polymerase makes DNA contacts upstream and downstream of CRP, there is generally a poor match to the consensus −35 sequence. Assays of YkgR-SPA synthesis in a Δcrp mutant confirmed that high levels of YkgR synthesis in minimal glycerol medium at 37°C are dependent on CRP (Fig. 6B). Although the ykgR-SPA transcript was barely detectable, the levels were higher for cells grown in minimal glycerol medium than those grown in minimal glucose medium, and this difference was CRP dependent (Fig. 6B).
FIG. 6.
CRP activation of ykgR. (A) Sequence of the ykgR promoter and coding region. The +1 site of transcription (position 312510 of the E. coli K-12 genome) is denoted with an arrow. A potential σ70 −10 sequence is indicated in bold, and the predicted CRP binding site is boxed. (B) Primer extension analysis of ykgR-SPA mRNA (top) and Western blot analysis of YkgR-SPA protein (bottom) levels in MG1655 and Δcrp cells grown in minimal glucose (glu) and minimal glycerol (gly) medium supplemented with 0.2% Casamino Acids and 0.0005% vitamin B1. M63 glucose and M63 glycerol cultures (inoculated from overnight cultures grown in the respective medium) were grown at 37°C to an OD600 of 0.3 to 0.4. Primer extension assays were conducted using 5 μg total RNA of each sample and an end-labeled oligonucleotide complementary to the ykgR ORF. Western blot analysis was performed as stated for Fig. 2. The asterisk denotes a band corresponding to the full-length SPA-tagged YkgR protein.
In addition to being activated by CRP, YkgR-SPA accumulation was strongly elevated upon heat shock (Fig. 7A). The levels of the ykgR-SPA mRNA were similarly induced upon a shift to 45°C in the wild-type strain, although no transcript could be detected without or with heat shock in the Δcrp mutant strain (data not shown). To test if ykgR heat shock induction occurs at a transcriptional or posttranscriptional level, we generated SPA fusion strains similar to those used to examine azuC expression. A transcriptional fusion was constructed in which the ykgR 5′-UTR was replaced on the chromosome by the pBAD24 MCS 5′-UTR and the ykgR ORF was replaced by the SPA tag sequence. For the translational fusion only the ykgR ORF was replaced by the SPA sequence. Heat shock led to similarly increased levels of the SPA tag from both fusions (Fig. 7B), suggesting that ykgR transcription is induced by heat shock. The genes of most heat shock-induced proteins are transcribed by the heat shock factor σ32 or the cell envelope stress regulator σE (1, 19). However, overexpression of neither σ32 nor σE from a plasmid led to YkgR-SPA induction (Fig. 7C). YkgR-SPA levels were also unchanged in ΔrseA cells, which lack the σE anti-sigma factor and thus have elevated levels of σE (data not shown), as well as in an rpoS mutant strain (Fig. 7C). These data indicate that ykgR induction is not regulated by these alternate σ factors, consistent with an absence of recognizable binding sites upstream of the ykgR transcriptional start site. In addition, the results suggest that ykgR heat shock induction is regulated by redundant or as-yet-unidentified mechanisms.
FIG. 7.
Heat shock induction of ykgR. (A) Primer extension analysis of ykgR-SPA mRNA (top) and Western blot analysis of YkgR-SPA protein (bottom) levels in MG1655 without and with heat shock. Thirty-milliliter LB, M63 glucose, or M63 glycerol cultures were inoculated with a dilution of overnight LB cultures and grown to an OD600 of 0.3 to 0.4 before being split into three 10-ml aliquots. Two aliquots were kept at 30°C while the other was incubated at 45°C. Cells were harvested before transfer (T0) and after 20 min (30°C or 45°C). (B) Heat shock induction of ykgR transcriptional and translational fusions. Extracts from strains containing the SPA-tagged ykgR allele (ykgR-SPA), a transcriptional fusion to the ykgR promoter (PykgR-5′+SPA), a translational fusion to the ykgR promoter and 5′-UTR (P + 5′ykgR-SPA), or a control transcriptional fusion to the azuC promoter (PazuC-5′+SPA) were probed for SPA expression in cells without or with heat shock. In the transcriptional fusions, the ykgR and azuC 5′-UTRs were replaced by the MCS 5′-UTR from pBAD24, and the ORFs were replaced by the SPA tag. In the translational fusion, the ykgR ORF was replaced by the SPA tag. Cultures grown overnight in LB were diluted into 30 ml LB and grown to an OD600 of 0.4 to 0.5 before being split into two 10-ml aliquots. One aliquot was kept at 30°C while the other was incubated at 45°C. (C) YkgR-SPA expression in wild-type cells and cells with altered sigma factor levels. YkgR-SPA levels were assayed in wild-type and ΔrpoS cells without and with heat shock, as well as in ykgR-SPA cells in which σH or σE synthesis was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to half of the sample. MG1655 and ΔrpoS cells grown overnight in LB were diluted into 30 ml LB and incubated at 30°C until the OD600 reached 0.4. Cultures were then split into two 10-ml aliquots. One set of samples was transferred to 45°C while the other was kept at 30°C. Cells were harvested before transfer (T0) and after 20 min of induction (30°C and 45°C). YkgR-SPA cells containing rpoH (pSAKTtrc) and rpoE (pCL245) overexpression plasmids were grown overnight in LB plus 100 μg/ml carbenicillin and were diluted into 30 ml of LB plus 100 μg/ml carbenicillin. At an OD600 of 0.4, cultures were split into two 10-ml aliquots, and sigma factor expression was induced in one set of samples by adding IPTG to 1 mM. Cells were harvested before induction (T0) and 20 min after induction (− and +, respectively). Primer extension assays were as conducted as described for Fig. 6. Western blot analysis was performed as stated for Fig. 2. A single asterisk denotes a band corresponding to the full-length SPA-tagged YkgR protein, and a double asterisk denotes a band corresponding to the SPA peptide.
Heat shock induction of YobF occurs at a posttranscriptional level.
The levels of the YobF-SPA protein were also strongly induced by heat shock (Fig. 2C). Primer extension analysis showed that the yobF-cspC mRNA was transcribed from two different promoters mapping 25 to 26 nt and 202 nt upstream of the yobF AUG (Fig. 8A; see also Fig. S2 in the supplemental material). However, in contrast to ykgR, Northern analysis of the yobF-cspC operon after heat shock showed that elevated temperature did not lead to increased levels of the mRNA (Fig. 8B). Together, these data suggest that YobF heat shock induction occurs at a posttranscriptional level.
FIG. 8.
Posttranscriptional heat shock induction of YobF. (A) Sequence of the yobF-cspC promoter and coding region. The +1 site of both the longer transcript (position 1905817 of the E. coli K-12 genome) and the shorter transcript (position 1996212 of the E. coli K-12 genome) are denoted by arrows. Possible σ70 binding sites are indicated in bold, and nucleotides that are predicted to base pair with the OxyS small RNA are denoted with dots. (B) yobF-cspC mRNA (top) and YobF-SPA protein (bottom) levels in cells exposed to heat shock. Samples were treated as for Fig. 7A. Cells were harvested before transfer (T0) and after 20 min at 30°C or 45°C. (C) YobF-SPA levels in a Δlon mutant strain. Again, cells were harvested from samples kept at 30°C or 45°C for 20 min or before transfer (T0). (D) YobF heat shock induction with and without hydrogen peroxide exposure in MG1655 and an ΔoxyS mutant. Heat shock induction was conducted as described for panel C, except that in some cases, hydrogen peroxide was added to the cells to a final concentration of 250 μM, 10 min prior to heat shock. Northern analysis was performed as described for Fig. 3. The prominent smaller RNA band runs at ∼600 nucleotides, consistent with the expected size of the shorter yobF-SPA transcript. Western blot analysis was performed as stated for Fig. 2. The asterisk denotes a band corresponding to the full-length SPA-tagged YobF protein.
YobF-SPA accumulation after heat shock could be due to a decrease in degradation. However, there were no changes in YobF-SPA levels in ΔclpP, ΔclpQ, and ΔclpY mutant strains, indicating that YobF is not a substrate for these proteases (data not shown). YobF is a substrate for the Lon protease, but YobF-SPA levels still increase upon heat shock in a Δlon mutant (Fig. 8C). The possibility that YobF is a substrate of yet another protease cannot be ruled out.
Another plausible explanation for the observed heat shock induction is occlusion of the ribosome binding site by the mRNA secondary structure, which melts at higher temperatures, as has been observed for 5′-UTRs that function as RNA thermometers (reviewed in reference 33). The yobF-cspC mRNA with either 5′ end is predicted to be highly structured, and this structure may inhibit translation of the YobF protein (see Fig. S3 in the supplemental material). In previous studies we noted a potential region of base pairing between the hydrogen peroxide-induced OxyS small RNA and yobF in a region overlapping the ribosome binding site (Fig. 8A) and showed that plasmid-expressed OxyS led to significantly decreased yobF mRNA levels (43). In the present study, we found that while endogenous OxyS did not affect YobF-SPA levels under normal conditions (data not shown), the small RNA did inhibit heat shock induction (Fig. 8D). If added immediately prior to heat shock, hydrogen peroxide exposure reduced the level of YobF-SPA accumulation in an OxyS-dependent manner. These data are consistent with the hypothesis that the 5′-UTR of the yobF transcript is generally occluded in a secondary structure during normal growth, that this second structure unfolds upon heat shock, and that heat shock-dependent unfolding of the yobF-cspC operon facilitates OxyS repression of YobF translation. Hydrogen peroxide exposure is associated with a decrease in the levels of yobF-cspC transcript in wild-type cells but not in the ΔoxyS mutant strain, consistent with the assumption that OxyS promotes some degradation of the yobF-cspC mRNA.
DISCUSSION
The number of small proteins is not known for the proteome of any organism. A survey of the literature shows the smallest proteins identified in most proteomics analyses in E. coli are around 10 kDa (21), in contrast to the 2- to 5-kDa small proteins described in this work. In a previous study, we showed that the E. coli K-12 genome encodes many more expressed small proteins than had previously been predicted. Given that even less is known about the regulation of small protein accumulation in E. coli and other organisms, we determined the levels of 51 confirmed and putative small proteins under a variety of growth conditions. We found that many show medium- and stress-dependent regulation.
Of the 51 proteins tested, 21 were induced under at least one of the conditions tested. Although these results show that the accumulation of many small proteins is subject to regulation in response to environmental conditions, it is important to note possible limitations of these experiments. Each of these small proteins is expressed as a chimeric protein with a C-terminal SPA tag that is larger than the endogenous protein. The presence of this tag could potentially inhibit normal regulation at the transcriptional, translational, or posttranslational level. The fact that we observed previously predicted regulation, such as low-oxygen induction of YbgT and YccB, and observed the same regulation with the endogenous and tagged ykgR and yobF-cspC transcripts (data not shown), suggests that the C-terminal SPA tag probably does not alter the native regulation of most of the small proteins. It is also possible that some regulation may have been missed because of the background levels of the SPA fragment we observed for a subset of the proteins (22). High levels of this fragment could mask changes in levels of the full-length protein in the initial dot blot assays. However, it is difficult to imagine a situation in which the SPA tag leads to new regulation not associated with the native protein, and it is therefore likely that the examples of regulation observed in this study accurately reflect that imposed on the endogenous proteins.
Transcription of some small protein genes is regulated by specific DNA binding proteins.
Information about the transcriptional regulation of E. coli operons encoding some of the small proteins is available. Of the small proteins examined in this study, four show regulation consistent with previously predicted transcriptional regulation of the corresponding operons. The synthesis of two small proteins, YbgT and YccB, which are encoded in cytochrome oxidase operons with ArcA-dependent activation (3, 8), was induced under low-oxygen growth conditions as would be expected. Interestingly, the appCB-yccB-appA operon has been suggested to be nonfunctional based on the inability of the operon to complement a cytochrome oxidase mutant for aerobic growth on succinate-containing medium (41). Rather than being nonfunctional, the dramatic induction of YccB levels during growth in low oxygen (∼30-fold) compared to aerobic conditions suggests that the appABC cytochrome oxidase may only be functioning under low-oxygen conditions, in contrast to the two other cytochrome oxidases in E. coli. The small protein IlvX, which is encoded in the ilvXGMEDA isoleucine biosynthesis operon regulated by Lrp (35), shows the expected increase in protein levels in cells grown in minimal medium (∼8-fold). Finally, the ykgMO operon was predicted to be regulated by the Zur transcription factor (34), a prediction that was substantiated by the observed effects of zinc and a Δzur deletion. A recent microarray study of genes induced by zinc depletion (17) identified ykgM as a zinc-repressed gene but did not report on ykgO, as the microarray did not include probes for the smaller gene. YtiA, a Bacillus subtilis ortholog of YkgM, has also recently been shown to be regulated by zinc through Zur (13).
In addition to confirming predicted regulation of small protein expression, we observed new transcriptional regulation. We showed that CRP regulates the synthesis of at least two small proteins: repressing azuC and activating ykgR. We also found that azuC transcription is induced in acidic medium and ykgR transcription is induced by heat shock but noted that previously characterized transcriptional regulators of the acid and heat shock responses were not responsible for the induction of these genes. Possibly there is redundancy in the transcriptional regulators needed for the induction. Alternately, azuC and ykgR may be induced by other regulators that were previously not associated with the acid and heat shock responses. A final possibility is that CRP activity is altered under acid and heat shock stress conditions and that this is responsible for the changes in AzuC and YkgR levels. In fact, the difference between AzuC-SPA levels in neutral and acidic media is reduced in the crp mutant. Little is known about the effects of acid and heat stress on cAMP levels and CRP activity, but it has been shown that the expression levels of other acid stress genes (gadA and gadBC) (5) and heat shock genes (hslS, hslT, and htpG) (16) are altered in crp mutant strains.
Stress induction of some small proteins occurs at a posttranscriptional level.
There was little information about possible transcriptional regulation for small proteins not encoded in operons or encoded in operons of unknown function. Many of these genes were only recently annotated and would have been missed in microarray analyses carried out using gene-specific probes. It is also possible that the accumulation of some proteins is regulated at the level of translation or protein stability and thus would have been missed in the surveys of transcriptional responses to stress. We obtained evidence that the heat shock induction of YobF occurs at the posttranscriptional level. One potential mechanism for the posttranscriptional heat shock induction of proteins is the melting of temperature-sensitive RNA stems that occlude the ribosome binding site at normal temperatures (45). Consistent with this model, the yobF-cspC mRNA leaders are predicted to fold into a secondary structure that could block ribosome binding (see Fig. S3 in the supplemental material). It is noteworthy that the predicted yobF ribosome binding site is unusually far removed from the AUG (13-nucleotide spacer), another factor that could impact the heat shock regulation of YobF synthesis. The findings that the yobF-cspC transcript is the target of the OxyS small RNA and that YobF-SPA is a substrate of the Lon protease support the conclusion that YobF accumulation is subject to significant posttranscriptional control.
We suggest that still other small proteins are subject to posttranscriptional regulation. M-Fold predictions (52) of the 5′-UTRs of at least one other heat shock-induced protein, YqeL, showed that this coding sequence may be preceded by structures reminiscent of a 4U-like motif, a temperature-sensitive RNA structure where the ribosome binding site is paired with four uridine nucleotides (46). If RNA melting contributed to YqeL induction and possibly other small proteins, this would allow for a rapid increase in protein synthesis in response to the stress. Another possibility is that other small heat shock-induced proteins besides YobF are protease targets and that they accumulate after heat shock because the proteases are overwhelmed with other unfolded protein substrates. An examination of the small protein levels in protease mutants will be valuable for discerning whether this regulation contributes to the heat shock induction.
Possible roles of small proteins in stress responses.
Nineteen of the proteins that were found to be induced under specific conditions are predicted to contain single transmembrane helices, and many of these have been shown to fractionate with the cell membrane (22). For these proteins, it is tempting to speculate that they could interact with the inner membrane under conditions of slow growth or stress and thus modulate the function of other transmembrane proteins, affect membrane permeability, or serve a stabilizing role in the membrane. Other stress-inducible proteins could be acting as chaperones or facilitate protein degradation upon stress exposure, roles that have been determined for IbpA and IbpB (32). Yet another possibility is that some of the proteins are of prophage origin. The heat shock-induced YthA protein is predicted to be encoded in the yjhB-yjhC-ythA operon, which is flanked by IS elements and shows no conservation outside of E. coli. Phage-related proteins often accumulate under stress conditions, presumably so that the prophage can become lytic (36). One might imagine that small hydrophobic proteins such as YthA descended from small transmembrane holin proteins used by phage to lyse the bacterium host.
In a separate but complementary study, mutants carrying bar-coded deletions of the genes encoding the small proteins were screened for sensitivity to cell envelope and acid stress in large-scale competition experiments (23). Surprisingly, there was little overlap between the regulation we observed and the phenotypes of the deletion strains. This discrepancy may be due to the fact that the conditions used in the competition assays were different from those used for the studies described here. Contrary to the short-term and relatively mild exposure to stress used for these expression assays, the survival assays described by Hobbs et al. consisted of a long-term exposure to SDS-EDTA and treatment with extreme acid (pH 1.8). It is possible that the stress-induced small proteins identified in the studies described here are not involved under more severe stress conditions as tested in the competition studies. Alternatively, some of the small proteins may have functions that are redundant with other stress response proteins. The limited convergence between the two approaches highlights the importance of carrying out multiple methodologies in characterizing these small proteins of unknown function.
The results presented here show that E. coli contains many stress-induced proteins that were missed using classical biochemical techniques. Given the lack of probes for these genes on most microarrays, the genes also were not assayed in whole-genome expression surveys to characterize the regulons of well-studied global transcriptional regulators such as CRP (16). Based on these and other findings, it is clear that small proteins and the genes that encode them need to be taken into consideration when designing future experiments. In addition, information about the accumulation of the small proteins could set the stage for further functional characterizations, pointing to growth conditions best suited for biochemical assays, such as copurification, as well as for phenotypic assays of mutant strains.
Supplementary Material
Acknowledgments
We thank S. Goodwin for conducting the primer extension analysis of the yobF operon, S. Gottesman, K. Moon, and V. Rhodius for sharing unpublished data, the S. Gottesman, C. Gross, and M. Maurizi labs for strains, A. Huerta for originally identifying the CRP binding site upstream of AzuC, and members of the Storz lab for helpful discussion and comments. We particularly thank B.-M. Koo and E. Gogol for sharing the unpublished strain CAG62093 and K. M. Thompson and N. Majdalani for sharing unpublished strain KMT249.
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and by postdoctoral fellowships from the Life Sciences Foundation (M.R.H.) and the National Research Council (B.J.P.).
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E. 2008. Regulation by destruction: design of the σE envelope stress response. Curr. Opin. Microbiol. 11:535-540. [DOI] [PubMed] [Google Scholar]
- 2.Alix, E., and A. B. Blanc-Potard. 2008. Peptide-assisted degradation of the Salmonella MgtC virulence factor. EMBO J. 27:546-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brøndsted, L., and T. Atlung. 1996. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J. Bacteriol. 178:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 5.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dassa, J., H. Fsihi, C. Marck, M. Dion, M. Kieffer-Bontemps, and P. L. Boquet. 1991. A new oxygen-regulated operon in Escherichia coli comprises the genes for a putative third cytochrome oxidase and for pH 2.5 acid phosphatase (appA). Mol. Gen. Genet. 229:341-352. [DOI] [PubMed] [Google Scholar]
- 10.De Lay, N., and S. Gottesman. 2009. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191:461-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fountoulakis, M., M. F. Takács, P. Berndt, H. Langen, and B. Takács. 1999. Enrichment of low abundance proteins of Escherichia coli by hydroxyapatite chromatography. Electrophoresis 20:2181-2195. [DOI] [PubMed] [Google Scholar]
- 12.Fozo, E. M., M. Kawano, Y. Kaya, K. Jones, K. E. Rudd, and G. Storz. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 70:1076-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel, S. E., and J. D. Helmann. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 191:6116-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galindo, M. I., J. I. Pueyo, S. Fouix, S. A. Bishop, and J. P. Couso. 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5:1052-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier, R., D. Douguet, B. Antonny, and G. Drin. 2008. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24:2101-2102. [DOI] [PubMed] [Google Scholar]
- 16.Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier, Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, A. I., S. Hunt, S. L. Stokes, N. Bramall, J. Bunch, A. G. Cox, C. W. McLeod, and R. K. Poole. 2009. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 284:18377-18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grainger, D. C., D. Hurd, M. Harrison, J. Holdstock, and S. J. Busby. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guisbert, E., T. Yura, V. A. Rhodius, and C. A. Gross. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 72:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, M. J., and S. Y. Lee. 2006. The Escherichia coli proteome: past, present, and future prospects. Microbiol. Mol. Biol. Rev. 70:362-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemm, M. R., B. J. Paul, T. D. Schneider, G. Storz, and K. E. Rudd. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 70:1487-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs, E. C., J. L. Astarita, and G. Storz. 2010. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J. Bacteriol. 192:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huerta, A. M., and J. Collado-Vides. 2003. σ70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333:261-278. [DOI] [PubMed] [Google Scholar]
- 25.Laskowska, E., A. Wawrzynów, and A. Taylor. 1996. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78:117-122. [DOI] [PubMed] [Google Scholar]
- 26.Law, E. C., N. J. Savery, and S. J. Busby. 1999. Interactions between the Escherichia coli cAMP receptor protein and the C-terminal domain of the alpha subunit of RNA polymerase at class I promoters. Biochem. J. 337:415-423. [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson, C. L., D. Swigon, K. S. Murakami, S. A. Darst, H. M. Berman, and R. H. Ebright. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14:10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lies, M., and M. R. Maurizi. 2008. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J. Biol. Chem. 283:22918-22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link, A. J., K. Robison, and G. M. Church. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18:1259-1313. [DOI] [PubMed] [Google Scholar]
- 30.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 31.Mandin, P., and S. Gottesman. 2009. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol. Microbiol. 72:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matuszewska, M., D. Kuczyńska-Wiśnik, E. Laskowska, and K. Liberek. 2005. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J. Biol. Chem. 280:12292-12298. [DOI] [PubMed] [Google Scholar]
- 32a.Moon, K., and S. Gottesman. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides.Mol. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 33.Narberhaus, F., T. Waldminghaus, and S. Chowdhury. 2006. RNA thermometers. FEMS Microbiol. Rev. 30:3-16. [DOI] [PubMed] [Google Scholar]
- 34.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. U. S. A. 100:9912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee, K. Y., B. S. Parekh, and G. W. Hatfield. 1996. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli. J. Biol. Chem. 271:26499-26507. [DOI] [PubMed] [Google Scholar]
- 36.Rokney, A., O. Kobiler, A. Amir, D. L. Court, J. Stavans, S. Adhya, and A. B. Oppenheim. 2008. Host responses influence on the induction of lambda prophage. Mol. Microbiol. 68:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejewski, A. D. Grossman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13:689-701. [DOI] [PubMed] [Google Scholar]
- 38.Rudd, K. E., I. Humphery-Smith, V. C. Wasinger, and A. Bairoch. 1998. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19:536-544. [DOI] [PubMed] [Google Scholar]
- 39.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salgado, H., S. Gama-Castro, M. Peralta-Gil, E. Díaz-Peredo, F. Sánchez-Solano, A. Santos-Zavaleta, I. Martínez-Flores, V. Jiménez-Jacinto, C. Bonavides-Martínez, J. Segura-Salazar, A. Martínez-Antonio, and J. Collado-Vides. 2006. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 34:D394-D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturr, M. G., T. A. Krulwich, and D. B. Hicks. 1996. Purification of a cytochrome bd terminal oxidase encoded by the Escherichia coli app locus from a Δcyo Δcyd strain complemented by genes from Bacillus firmus OF4. J. Bacteriol. 178:1742-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subrahmanyam, C. S., J. D. Noti, and H. E. Umbarger. 1980. Regulation of ilvEDA expression occurs upstream of ilvG in Escherichia coli: additional evidence for an ilvGEDA operon. J. Bacteriol. 144:279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tjaden, B., S. S. Goodwin, J. A. Opdyke, M. Guillier, D. X. Fu, S. Gottesman, and G. Storz. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 34:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wadler, C. S., and C. K. Vanderpool. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U. S. A. 104:20454-20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldminghaus, T., A. Fippinger, J. Alfsmann, and F. Narberhaus. 2005. RNA thermometers are common in α- and γ-proteobacteria. Biol. Chem. 386:1279-1286. [DOI] [PubMed] [Google Scholar]
- 46.Waldminghaus, T., N. Heidrich, S. Brantl, and F. Narberhaus. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65:413-424. [DOI] [PubMed] [Google Scholar]
- 47.Wang, R., K. R. Braughton, D. Kretschmer, T. H. Bach, S. Y. Queck, M. Li, A. D. Kennedy, D. W. Dorward, S. J. Klebanoff, A. Peschel, F. R. DeLeo, and M. Otto. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510-1514. [DOI] [PubMed] [Google Scholar]
- 48.Wawrzynow, A., J. L. Theibert, C. Murphy, I. Jona, A. Martonosi, and J. H. Collins. 1992. Sarcolipin, the “proteolipid” of skeletal muscle sarcoplasmic reticulum, is a unique, amphipathic, 31-residue peptide. Arch. Biochem. Biophys. 298:620-623. [DOI] [PubMed] [Google Scholar]
- 49.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 50.Yamanaka, K., W. Zheng, E. Crooke, Y. H. Wang, and M. Inouye. 2001. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39:1572-1584. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.