Abstract
J proteins are structurally diverse, obligatory cochaperones of Hsp70s, each with a highly conserved J domain that plays a critical role in the stimulation of Hsp70's ATPase activity. The essential protein, Cwc23, is one of 13 J proteins found in the cytosol and/or nucleus of Saccharomyces cerevisiae. We report that a partial loss-of-function CWC23 mutant has severe, global defects in pre-mRNA splicing. This mutation leads to accumulation of the excised, lariat form of the intron, as well as unspliced pre-mRNA, suggesting a role for Cwc23 in spliceosome disassembly. Such a role is further supported by the observation that this mutation results in reduced interaction between Cwc23 and Ntr1 (SPP382), a known component of the disassembly pathway. However, Cwc23 is a very atypical J protein. Its J domain, although functional, is dispensable for both cell viability and pre-mRNA splicing. Nevertheless, strong genetic interactions were uncovered between point mutations encoding alterations in Cwc23's J domain and either Ntr1 or Prp43, a DExD/H-box helicase essential for spliceosome disassembly. These genetic interactions suggest that Hsp70-based chaperone machinery does play a role in the disassembly process. Cwc23 provides a unique example of a J protein; its partnership with Hsp70 plays an auxiliary, rather than a central, role in its essential cellular function.
Hsp70-based machineries constitute a key component of the cell's chaperone network, playing a central role in many processes, including de novo protein folding, protein translocation across membranes, and remodeling of protein complexes (6, 13). By their ability to bind to short, exposed, hydrophobic stretches of polypeptide, Hsp70s serve as the core of this protein folding machinery. However, Hsp70s cannot function alone. J proteins (often referred to as Hsp40s) are their obligatory partners, serving to stimulate Hsp70's ATPase activity and thereby stabilizing interaction with client proteins (19). J proteins are very diverse but, by definition, contain an ∼70-amino-acid “J domain” that interacts directly with the Hsp70 ATPase domain (12, 17). It is well established that the J domain is critical for function of the Hsp70-based chaperone machinery, since single amino acid alterations in its highly conserved HPD motif disrupt function, both in vitro and in vivo, without affecting domain structure (9, 14). The Saccharomyces cerevisiae genome encodes 22 J proteins. Of these, 13 are found in the cytosol and/or nucleus. One, Cwc23, is the focus of this report. Deletion of CWC23 causes inviability in some strain backgrounds, although adaptation or suppression allows slow growth in others (31, 34). The growth defect of cwc23Δ cells cannot be rescued by overexpression of any of the other 12 J proteins (31), indicating that Cwc23 carries out a specialized cellular function. Cwc23 has been linked to pre-mRNA splicing via both genome-wide and spliceosomal component-directed physical interaction studies (7, 25).
Pre-mRNA splicing is a highly precise and stepwise process whereby noncoding introns are removed from nascent transcripts. The splicing reaction is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex composed of five small nuclear RNAs and at least 100 different proteins (5, 24, 38). The widely held model of the splicing pathway posits that the spliceosome is not a monolithic, stable complex but rather a series of complexes that must be assembled de novo for each splicing event and subsequently disassembled upon completion. Both the assembly and the disassembly steps of pre-mRNA splicing require a series of macromolecular rearrangements involving changes in RNA-RNA, RNA-protein, and protein-protein interactions. A positive “two-hybrid” interaction between Cwc23 and Ntr1, an essential protein required for efficient spliceosomal disassembly, was recently reported (26). As part of the “NTR complex,” Ntr1 recruits the DExD/H-box ATPase Prp43, which is critical for disassembly, to the spliceosome (4, 35). The interaction surface between Prp43 and Ntr1 involves a region of Ntr1 termed the G patch, a motif found in many proteins and known to be involved in both protein-protein and protein-RNA interactions (1). Given the links between Cwc23 and RNA splicing, we further investigated whether Cwc23 plays a role in this process. Indeed, a partial loss-of-function mutation in CWC23 causes profound global effects on splicing, demonstrating a role of Cwc23 in this process. However, the J domain of Cwc23 is not normally required for either cell viability or pre-mRNA splicing. However, when the interaction between Ntr1 and Prp43 is compromised, Cwc23's J domain becomes essential, indicating a role for it, and thus Hsp70-based chaperone machinery, in pre-mRNA splicing.
MATERIALS AND METHODS
Construction of plasmids.
A DNA fragment containing 758 bp upstream and 545 bp downstream of start and stop codon of CWC23 was PCR amplified from genomic DNA and cloned in pRS316 (URA3 CEN), pRS314 (TRP CEN) and pRS313 (HIS3 CEN) vectors as a BamHI/SacII fragment. To obtain approximately normal expression levels of Cwc231-225, the corresponding DNA fragment was cloned under the control of the alcohol dehydrogenase (ADH) promoter in the CEN-based vectors pRS413, pRS414, and pRS415 (23). All mutations, truncations, and internal deletions were made by quick change or sewing PCR using appropriate primer pairs and standard methods. For overexpression, the coding sequence for full-length Cwc23 was PCR amplified and cloned into the SpeI/XhoI sites of the 2μm plasmid, pRS424, which contains the GPD promoter (23). The Cwc231-225 coding sequence was also cloned into the same sites in CEN TEF- and CEN GPD-based vectors (23). Jjj1-J domain (amino acids 1 to 128) overexpression was carried out essentially as described previously (31), except the coding fragment was subcloned from pRS424 into the analogous vector having a LEU2 marker, pRS425. By PCR sewing using overlapping primers, DNA encoding the J domain of Cwc23 was fused to a random stretch of 32 codons that preceded a 3×HA tag and then cloned into the SpeI/XhoI sites of pRS425. For the two-hybrid experiment, the complete Ntr1 coding sequence was PCR amplified and cloned in the GAL4 binding domain plasmid, pGBDU-C1, whereas CWC23 fragments were cloned into the BamHI/EcoRI sites of a GAL4 activation domain plasmid pGAD-C1 (16). Constructs for bacterial expression were made by PCR amplifying CWC23 fragments with appropriate primers such that all contained His6 tags at their C termini. The fragments were cloned into the NdeI/BamHI sites in pET3a (Novagen).
Genetic methods.
Most yeast strains used have the W303 genetic background (10). Those generated here are listed in Table 1. The starting strain for many of these is Y1818 cwc23Δ:KanMx [pRS316-Cwc23]. Plasmids carrying wild-type (wt) or mutant CWC23 alleles were transformed into Y1818, and transformants were selected on appropriate dropout media. The functionality of different CWC23 alleles was tested by checking the ability of the cells to lose the URA3-based pRS316-Cwc23 plasmid by spotting on 5-fluoroorotic acid (5-FOA) plates and incubation at 30°C for 3 days.
TABLE 1.
Yeast strains generated in this study
| Strain | Description |
|---|---|
| Y1818 | Δcwc23:KanMx [316-Cwc23] |
| Y2178 | Δcwc23:ADE2 [316-Cwc23] |
| Y2236 | Δcwc23:KanMx [314-Cwc23ΔJ] Δapj1:HIS3 |
| Y2237 | Δcwc23:KanMx [314-Cwc23ΔJ] Δydj1:HIS3 |
| Y2238 | Δcwc23:KanMx [314-Cwc23ΔJ] Δjjj1:URA3 |
| Y2239 | Δcwc23:KanMx [314-Cwc23ΔJ] Δxdj1:LEU2 |
| Y2240 | Δcwc23:KanMx [314-Cwc23ΔJ] Δcaj1:LEU2 |
| Y2241 | Δcwc23:KanMx [313-Cwc23] Δsis1:LEU2 [Tetr-Sis1] |
| Y2242 | Δcwc23:KanMx [313-Cwc23ΔJ] Δsis1:LEU2 [Tetr-Sis1] |
| Y2244 | Δcwc23:KanMx [314-Cwc23] Δntr1:natR [p360-Ntr1] |
| Y2245 | Δcwc23:KanMx [314-Cwc23 H50Q] Δntr1:natR [p360-Ntr1] |
| Y2246 | Δcwc23:KanMx [314-Cwc23ΔJ] Δntr1:natR [p360-Ntr1] |
| Y2251 | Δcwc23:ADE2 [316-Cwc23] Δprp43:KanMx [p358-Prp43] |
| Y2253 | Δcwc23:ADE2 [316-Cwc23] Δprp43:KanMx [p358-Prp43-Y402A] |
| Y2305 | Δcwc23:ADE2 [316-Cwc23] Δntc20:KanMx |
| Y2306 | Δcwc23:ADE2 [316-Cwc23] Δisy1:KanMx |
| Y2307 | Δcwc23:ADE2 [316-Cwc23] Δlea1:KanMx |
| Y2308 | Δcwc23:ADE2 [316-Cwc23] Δmsl1:KanMx |
| Y2309 | Δcwc23:ADE2 [316-Cwc23] Δmud1:KanMx |
| Y2310 | Δcwc23:ADE2 [316-Cwc23] Δmud2:KanMx |
Yeast strains Y2236, Y2237, Y2238, Y2239, and Y2240 were constructed to investigate the synthetic genetic interaction between cwc23ΔJ and deletions of genes encoding nonessential cytosolic/nuclear J proteins (Apj1, Ydj1, Jjj1, Xdj1, and Caj1) by mating single J protein deletion strains as described previously (31) with a cwc23Δ:KanMx strain expressing Cwc23ΔJ from pRS314 plasmid (constructed as described in the previous paragraph), after losing the URA-based wt CWC23 plasmid. Subsequently, the diploids were sporulated, and tetrads were dissected to obtain the haploid strains. Y2241 and Y2242 strains were made to study the genetic interaction of cwc23ΔJ with the essential SIS1 gene by allowing repression of Sis1 synthesis by the addition of doxycycline. sis1Δ:LEU2 [Tetr-Sis1] (3) was crossed with cwc23Δ:KanMx expressing either wt Cwc23 or Cwc23ΔJ from a HIS3 CEN-based plasmid (pRS313), followed by tetrad dissection to obtain the haploid strain.
ntr1Δ and prp43Δ strains were obtained from Beate Schwer (33). Y2244, Y2245, and Y2246, used to assess synthetic growth defects with NTR1 mutants, were made by crossing cwc23Δ cells expressing wt Cwc23, Cwc23ΔJ, or Cwc23H50Q from a pRS314 plasmid (obtained as described above) with ntr1Δ carrying wt NTR1 on a URA3 CEN plasmid (p360-Ntr1). Haploid strains were obtained by tetrad dissection. Genetic interactions between NTR1 and CWC23 mutants were investigated by transforming pRS413-based plasmids (HIS3 CEN) with wt or G-patch mutant NTR1 genes (encoding the alterations L68A, Y74A, or L80A, which were described previously [33]) into Y2244, Y2245, and Y2246. To investigate whether the expression of a J domain fragment could complement the synthetic growth defect between J domain mutants of CWC23 and G-patch NTR1 mutants in trans, J domain-encoding fragments of CWC23 (Cwc231-225 [CEN ADH] or Cwc23-J [2μm GPD]) or another cytosolic J protein gene JJJ1 (Jjj11-128 [2μm GPD]) were transformed into Y2244, Y2245, and Y2246. The Y2178 strain (cwc23Δ:ADE2 [pRS316-Cwc23]), was constructed by swapping the KanMx cassette in the heterozygous diploid cwc23Δ:KanMx strain with the NotI-linearized ADE2 disruption cassette plasmid (37). The resulting strain was transformed with the pRS316-Cwc23 plasmid and sporulated, and tetrads were dissected to yield Y2178. To test a synthetic genetic interaction with PRP43, yeast strains Y2251 and Y2253 were constructed by crossing Y2178 with prp43Δ cells harboring either p358-Prp43 or p358-Prp43Y402A (TRP CEN) plasmids (33). Haploids having the deletion alleles of both CWC23 and PRP43 were obtained by tetrad dissection. Finally, Y2251 and Y2253 were transformed with pRS313 (HIS3 CEN) plasmids expressing wt Cwc23, Cwc23ΔJ, or Cwc23H50Q. Transformants were selected on dropout media, and synthetic genetic interactions between CWC23 and PRP43 mutants were scored after plating cells on 5-FOA medium. The effect of expression of a J domain containing fragment in trans was investigated by transforming Cwc231-225 (CEN ADH), Cwc23-J (2μm GPD), or Jjj11-128 (2μm GPD) into Y2251 and Y2253 expressing Cwc23ΔJ.
Strains Y2305, Y2306, Y2307, Y2308, Y2309, and Y2310 (see Table 1) were made to study the genetic interaction between mutations in CWC23 and other spliceosomal genes. Homozygous diploid strains from the yeast knockout collection (40) for ntc20, isy1, lea1, msl1, mud1, or mud2 were sporulated. Haploids obtained by tetrad dissection were mated with Y2178. Double deletions of CWC23 and one of the six genes, obtained by tetrad dissection, were transformed with pRS314 or pRS314 harboring wt Cwc23 or Cwc23ΔJ.
Protein expression and purification.
His-tagged Ssa1 and Ssb1 were purified as described previously (15, 28). Cwc231-225-His6 was expressed from pET3a plasmid in C41(DE3) cells (22). Cells were induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C for 8 h. The cell pellet was resuspended and lysed in the Ni-NTA binding buffer (25 mM HEPES [pH 7.5], 300 mM KCl, 25 mM imidazole, 5% glycerol) supplemented with protease inhibitor cocktail (Roche). The supernatant was loaded onto a Ni-NTA agarose column (Novagen), washed with several volumes of Ni-NTA buffer, and eluted with 300 mM imidazole in Ni-NTA binding buffer. Eluted protein was further purified by using HiPrep 16/60 Sephacryl S-200 HR (GE Healthcare) equilibrated with the Gel filtration buffer (25 mM HEPES [pH 7.5], 100 mM KCl, 10 mM magnesium acetate, 5% glycerol). The purified protein was concentrated by using a Centriprep YM-10 (Millipore), divided into aliquots, snap-frozen in liquid nitrogen, and stored at −80°C. For antibody production, pET3a-Cwc2381-171-His6 was used. Eluted protein was dialyzed overnight against 10% glycerol at 4°C. Polyclonal antibodies to this protein were commercially generated at Harlan (Madison, WI).
ATPase assay.
Single-turnover ATPase assays of Ssa1 and Ssb1 were carried out as described previously (15, 21). Isolated Hsp70-[α-32P]ATP complexes were incubated with equimolar amounts of Cwc231-225 or Cwc231-225/H50Q at 25°C, and the samples were removed for detection of ATP and ADP. The fractions of ATP hydrolyzed to ADP over the time courses were quantified by PhosphorImager analysis (Molecular Dynamics) and fit to one-phase exponential decay by using Prism 4.0 (GraphPad Software).
Splicing assays.
To examine pre-mRNA splicing defects in the cwc23Δ strain by Northern blot analysis, yeast cells from wt and cwc23Δ microcolonies were subcultured in rich media at 30°C for 2 days. Total cellular RNA was isolated by using acid phenol (32). Equivalent cells (optical density at 600 nm = 1.0) were spun down at 2,000 × g for 5 min at room temperature in a 2-ml microcentrifuge tube. The cells were washed once with chilled distilled water. The cells were resuspended in 400 μl of TES buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 0.5% sodium dodecyl sulfate [SDS]) and 400 μl of acidic phenol-chloroform-isoamyl alcohol (25:24:1 [pH 4.2]). The cells were lysed by adding 400-μl equivalents of 0.5-mm glass beads, followed by incubation at 65°C for 1 h with intermittent vortexing. The tubes were incubated on ice for 5 min and then centrifuged at 16,000 × g at 4°C. The upper aqueous phase containing the total RNA was precipitated by addition of 1/10 volume of sodium acetate (pH 5.2) and 2.5 volumes of chilled ethanol. Total RNA was spun down for 10 min at 16,000 × g at 4°C. Finally, the RNA pellet was washed with 70% ethanol, air dried, and dissolved in 50 μl of RNase-free distilled water. For use with a full-length actin probe, 5 μg of total RNA was resolved on a denaturing morpholinepropanesulfonic acid-formaldehyde 1% agarose gel and transferred by capillary to a nylon membrane (GE Healthcare). For use with actin intron probe, 20 μg of RNA was resolved on a denaturing morpholinepropanesulfonic acid-formaldehyde 1.5% agarose gel. Radiolabeled PCR fragment corresponding to the full-length or ACT1 intron DNA was used as a probe. Radiolabeling was done by using the Prime-A gene labeling system (Promega, Madison, WI). Hybridization was carried out in ULTRAhyb solution (Ambion) at 42°C, overnight. After the washings, the bands were visualized by using a PhosphorImager.
To monitor pre-mRNA splicing defects on a global scale, splicing-sensitive whole genome microarrays were used. Cultures were grown as previously described (29). Total RNA was isolated as described previously, but for each microarray cDNA was prepared from 20 μg of total RNA in a 50-μl reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.5 mM TTP, 0.01 mM 5-(3-aminoallyl)-dUTP, 12.5 μg of dN9 primer, and 5 ng of M-MLV RT. Labeled cDNA samples were hybridized on custom designed Agilent 8x15K microarrays. These microarrays contain probes targeting over 6,000 yeast genes. For intron-containing genes, the microarrays also contain probes that target regions of the introns, as well as the junction between neighboring exons (29). Microarray design details can be found at NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GPL8154. Samples were hybridized according to manufacturer's protocols at 60°C for 17 h. Microarrays were washed for one minute in 6× SSPE (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA), 0.005× Sarkosyl, and then briefly washed in 0.06× SSPE-0.005× Sarkosyl prior to scanning. Images were obtained by using an Axon 4000B scanner, and data were extracted from the images by using GenePix 6.0 software. Raw data were processed by using Bioconductor to implement the Loess normalization function. After normalization, replicate values were averaged to determine the behavior of each feature on the array. Each experiment was performed as a dye-flipped replicate, with the composite behavior being presented in the figures and text. Both raw and processed data are available from GEO.
Other methods.
Total protein for immunoblot analysis was isolated by treating cells with 0.1 N NaOH for 7 min, followed by resuspension in 1× SDS sample buffer (62.5 mM Tris-HCl [pH 6.8], 5% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.01% bromophenol blue) and incubation for 5 min in a boiling water bath. Anti-Cwc23 polyclonal antibody was affinity purified using Cwc2381-171 as described previously (20). Then, 5 mg of affinity-purified Cwc2381-171 was resolved by SDS-12.5% polyacrylamide gel electrophoresis (PAGE). Protein was transferred to a nitrocellulose membrane by electroblotting. The membrane was stained with Ponceau S for 5 min and destained with 5% acetic acid. The section of the membrane containing the protein band was excised, washed three times (for 5 min each time) with TBS buffer (10 mM Tris-HCl [pH 7.5], 0.9% NaCl), and then incubated with 10 ml of anti-Cwc23 polyclonal antibody for 1 h at room temperature. The membrane was then washed four times, for 5 min each time, with TBS buffer and then briefly in distilled water. Purified antibody was eluted with 3 ml of cold 100 mM glycine-HCl (pH 2.5) for 10 min. Eluted antibody was transferred to a fresh tube and neutralized with 1 M Tris-HCl (pH 8.5). Cwc23 expressed from either its native promoter or an ADH promoter on a CEN-based plasmid was detected by the more sensitive ECL Advance Western detection kit (GE Healthcare). Otherwise, a Western Lightening Plus ECL kit (Perkin-Elmer) was used.
The fungal orthologs of Cwc23 were identified at http://www.broad.mit.edu/regev/orthogroups/html/6/6/OG3966.html. Higher eukaryotic homologs were identified by using amino acids 58 to 289 of Cwf23 (Cwc23 ortholog in S. pombe), which lacks the J domain, as a query sequence in BLASTp at the National Center for Biotechnology Information. The amino acids corresponding to the J domains of selected Cwc23 orthologs were aligned by using the CLUSTAL W mode of MegAlign module of DNASTAR (Lasergene 8). RNA recognition motif in Cwc23 orthologs was identified by searching the respective protein sequences at Conserved Domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and Pfam (http://pfam.sanger.ac.uk/search).
RESULTS
Cwc23's C terminus, but not its N-terminal J domain, is essential for cell viability.
The N-terminal 80-amino-acid segment of S. cerevisiae Cwc23 (Fig. 1A), as well as orthologs identified in other fungal species and in higher eukaryotes (see Fig. S1 in the supplemental material), are highly similar in sequence to known J domains. To determine whether the J domain of Cwc23 is essential for its in vivo function, a mutation encoding a histidine-to-glutamic-acid substitution in the invariant HPD motif was made. This particular mutation was selected because it has been demonstrated to severely affect the ability of several J proteins to stimulate the ATPase activity of their partner Hsp70 and consequently their in vivo function (9). Surprisingly, not only was the cwc23H50Q strain viable (Fig. 1B, panels 1 and 2), it grew indistinguishably from wild-type (wt) (Fig. 1B, panels 3 to 5). To address the concern that the H50Q mutation might not completely abolish J domain function, we constructed cwc23ΔJ, with a deletion of the 80 codons of the N-terminal J domain. Remarkably, cwc23ΔJ cells also grew indistinguishably from wt cells (Fig. 1B), establishing that the J domain region is not required for Cwc23's essential function.
FIG. 1.
The J domain of Cwc23 is dispensable. (A) Cwc23 has a conserved J domain. A sequence alignment of the predicted J domains of the indicated J proteins of S. cerevisiae is shown. The conserved histidine-proline-aspartic acid (HPD) tripeptide motif is marked (***). (B) The J domain is not critical for growth. In panels 1 and 2, equal numbers of cwc23Δ cells harboring two plasmids—URA3 CEN-Cwc23 and a vector containing no insert (−), CWC23 (WT), cwc23H50Q (H50Q), or cwc23ΔJ (ΔJ)—were spotted on medium without (−) or with (+) 5-FOA to select for cells having lost the URA3 CEN-Cwc23 plasmid, as indicated. In panels 3 to 5, 10-fold serial dilutions of equivalent numbers of cwc23Δ cells expressing either wt Cwc23 (WT), Cwc23H50Q (H50Q), or Cwc23ΔJ (ΔJ) were spotted on selective medium and incubated at the indicated temperatures for 3 days. (C) Cwc23's J domain is functional. For the left panel, 10-fold serial dilutions of an equivalent number of ydj1Δ cells expressing either wt Ydj1 from its own promoter (Ydj1), or Cwc23 (WT), or Cwc23H50Q (H50Q) from the GPD promoter or harboring only an empty vector (−) were spotted on selective medium and incubated at 30°C for 3 days. For the right panel, total protein was isolated from strains indicated in the left panel and subjected to SDS-PAGE, electroblotted, and probed with antibodies specific for Cwc23 or, as a loading control, Ssc1 (row C). (D) Cwc23 stimulates the ATPase activity of the Hsp70, Ssa1. Affinity-purified Cwc231-225-His6 (WT) or the J domain mutant (H50Q) was incubated with equimolar amounts of Ssa1-[α-32P]ATP (left panel) or Ssb1-[α-32P]ATP (right panel). Reactions were stopped after the indicated times and subjected to chromatography. ATP hydrolysis was quantified, and the data were fit to a one-phase exponential decay equation.
The dispensability of the J domain region of Cwc23 raises the question of whether it is actually a functional J domain. To address this question, we carried out two experiments. First, we made use of our previous observation that Cwc23, when expressed from the strong GPD promoter, substantially rescued the severe growth defects of cells lacking Ydj1, the most abundant cytosolic/nuclear J protein (31). However, Cwc23H50Q was completely ineffective in rescuing the growth defect of ydj1Δ cells (Fig. 1C), even though it was expressed at levels similar to that of wt Cwc23 (Fig. 1C, right panel). Second, we tested the effect of the H50Q alteration on the ability of Cwc23 to stimulate the ATPase activity of Hsp70. Since this was the first time Cwc23 had been purified, we first tested whether Cwc23 was competent to stimulate the ATPase activity of either of the two classes of cytosolic/nuclear Hsp70s, Ssa and Ssb. Ssa1 and Ssb1 were loaded with [32P]ATP, and the ability of a 225-amino-acid fragment of Cwc23 to stimulate the basal ATPase activities was monitored. Although little effect on Ssb1's ATPase activity was observed (Fig. 1D, right), the ATPase activity of Ssa1 was stimulated ∼7-fold when present in a stoichiometric amount (Fig. 1D, left). This level of stimulation was similar to that of Ydj1, an established J protein partner of Ssa (15). The H50Q alteration virtually eliminated this stimulatory ability (Fig. 1D, left). These results support the idea that Cwc23 is a functional J protein, which, like Ydj1 and many other cytosolic J proteins, can partner with Ssa. Nevertheless, inactivation of the J domain does not result in a loss of Cwc23's essential cellular function.
Truncation of Cwc23's C terminus, but not its J domain, causes global defects in pre-mRNA splicing.
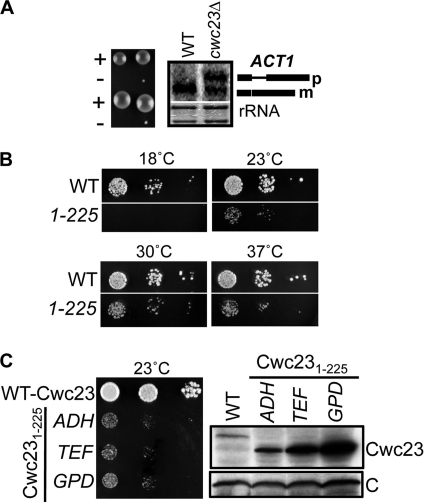
Since Cwc23 was found physically associated with the spliceosome, we sought to determine whether Cwc23 is indeed involved in pre-mRNA splicing. Earlier, we reported CWC23 to be an essential gene, since viable colonies could not be obtained using the standard selection on 5-FOA containing medium for mutants whose viability is maintained by a wt gene in a URA3-based plasmid (31) (Fig. 1B). However, as previously reported (34), dissection of a cwc23Δ heterozygous diploid strain occasionally yielded cwc23Δ microcolonies (Fig. 2A, left panel). Total RNA from primary cultures of the cwc23Δ microcolonies and from wt cells was used for hybridization analysis using radiolabeled full-length actin (ACT1) genomic DNA probe. Consistent with a role for Cwc23 in splicing of actin pre-mRNA, two bands were observed in the cwc23Δ sample: one at the position of the mature mRNA found in the wt sample and the other at the position expected for the larger unspliced pre-mRNA species (Fig. 2A, right panel).
FIG. 2.
Cwc23 plays a role in pre-mRNA splicing. (A) Cwc23 is critical for the splicing of actin pre-mRNA. For the left panel, spores from individual asci of cwc23Δ:KanMx heterozygous diploid cells were separated on solid rich media and then incubated at 30°C for 5 days. Two sample tetrads are shown. The presence (+) or absence (−) of wt CWC23 is indicated. For the right panel, equal amounts of total RNA from wt and cwc23Δ cells were resolved in a denaturing agarose gel, transferred to nylon membrane, and probed with radiolabeled DNA encompassing the full-length actin gene (ACT1). Bands were visualized utilizing a PhosphorImager (top panel). As a loading control, the membrane was stained with methylene blue to visualize rRNA (bottom panel). (B) The C-terminal region of Cwc23 is functionally important. Tenfold serial dilutions of equal numbers of cwc23Δ cells expressing either Cwc23 (WT) or Cwc231-225 (225) were spotted on minimal medium plates and incubated at the indicated temperatures. (C) Overexpression of Cwc231-225 does not significantly affect the growth phenotype. For the left panel, equivalent numbers of cwc23Δ cells expressing Cwc23 from its own promoter (WT-Cwc23) or Cwc231-225 from the ADH, TEF, or GPD promoter were spotted on solid minimal medium and incubated for 3 days. For the right panel, total protein extracted from the samples on the left was subjected to SDS-PAGE and electroblotted. Membranes were probed with anti-Cwc23 antibodies and, as a loading control, Ssc1 (row C).
Because the haploid cwc23Δ strains commonly accumulated suppressing mutations that could complicate analysis of Cwc23 function, we chose to continue our investigation by searching for partial loss-of-function mutants of CWC23 by truncating the 3′ end of the gene. Although cwc231-200 was inviable (data not shown), cwc231-225, which encodes a protein lacking the C-terminal 58 amino acids, was viable, growing more poorly than wt cells between 23 and 37°C but not forming colonies at 18°C (Fig. 2B). We noted that cells expressing 5- to 10-fold higher levels of Cwc231-225 grew no better than cells expressing nearly normal levels of the mutant protein, supporting our conclusion that Cwc231-225 is not simply a hypomorphic allele but rather is functionally defective (Fig. 2C).
To determine whether pre-mRNA splicing was globally affected when Cwc23 function was compromised, whole-genome splicing microarrays were used. For each intron-containing gene a minimum of three primers, targeting three regions of each pre-mRNA, were present on the microarray: the intron to probe pre-mRNA levels, the exon-exon junction to probe mature mRNA levels, and an exonic region to assess changes in total mRNA levels. By simultaneously considering the pre-mRNA, mature mRNA, and total RNA levels in mutant relative to wt cells, differences in splicing efficiency can be assessed. As seen in Fig. 3A, significant levels of unspliced pre-mRNA were detected for most intron-containing genes in cells expressing cwc231-225 whether grown at the permissive temperature of 30°C or after a shift to the nonpermissive temperature of 16°C for 30 min. When we consider both the number of transcripts affected and the magnitude of pre-mRNA accumulation, the splicing defects seen in the cwc231-225 mutant are similar to those seen in a strain containing a temperature-sensitive allele of the essential spliceosomal protein Prp43, prp43-Y402A (33) (Fig. 3A). Although no determination can be made regarding the mechanistic role of a protein based solely on the identity of the pre-mRNAs whose splicing is impacted in an experiment such as this (29), these data nevertheless make it clear that Cwc23 plays a prominent, global role in pre-mRNA splicing based on both the widespread defect and the level of pre-mRNA accumulation seen in the cwc231-225 strain.
FIG. 3.
cwc231-225 is defective in pre-mRNA splicing, but cwc23ΔJ is not. (A) Global analyses of pre-mRNA splicing. For each of 301 intron-containing genes, the behaviors of the total mRNA, pre-mRNA, and mature mRNA were determined (indicated as T, P, and M, respectively, above each lane; see the diagram at top). Each horizontal line describes the behavior of a single intron-containing gene. Ratio values were calculated for each of the strain comparisons. The data are presented as false-colored representations of the log2 value for each feature. For the cwc231-225 experiments, samples were first collected from both mutant and wt strains while growing at 30°C. An additional sample was collected after both strains had been shifted to 16°C for 30 min. For the prp43Y402A experiment, both mutant and wt strains were grown at 30°C then shifted to 37°C for 30 min prior to sample collection. For the Cwc23ΔJ experiment, samples were collected from both mutant and wt strains growing at 30°C. (B) cwc231-225 accumulates actin lariat-introns. Equal amounts of total RNA isolated from cwc23Δ cells expressing wt Cwc23 (WT), Cwc23ΔJ (ΔJ), Cwc231-225 (1-225) were resolved on denaturing agarose gels, transferred to nylon membrane, and probed with either full-length (left) or intronic (right) actin DNA. Bands were visualized utilizing a PhosphorImager (top panel). p, precursor; m, mature; i, intron lariat. As a loading control, the membrane was stained with ethidium bromide to visualize rRNA (bottom panel).
Cwc23 interacts with Ntr1, an essential spliceosome disassembly factor (4, 35), suggesting that it might also be important for the disassembly process. A hallmark of mutants in this pathway is that the excised lariat introns, which are normally rapidly degraded, become significantly stabilized (2). The accumulation of these lariat-bound complexes sequesters essential spliceosomal complexes, indirectly leading to a global defect in pre-mRNA splicing. To directly test this idea, we probed RNA isolated from cwc231-225, cells with an intron-specific probe, as well as the more standard probe encompassing the entire actin gene. cwc231-225 cells had high levels of both actin pre-mRNA and intron lariat compared to wt cells (Fig. 3B), indicating a critical involvement of Cwc23 in the splicing disassembly pathway.
Having obtained evidence that Cwc23 plays a role in disassembly of the spliceosome, we addressed whether the J domain was important for splicing. We observed virtually no pre-mRNA splicing defects in the cwc23ΔJ strain using microarray analysis (Fig. 3A). In addition, the level of the intron lariat in cwc23ΔJ cells was indistinguishable from that present in wt cells (Fig. 3B). Together, our results confirm that although the C terminus of Cwc23 is required for both cell viability and efficient pre-mRNA splicing, the J domain is dispensable for both.
No compensation by other J proteins in the absence of Cwc23's J domain.
Since the J domain is critical for the function of all yeast J proteins tested thus far, we wanted to determine whether the lack of an observable effect upon deletion of Cwc23's J domain was attributable to compensation, in trans, by other J proteins. Besides Cwc23, six other J proteins (Apj1, Xdj1, Caj1, Sis1, Jjj1, and Ydj1) are known to be at least partially localized to the nucleus or involved in nuclear function (www.yeastgenome.org). Thus, we constructed a set of double mutants, each having cwc23ΔJ and a complete deletion of one of the six J protein genes listed above. Four of the double mutant strains (the apj1Δ cwc23Δ, xdj1Δ cwc23Δ, caj1Δ cwc23Δ, and jjj1Δ cwc23Δ strains) expressing cwc23ΔJ grew indistinguishably from the single J protein gene deletions. In addition, no defects in the splicing of actin pre-mRNA were observed (data not shown). In the case of cells lacking Ydj1, a very subtle but reproducible synthetic growth defect was observed at 18°C (see Fig. S2A in the supplemental material). However, accumulation of actin pre-mRNA was not detected at 23°C or after a shift to 18, 37, or 42°C (see Fig. S2B in the supplemental material). We conclude that the slight synthetic growth defect at 18°C is not related to pre-mRNA splicing. Rather, we think that Cwc23, at its normal low levels, is slightly compensating for the absence of Ydj1, a finding consistent with the robust compensation observed when Cwc23 is expressed at higher levels (Fig. 1C). Since Sis1 is essential, we exploited the fact that levels of Sis1 can be expressed at ca. 10% of normal levels by placing Sis1 under the control of the Tetr repressible promoter to test for a genetic interaction between SIS1 and cwc23ΔJ (3). Cells with lowered levels of Sis1 and either wt Cwc23 or Cwc23ΔJ grew indistinguishably. Likewise, no defect in the splicing of actin pre-mRNA was detected (data not shown).
As a complementary approach, we queried our microarray data to determine whether cytosolic HSP70s and other J protein genes might be upregulated in CWC23 mutant strains (8, 9, 39). We plotted the geometric mean intensity versus the log ratio value for each of the ∼6,000 unique mRNA probes included on our microarray analyses of cwc231-225 and cwc23ΔJ strains compared to a wt strain (see Fig. S2C in the supplemental material). CWC23 mRNA was significantly increased in cwc231-225, presumably because it is expressed from the ADH promoter. Even though it is under the control of its endogenous promoter, a modest increase in Cwc23ΔJ mRNA was observed, perhaps because it is expressed from a plasmid. Strikingly, none of the other 21 J protein genes showed any significant difference in expression compared to the wt strain. Likewise, none of the Ssa or Ssb Hsp70 genes showed any significant change in expression. In sum, we obtained no evidence in either our genetic or our biochemical analyses that any other J domain is functionally substituting in trans when Cwc23's J domain is absent or nonfunctional. Rather, our data support the idea that under typical laboratory conditions the J domain of Cwc23, and thus Hsp70 machinery function, is dispensable.
Cwc23's J domain is critical when interaction between Cwc23 and Ntr1 is compromised.
Since the Hsp70:J protein chaperone machinery is known to modulate protein-protein interactions, we wanted to determine whether Cwc23's J domain is important when the function of a protein with which Cwc23 interacts, Ntr1 (26), is compromised. First, we carried out a yeast two-hybrid experiment to exclude the possibility that the J domain of Cwc23 is important for this interaction. Full-length CWC23 and fragments encoding amino acids 1 to 83 (J domain), 1 to 225, 81 to 283 (Cwc23ΔJ), and 226 to 283 (the C-terminal 58 amino acids) were cloned into a GAL4 activation domain plasmid, and the fusion proteins were coexpressed with a fusion of the GAL4 DNA-binding domain and Ntr1. Since the HIS3 gene was under the control of GAL4 in the tester strain, growth in the absence of histidine is dependent on interaction between Cwc23 and Ntr1. As expected from previous work (26), the full-length Cwc23 fusion allowed robust cell growth (Fig. 4A). Cells expressing the Cwc23ΔJ fusion grew as well as those expressing full-length protein, while those expressing a fusion of the N-terminal 81 amino acids did not, indicating that the J domain is not important for interaction with Ntr1. On the other hand, cells expressing the Cwc231-225 fusion grew extremely poorly in the absence of histidine (Fig. 4A), indicating the importance of the C-terminal 58 amino acids of Cwc23 for interaction with Ntr1. However, a fusion of the C-terminal 58 amino acids did not support growth. These results are consistent with the idea that the C-terminal 58 amino acids of Cwc23 is important, but not sufficient, for interaction with Ntr1, and that the J domain does not play a role in the interaction.
FIG. 4.
The J domain of Cwc23 is critical when the G patch of Ntr1 is altered. (A) The C-terminal region is critical for interaction with Ntr1. A two-hybrid analysis was performed. Ntr1 fused to the GAL4 binding domain (BD) and/or Cwc23 proteins fused to the GAL4 activation domain (AD) was expressed in a two-hybrid tester strain (see Materials and Methods). Equal numbers of cells were spotted on nonselective or selective medium. Plates were incubated at 30°C for 2 days. Growth on selective medium is indicative of a positive two-hybrid interaction. (B) Equal numbers of ntr1Δ cwc23Δ URA3-Ntr1 cells harboring CWC23 (WT), cwc23ΔJ (ΔJ), or cwc23H50Q (H50Q) on a TRP-based plasmid and either a HIS3-based empty plasmid (−) or the same plasmid carrying NTR1 (WT), ntr1L68A (L68A), ntr1Y74A (Y74A), or ntr1L80A (L80A) were spotted either on a plate with (+) or without (−) 5-FOA and incubated at 30°C for 3 days. The white space indicates removal of strains not relevant to the present study. (C) The J domain is critical in the absence of C-terminal 58 amino acids. Equal numbers of cwc23Δ cells harboring two plasmids (URA3-Cwc23 and a vector containing no insert [−], CWC23, or the indicated cwc23 mutant genes) were spotted on medium with (+) or without (−) 5-FOA. Plates were incubated at 30°C for 3 days. Cwc23H50Q (H50Q), Cwc23ΔJ (ΔJ), Cwc231-225 (1-225), Cwc2381-225 (81-225), and Cwc231-225/H50Q (1-225/H50Q) are indicated. (D) Total protein extracted from strains, indicated in panel C was subjected to SDS-PAGE, electroblotted to a membrane, and probed with antibody specific for Cwc23 and, as a loading control, Ssc1 (row C, bottom panel). Cwc2381-225 (81-225), and Cwc231-225/H50Q (1-225 H50Q) were assessed prior to transfer to 5-FOA, since they are inviable in the absence of wt Cwc23. A dotted line denotes the position of a segment of the gel which contained samples that were irrelevant to the present study and thus removed.
Since the absence of the J domain did not compromise the interaction between Cwc23 and Ntr1, we proceeded to investigate whether mutations in NTR1 have synthetic genetic interactions with CWC23 J domain mutations. We tested three NTR1 point mutations, L68A, Y74A, and L80A, each of which alters the G patch, causing significant defects in interaction with Prp43 in vitro but no obvious defect in vivo (33, 35). We used the plasmid shuffling technique, plating strains with different combinations of NTR1 and CWC23 alleles, as well as a wt NTR1 gene on a URA3-based plasmid. Cells were plated on 5-FOA-containing plates to select for those having lost the URA3-based plasmid and thus the wt NTR1 gene it contained. As expected, the control strains expressing either the wt NTR1 or CWC23 genes grew well on the 5-FOA plates. However, we were unable to recover yeast strains expressing any of the Ntr1 variants in combination with Cwc23ΔJ (Fig. 4B). We reasoned that this lack of function could be the result of indirect effects on protein conformation, rather than loss of J domain function. Therefore, we combined the H-to-Q substitution mutation in CWC23 described above that renders J domains nonfunctional with G-patch NTR1 mutations. No double mutants containing both cwc23H50Q and ntr1 mutants were recovered on 5-FOA plates, suggesting that cells cannot tolerate defects in both the J domain of Cwc23 and the G patch of Ntr1.
Since we demonstrated that the C terminus, but not the J domain, is important for Cwc23's interaction with Ntr1, we performed the analogous intragenic synthetic genetic interaction test, combining the N-terminal and C-terminal truncations, generating cwc2381-225. Cwc2381-225 was stably expressed (Fig. 4D). However, no viable cwc2381-225 cells were obtained (Fig. 4C). Consistent with this result, cwc231-225, H50Q was also unable to rescue the viability of a cwc23Δ strain (Fig. 4C). We conclude that J domain function becomes critical when the function of the C-terminal region, which is important for interaction with Ntr1, is compromised.
Specificity of synthetic genetic interaction of CWC23 J domain mutations.
Both circumstances in which Cwc23's J domain was found to be important involved Ntr1: (i) when Cwc23's interaction with Ntr1 was compromised by the alteration of Cwc23 itself and (ii) when the Ntr1-Prp43 interaction was compromised by the alteration of Ntr1. We pursued the idea that J domain function was important when interaction among the components of the NTR complex was compromised by testing other synthetic genetic interactions. First, we tested a PRP43 mutation, prp43Y402A, an allele of PRP43 known to affect interaction with Ntr1 (33). Again, we used the plasmid shuffling technique, plating strains on 5-FOA-containing plates. As expected, prp43Y402A cells expressing full-length Cwc23 grew indistinguishably from wt cells. However, yeast strains expressing prp43Y402A and cwc23ΔJ or cwc23H50Q could not be recovered at 18 and 23°C. Double mutants containing prp43Y402A and either cwc23ΔJ or cwc23H50Q were recovered at 30°C but grew slowly at that temperature (Fig. 5A and data not shown).
FIG. 5.
The J domain of Cwc23 is important specifically when interactions between NTR complex components are compromised. (A) The J domain of Cwc23 is important in the presence of Prp43Y402A. Equal numbers of prp43Δ cwc23Δ URA3-Cwc23 cells harboring a PRP43 (WT) or prp43Y402A plasmid and either a HIS3-based empty plasmid (−), CWC23 (WT), cwc23ΔJ (ΔJ), or cwc23H50Q (H50Q) were spotted onto media with (+) or without (−) 5-FOA and incubated at 30°C for 2 days or 23°C for 3 days. (B) Expression of J domain-containing fragments in trans did not restore growth of prp43Y402A cells expressing J domain mutant genes. prp43Δ cwc23Δ URA3-Cwc23 cells harboring either PRP43 (WT) or prp43Y402A plasmids were transformed with an empty HIS3-based vectors (−) or the same vector containing CWC23 (WT), cwc23H50Q (H50Q), or cwc23ΔJ (ΔJ) in combination with either an empty LEU2-based (−) vector or the same vector expressing one of three J domain containing fragments: Cwc231-225 (1-225) from the ADH promoter, or the J domain fragments of Jjj1 or Cwc23 driven by the GPD promoter (Jjj1-J and Cwc23-J, respectively). Transformants were spotted onto media with or without 5-FOA and incubated at 23°C for 3 days.
To more broadly explore the relationship between Cwc23 and splicing, we next looked for synthetic genetic interactions with components of the spliceosomal machinery that are known to function at steps other than disassembly. We examined interactions with deletions of six nonessential components, including the early-acting U1 component MUD1, the commitment complex factor MUD2, the U2-associated components LEA1 and MSL1, and finally ISY1 and NTC20, which are components of the NTC (for Prp nineteen complex) that is added during spliceosomal activation (5). Notably, the NTR (for nineteen-complex related) components Ntr1 and Ntr2 have been shown to weakly interact with the NTC, suggesting that they could interact with the spliceosome well prior to the disassembly step (35). Nevertheless, none of the genes we tested showed synthetic growth defects with cwc23ΔJ (see Fig. S3 in the supplemental material), indicating that the severe synthetic growth defects observed with the NTR1 and PRP43 mutations are specific.
The J domain and C-terminal regions of Cwc23 cannot function in trans.
The results described above suggest the possibility that Cwc23 carries out two independent functions: the C-terminal region being critical for an interaction with Ntr1 and the N-terminal J domain for an interaction with Hsp70. We therefore examined, again using the 5-FOA selection technique, whether the expression of Cwc231-225 could suppress the synthetic genetic defects seen in double mutant containing prp43Y402A and either cwc23H50Q or cwc23ΔJ. The prp43Y402A mutation was chosen because the synthetic growth defects with J domain mutations were the least severe of the PRP43 and NTR1 mutants discussed above. As seen in Fig. 5B, the inclusion of cwc231-225 was insufficient for rescue. Similar results were obtained when examining the synthetic interaction seen with strains containing ntr1L80A, a mutation in the G patch of Ntr1 (see Fig. S4 in the supplemental material).
Since Cwc23 is of low abundance in cells, we also wanted to test whether higher levels of expression of a J domain could overcome the synthetic growth defect when in trans with the CWC23 J domain mutants. Plasmids were constructed that allowed for the overexpression of J domain fragments of either Cwc23 or the J protein Jjj1 from the GPD promoter. Overexpression of the J domains also failed to rescue the synthetic growth defects of strains combining J domain mutations in CWC23 with prp43Y402A (Fig. 5B). In sum, we conclude that the two domains of Cwc23 must be present as a single polypeptide to perform their function(s).
DISCUSSION
Here we report evidence supporting an important role for Cwc23 in the disassembly of the spliceosome. In addition, Cwc23 was revealed to be a J protein with unusual, if not unique, characteristics. Although it plays a critical role in pre-mRNA splicing, its J domain is not normally required for this function. Unlike the situation with many J proteins, the J domain plays only an auxiliary role; its involvement in splicing is revealed when the function of other proteins (i.e., Ntr1 or Prp43) involved in spliceosomal disassembly are compromised.
Cwc23's role in pre-mRNA splicing: disassembly of the spliceosome.
Several lines of evidence point to an essential role for Cwc23 in the pre-mRNA splicing pathway. First, a partial loss-of-function CWC23 mutant affects both cell fitness and global pre-mRNA splicing. Second, this mutation, a truncation of the codons encoding the 58 C-terminal amino acids, affects Cwc23's physical interaction with Ntr1, a known essential splicing factor (35). More specifically, available data point to a critical role for Cwc23 in the spliceosome disassembly step of the pathway. CWC23 mutants accumulate a product of the splicing reaction, the lariat-intron, which is known to be stabilized upon failure to disassemble the spliceosome (2). Indeed, extensive evidence accumulated over a decade supports a role of Prp43 in catalyzing the final step of splicing, the dissociation of the lariat-intron, and the disassembly of the spliceosome (38). Thus, the synthetic genetic interactions between CWC23 mutations and NTR1 or PRP43 mutations, but not mutations in genes involved in other steps in the pathway, also serve to place Cwc23 at the disassembly step.
Cwc23: an essential J protein with a nonessential J domain.
The most surprising result of the present study is the lack of a phenotype upon deletion of the J domain of Cwc23; there is no obvious defect in either cell fitness or pre-mRNA splicing. To our knowledge, Cwc23 is the only example of an essential J protein with a nonessential J domain. Usually, alteration of the J domain leads to the same phenotypic effect as deletion of the entire protein. Indeed, in some cases much of a J protein can be deleted with little or no affect, as long as the J domain itself is left intact (18, 41). An extreme example of this is provided by Ydj1, a 409-amino-acid J protein with a well-characterized client protein-binding domain. Although a complete deletion of Ydj1 has severe effects on cell growth, expression of the J domain alone, at the level that Ydj1 is normally present, permits quite robust growth (31). On the other hand, when dramatic effects of mutations outside the J domain have been found, the consequences are typically no more severe than those that only affect J domain mutation. For example, deletion of the region necessary for binding of the J protein Zuo1 to the ribosome results in the same phenotype as disruption of only the J domain (43).
To our knowledge, only two examples in which the J domain of a protein appears to be of less biological importance than other domains have been reported previously: P58IPK and RSP16. Neither case is as striking as that of Cwc23. In the case of P58IPK, its J domain-independent function appears to be in a different cellular compartment from its J domain-dependent function. In the cytosol, where it is present in low abundance, it acts as an inhibitor of interferon-induced protein kinase (PKR), a J domain-independent activity important for productive infection by influenza virus (42). However, P58IPK is predominantly found in the endoplasmic reticulum lumen, where it plays an important role as a bona fide J protein cochaperone of Hsp70 (27). RSP16 plays a role in flagellar stroke movement in Chlamydomonas. A fragment lacking the J domain appears to function as well as full-length protein in regulation of flagellar beating (44), although its exact function in this process is not known. The functionality of the J domain of RSP16 has yet to be demonstrated. RSP16 does have the defining HPD motif, but its orthologs from human, zebrafish, mouse, and mosquito do not (44).
Function of Cwc23's J domain.
Both our in vitro and in vivo results establish that S. cerevisiae Cwc23 has a functional J domain. It has the capacity to stimulate the weak ATPase activity of the Hsp70, Ssa1, and substantially rescue the severe growth defect of ydj1Δ cells. Both abilities are eliminated by the same single amino acid alteration in the J domain. However, the J domain of Cwc23 is clearly dispensable for cell viability and efficient pre-mRNA splicing. Nevertheless, the severe synthetic genetic interactions between mutations that alter the J domain of Cwc23 in ways known to disrupt functional interaction with Hsp70 and NTR1 or PRP43 mutations that affect the Ntr1:Prp43 interaction suggests that the J domain function of Cwc23 becomes critical when the interaction between Ntr1 and Prp43 is compromised. We suspect that the J domain is important under some environmental condition that we did not reproduce in the laboratory.
The questionable functionality of the J domain of RSP16 orthologs in higher eukaryotes raises the issue of the conservation of Cwc23 as a J protein. However, unlike RSP16 discussed above, Cwc23 orthologs in higher eukaryotes do contain an HPD motif, suggesting a conservation of J domain function. Interestingly, S. cerevisiae, compared to most eukaryotes, has evolved a simplified splicing machinery, since it lacks alternative splicing and overall has few intron-containing genes (5, 30). Perhaps the J domains of Cwc23 orthologs play a more prominent role in organisms with more complex splicing machineries. That Cwc23 orthologs identified in higher eukaryotes have an RNA recognition motif (i.e., RRM; data not shown), a motif known to mediate protein-RNA interactions (11) that is absent from the fungal proteins, is in keeping with this line of logic.
What is the relationship between the role of Cwc23's J domain, and thus Hsp70, and that of the C-terminal region of Cwc23? Are their functions totally independent or at least partially inter-related? Although we cannot answer these questions with certainty, the results reported here provide some insight. The function of the two domains cannot be completely independent, since the J domain cannot carry out its roles when expressed as an independent polypeptide. Interaction of Cwc23 with Ntr1 may serve to tether the J domain to a particular location, enormously increasing its local concentration, and efficiently allowing recruitment of Hsp70. However, on the other hand, the C-terminal domain must be playing a role beyond simply tethering the J domain since this domain is essential and Cwc23's J domain is not.
Recent biochemical results indicate that the interaction between Prp43 and Ntr1 is critical in the disassembly step, with Ntr1 not only tethering Prp43 to the spliceosome but also able to activate its helicase activity (4, 33, 36). An involvement of Cwc23 in modulating the interaction between Prp43 and the NTR complex, increasing the efficiency of spliceosome disassembly, is an intriguing possibility. Participation in the remodeling of protein complexes is a known role for Hsp70-J protein systems. Regardless of the exact role of chaperone system in spliceosome disassembly, Cwc23 may provide an extreme example of an “underappreciated class” of J proteins whose biological role is defined more by their non-J domain regions than by their J domains, that is, their traditional Hsp70-related functions. Understanding how such J proteins evolved may afford important insight into the diversity of J protein function and how chaperone systems may be recruited to fine-tune protein complex remodeling.
Supplementary Material
Acknowledgments
We thank David Brow (University of Wisconsin-Madison) and Beate Schwer (Weill Cornell Medical College) for yeast strains, plasmids, and helpful discussions. We also thank members of the Craig lab for critical comments on this work.
This study was supported by National Institutes of Health grant GM31107 (E.A.C.).
Footnotes
Published ahead of print on 12 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24:342-344. [DOI] [PubMed] [Google Scholar]
- 2.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aron, R., T. Higurashi, C. Sahi, and E. A. Craig. 2007. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 26:3794-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon, K. L., T. Auchynnikava, G. Edwalds-Gilbert, J. D. Barrass, A. P. Droop, C. Dez, and J. D. Beggs. 2006. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell. Biol. 26:6016-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 6.Bukau, B., J. Weissman, and A. Horwich. 2006. Molecular chaperones and protein quality control. Cell 125:443-451. [DOI] [PubMed] [Google Scholar]
- 7.Collins, S. R., P. Kemmeren, X. C. Zhao, J. F. Greenblatt, F. Spencer, F. C. Holstege, J. S. Weissman, and N. J. Krogan. 2007. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics 6:439-450. [DOI] [PubMed] [Google Scholar]
- 8.Craig, E., T. Ziegelhoffer, J. Nelson, S. Laloraya, and J. Halladay. 1995. Complex multigene family of functionally distinct Hsp70s of yeast. Cold Spring Harbor Symp. Quant. Biol. 60:441-449. [DOI] [PubMed] [Google Scholar]
- 9.Craig, E. A., P. Huang, R. Aron, and A. Andrew. 2006. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156:1-21. [DOI] [PubMed] [Google Scholar]
- 10.Fan, H. Y., K. K. Cheng, and H. L. Klein. 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics 142:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grainger, R. J., and J. D. Beggs. 2005. Prp8 protein: at the heart of the spliceosome. RNA 11:533-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene, M. K., K. Maskos, and S. J. Landry. 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA 95:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 14.Hennessy, F., W. S. Nicoll, R. Zimmermann, M. E. Cheetham, and G. L. Blatch. 2005. Not all J. domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, P., M. Gautschi, W. Walter, S. Rospert, and E. A. Craig. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497-504. [DOI] [PubMed] [Google Scholar]
- 16.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, J., E. G. Maes, A. B. Taylor, L. Wang, A. P. Hinck, E. M. Lafer, and R. Sousa. 2007. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28:422-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. L., and E. A. Craig. 2000. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 20:3027-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laufen, T., M. P. Mayer, C. Beisel, D. Klostermeier, A. Mogk, J. Reinstein, and B. Bukau. 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA 96:5452-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Q., P. D'Silva, W. Walter, J. Marszalek, and E. A. Craig. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300:139-141. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Buesa, P., C. Pfund, and E. A. Craig. 1998. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc. Natl. Acad. Sci. USA 95:15253-15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 24.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25:1147-1149. [DOI] [PubMed] [Google Scholar]
- 25.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22:2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandit, S., B. Lynn, and B. C. Rymond. 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. USA 103:13700-13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrova, K., S. Oyadomari, L. M. Hendershot, and D. Ron. 2008. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27:2862-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfund, C., P. Huang, N. Lopez-Hoyo, and E. A. Craig. 2001. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12:3773-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleiss, J. A., G. B. Whitworth, M. Bergkessel, and C. Guthrie. 2007. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 5:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy, S. W., and W. Gilbert. 2006. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7:211-221. [DOI] [PubMed] [Google Scholar]
- 31.Sahi, C., and E. A. Craig. 2007. Network of general and specialty J. protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 104:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Tanaka, N., A. Aronova, and B. Schwer. 2007. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 21:2312-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tizon, B., A. M. Rodriguez-Torres, and M. E. Cerdan. 1999. Disruption of six novel Saccharomyces cerevisiae genes reveals that YGL129c is necessary for growth in non-fermentable carbon sources, YGL128c for growth at low or high temperatures and YGL125w is implicated in the biosynthesis of methionine. Yeast 15:145-154. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, R. T., R. H. Fu, F. L. Yeh, C. K. Tseng, Y. C. Lin, Y. H. Huang, and S. C. Cheng. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai, R. T., C. K. Tseng, P. J. Lee, H. C. Chen, R. H. Fu, K. J. Chang, F. L. Yeh, and S. C. Cheng. 2007. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 27:8027-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voth, W. P., Y. W. Jiang, and D. J. Stillman. 2003. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20:985-993. [DOI] [PubMed] [Google Scholar]
- 38.Wahl, M. C., C. L. Will, and R. Luhrmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701-718. [DOI] [PubMed] [Google Scholar]
- 39.Walsh, P., D. Bursac, Y. C. Law, D. Cyr, and T. Lithgow. 2004. The J-protein family: modulating protein assembly, disassembly, and translocation. EMBO Rep. 5:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 41.Yan, W., and E. A. Craig. 1999. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 19:7751-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan, W., M. J. Gale, Jr., S. L. Tan, and M. G. Katze. 2002. Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone P58(IPK) does not require J. domain function. Biochemistry 41:4938-4945. [DOI] [PubMed] [Google Scholar]
- 43.Yan, W., B. Schilke, C. Pfund, W. Walter, S. Kim, and E. A. Craig. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17:4809-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, C., H. A. Owen, and P. Yang. 2008. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9+2 flagella. J. Cell Biol. 180:403-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.