Abstract
Saccharomyces cerevisiae cells lacking the cytochrome c oxidase (CcO) assembly factor Coa2 are impaired in Cox1 maturation and exhibit a rapid degradation of newly synthesized Cox1. The respiratory deficiency of coa2Δ cells is suppressed either by the presence of a mutant allele of the Cox10 farnesyl transferase involved in heme a biosynthesis or through impaired proteolysis by the disruption of the mitochondrial Oma1 protease. Cox10 with an N196K substitution functions as a robust gain-of-function suppressor of the respiratory deficiency of coa2Δ cells but lacks suppressor activity for two other CcO assembly mutant strains, the coa1Δ and shy1Δ mutants. The suppressor activity of N196K mutant Cox10 is dependent on its catalytic function and the presence of Cox15, the second enzyme involved in heme a biosynthesis. Varying the substitution at Asn196 reveals a correlation between the suppressor activity and the stabilization of the high-mass homo-oligomeric Cox10 complex. We postulate that the mutant Cox10 complex has enhanced efficiency in the addition of heme a to Cox1. Coa2 appears to impart stability to the oligomeric wild-type Cox10 complex involved in Cox1 hemylation.
Cytochrome c oxidase (CcO) is the terminal enzyme of the energy-transducing respiratory chain in mitochondria. Eukaryotic CcO consists of 12 to 13 subunits, with three mitochondrion-encoded subunits (Cox1 to Cox3) forming the core enzyme embedded within the mitochondrial inner membrane (IM). The remaining nucleus-encoded subunits pack on the periphery of the catalytic core (35). The assembled holoenzyme is further organized into supercomplexes with the bc1 cytochrome c reductase (8). The core CcO subunits contain three copper atoms and two modified heme cofactors (34). Cox1 contains both heme moieties, one of which exists as a heterobinuclear center with copper. Cox2 contains the other two Cu ions within a binuclear copper center. Two modifying enzymes, Cox10 and Cox15, form the modified heme moiety in CcO. Cox10 catalyzes the conversion of heme b into heme o by the addition of a hydroxyethyfarnesyl group (12). Cox15 converts heme o into heme a through the oxidation of a C8 pyrrole methyl moiety (11). Cox15, heme a synthase, works in conjunction with ferredoxin and ferredoxin reductase (4).
The biogenesis of CcO, occurring within the IM, commences with the mitochondrial synthesis of the Cox1 subunit, followed by stepwise insertion of the heme and copper redox cofactors and the addition of the remaining subunits. The translation of Cox1 on mitoribosomes occurs in juxtaposition to the IM and is mediated in yeast by the IM-tethered translational activators Pet309 and Mss51 (21, 22, 27, 32). Mdm38 and Mba1 facilitate recruitment of mitoribosomes for Cox1 translation (26, 30). Nascent Cox1 appears to be inserted into the IM by the Oxa1 translocase (16). Newly synthesized Cox1 is escorted through the maturation process by IM-associated protein complexes containing Mss51, Cox14, Coa1, and Shy1. The initial Cox1 assembly intermediate involves a complex with Mss51 and Cox14 (3, 27). Cox14 is not important for the translational initiation function of Mss51, since cox14Δ cells synthesize Cox1 but are stalled in its maturation presumably due to the failure to form the newly synthesized Cox1-Mss51 complex (3, 27). Coa1 appears to coordinate the transition of newly synthesized Cox1 from the Mss51-Cox14 complex to a later intermediate involving Shy1 that likely functions in the heterobimetallic heme-copper center (23, 28). Cells lacking Coa1 or Shy1 synthesize Cox1, but maturation is largely stalled prior to cofactor insertion, leading to Cox1 degradation. Limited assembly of CcO occurs in either deficient strain, but the residual CcO is insufficient to promote respiratory growth.
Clues to the CcO assembly pathway have been gleaned by the isolation of genetic suppressors of respiration-deficient strains (1). Overexpression of MSS51 in shy1Δ cells yields respiratory growth, most likely through elevated Cox1 translation (2). Mss51 was proposed to be sequestered in a nonproductive complex in shy1Δ cells, thereby limiting its level for Cox1 translation initiation (3). Overexpression of Hap4, the catalytic subunit of the Hap2/3/4/5 transcription complex, suppresses the CcO deficiency of shy1Δ cells through induced expression of nucleus-encoded subunits of CcO (10). Enhanced levels of Cox5a and Cox6 may stabilize Cox1 in shy1Δ cells, enabling progression to later stages of CcO assembly. A series of extragenic suppressors of the coa1Δ cell respiratory defect was isolated (28). High-copy-number MSS51 was an efficient suppressor of coa1Δ cells, and three weaker suppressors were high-copy-number COX10, MDJ1, and COA2. As described, Cox10 is involved in the biosynthesis of heme a, Mdj1 is a DnaJ cochaperone with Hsp70, and Coa2 is a newly identified CcO assembly factor. Strong synergism was observed with MSS51 in combination with either COX10 or COA2 in the suppression of both coa1Δ and shy1Δ cells (29).
Coa2 is a small matrix-localized protein that functions downstream of the Mss51-dependent step in Cox1 maturation (29). The observed transient interaction of Coa2 and Shy1 suggested a role in the ShyI-related step in Cox1 maturation. One dramatic phenotype of coa2Δ cells is the rapid degradation of newly synthesized Cox1 (29). High-copy-number Mss51, which efficiently suppresses the respiratory defect of coa1Δ and shy1Δ cells, had no effect in coa2Δ cells, although high-copy-number Cox10 was weakly effective. During these studies, we observed spontaneous mutant clones of coa2Δ cells with robust respiratory growth. In the present study we report the characterization of a mutant allele of COX10 that is a dominant suppressor of coa2Δ cells. We show that the suppressor activity of mutant Cox10 is dependent on its catalytic activity and suggest that the mutant Cox10 facilitates heme a addition into Cox1, a step that is compromised in coa2Δ cells. Coa2 per se is not essential for hemylation of Cox1, since CcO biosynthesis can be restored in coa2Δ cells by depleting cells of the metallopeptidase Oma1. The present work provides novel insight into the function of Cox10, Coa2, and Oma1.
MATERIALS AND METHODS
Yeast strains and vectors.
The Saccharomyces cerevisiae yeast strains used in this study are listed in Table 1. The hemagglutinin (HA)-tagged Cox1 strain was a generous gift from Xochitl Perez-Martinez. Strains with open reading frame (ORF) deletions generated for this work were created by homologous recombination of disruption cassettes using either KanMX4 or Candida albicans URA3. Strains with an integrated 3′-13Myc or 3′-3HA epitope tag were generated as described previously (20). All strains generated for this work were confirmed by PCR. The COX10 ORF was cloned into plasmid pRS426 and pRS416 under the control of its own promoter and terminator (450 base pairs upstream and downstream of the ORF). The N196K mutation was made in both the high- and low-copy-number vectors using site-directed mutagenesis. The COX10 ORF with a 3′-13Myc tag was cloned from genomic DNA of the DY5113 COX10-13Myc strain into pRS416 and pRS415 vectors under the control of its own promoter and the ADH1 terminator. The N196K, N196A, N196D, R212,216A, and H317A mutations were generated by site-directed mutagenesis. Successive rounds of site-directed mutagenesis created the double and triple mutations. The COX15 ORF with a 3′-6His epitope tag was cloned into pRS423 under the control of the MET25 promoter and CYC1 terminator. The YAH1 ORF was cloned into pRS425 under the control of its own promoter (450 base pairs upstream of the ORF) and the CYC1 terminator. Sequencing was used to confirm all cloning and site-directed mutagenesis products in the vectors created. The MSS51, COA2, and YTA12 (E614Q) vectors used have been previously described (29). Yeast strains were transformed using lithium acetate. Culture conditions for the yeast strains were either rich medium (YP) or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection.
TABLE 1.
S. cerevisiae yeast strains used in this study
| Strain | Genotype | Source or reference(s) |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| BY4741 coa2Δ | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 coa2::kanMX4 | 29 |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 | Invitrogen |
| BY4743 cox10Δ | BY4743 cox10::kanMX4 | Invitrogen |
| BY4743 coa1Δ | BY4743 coa1::kanMX4 | Invitrogen |
| BY4743 shy1Δ | BY4743 shy1::kanMX4 | Invitrogen |
| W303 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1, ura3-1 | |
| W303 coa2Δ | W303 coa2::kanMX4 | 29 |
| W303 cox10Δ | W303 cox10::HIS3 | 25 |
| W303 coa1Δ | W303 coa1::kanMX4 | 23, 28 |
| W303 cox4Δ | W303 cox4::CaURA3 | 13 |
| W303 cox11Δ | W303 cox11::HIS3 | 7 |
| W303 oma1Δ | W303 oma1::CaURA3 | This work |
| W303 coa2Δyta12Δ | W303 coa2::CaURA3 yta12::kanMX4 | 29 |
| W303 coa2Δyme1Δ | W303 coa2::CaURA3 yme1::kanMX4 | This work |
| W303 coa2Δoma1Δ | W303 coa2::kanMX4 oma1::CaURA3 | This work |
| W303 coa2Δcox15Δ | W303 coa2:: kanMX4 cox15::CaURA3 | This work |
| DY5113 (W303) | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ, ura3-1 | |
| COX10-3HA | DY5113 COX10-3HA::TRP1 | This work |
| COX10-13Myc | DY5113 COX10-13Myc::TRP1 | This work |
| COX10-13Myc mss51Δ | DY5113 COX10-13Myc::TRP1 mss51::kanMX4 | This work |
| COX10-13Myc cox14Δ | DY5113 COX10-13Myc::TRP1 cox14::CaURA3 | This work |
| COX10-13Myc coa1Δ | DY5113, COX10-13Myc::TRP1, coa1::CaURA3 | This work |
| COX10-13Myc shy1Δ | DY5113 COX10-13Myc::TRP1 shy1::CaURA3 | This work |
| COX10-13Myc sco1Δ | DY5113 COX10-13Myc::TRP1 sco1::kanMX4 | This work |
| COX10-13Myc cox1Δ | MATα lys2 leu2-3,112 arg8::hisG ura3-52 cox1Δ::ARG8m COX10-13Myc::kanMX | This work |
| COX15-13Myc | DY5113 COX15-13Myc::TRP1 | This work |
| SHY1-13Myc | DY5113 SHY1-13Myc::TRP1 | 29 |
| SHY1-13Myc cox11Δ | DY5113 SHY1-13Myc::TRP1 cox11::HIS3 | This work |
| COA1-13Myc | DY5113 COA1-13Myc::HIS3 | 23, 28 |
| COX1-3HA | MATaarg8::hisG leu2-3,112 lys2 ura3-52 COX1-3HA D273-10B | X. Perez-Martinez |
Mitochondrial purification and assays.
Intact mitochondria were isolated from yeast as previously described (9). The standard Bradford assay was used to determine total mitochondrial protein concentration (5). Total heme was extracted from 1 to 2 mg of purified mitochondria with 0.5 ml acetone containing 2.5% HCl as described previously (4). The pH of the extract was adjusted to 4.0 by the addition of 1 μl formic acid and titration with 5 M KOH (elution times are affected by the pH). The sample was clarified by centrifugation at 13,000 rpm for 5 min, and 1 ml was injected onto a 4.6- by 250-mm SunFire C18 5-μm column (Waters). Hemes were eluted from the column at a flow rate of 1 ml/min using a 30 to 50% gradient of acetonitrile containing 0.05% trifluoroacetic acid for the first 3 ml and a 50 to 80% gradient for the next 42 ml. The elution of heme compounds was monitored at 400 nm; purified heme a, heme o, and hemin were used as standards to determine elution times. Heme a was a generous gift from Winslow Caughey.
Blue native PAGE.
Blue native PAGE (BN-PAGE) was performed essentially as described previously (38) with 1% digitonin. After incubation on ice for 15 min and centrifugation (20,000 × g for 15 min at 2°C), supernatants were mixed with sample buffer (5% Coomassie brilliant blue G250, 0.5 M 6-aminocaproic acid, pH 7.0) and then loaded on a gradient polyacrylamide gel. Separated complexes were detected by immunoblotting on a polyvinylidene difluoride (PVDF) membrane. The high-mass marker proteins were obtained from GE Healthcare.
Immunoblotting.
Mitochondrial protein samples were separated on 12% polyacrylamide gels and transferred to nitrocellulose. Proteins were visualized using either enhanced chemiluminescence (ECL) reagents with horseradish peroxidase-conjugated secondary antibodies or the Odyssey infrared imaging system (Li-Cor Biosciences) with fluorescent secondary antibodies (anti-mouse IRDye 800 and anti-rabbit IRDye 680; Li-Cor Biosciences). Anti-Myc and anti-HA rabbit polyclonal antisera was purchased from Santa Cruz, anti-Myc and anti-HA mouse monoclonal antibodies were from Roche, antiporin was from Molecular Probes, and anti-Cox2 was from Mitosciences. Alex Tzagoloff generously provided antiserum to F1 ATP synthase, and Thomas Langer provided antisera to Phb1 and Phb2.
IP.
Immunoprecipitations (IPs) were performed essentially as described previously (29) except that the IP buffer used was 20 mM Tris (pH 7.4), 50 mM sodium chloride, 1 mM phenylmethylsulfonyl fluoride, and 1% digitonin. Anti-HA-agarose and anti-Myc-agarose conjugates were purchased from Santa Cruz. The clarified lysate, unbound, final wash, and elution fractions were analyzed by immunoblotting.
Additional assays.
The respiratory competency of strains was determined by growth tests on plates containing 2% glucose or 2% glycerol-2% lactate as a carbon source. Yeast cells were grown overnight in liquid cultures in selective medium containing 2% raffinose-0.2% glucose and adjusted to an optical density at 600 nm (OD600) of 0.5, and serial dilutions were spotted onto the plates and incubated at 30°C for 2 days (glucose plates) or 4 to 6 days (glycerol-lactate plates). The oxygen consumption of cells grown to stationary phase was determined on a 5300A biological oxygen monitor (Yellow Springs Instrument Co.). The rate of oxygen consumption (% O2/second/OD600 unit) was calculated from the linear response (17). For in vivo mitochondrial labeling with [35S]methionine, cells were grown overnight in selective medium containing 2% raffinose-0.2% glucose and then reinoculated into YP-2% galactose to grow to an OD600 of 1. The labeling and preparation of the samples for 12% SDS-PAGE have been described previously (2). Gels were dried, and radiolabeled proteins were visualized by exposing autoradiographic films at −80°C. Sensitivity of yeast strains to hydrogen peroxide was determined as previously described (19).
Construction of the low-copy-number library.
Genomic DNA purified from the coa2Δ suppressor strain containing the COX10-N196K mutation was partially digested with Sau3A. DNA fragments were then purified from an agarose gel and ligated with plasmid pRS416 that was digested to completion with BamHI. The resulting number of transformants was sufficient for severalfold coverage of the yeast genome.
RESULTS
Isolation of a coa2Δ dominant suppressor.
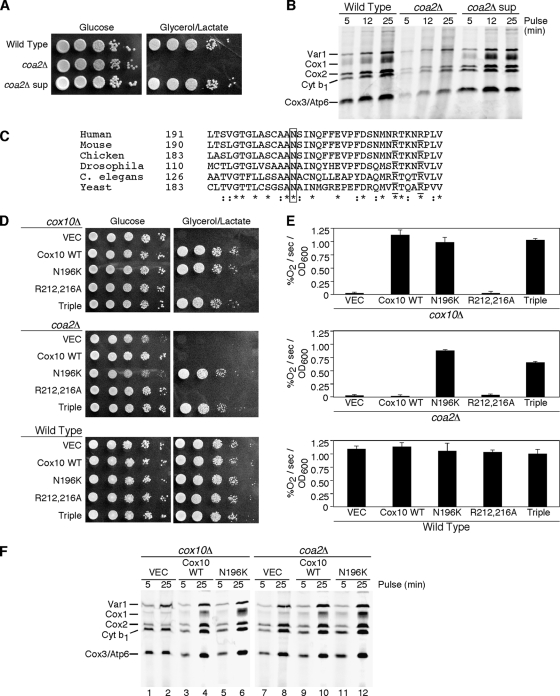
Coa2 is required for the assembly of CcO in yeast (29). Cells lacking Coa2 fail to propagate on glycerol/lactate growth medium (29) (Fig. 1A). With prolonged incubation on glycerol/lactate medium, spontaneous mutants emerged that showed robust growth upon subsequent replating. The frequencies of appearance of spontaneous mutants were 1 in 107 colonies for coa2Δ cells in the BY4741 background and 1.7 in 106 colonies for coa2Δ cells in the W303 background. Recovery of one spontaneous suppressor clone showed that upon subsequent plating, respiratory growth resembled that of wild-type (WT) cells (Fig. 1A). coa2Δ null cells exhibit a marked attenuation of newly synthesized Cox1 in an in vivo translation assay, and this attenuation was shown to arise from enhanced degradation rather than impaired translation (29). The suppressor mutant showed normal Cox1 levels in the in vivo translation assay (Fig. 1B). To assess whether the suppressing mutation was recessive or dominant, the suppressor isolate was crossed with the parent coa2Δ strain of opposite mating type. The resulting diploid strain was found to be respiration competent in showing WT growth on glycerol/lactate medium. The diploid was sporulated, and tetrad dissection revealed that the suppressing phenotype segregated 2:2, suggesting that a single mutant gene was responsible for the phenotype.
FIG. 1.
Identification of the coa2Δ suppressor mutant as N196K Cox10. (A) W303 wild type (WT), coa2Δ, and isolated coa2Δ suppressor strains were grown in yeast extract-peptone-dextrose, serially diluted, and spotted on yeast extract-peptone-2% glucose and yeast extract-peptone-2% glycerol-2% lactate. Plates were grown at 30°C. (B) In vivo labeling of mitochondrial translation products. The strains described for panel A were pulsed with [35S]methionine for 5, 12, and 25 min at 30°C. The samples were separated by 12% SDS-PAGE, and the gel was dried and then exposed to autoradiographic film. (C) Alignment of the peptide sequences of Cox10 from various species using ClustalW. The box highlights the conserved N196 residue. Arg212 and Arg216, residues required for activity, are underlined. Residues with conserved substitutions across species are indicated by colons, and identical residues across species are indicated by asterisks. (D) BY cox10Δ, coa2Δ, and wild-type cells transformed with centromeric vectors expressing WT Cox10, N196K Cox10, R212A,R216A Cox10, and N196K,R212A,R216A Cox10 (Triple) were grown in SC-2% raffinose-0.2% glucose selective medium, serially diluted, and spotted on SC-2% glucose and SC-2% glycerol-2% lactate. The plates were incubated at 30°C. Similar results were observed for cells transformed with episomal vectors. (E) The cells described for panel D were grown in liquid SC-1% glucose overnight, and oxygen consumption (% O2/second/OD600 unit) was measured. The data represent the averages of four independent repeats, and the error bars represent standard errors of the means. (F) In vivo labeling of mitochondrial translation products. W303 cox10Δ cells (lanes 1 to 6) and W303 coa2Δ cells (lanes 7 to 12) transformed with episomal vectors expressing WT Cox10 and N196K Cox10 were pulsed with [35S]methionine for 5 and 25 min. The samples were analyzed as described for panel B.
To clone the dominant mutant gene, a genomic library was constructed from the genomic DNA of the suppressor coa2Δ clone in a centromeric vector. Recombinant plasmid DNA was used to transform coa2Δ parent cells, and transformants that grew on glycerol/lactate medium were isolated. The respiratory growth was shown to be dependent on the library vector, as shedding the vector by growth in 5-fluorortate abrogated respiratory growth. The plasmid rescued from the respiration-competent coa2Δ cells was used to retransform coa2Δ cells, and respiratory competence was recovered. DNA sequencing revealed the suppressor gene to be a mutated COX10 causing an N196K amino acid change. Asn196 is a highly conserved residue in diverse Cox10 orthologs (Fig. 1C).
The N196K mutation was introduced in a low-copy-number plasmid containing COX10 and transformed into coa2Δ or cox10Δ cells (Fig. 1D). The mutant Cox10 restored respiratory growth in coa2Δ cells, demonstrating that the mutation was sufficient to confer the suppressor phenotype. WT COX10 is only a weak suppressor in coa2Δ cells when expressed in high copy number, a situation similar to its weak suppressor activity in coa1Δ cells (28, 29). The observed respiratory growth of cox10Δ cells harboring the N196K mutant allele showed that the substitution did not impair the catalytic activity of Cox10. This conclusion was substantiated by the observation that oxygen consumption by cox10Δ cells containing either WT Cox10 or the N196K mutant protein was similar (Fig. 1E). No dominant negative effects were observed in transformants of WT cells with the mutant Cox10 regardless of whether it was expressed from a low- or high-copy-number vector.
The presence of the mutant Cox10 in coa2Δ cells restored Cox1 levels in the in vivo mitochondrial translation assay (Fig. 1F) better than the WT Cox10 protein. Likewise, mutant Cox10 restored Cox1 levels in cox10Δ cells.
N196K Cox10 is a coa2Δ-specific suppressor.
N196K Cox10 has a gain-of-function activity in coa2Δ cells. To substantiate this activity, we engineered the mutation in a cox10 allele encoding a double substitution of two functionally important conserved Arg residues implicated in the binding of the pyrophosphate moiety of the farnesyl pyrophosphate substrate of the Cox10-related CyoE heme o synthase of Escherichia coli (31). The double-Arg Cox10 mutant (R212,216A) was nonfunctional in cox10Δ cells (Fig. 1D) and failed to suppress either coa1Δ cells (28) or coa2Δ cells despite the presence of the mutant protein by immunoblotting (data not shown). However, the triple mutant (N196K, R212,216A) retained the suppressor activity in coa2Δ cells and, surprisingly, restored Cox10 function in cox10Δ cells (Fig. 1D and E). Thus, the N196K substitution has a dominant effect on the Cox10 suppressor activity.
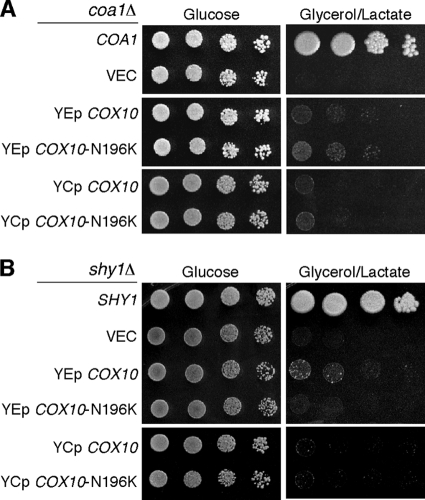
We previously demonstrated that high-copy-number COX10 was a weak suppressor of coa1Δ and shy1Δ cells (28). To assess whether the N196K mutant Cox10 exhibited gain-of-function suppressor activity in these cells, low- and high-copy-number vectors carrying either WT or mutant COX10 were transformed in coa1Δ and shy1Δ cells, and respiratory growth was assessed (Fig. 2). The WT and mutant genes were equally weak suppressors in these CcO assembly mutants. In addition, no suppression by the N196K Cox10 was seen in cox14Δ cells (data not shown). Thus, the N196K Cox10 has a unique suppressor function in coa2Δ cells.
FIG. 2.
N196K Cox10 does not alter suppression of the respiratory defect of coa1Δ or shy1Δ cells. (A) BY coa1Δ cells transformed with either episomal (YEp) or centromeric (YCp) vectors expressing WT COX10 or COX10 (N196K) were grown in SC-2% raffinose-0.2% glucose selective medium, serially diluted, and spotted on SC-2% glucose and SC-2% glycerol-2% lactate agar plates. The plates were incubated at 30°C. (B) BY shy1Δ cells with either YEp or YCp vectors expressing WT COX10 or COX10 (N196K) were treated as described for panel A.
The N196K mutant Cox10 may exert its specific suppressor function in coa2Δ cells by one of two mechanisms. The first scenario is that the mutant Cox10 has a gain-of-function role in directly stabilizing newly synthesized Cox1, thereby impairing proteolytic degradation. The second candidate mechanism is that mutant Cox10 has an enhanced efficiency in heme a site formation in Cox1. The marked instability of Cox1 seen in coa2Δ cells may arise from an impaired addition of heme a, since the heme a site stabilizes the Cox1 helical bundle. Studies were conducted to discern between these candidate mechanisms of suppression.
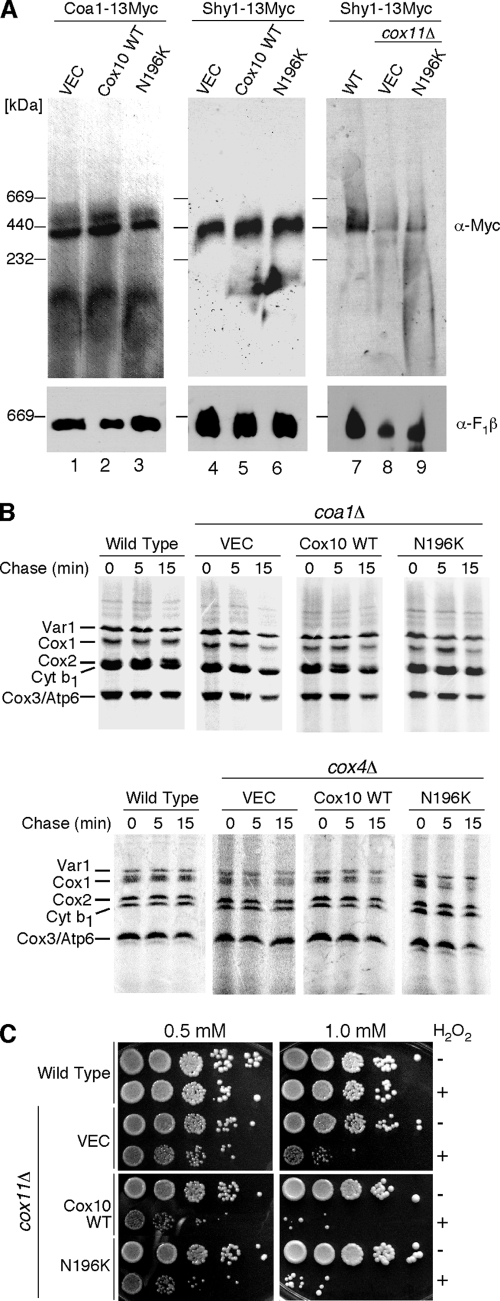
N196K Cox10 has no general role in the stabilization of Cox1.
The first postulate is that the N196K mutant Cox10 has a gain-of-function role in stabilizing Cox1 within early assembly intermediates. One marked phenotype of coa2Δ cells is the rapid degradation of Cox1 observed with in vivo mitochondrial translation assays (29) (Fig. 1F). We conducted three studies to test this first postulate. First, Coa1 and Shy1 participate in early Cox1 assembly intermediates downstream of the Mss51-Cox14 complex and have previously been shown to bind Cox1 (23, 28). To assess whether N196K mutant Cox10 had an effect on the abundance of these Cox1 assembly complexes, mitochondria isolated from cells chromosomally epitope tagged at the COA1 and SHY1 loci were fractionated on BN-PAGE (Fig. 3A). The presence of N196K Cox10 had no effect on the abundance of Cox1-containing high-mass Coa1 and Shy1 complexes. The high-mass Shy1 complex is attenuated in numerous CcO assembly mutants, such as cox11Δ cells (Fig. 3A, lane 8). The N196K mutant Cox10 had no stabilizing effect on the Cox1-containing ∼450-kDa Shy1 complex in these cells (Fig. 3, lane 9). Second, the N196K mutant Cox10 fails to stabilize Cox1 in two CcO assembly mutants (coa1Δ and cox4Δ cells) in which newly synthesized Cox1 is proteolytically unstable (Fig. 3B). Cox1 is synthesized in coa1Δ and cox4Δ cells as seen in an in vivo mitochondrial translation assay, but it is degraded during the chase phase of the assay. The presence of either WT Cox10 or the N196K mutant failed to retard Cox1 degradation in either mutant strain. Third, mutant Cox10 lacks any Cox1 stabilization function as determined using a hydrogen peroxide sensitivity assay. Cells lacking Cox11 are sensitive to hydrogen peroxide due to the accumulation of a stalled heme a3-Cox1 assembly intermediate (19). We reported previously that overexpression of either COX10 or COA2 exacerbated the sensitivity of cox11Δ cells to peroxide, most likely through stabilizing the heme a3-Cox1 assembly intermediate (29). The N196K mutant Cox10 did not induce any further peroxide hypersensitivity to cox11Δ cells relative to WT Cox10 (Fig. 3C). These three studies argue against mutant Cox10 having any general Cox1 stabilization function and further support the specific suppression of coa2Δ cells.
FIG. 3.
Stabilization of Cox1 by N196K Cox10 is specific to coa2Δ cells. (A) Mitochondria (50 μg protein) isolated from strains containing a genomically tagged 13-Myc COA1 gene (lanes 1 to 3) or SHY1 gene (lanes 4 to 6) and expressing either WT Cox10 or N196K Cox10 were solubilized in buffer containing 1% digitonin. Lysates were loaded onto a continuous 5 to 13% gradient gel, and protein complexes were separated by BN-PAGE. Complexes were analyzed by immunoblotting with anti-Myc antibody. Molecular mass markers are indicated, and antibody to the F1β subunit of monomeric complex V was used as a loading control. Mitochondria (50 μg protein) isolated from WT, cox11Δ, or cox11Δ cells expressing N196K Cox10 and containing an endogenously tagged SHY1-13Myc (lanes 7 to 9) were solubilized, separated by BN-PAGE, and analyzed by immunoblotting as in the first two panels. (B) In vivo labeling of mitochondrial translation products. W303 WT, coa1Δ, and cox4Δ cells transformed with WT Cox10 or N196K Cox10 were used in the translation assay as described for Fig. 1B. (C) cox11Δ cells transformed with YCp Cox10 or YCp N196K Cox10 vectors and W303 WT cells were grown to mid-exponential phase and incubated with (+) or without (−) the indicated concentrations of H2O2 for 2 h at 30°C. Serial dilutions were spotted onto yeast extract-peptone-dextrose plates and incubated for 36 to 48 h at 30°C. Similar results were obtained for YEp COX10 and N196K COX10 in the cox11Δ background.
Suppressor activity of N196K Cox10 is dependent on its catalytic activity.
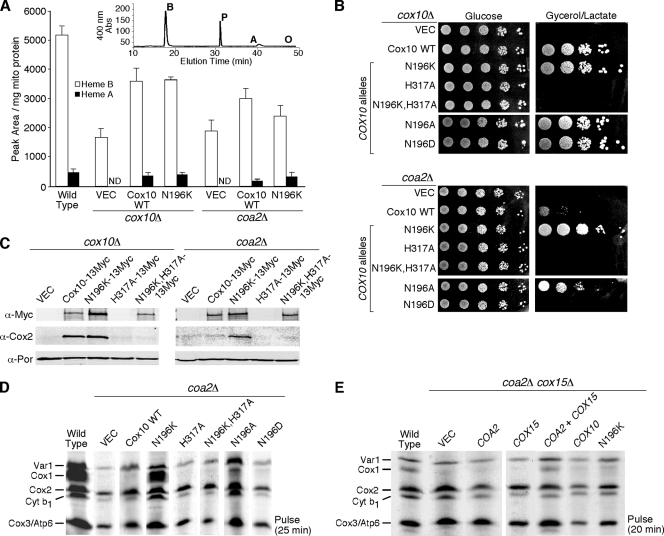
The second candidate mechanism of suppression by the N196K mutant Cox10 is related to its role in heme a biosynthesis. Suppression of coa2Δ cells by the mutant Cox10 may arise from more efficient hemylation of Cox1. If Coa2 functions at a point in which the heme a site is formed, the absence of Coa2 may destabilize the heme a site occupancy, leading to an altered Cox1 conformation that is susceptible to proteolytic degradation. To test whether N196K Cox10 had enhanced catalytic activity, we evaluated whether strains harboring the mutant had elevated levels of heme o (Fig. 4A). The mutant and WT Cox10 proteins were expressed in either cox10Δ or coa2Δ cells. Mitochondria purified from each transformant were quantified for various hemes using reverse-phase high-pressure liquid chromatography (HPLC) after organic extraction (Fig. 4A, inset). Comparable levels of heme a were evident in cox10Δ cells containing either the WT or the N196K Cox10 mutant (80 and 85% of WT level, respectively), but no heme o was evident in either extraction. The detection limit for heme o in our assay is 10% of heme a levels. Likewise, coa2Δ cells harboring either the WT or the N196K Cox10 mutant had no detectable levels of heme o.
FIG. 4.
Catalytically active Cox10 and heme a generation are required for suppressor activity by N196K Cox10. (A) Heme was extracted from mitochondria (1.5 to 2 mg protein) isolated from WT cells, cox10Δ cells expressing YCp WT Cox10 or N196K Cox10, and coa2Δ cells expressing YCp WT Cox10 or Cox10-N196K and separated by reverse-phase high-performance liquid chromatography. The inset shows a representative chromatogram from WT mitochondria. The peaks corresponding to heme b (B), heme a (A), and protoporphyrin (P) and the expected elution time of heme o (O) are indicated. The white bars indicate the area under the heme b peak normalized to mg of mitochondrial protein, and the black bars represent the area under the heme a peak also normalized to mg of mitochondrial protein. The data represents the averages of three independent repeats, and the error bars represent the standard errors of the means. ND, not detectable. (B) cox10Δ and coa2Δ cells expressing WT Cox10, N196K Cox10, H317A Cox10, N196K-H317A Cox10, N196A Cox10, and N196D Cox10 were grown in SC-2% raffinose-0.2% glucose selective medium, serially diluted, and drop tested on SC-2% glucose or yeast extract-peptone-2% glycerol-2% lactate. The plates were incubated at 30°C for 2 days (glucose) or 4 days (glycerol/lactate). (C) Purified mitochondria (30 μg protein) from cox10Δ and coa2Δ cells expressing WT Cox10-13Myc, N196K Cox10-13Myc, H317A Cox10-13Myc, and N196K-H317A Cox10-13Myc were separated by SDS-PAGE and analyzed by immunoblotting using anti-Myc, anti-Cox2, and anti-Por1 (loading control) antibodies. (D) In vivo mitochondrial translation. W303 coa2Δ cells expressing WT Cox10, N196K Cox10, H317A Cox10, N196K-H317A Cox10, N196A Cox10, and N196D Cox10 were pulsed with [35S]methionine as for Fig. 1B. (E) Analysis of mitochondrial translation products in W303 coa2Δ cox15Δ cells expressing COA2, COX15, WT COX10, and COX10 (N196K). The cells were pulsed with [35S]methionine for 20 min at 30°C and analyzed as for Fig. 1B.
His317 of Cox10 is a highly conserved residue and is implicated in catalytic activity (24). To test whether the suppressor activity of the N196K Cox10 mutant was dependent on the catalytic activity of the enzyme, an H317A substitution was engineered in WT and N196K Cox10 and the resulting mutants were tested for function in cox10Δ cells or suppressor activity in coa2Δ cells (Fig. 4B). The H317A Cox10 mutant failed to support respiratory growth when expressed in cox10Δ cells, as was true with the double H317A,N196K mutant. Immunoblotting revealed that the single H317A mutant was unstable when expressed in cox10Δ cells, but the double H317A,N196K mutant was stably expressed (Fig. 4C). In addition to the respiratory deficiency of cox10Δ transformants with the double H317A,N196K mutant, steady-state levels of Cox2 were also attenuated as seen in the parent cox10Δ cells (Fig. 4C). Expression of the mutants in coa2Δ cells revealed that the H317A,N196K double mutant failed to suppress the respiratory deficiency of coa2Δ cells. Whereas expression of the N196K mutant Cox10 in coa2Δ cells resulted in stable Cox1 levels seen in the mitochondrial translation assay, expression of the H317A,N196K double mutant showed a rapid degradation of Cox1 similar to that seen in untransformed coa2Δ cells (Fig. 4D). To corroborate the evidence that heme a biosynthesis was essential for the suppressor activity of the N196K mutant Cox10, we tested whether the N196K mutant Cox10 retained suppressor activity in stabilizing Cox1 in the mitochondrial translation assay in cells lacking Cox15, the second enzyme required in heme a biosynthesis. The N196K mutant Cox10 was expressed in coa2Δ cox15Δ cells (Fig. 4E). In the mitochondrial translation assay, no Cox1 was observed during the pulse despite the presence of the N196K Cox10. Thus, the catalytic activity of Cox10 and the synthesis of heme a are essential for the suppression of respiratory function and Cox1 stabilization in coa2Δ cells.
We addressed the specificity of the Asn196 substitution for the gain-of-function suppressor activity in coa2Δ cells. Two mutations were introduced into COX10, yielding N196A and N196D substitutions, and these mutants were expressed in cox10Δ or coa2Δ cells (Fig. 4B). None of the substitutions at Asn196 altered the function of Cox10, since all cox10Δ transformants grew well on glycerol/lactate medium. However, the N196D mutant Cox10 lacked suppressor activity in coa2Δ cells, and the N196A Cox10 yielded only partial restoration of respiratory growth. Thus, the charge at position 196 appears to be important for suppressor activity. The glycerol/lactate growth results matched the stabilization of Cox1 in the mitochondrial translation assay (Fig. 4D).
Suppressor activity of N196K Cox10 correlates with the abundance of its high-mass complex.
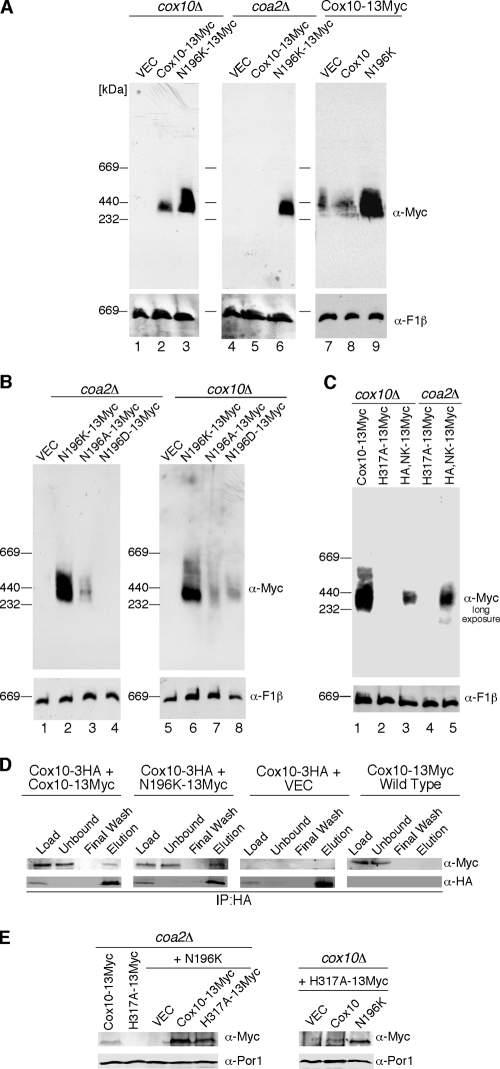
The N196K Cox10 was reproducibly more abundant relative to the WT protein when expressed in cox10Δ and coa2Δ cells (Fig. 4C) or WT cells (data not shown). To gain further insights into Cox10 function, we carried out BN-PAGE on Myc-tagged Cox10 (Fig. 5A). Cox10 forms a high-mass complex when expressed either from a vector or chromosomally (Fig. 5A, lanes 2 and 7). Surprisingly, the N196K mutant protein forms a markedly more abundant high-mass complex than the WT protein (Fig. 5A, lanes 3 and 6). The presence of the untagged N196K mutant Cox10 in WT cells containing a chromosomally tagged COX10 also yields a marked enhancement in the abundance of the high-mass complex (Fig. 5A, lanes 8 and 9). Importantly, the high-mass Cox10 complex is gone in coa2Δ cells (Fig. 5A, lane 5) despite the presence of the protein seen by SDS-PAGE (Fig. 4C). The N196K mutant protein forms an abundant complex in coa2Δ cells (Fig. 5A, lane 6). BN analyses were carried out on mutant Cox10 alleles with various Asn196 substitutions. The N196D mutant, which lacked suppressor activity (Fig. 4B), also lacked any appreciable BN complex (Fig. 5B, lane 4), although the protein was equally present as determined by steady-state SDS-PAGE analysis. The N196A mutant, which had limited suppressor activity, formed a high-mass complex that was less abundant than the N196K mutant protein. Thus, a correlation exists between the abundance of the Cox10 complex and its suppressor activity. The N196D and N196A mutant proteins formed a high-mass complex of similar abundance when expressed in cox10Δ cells (Fig. 5B, lanes 7 and 8).
FIG. 5.
Assessing the Cox10 high-molecular-weight complex. (A) Purified mitochondria (100 μg protein) from cox10Δ and coa2Δ cells expressing WT Cox10-13Myc and N196K Cox10-13Myc (lanes 1 to 6) were solubilized in buffer containing 1% digitonin. Lysates were loaded onto a 5 to 13% gradient gel, and complexes were separated by BN-PAGE. Lanes 7 to 9 show the BN-PAGE analysis of mitochondria (100 μg protein) isolated from a genomically tagged COX10-13Myc strain expressing WT Cox10 or N196K Cox10 (both untagged). The Cox10-13Myc complex was detected by immunoblotting using anti-Myc antibody in all panels, and monomeric complex V was detected using anti-F1β antibody as a loading control. (B) Purified mitochondria (100 μg proteins) from both coa2Δ and cox10Δ cells expressing N196K Cox10-13Myc, N196A Cox10-13Myc, and N196D Cox10-13Myc were analyzed by BN-PAGE as described for panel A. (C) Isolated mitochondria from cox10Δ cells expressing WT Cox10-13Myc, H317A Cox10-13Myc, and H317A,N196K Cox10-13Myc (HA,NK-13Myc), and from coa2Δ cells expressing H317A Cox10-13Myc and HA,NK-13Myc were analyzed by BN-PAGE as described for panel A. (D) Purified mitochondria (500 μg protein) from endogenously tagged COX10-3HA cells, COX10-3HA cells expressing WT Cox10-13Myc or N196K Cox10-13Myc, and endogenously tagged COX10-13Myc cells were solubilized in IP buffer, and clarified extracts were immunoprecipitated with rabbit polyclonal anti-HA beads overnight. The fractions for each co-IP were analyzed by SDS-PAGE and immunoblotting using anti-Myc and anti-HA antibodies. Load, 2% of the soluble extract; unbound, 2% of the soluble extract after incubation with the HA-conjugated beads; wash, 50% of the last wash concentrated by trichloroacetic acid precipitation; and elution, 50% of the bead elution. IP using mouse monoclonal Myc-conjugated beads provided similar results. (E) Mitochondria (30 μg protein) purified from coa2Δ cells expressing either WT Cox10-13Myc or H317A Cox10-13Myc and these two plasmids expressed in trans with untagged N196K Cox10 were analyzed for steady-state levels of Cox10 by SDS-PAGE and immunoblotting using anti-Myc antibody and Por1 as a loading control. Similarly, mitochondria (30 μg protein) purified from cox10Δ cells expressing H317A Cox10-13Myc in trans with untagged WT Cox10 or N196K Cox10 was analyzed as described.
As mentioned, the H317A mutation in Cox10 abrogates any suppressor activity of the N196K allele. However, the inactive N196K,H317A mutant Cox10 still forms a low-abundance complex in both coa2Δ and cox10Δ cells (Fig. 5C). Thus, formation of the high-mass Cox10 complex is not dependent on a catalytically active enzyme.
The enhanced Cox10 BN complex seen in chromosomally tagged Cox10 cells expressing a low-copy-number untagged N196K Cox10 from a vector suggests that the ∼370-kDa complex may be a Cox10 oligomer. Two additional studies corroborate the existence of a homo-oligomer complex. First, cells expressing chromosomally HA-tagged Cox10 and vector Myc-tagged Cox10 were tested for co-IP (Fig. 5D). Precipitation of HA-Cox10 resulted in co-IP of a fraction of Myc-Cox10, especially in the cells with the N196K Cox10. Second, coexpression of the unstable H317A Cox10 mutant (Fig. 4C) with the N196K mutant led to a pronounced stabilization of the unstable H317A Cox10 in either coa2Δ or cox10Δ cells (Fig. 5E). The cotransformants of both strains were fully competent in growth on glycerol/lactate medium (data not shown). The stabilized H317A Cox10 did not have any dominant negative effects.
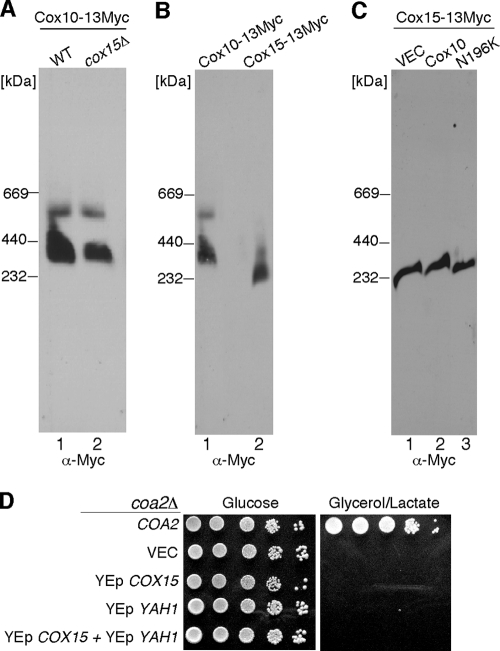
We sought to assess whether other components were present in the high-mass Cox10 complex by taking a candidate protein approach. Three candidates are Coa2, Cox15, and Cox1. No stable interaction between Coa2 and Cox10 was observed in co-IP studies. Cox10 functions in heme a biosynthesis together with Cox15. Since bacterial Cox10 and Cox15 orthologs were reported to associate within a complex (6), we tested whether the Cox10 complex was abrogated in cells lacking Cox15 (Fig. 6A). COX15 was deleted in cells chromosomally tagged at the COX10 locus. The Cox10 BN complex persisted in cox15Δ cells, suggesting that Cox15 is not a stable component of the high-mass Cox10 complex. This conclusion is substantiated by the observation that Cox10 and Cox15 form distinct high-mass complexes on BN-PAGE that migrate at different sizes (Fig. 6B). The N196K Cox10 mutant had no effect on the abundance of the high-mass Cox15 BN complex (Fig. 6C), suggesting that the mutant Cox10 has a specific enhancement effect on the Cox10 BN complex.
FIG. 6.
Cox10 and Cox15 form distinct high-molecular-weight complexes. (A) Mitochondria (100 μg protein) isolated from WT or cox15Δ cells containing an endogenously tagged 13-Myc COX10 gene were solubilized in buffer containing 1.5% digitonin. The lysates were run on a continuous 5 to 13% gradient gel, and protein complexes were separated by BN-PAGE. The Cox10-13Myc complex was detected by immunoblotting with anti-Myc antibody. Molecular mass markers are indicated. (B) Purified mitochondria from cells containing either a genomically tagged 13-Myc COX10 gene or 13-Myc COX15 gene were solubilized (100 μg and 30 μg mitochondrial protein, respectively) and analyzed by BN-PAGE as described for panel A. (C) Mitochondria (35 μg protein) isolated from strains containing a genomically tagged 13-Myc COX15 gene expressing either WT Cox10 or Cox10-N196K were solubilized and analyzed by BN-PAGE as described for panel A. The Cox10-13Myc and Cox15-13Myc complexes were detected by immunoblotting with anti-Myc antibody. (D) coa2Δ cells transformed with COA2, YEp COX15, and/or YEp YAH1 were grown in SC-2% raffinose-0.2% glucose selective medium, serially diluted, and drop tested on SC-2% glucose and SC-2% glycerol-2% lactate. Plates were at 30°C for 2 days (glucose) or 5 days (glycerol/lactate).
Cells stalled in CcO biogenesis show limited levels of heme o. Overexpression of COX15 along with the YAH1 ferredoxin was shown to elevate heme a levels by greater than 50-fold (4). To assess whether high heme a levels were sufficient to suppress the respiratory deficiency of coa2Δ cells, mutant cells containing both genes on high-copy-number vectors were tested for growth on glycerol/lactate (Fig. 6D). Elevated levels of both proteins failed to impart respiratory growth. Thus, the robust suppressor activity of N196K Cox10 relates to a function of Cox10 in addition to its catalytic role in heme o formation.
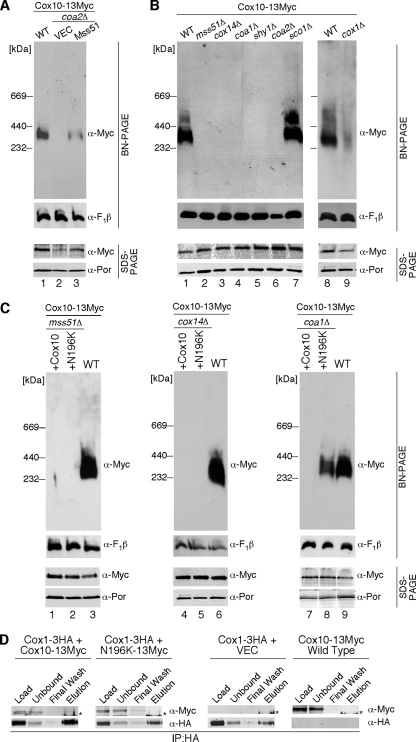
The high-mass complexes of Mss51, Coa1, and Shy1 contain Cox1, so Cox1 is a third candidate component of the ∼370-kDa Cox10 complex. Although no general Cox1 stabilization effect was observed with the N196K mutant Cox10, Cox10 may interact with Cox1 during the insertion of heme a. The lack of the high-mass Cox10 complex in coa2Δ cells may arise from Cox1 degradation. We tested whether the high-mass Cox10 complex was restored in coa2Δ cells overexpressing Mss51 to stimulate Cox1 translation. As can be seen in Fig. 7A (lane 3), elevated levels of Mss51 partially restored steady-state levels of the high-mass Cox10 BN complex. This result implies a link between Cox1 and the Cox10 complex. To test the involvement of early Cox1 assembly intermediates in the oligomeric Cox10 complex, cells containing chromosomally Myc-tagged COX10 were used to construct deletions in MSS51, COX14, COA1, SHY1, or SCO1. The Cox10 BN complex was abrogated in all mutant strains other than sco1Δ cells (Fig. 7B). Cells lacking Mss51 fail to translate full-length Cox1, but cells lacking either Cox14 or Coa1 synthesize Cox1; however, the protein is unstable (28). The oligomeric Cox10 complex is largely attenuated in cox1Δ cells, but a residual complex persists. To assess whether the oligomeric complex of N196K mutant Cox10 is dependent on Cox1, WT and mutant Cox10 were expressed in cells lacking Mss51, Cox14, or Coa1 (Fig. 7C). Whereas neither form of Cox10 was competent to form stable oligomeric complexes in mss51Δ and cox14Δ cells, the oligomeric N196K mutant Cox10 complex was seen in coa1Δ cells. Levels of newly synthesized Cox1 are comparable in cox14Δ and coa1Δ cells, yet Cox10 is responsive to Cox1 levels only in coa1Δ cells. No direct stable interaction of Cox10 and Cox1 is observed. HA-tagged Cox1 did not co-IP with Myc-tagged Cox10 (Fig. 7D), and two-dimensional BN-PAGE failed to show comigration of Cox10 and Cox1 (data not shown). Although a co-IP of Cox10 and Cox1 was not observed, we did reproduce the known interactions of Coa1 with Cox1 and of Shyl with Cox1 (data not shown). Thus, formation of the high-mass Cox10 complex is linked to Cox1 translation and formation of the Mss51/Cox14-containing Cox1 complex, although no direct interaction between Cox10 and Cox1 exists that is stable to co-IP.
FIG. 7.
Cox1 is not a stable component of the Cox10 high-molecular-weight complex. (A) Isolated mitochondria (100 μg protein) from WT, coa2Δ, and coa2Δ cells expressing YEp MSS51 containing an endogenously tagged 13-Myc COX10 gene were solubilized and analyzed by BN-PAGE as described previously. Detection of the F1β subunit of monomeric complex V was used as a loading control. The lower panel represents purified mitochondria (30 μg protein) from the same strains analyzed under denaturing (SDS-PAGE) conditions. Anti-Myc antibody was used to detect Cox10-13Myc, and Por1 was used as a loading control. (B) Mitochondria purified from WT, mss51Δ, cox14Δ, coa1Δ, shy1Δ, coa2Δ, sco1Δ, and cox1Δ strains containing chromosomally tagged COX10-13Myc were analyzed by both native (BN, 100 μg mitochondrial protein) and denaturing (SDS, 30 μg mitochondrial protein) PAGE using anti-Myc antibody. (C) Mitochondria (100 μg protein) from chromosomally tagged COX10-13Myc strains with mss51Δ (lanes 1 and 2), cox14Δ (lanes 4 and 5), or coa1Δ (lanes 7 and 8) expressing WT Cox10 or mutant N196K Cox10 were analyzed by BN-PAGE and SDS-PAGE as described previously. (D) Cox1 and Cox10 do not have a stable interaction. Mitochondria (350 μg protein) from COX1-3HA cells, COX1-3HA cells expressing YCp WT Cox10, or Cox10-N196K and COX10-13Myc cells were solubilized in IP buffer, and clarified extracts were immunoprecipitated with rabbit polyclonal anti-HA beads. All co-IP samples were analyzed by SDS-PAGE and immunoblotting as indicated. Details are as described for Fig. 6B. The asterisk indicates the rabbit immunoglobulin G heavy chain detected with the secondary antibody used in the analysis. Similar co-IP results were obtained using mouse monoclonal Myc-conjugated beads.
An unbiased proteomic approach was taken to identify components in the high-mass Cox10 BN complex. Digitonin-solubilized mitochondrial supernatant isolated from chromosomally tagged Cox10 cells was used for immunoprecipitation (IP) by anti-Myc agarose beads. Resolution of the IP on SDS-PAGE followed by mass spectrometry of the silver-stained bands revealed Cox10 and prohibitin subunits (Phb1 and Phb2) and the mAAA subunits (Yta10 and Yta12). Cox1 was not observed. The prohibitin and mAAA components are apparently nonspecific IP products, as prohibitin subunits also were recovered with control beads and mAAA associates with the prohibitin complex (33).
Cox1 degradation in coa2Δ cells by the Oma1 metallopeptidase.
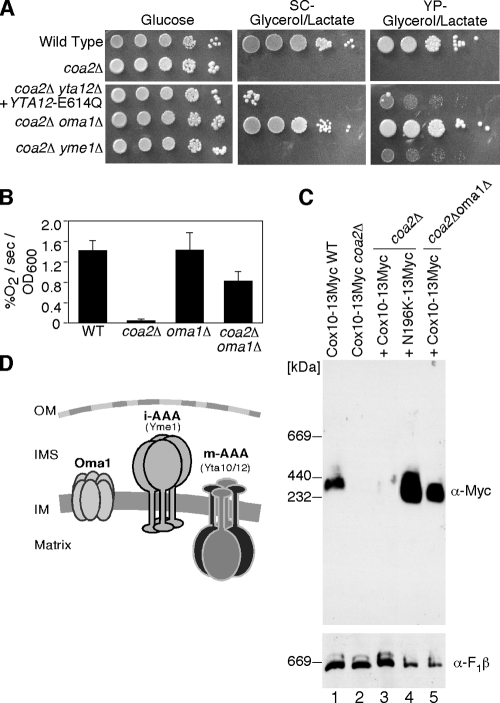
One dramatic effect of the N196K Cox10 mutant is the abrogation of the facile degradation of Cox1 in coa2Δ cells. The final question addressed is whether full respiratory activity could be restored in coa2Δ cells if Cox1 degradation was impaired. The mAAA is the most obvious candidate IM protease, although our previous studies with coa2Δ cells containing a compromised mAAA protease showed only weak respiratory growth (29). Two other protease systems besides mAAA reside within the mitochondrial IM, iAAA (Yme1) and Oma1 (Fig. 8D). The iAAA has its catalytic activity facing the intermembrane space (IMS) side of the IM and has a role in Cox2 degradation (14, 15). Oma1 is a conserved metallopeptidase that functions with the mAAA protease in the degradation of misfolded polytopic IM proteins (18). To assess whether the other IM-associated proteases contribute to the rapid Cox1 degradation seen in coa2Δ cells, we constructed double-deletion strains in which OMA1 or YME1 was disrupted in the coa2Δ background (Fig. 8A). The absence of Yme1 failed to markedly enhance respiratory growth of coa2Δ cells, but robust respiratory growth was achieved by the deletion of OMA1. Respiratory activity as measured by oxygen consumption was restored to ∼50% of the WT level in coa2Δ oma1Δ cells (Fig. 8B), and Cox1 protein levels were restored in the in vivo mitochondrial translation assay (data not shown). The high-mass BN Cox10 complex was also restored in the double-null cells (Fig. 8C). Thus, respiratory function can be restored in coa2Δ cells either by the suppressor activity of mutant Cox10 or by selectively impaired proteolysis. A modest synergism exists between the two mechanisms. Respiratory growth of coa2Δ oma1Δ cells is enhanced by the expression of the N196K mutant Cox10 (data not shown).
FIG. 8.
Suppression of the coa2Δ respiratory defect by deletion of OMA1. (A) WT cells, coa2Δ cells, coa2Δ yta12Δ cells transformed with YCp Yta12(E614Q), coa2Δ oma1Δ cells, and coa2Δ yme1Δ cells were grown in SC-2% raffinose-0.2% glucose, serially diluted, and spotted onto SC-2% glucose, SC-2% glycerol-2% lactate, and yeast extract-peptone-2% glycerol-2% lactate. The plates were incubated at 30°C for 2 days (glucose) or 6 days (glycerol-lactate). (B) WT, coa2Δ, oma1Δ, and coa2Δ oma1Δ cells were grown overnight in SC-1% glucose, and oxygen consumption (% O2/second/OD600 unit) was measured. The data represent the averages of three independent repeats, and the error bars represent standard errors of the means. (C) Purified mitochondria (100 μg protein) from WT or coa2Δ cells expressing an endogenously tagged COX10-13Myc, coa2Δ cells expressing WT Cox10-13Myc or N196K Cox10-13Myc, or coa2Δ oma1Δ cells expressing N196K Cox10-13Myc were solubilized in buffer containing 1% digitonin. Lysates were run on a continuous 5 to 13% gel, and complexes were separated by BN-PAGE. Cox10-13Myc was detected by immunoblotting with anti-Myc antisera, and monomeric complex V detected with anti-F1β antisera was used as a loading control. (D) A schematic of mitochondrial IM proteases. The mAAA protease is composed of Yta10 and Yta12, the iAAA protease is a homo-oligomer of Yme1, and Oma1 is proposed/speculated to be a hexamer based on previously published data (18).
DISCUSSION
Cells lacking Coa2 are stalled in CcO biogenesis and exhibit a rapid degradation of newly synthesized Cox1, leading to impaired CcO biogenesis. The respiratory deficiency of coa2Δ cells is suppressed either by the presence of a mutant allele of the Cox10 farnesyl transferase or by impairing proteolysis of newly synthesized Cox1 through the disruption of the Oma1 IM protease. The N196K mutant Cox10 is a gain-of-function and specific suppressor of coa2Δ cells. Whereas high-copy-number WT Cox10 has limited suppressor activity in restoring respiratory growth to coa1Δ and shy1Δ cells, the mutant Cox10 was no more effective than the WT protein in those CcO assembly mutants. The N196K substitution did not attenuate Cox10 catalytic activity, since the mutant protein was fully functional when expressed in cox10Δ cells. The mutant Cox10 restores near-WT levels of oxygen consumption in coa2Δ cells and Cox1 levels in the mitochondrial translation assay.
To assess the mechanism of suppression by the N196K Cox10 in coa2Δ cells, we showed that the mutant Cox10 lacked any general Cox1 chaperone activity. The mutant Cox10 failed to stabilize newly synthesized Cox1 in two CcO assembly mutants, coa1Δ and cox4Δ cells. The mutant and WT Cox10 were similar in having only a modest effect in enhancing the hydrogen peroxide sensitivity of cox11Δ cells arising from the accumulation of a prooxidant heme a3-Cox1 assembly intermediate. If the mutant Cox10 had a general stabilizing effect on Cox1, an effect would be evident in these assays. The lack of a general stabilization effect on Cox1 was substantiated by the inability to detect any stable interaction between Cox10 and Cox1.
The suppressor function of the N196K Cox10 requires the catalytic function of Cox10. An H317A mutation that abrogates Cox10 catalysis also blocks suppressor activity of the N196K Cox10. Likewise, the stabilization of Cox1 in the pulse phase of the mitochondrial translation assay imparted by the N196K Cox10 is dependent on a functional Cox15. Heme a biosynthesis is critical for the suppressor activity, although high-copy-number coexpression of Cox15 and the Yah1 ferredoxin, which is known to stimulate heme a formation in CcO mutants (4), lacked suppressor activity.
Cox10 forms a high-mass ∼370-kDa oligomeric complex as visualized with BN gels. The abundance of the Cox10 high-mass complex seen on BN gels correlates with its suppressor activity. The complex is highly abundant in cells harboring the N196K mutant relative to the WT protein. The complex is intermediate in abundance in cells with a N196A Cox10 substitution, which has weaker suppressor activity than the N196K mutant protein. Suppressor activity and formation of the high-mass Cox10 complex are abrogated with an N196D substitution. Formation of the high-mass Cox10 complex is not dependent on a catalytically active enzyme as shown with the N196K,H317A mutant Cox10. Whereas the N196K Cox10 mutant has a marked effect on the abundance of the Cox10 BN complex, the mutant protein has no effect on the abundance of the high-mass Cox1-containing Coa1 or Shy1 complexes. Thus, the effect of the N196K mutant is restricted to the Cox10 complex.
Heme a biosynthesis involves both Cox10 and Cox15. Cox15 is present at nearly 10-fold-higher levels than Cox10 in yeast (37). Therefore, Cox10 may be the limiting factor in generation of heme a. We postulate that Cox10 has a key role in Cox1 hemylation besides its catalytic role in forming heme o. Cox10 appears to sense newly synthesized Cox1, although Cox10 does not stably interact with Cox1. Formation of the Cox10 oligomeric complex is dependent on Cox1 at an early step in Cox1 maturation. Cells lacking Cox14 fail to form the high-mass Mss51/Cox14-containing Cox1 complex (3, 30) and are unable to form the Cox10 oligomeric complex despite having WT levels of newly synthesized Cox1. In contrast, cells lacking Coa1 retain a WT Mss51/Cox14 complex and show an oligomeric Cox10 complex with the N196K mutant Cox10 but not the WT protein. Thus, formation of the oligomeric Cox10 complex with the WT protein appears to be dependent on the Coa1-containing Cox1 complex, but the mutant protein can oligomerize in the absence of Coa1. Coa1 may have a role in promoting Cox10 oligomer formation. An attractive postulate is that the Cox10 catalytic function is dependent on oligomerization, such that Cox10 activity is attenuated in cells lacking Coa1 or the upstream Mss51/Cox14 factors. Previously it was reported that heme o formation was dependent on the presence of Cox15 (4), although the mechanism by which Cox15 modulates Cox10 remains unknown. Formation of the potentially deleterious heme a moiety may be temporally coordinated with formation of a Cox1 assembly intermediate. This may be one mechanism to minimize the deleterious effects of stalled Cox1 assembly intermediates. Induced heme o synthesis may rapidly lead to heme a production by the abundant Cox15 followed by heme a insertion into Cox1. No information exists on whether heme a insertion is directly facilitated by Cox15 or another protein.
The marked instability of Cox1 seen in coa2Δ cells may arise in part from an impaired addition of heme a. Cox1 spans the IM with 12 transmembrane (TM) helices organized in six successive helical pairs forming a closed bundle (34). The heme a farnesyl tail is packed between helices 1, 11, and 12. The axial His ligands for heme a ligation come from His62 in helix 2 and His378 in helix 10. Thus, heme a binding is expected to stabilize the Cox1 helical bundle. Impaired formation of the heme a-Cox1 complex would therefore be expected to destabilize Cox1, leading to rapid degradation. The N196K Cox10 may specifically overcome the impaired hemylation of Cox1 in coa2Δ cells, and this may explain why the mutant Cox10 has no gain-of-function activity in other CcO assembly mutants.
The N196K substitution in Cox10 has a dramatic effect on the Cox10 function. Whereas a double Arg substitution in Cox10 abrogates function, activity is restored by the addition of the N196K substitution. The two Arg residues (R212 and R216) are present in a matrix-facing loop of Cox10 between TM helices 2 and 3, and Asn196 lies at the end of TM2 near the matrix boundary. The introduced Lys in the N196K substitution may restore an important positive charge needed in catalysis. The conserved Arg residues are implicated in the binding of the pyrophosphate moiety of the farnesyl pyrophosphate substrate of the Cox10-related CyoE heme o synthase of E. coli (31). Further mechanistic details on this effect are lacking, since no structural information on Cox10 is available.
Curiously, an identical mutation corresponding to N196K was reported in human Cox10 (N204K) from two related patients with a CcO deficiency disorder (36). Lymphocytes harboring the mutant human protein showed residual CcO activity, and the mutant protein restored respiratory growth in cox10Δ yeast cells when overexpressed but only limited growth on glycerol medium in the absence of overexpression. In contrast to the yeast N196K Cox10, the human N204K mutant Cox10 was only a weak suppressor of coa2Δ cells (data not shown).
The second major mechanism of suppression of the respiratory defect in coa2Δ cells is through depletion of the Oma1 metallopeptidase. The depletion of Oma1 restored the WT Cox10 BN complex and restored Cox1 levels seen in the mitochondrial translation assay. Oma1 was shown to function with the mAAA protease in the degradation of a misfolded polytopic protein, Oxa1 (18). The catalytic center of Oma1 appears to reside on the matrix side of the IM, although it mediated an Oxa1 cleavage site also on the IMS side of the IM. The present work clearly documents that Oma1 has a significant role in the Cox1 instability in coa2Δ cells. The likely Oma1 substrate is Cox1 and not Cox10, since overexpression of MSS51 partially restores the BN Cox10 complex in coa2Δ cells yet fails to promote respiratory growth. In addition, only the BN Cox10 complex is depleted in coa2Δ cells; steady-state Cox10 levels remain normal. Thus, the observed depletion of the high-mass Cox10 complex may arise merely from Cox1 degradation of the Coa1-containing Cox1 complex. Impaired hemylation of Cox1 may lead to a misfolded Cox1 that is an efficient substrate for Oma1. We previously demonstrated that weak respiratory growth in coa2Δ cells is induced in cells with an attenuated mAAA. The reported proteolytic degradation of the misfolded Oxa1 was also facilitated by a combination of Oma1 and the mAAA protease (18). Future studies will address the substrate specificity of Oma1 and whether Oma1 depletion has similar effects on other CcO assembly mutants. Oma1 is conserved in metazoans, so future studies will also address its role in Cox1 degradation in human CcO deficiency mutants.
The two mechanisms of suppression of coa2Δ cells have implications for the physiological function of Coa2 in CcO biogenesis. The robust suppressor function of N196K Cox10 suggests that Coa2 functions in part in a step related to heme a addition to Cox1. Cells lacking Coa2 fail to form the oligomeric Cox10 complex as well as the Cox1 assembly intermediates containing Coa1 and Shy1. Coa2 may impart stability to the oligomeric Cox10 complex. It is unlikely that Coa2 imparts stability to the Coa1-containing Cox1 complex, as Coa2 and Coa1 do not interact. In contrast, Coa2 forms a transient interaction with Shy1 and may contribute to its stability (29) as well as the Cox10 complex. Shy1 stabilizes the Cox1 assembly intermediate in which the CuB-heme a3 bimetallic center is formed (29). In the absence of Coa2, formation of the heme a and CuB-heme a3 centers occurs to a limited extent, leading to low levels of the prooxidant heme a3-Cox1 intermediate that confers weak hydrogen peroxide sensitivity. Thus, Coa2 is not essential for formation of the redox centers. Future studies will address whether Coa1 and/or Coa2 mediates Cox10 oligomerization.
A second function of Coa2 involves the apparent stabilization of Cox1 during its maturation. This may arise either from a direct effect on Cox1 or indirectly by modulating a protease such as Oma1. Newly synthesized Cox1 is less stable in coa2Δ cells than in cox10Δ cells. Although Cox1 appears to be rapidly degraded in coa2Δ and cox10Δ cells (Fig. 1F), two other studies reported the appearance of newly synthesized Cox1 in mitochondrial translation assays performed on cox10Δ cells (3, 25). The differences in Cox1 levels seen during the pulse phase of the translation assay in the two null strains must relate to enhanced proteolysis in coa2Δ cells. Coa2 may be a negative regulator of a protease such as Oma1. Future studies will directly assess the role of Coa2 in IM proteolysis.
Acknowledgments
We acknowledge the assistance of Greg Keller and Talina Watts in the construction of certain yeast strains and plasmids.
This work was supported by grant ES03817 from the National Institute of Environmental Health Sciences, NIH, to D.R.W. M.B. was supported by predoctoral training grant T32 DK007115 and a University of Utah Graduate Research Fellowship.
Footnotes
Published ahead of print on 19 October 2009.
REFERENCES
- 1.Barrientos, A., K. Gouget, D. Horn, I. C. Soto, and F. Fontanesi. 2009. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta 1793:97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrientos, A., D. Korr, and A. Tzagoloff. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 21:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barros, M. H., and A. Tzagoloff. 2002. Regulation of the heme a biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516:119-123. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, N. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brown, B. M., Z. Wang, K. R. Brown, J. A. Cricco, and E. L. Hegg. 2004. Heme O synthase and heme A synthase from Bacillus subtilis and Rhodobacter sphaeroides interact in Escherichia coli. Biochemistry 43:13541-13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, H. S., G. N. George, and D. R. Winge. 2002. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I) binding protein. J. Biol. Chem. 277:31237-31242. [DOI] [PubMed] [Google Scholar]
- 8.Cruciat, C. M., S. Brunner, F. Baumann, W. Neupert, and R. A. Stuart. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 275:18093-18098. [DOI] [PubMed] [Google Scholar]
- 9.Diekert, K., A. I. De Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37-51. [DOI] [PubMed] [Google Scholar]
- 10.Fontanesi, F., C. Jin, A. Tzagoloff, and A. Barrientos. 2008. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 17:775-788. [DOI] [PubMed] [Google Scholar]
- 11.Glerum, D. M., I. Muroff, C. Jin, and A. Tzagoloff. 1997. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 272:19088-19094. [DOI] [PubMed] [Google Scholar]
- 12.Glerum, D. M., and A. Tzagoloff. 1994. Isolation of a human cDNA for heme A:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proc. Natl. Acad. Sci. U.S.A. 91:8452-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glerum, D. M., and A. Tzagoloff. 1997. Submitochondrial distributions and stabilities of subunits 4, 5, and 6 of yeast cytochrome oxidase in assembly defective mutants. FEBS Lett. 412:410-414. [DOI] [PubMed] [Google Scholar]
- 14.Graef, M., and T. Langer. 2006. Substrate specific consequences of central pore mutations in the i-AAA protease Yme1 on substrate engagement. J. Struct. Biol. 156:101-108. [DOI] [PubMed] [Google Scholar]
- 15.Graef, M., G. Seewald, and T. Langer. 2007. Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell. Biol. 27:2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horng, Y.-C., S. C. Leary, P. A. Cobine, F. B. J. Young, G. N. George, E. A. Shoubridge, and D. R. Winge. 2005. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280:34113-34122. [DOI] [PubMed] [Google Scholar]
- 18.Kaser, M., M. Kambacheld, B. Kisters-Woike, and T. Langer. 2003. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 278:46414-46423. [DOI] [PubMed] [Google Scholar]
- 19.Khalimonchuk, O., A. Bird, and D. R. Winge. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282:17442-17449. [DOI] [PubMed] [Google Scholar]
- 20.Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 21.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14:4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manthey, G. M., B. D. Przybyla-Zawislak, and J. E. McEwen. 1998. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 255:156-161. [DOI] [PubMed] [Google Scholar]
- 23.Mick, D. U., K. Wagner, M. van der Laan, A. E. Frazier, I. Perschil, M. Pawlas, H. E. Meyer, B. Warscheid, and P. Rehling. 2007. ShyI couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogi, T. 2009. Probing structure of heme A synthase from Bacillus subtilis by site-directed mutagenesis. J. Biochem. 145:625-633. [DOI] [PubMed] [Google Scholar]
- 25.Nobrega, M. P., F. G. Nobrega, and A. Tzagoloff. 1990. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 265:14220-14226. [PubMed] [Google Scholar]
- 26.Ott, M., M. Prestele, H. Bauerschmitt, S. Funes, N. Bonnefoy, and J. M. Herrmann. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25:1603-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierrel, F., M. L. Bestwick, P. A. Cobine, O. Khalimonchuk, J. A. Cricco, and D. R. Winge. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierrel, F., O. Khalimonchuk, P. A. Cobine, M. Bestwick, and D. R. Winge. 2008. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28:4927-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preuss, M., K. Leonhard, K. Hell, R. A. Stuart, W. Neupert, and J. M. Herrmann. 2001. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 153:1085-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saiki, K., T. Mogi, H. Hori, M. Tsubaki, and Y. Anraku. 1993. Identification of the functional domains in heme O synthase. Site-directed mutagenesis studies on the cyoE gene of the cytochrome bo operon in Escherichia coli. J. Biol. Chem. 268:26927-26934. [PubMed] [Google Scholar]
- 32.Siep, M., K. van Oosterum, H. Neufeglise, H. van der Spek, and L. A. Grivell. 2000. Mss51p, a putative translational activator of cytochrome c oxidase subbunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet. 37:213-220. [DOI] [PubMed] [Google Scholar]
- 33.Steglich, G., W. Neupert, and T. Langer. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 19:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Hakashima, R. Yaono, and S. Yoshikawa. 1995. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8A. Science 269:1069-1074. [DOI] [PubMed] [Google Scholar]
- 35.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguichi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272:1136-1144. [DOI] [PubMed] [Google Scholar]
- 36.Valnot, I., J.-C. Von Kleist-Retzow, A. Barrientos, M. Gorbatyuk, J.-W. Taanman, B. Mehaye, P. Rustin, A. Tzagoloff, A. Munnich, and A. Rotig. 2000. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 9:1245-1249. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Z., Y. Wang, and E. L. Hegg. 2009. Regulation of the heme A biosynthetic pathway: differential regulation of heme A synthase and heme O synthase in Saccharomyces cerevisiae. J. Biol. Chem. 284:839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]