Abstract
An important difference between placental mammals and marsupials is the maturity of the fetus at birth. Placental mammals achieved this maturity by developing a complex and invasive placenta to support and prolong internal development. The exact genomic modifications that facilitated the evolution of this complex structure are unknown, but the emergence of genomic imprinting within mammalian lineages suggests a role for gene dosage. Here we show that a maximally altered placental structure is achieved by a single extra dose of the imprinted Phlda2 gene characterized by a dramatically reduced junctional zone and a decrease in stored glycogen. In addition, glycogen cells do not migrate into the maternal decidua in a timely fashion, but instead, Tpbpa-positive cells progressively mislocalize into the labyrinth. These defects are linked to a progressive restriction of embryonic growth from embryonic day 16.5. This work has identified a critical role for the imprinted Phlda2 gene in regulating glycogen storage in the eutherian placenta and implies that imprinting has provided a mechanism to boost nutrient supply to the fetus late in gestation, when the fetus is placing the highest demands on maternal resources, to enhance growth.
Distinct to mammals, embryonic growth is dependent on the ability of the mother to support in utero growth. The choriovitelline placenta initially provides access to maternal nutrients, and, as the demands of fetal growth increase, monotremes and marsupials remain dependent on the yolk sac placenta but eutherian mammals switch to an elaborate chorioallantoic placenta (22, 43). Very few genes are expressed uniquely in the placenta. The majority have arisen from existing genes by means of placenta-specific promoters, from the duplication of large gene families, or through the adoption of functions associated with endogenous retroviruses and retroelements (42). A surprising number of imprinted gene knockout models exhibit placental defects (19), suggesting gene dosage as another mechanism important in the evolution of the fetoplacental unit. Approximately 0.3% of autosomal genes are imprinted in eutherian mammals, while a subset of these genes are imprinted in marsupials with no evidence of imprinting in other vertebrates (1, 31, 32, 37, 39, 51, 54, 56, 58). Thus, the emergence of genomic imprinting coincides with the appearance of extraembryonic support, and, as the demands for this support have increased, the number of imprinted genes co-opted by the imprinting mechanism has increased (30), also suggesting the involvement of these unique genes in placental development.
The mouse placenta is organized into the histologically distinct labyrinth zone, junctional zone, giant cell layer, and maternal decidua (9-11, 27, 45, 49). The giant cells are thought to modify the maternal uterine vasculature, promoting maternal blood flow toward the implantation site, while in the labyrinth zone exchange takes place between the maternal and fetal circulation. The junctional zone, also known as the spongiotrophoblast layer, provides a source of pregnancy-related hormones (9, 35), but, although this layer is absolutely required for embryonic survival (25, 26), its function is less well understood. It is composed of two major cell types, spongiotrophoblast and glycogen cells, which both express trophoblast-specific protein alpha (Tpbpa), with the glycogen cells additionally accumulating glycogen within their cytoplasm from embryonic day 12.5 (E12.5) (5, 9). An unusual feature of glycogen cells is their migration into the maternal decidua late in gestation, where they may function to provide a rapidly mobilizable energy source during late pregnancy and parturition. Despite the amazing variety in the forms and types of eutherian placenta, easily detectable stores of glycogen are a common feature (8).
Imprinted genes located at mouse distal chromosome 7 play an important role in regulating embryonic and placental growth (16, 38). With regard to imprinting, this chromosomal region can be separated mechanistically into two distinct domains (7). Each domain contains one key gene that directly modulates embryonic growth. The IC1 domain contains the gene for the potent embryonic growth factor insulin-like growth factor 2 (Igf2) (13, 14). Global loss of expression of Igf2 directly limits embryonic growth, while Igf2 deficiency localized to the placenta indirectly restricts embryonic growth (12). The predicted consequence of imprinting Igf2 (reduced dosage) would be to limit embryonic growth. Cyclin-dependent kinase inhibitor 1C (Cdkn1c) is the major regulator of embryonic growth within the adjacent IC2 domain (2). In contrast to Igf2, imprinting of Cdkn1c would be predicted to enhance embryonic growth. Pleckstrin homology-like domain family A member 2 (Phlda2) and achaete-scute complex homolog 2 (Ascl2) also map to the IC2 region (24, 41) but primarily play a role in extraembryonic development. Ascl2 deficiency results in embryonic lethality at midgestation due to placental failure, but tetraploid rescue experiments exclude a direct role in regulating embryonic growth or adult development (25, 26, 53). Phlda2 is also predominantly expressed in the placenta from the maternal allele being expressed in syncytiotrophoblast layers II and III of the labyrinth (15, 21, 41). Phlda2 deficiency results in placentomegaly with a specific increase in the area of the junctional zone but with no overt consequence for embryonic growth or adult development (20).
A mouse model of loss of imprinting of the IC2 domain, in which several imprinted genes are overexpressed, shows placental stunting (17) and a reduction of the junctional zone (46). We previously showed, indirectly, that Phlda2 rescues the volume of the junctional zone by normalizing Phlda2 expression in these Kvdmr1+/− mice. We also showed that excess dosage of the region spanning Phlda2 and a second imprinted gene, the solute carrier family 22 member 18 gene (Slc22a18), restricts placental growth and noted a subtle and late embryonic growth restriction phenotype (46). In our transgenic model the two imprinted genes were overexpressed at high levels from three copies of a bacterial artificial chromosome (BAC), suggesting misregulated expression. Given the importance of the junctional zone in embryonic viability and the potential role of PHLDA2 in human intrauterine growth restriction (IUGR) (36), we sought to perform a more detailed characterization of the consequence of excess expression of Phlda2 and Slc22a18 in three independent transgenic lines with increasing doses of the transgene and in two genetic backgrounds. Using a single-copy transgene, we asked whether normalizing Phlda2 expression rescued the identified phenotypes, which included a unique mislocalization defect. Finally, we characterized embryonic growth from E13.5 to E18.5 in two independent lines. We identify critical roles for Phlda2 in regulating glycogen storage and in coordinating the location of spongiotrophoblast and glycogen cells late in gestation.
MATERIALS AND METHODS
Mouse strains and genotyping.
All animal studies and breeding were approved by the Universities of Cardiff ethical committee and performed under a UK Home Office project license (R.M.J.). Lines 10-15, 5D3, and Phlda2+/− were previously described (20, 29). Transgenic line 10-10 was newly generated as described for line 10-15 (29). In addition to Phlda2 and Slc22a18, the BAC transgene spans a third gene, Cdkn1c, which is inactivated by insertion of lacZ in lines 10-15 and 10-10 but not in line 5D3. The BAC does not imprint (29), and experimental material was generated by male transmission. A small portion of the tail tip or ear pinnae was removed between postnatal day 7 (P7) and P14 and genotyped as described previously (20, 29).
Weighing studies.
Embryonic and placental wet weights were taken at the stated time points after the formation of a discernible plug. Genotyping data was obtained from yolk sac DNA as described or by staining for the lacZ reporter as described previously (20, 29).
mRNA analysis.
Quantitative PCR of reverse-transcribed RNA (QRT-PCR) was performed as described previously (2). For the additional primers used see Table S3 in the supplemental material. For genes with two primer sets, fold changes represent the mean result for both primer sets. Microarray analysis was performed as described previously (36), with three transgenic and three control E16.5 placental samples.
In situ hybridization and histological and immunohistochemical analyses.
Placentas were fixed overnight in phosphate-buffered 4% paraformaldehyde (PFA) and paraffin embedded, and 10-μm sections were taken. Hematoxylin-and-eosin (H&E) staining, riboprobe preparation, and in situ hybridizations were performed as described previously (29). Cdkn1c cDNA was prepared as described previously (29), Ascl2 was obtained from Azim Surani, Prl3d and Prl3b1 cDNAs were obtained from Louis Lefebvre, Flk-1 was subcloned from a cDNA gifted by David Bates, and Gcm1 and Hand1 cDNAs were obtained from Takahiro Arima. A 546-bp Phlda2 cDNA fragment was cloned using primers 5′-ACAGCCTGTTCCAGGTATGG-3′ and 5′-GGTTGGAAGCAGGTAACCAA-3′, and an 858-bp Slc22a18 cDNA was cloned using primers 5′-AGAGCGGCACTCTCACTCTC-3′ and 5′-GGTCAATGGTTGCACAGATG-3′. Immunohistochemistry was as described previously (21). For periodic acid-Schiff (PAS) staining slides were deparaffinized, rehydrated, and treated with 1% (wt/vol) periodic acid (Sigma) in distilled H2O for 5 min at room temperature. Slides were washed in running water, incubated in Schiff's reagent (Sigma) for 10 min at room temperature, rinsed in running tap water, counterstained with Gill's no. 3 hematoxylin (Sigma), dehydrated, and mounted by reversing the rehydration steps.
Cell proliferation by BrdU incorporation.
Pregnant females were injected with 100 μg/g bromodeoxyuridine (BrdU; Amersham) 3 h before dissection of embryos. Placentas were fixed for 2 h in 4% PFA at 4°C and paraffin embedded, and 5-μm sections were prepared. Slides were dewaxed in xylene and rehydrated through graded ethanols, submerged in 1× citrate buffer (LabVision), and heated at full power in a microwave for 15 min. Slides were cooled and blocked for 20 min in Peroxidase Block (Envsion) and 30 min in 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Slides were incubated overnight at 4°C with anti-BrdU antibody (BD) diluted 1:150 in 1% BSA in PBS. Antibody solution was washed off in PBS, and slides were incubated with horseradish peroxidase-labeled polymer (Envision) for 1 h at room temperature. A 3,3′-diaminobenzidine chromogen and substrate mixture was applied to the slides and allowed to develop for 10 min. Slides were counterstained in Mayer's hemalum, dehydrated, cleared, and mounted in DPX mounting medium.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay.
Apoptosis was examined on midline paraffin-embedded placenta sections at E10.5 and E12.5 using the DeadEnd Colorimetric TUNEL system (Promega) according to the manufacturer's instructions.
Biochemical determination of placental glycogen concentration.
Glycogen was extracted from whole placenta according to the method of Lo et al. (34) and resuspended in 1 ml of H2O. Glycogen extract was diluted 1:60 in H2O, and the glycogen concentration was determined using a glycogen assay kit (BioAssay Systems) according to the manufacturer's instructions.
Statistical analyses.
Statistical significance (probability values) was determined using the Student t test (two-tailed distribution and two-sample unequal variance) or the Mann-Whitney U test. The significance of the difference in observed over the expected appearance of a particular genotype was determined using the chi-square test.
RESULTS
A twofold increase in Phlda2 expression produces maximal placental stunting.
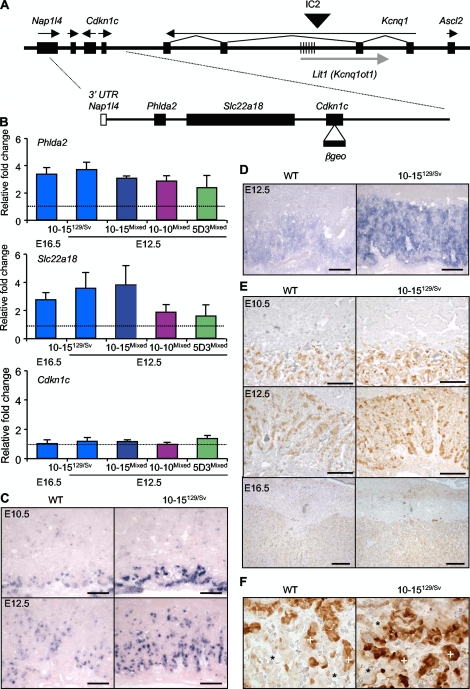
In our previous study three copies of a modified BAC transgene which spanned two intact genes, Phlda2 and Slc22a18 (Fig. 1A), caused a severe placental growth restriction phenotype (46). Phlda2 and Slc22a18 transcript levels did not correlate with the number of transgene copies, suggesting that the phenotype might be due to highly misregulated expression. Breeding this three-copy line for a further eight generations into the 129/Sv genetic background stabilized the expression of Phlda2 and Slc22a18 to approximately 4 times the endogenous level at E12.5 and E16.5 (Fig. 1B). Similar results were obtained in a mixed 129/Sv × C57BL/6 background (Fig. 1B). A newly generated line, 10-10, carrying the same modified transgene expressed Phlda2 at 2.8 times and Slc22a18 at 1.8 times the wild-type levels, and a third line 5D3, which carried a single copy of an unmodified version of the BAC transgene (2), expressed Phlda2 and Slc22a18 at 2.3 and 1.6 times the wild-type levels, respectively (Fig. 1B). The Cdkn1c placental enhancers are not encompassed by the BAC (20, 29), and Cdkn1c was not expressed from the transgene in the placenta in any line (Fig. 1B), thus uniquely isolating the consequences of excess Phlda2 and Slc22a18 in the placenta.
FIG. 1.
Increasing dosages of Phlda2 and Slc22a18 with precise temporal and spatial expression. (A) Schematic of the BAC transgene. The top line is a genomic map of the IC2 region on distal mouse chromosome 7. The hatched region marks KvDMR1, which is methylated only on the maternal allele. Arrows indicate the direction of transcription. Below is a map of the 85-kb transgene (BAC144D14). Filled boxes show the positions of the intact genes. In addition to Phlda2, Slc22a18, and Cdkn1c, the transgene includes the 3′ untranslated region (UTR) of Napl14 (white box) but not the 5′ untranslated region. The modified BAC used to generate lines 10-15 and 10-10 has a β-galactosidase-neomycin (βgeo) fusion gene inserted into Cdkn1c, indicated by the arrow and filled box. (B) Placental QRT-PCR results for Phlda2 (top), Slc22a18 (middle), and Cdkn1c (bottom). Data at E12.5 and E16.5 from the three-copy line 10-15 in a pure 129/Sv (>99% 129/Sv) or a mixed (129/Sv × C57BL/6) genetic background demonstrates the stabilization of expression of Phlda2 and Slc22a18 to approximately 4 times the endogenous level. Data from the single-copy line 5D3 and the newly generated line 10-10 in a mixed genetic background at E12.5 demonstrate approximately two- and threefold increased expression of Phlda2 and Slc22a18. Consistent with the absence of Cdkn1c placental enhancers on the transgene, Cdkn1c expression was comparable in wild-type (WT) and transgenic placentas in all three lines. (C) In situ hybridization with a Phlda2 riboprobe reveals similar spatial localizations of mRNA in control and transgenic 10-15129/Sv placentas at E10.5 and E12.5. (D) In situ hybridization with a Slc22a18 riboprobe reveals similar spatial localizations of mRNA in control and transgenic 10-15129/Sv placentas at E12.5. (E) Localization of Phlda2 protein at E10.5, E12.5, and E16.5 in both control and transgenic placentas with a similar pattern. Scale bar, 200 μm (×4 magnification). (F) Phlda2 protein localization at ×40 magnification. Asterisks indicate the large but inconspicuous type I trophoblast cells lining the maternal vessels, which are not stained and do not express Phlda2 in either wild-type or transgenic samples. Plus signs indicate the Phlda2-positive clusters of type II cells in the middle layer of the trilaminar trophoblast, and the type III trophoblast cells lining the fetal vessels (not marked) are weakly positive for Phlda2.
Phlda2 and Slc22a18 mRNAs were similarly localized to the labyrinth in control and transgenic placentas, with increased signal intensity in transgenic placentas reflecting the increased dosage of both genes (Fig. 1C and D). The spatial distribution of the Phlda2 mRNA partially mirrored that of Slc22a18, suggesting that they may share enhancers which, unlike the placental enhancers for Cdkn1c, were present within the transgene. The Phlda2 protein in the transgenic placentas correctly localized to the ectoplacental cone (Fig. 1E) and specifically to type II (strong staining) and type III (weak staining) trophoblast cells in the trilaminar labyrinthine layer (Fig. 1E and F). Type I trophoblast cells were Phlda2 negative in both the control and the transgenic placentas (Fig. 1F), demonstrating that the BAC transgene recapitulated both the gross and the microscopic Phlda2 expression profiles. This allelic series is therefore a useful tool for defining the phenotypic consequences of increasing the dosage of Phlda2 and Slc22a18 from the normal single dose to approximately two (line 5D3), three (line 10-10), and four (line 10-15) doses while maintaining the correct spatial and temporal pattern of expression of these genes in the placenta.
In our previous study transgenic placentas with >10-fold overexpression of Phlda2 and Slc22a18 were 25% and 17% lighter at E14.5 and E16.5, respectively. We examined placental wet weights in the pure 129/Sv background, where expression was 4 times the endogenous level from E12.5 to E18.5 (Fig. 2A). Transgenic placentas were 15.4% lighter than wild-type placentas at E12.5, and the difference increased to a maximal 26.2% at E13.5. Wild-type placentas reached their maximum weight at E14.5, exhibiting an increase of approximately 50% between E12.5 and E14.5, with the majority of this weight gain occurring from E13.5. Wild-type placental weight did not significantly alter between E14.5 and E18.5, but the weight of transgenic placentas increased by approximately 10% between E16.5 and E18.5 such that the weight difference at E18.5 (9%) was considerably less than at E14.5 (15%).
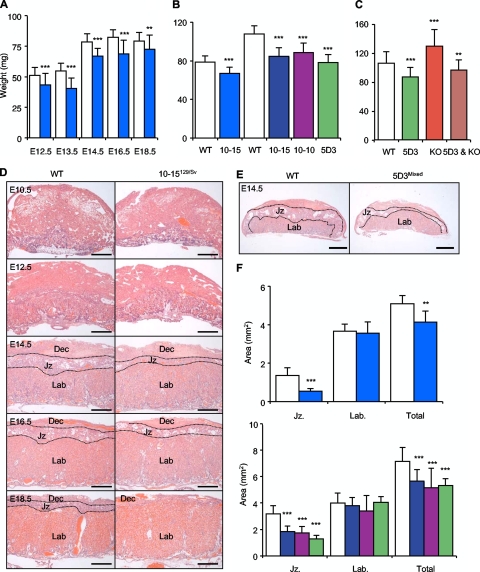
FIG. 2.
A double dose of Phlda2 induces the maximal reduction in the junctional zone irrespective of the genetic background. (A) Placental wet weights for the three-copy transgenic line 10-15 in a pure 129/Sv genetic background (see Table S1 in the supplemental material). Light blue columns, 10-15129/Sv transgenics. (B) Placental wet weights in a mixed (129/Sv × C57BL/6) genetic background. Line 10-15, line 10-10, and line 5D3 transgenic placentas show similar degrees of placental stunting at E14.5 (and E18.5; see Table S2 in the supplemental material). (C) Placental wet weights in a C57BL/6 genetic background for single-copy line 5D3, Phlda2−/+ null, and double-transgenic (5D3 and Phlda2−/+) placentas. (D) H&E staining for the three-copy transgenic line 10-15129/Sv from E10.5 to E18.5 illustrating a progressive loss of the junctional zone. Jz, junctional zone; Lab, labyrinth; Dec, maternal decidua. Scale bar, 500 μm. (E) H&E staining for the single-copy transgenic line 5D3Mixed at E14.5 illustrating a similar restriction of the junctional zone. Scale bar, 500 μm. (F) Junctional zone and labyrinth areas at the placental midline at E14.5. The line 10-15129/Sv (three copy, filled columns) mean junctional zone area of transgenic placentas is 59.9% less than the wild-type (WT) area with no significant effect on labyrinth area and a combined area reduction of 19%. Data for the mixed background are shown below. The line 10-15Mixed (dark blue columns) junctional zone area is 57% of the wild-type area with no significant effect on the labyrinth area and a combined area reduction of 21%. The line 10-10Mixed (purple columns) junctional zone area is 54.9% of the wild-type area with no significant effect on the labyrinth area and a combined area reduction of 25%. The line 5D3Mixed (single copy, green columns) junctional zone area is 60% less with no significant effect on the labyrinth area and a combined area reduction of 30% (see Table S2 in the supplemental material).
In order to make a precise comparison of placental stunting between 10-15129/Sv and the other transgenic lines that were not in the 129/Sv background, we first sorted to exclude the possibility of strain differences in the severity of placental stunting by examining placental weights in a mixed genetic background (129/Sv × C57BL/6). The overall weights of both the wild-type and 10-15 transgenic placentas were greater in the mixed background than in the pure 129/Sv background, but the degrees of stunting were similar (Fig. 2B). Lines 10-10Mixed and 5D3Mixed were stunted to similar extents (Fig. 2B; see Table S1 in the supplemental material). Thus, a single extra dose of the transgene was sufficient to induce the severest placental stunting, and normalizing the Phlda2 dosage in this single-copy line by introducing a targeted deletion of the maternal Phlda2 allele restored placental weights to the range of the wild type (106.0 ± 15.9 versus 96.8 ± 13.8 mg; wild type = 53, Phlda2−/+ plus 5D3 = 52; P = 0.002), excluding a major role for Slc22a18 in placental stunting (Fig. 2C).
Reduction in placental weight reflects a specific loss of the junctional zone.
Kvdmr1+/− mice, which overexpress Phlda2 in addition to Cdkn1c, Ascl2, and five other genes, exhibit a reduction in the junctional zone to labyrinth ratio at E14.5. Histological examination revealed a similar deficiency of the junctional zone in line 10-15129/Sv (Fig. 2D). There was a 2.3-fold reduction in the volume ratio of the junctional zone to the labyrinth at E14.5 (0.37 ± 0.056 versus 0.16 ± 0.025; P = 0.029) (see Table S2 in the supplemental material). The volume ratio was similar to the area ratio when measurements were made at the placental midline at E14.5 (2.4-fold reduction; 0.38 ± 0.13 versus 0.16 ± 0.05; P = 4.4 × 10−4) (Fig. 2C; see Table S2 in the supplemental material) and at E16.5 (2.4-fold reduction; 0.28 ± 0.06 versus 0.12 ± 0.04; three litters; n = 21; P = 5.67 × 10−6) (see Table S2 in the supplemental material). Consistent with the weight data the area of junctional zone loss was comparable (60% of the wild-type area) in the mixed genetic background for all three lines at E14.5 with no significant loss of the labyrinth zone (Fig. 2E and F; see Table S2 in the supplemental material).
Strain differences in the ratio of the junctional zone to the labyrinth zone.
We noted an absolute difference in placental wet weight between pure wild-type 129/Sv placentas and those from the 129/Sv × C57BL/6 cross, with the later being 38% heavier at E14.5 (78.7 ± 12.5 versus 108.6 ± 9.2 mg; wild-type placentas from 19 litters; n = 73; P = 2.10 × 10−18) and 45% heavier at E18.5 (79.6 ± 6.9 versus 115.4 ± 7.2 mg; wild-type placentas from 22 litters; n = 82; P = 7.22 × 10−37). The increased weight of placentas in a mixed genetic background appeared to be due entirely to the presence of a greater junctional zone area, which was 2.34 times greater in placentas in the mixed genetic background than in those in the 129/Sv background (1.36 ± 0.40 versus 3.17 ± 0.61 mm2; wild-type placentas from 11 litters; n = 37; P = 3.29 × 10−6). In contrast, there was no significant difference in the area of the labyrinth region (3.66 ± 0.36 versus 3.85 ± 0.79 mm2; P = 0.104). The difference in the combined area of the junctional zone and labyrinth region was comparable to the weight difference between the two genetic backgrounds, with a 37% increased area in mixed genetic background placentas (5.10 ± 0.41 versus 7.00 ± 1.04 mm2; P = 7.17 × 10−5).
Phlda2 limits glycogen storage and drives a progressive mislocalization of Tpbpa-positive cells into the labyrinth.
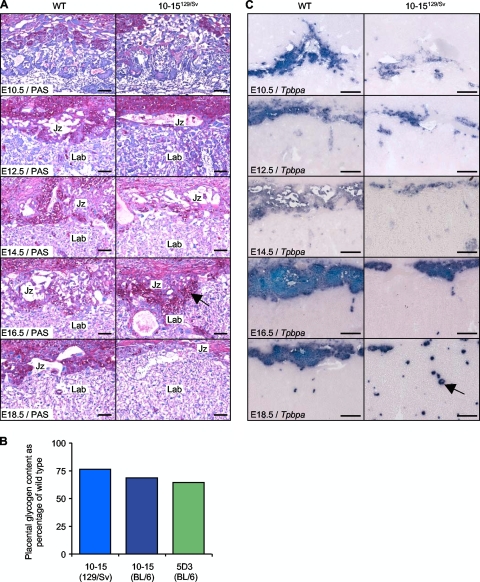
The distribution of glycogen was studied by PAS staining. PAS-positive cells were observed in the junctional zone at E12.5 in both control and transgenic placentas, but staining was markedly reduced at this time point and throughout gestation in the transgenic placenta (Fig. 3A), the reciprocal of Phlda2 deficiency. We also noted a second critical difference in glycogen staining. While the glycogen cells of wild-type placentas predominantly appeared to have migrated into the maternal decidua at E16.5, glycogen staining of transgenic placentas persisted in the junctional zone. A biochemical determination of glycogen was performed at E14.5 for line 10-15 in the 129/Sv and C57BL/6 backgrounds and for the single-copy line 5D3, which revealed a 25 to 35% loss of stored glycogen (Fig. 3B).
FIG. 3.
Mislocalization defects revealed by PAS staining and in situ hybridization analysis of Tpbpa. (A) PAS staining of midline placental sections from E10.5 to E18.5 reveals reduced glycogen staining in the transgenic junctional zone until E16.5. Glycogen staining locates to the maternal decidua in control placenta but not transgenic placenta at E16.5, as indicated by the black arrow. Scale bar, 100 μm. (B) Direct biochemical determination of stored glycogen at E14.5 shown as a percentage of wild-type (WT) values. Actual values: 10-15129/Sv, 5.13 ± 1.37 versus 3.92 ± 0.99 mg/g placenta (n = 25, P = 0.0148); 10-15Mixed, 8.92 ± 2.12 versus 6.13 ± 1.62 mg/g placenta (n = 30, P = 3.28 × 10−4); 5D3BL6, 11.44 ± 2.37 versus 7.39 ± 1.27 mg/g placenta (n = 29, P = 5.96 × 10−10). (C) Hybridization of adjacent midline placental sections from E10.5 to E18.5 with the junctional zone antisense riboprobe Tpbpa. Mislocalized patches of Tpbpa-positive cells within the labyrinth are indicated by the black arrow. Scale bar, 200 μm.
In situ hybridization for the ectoplacental cone and junctional zone marker Tpbpa (33) revealed a second mislocalization defect (Fig. 3C). Tpbpa staining was correctly localized to cells of the ectoplacental cone at E10.5 although staining was markedly reduced. A similar reduction in signal was observed at E12.5, with the junctional zone appearing as a more compact region in transgenic placentas. At E18.5 the junctional zone appeared as a distinct zone in wild-type placentas, with a clear boundary existing between this and the labyrinth region. In transgenic placentas, however, this boundary between the two zones was completely absent, with only a few Tpbpa-positive cells localized in the anticipated area. The majority of Tpbpa-positive cells were instead dispersed throughout the labyrinth region of transgenic placentas (Fig. 3C).
Excluding a role for Slc22a18 in regulating glycogen storage.
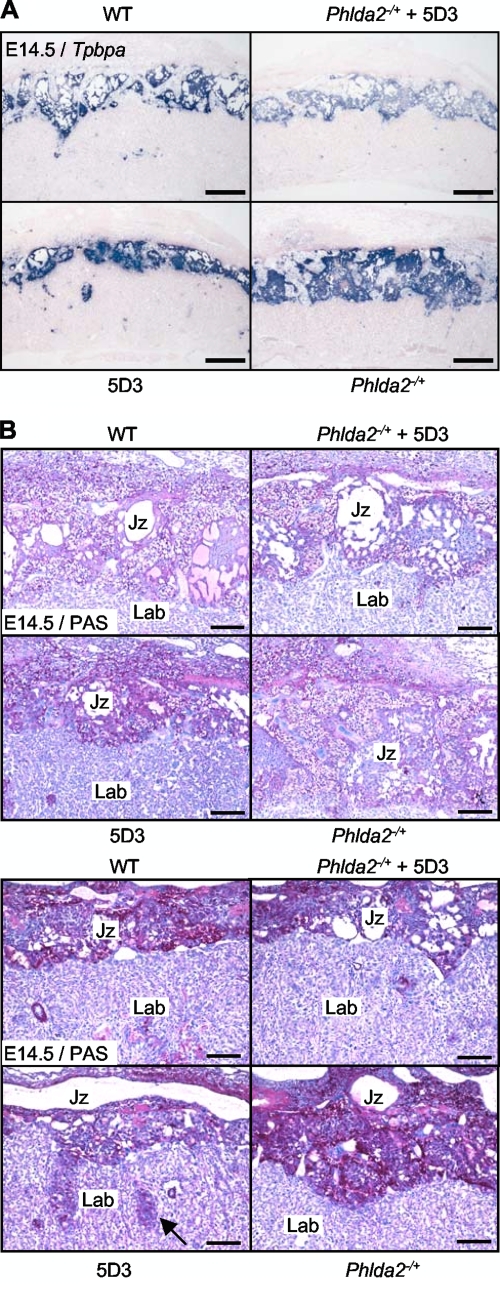
The junctional zone was restored by normalizing the Phlda2 dosage in the KvDMR1+/− placenta (46). In a similar experiment using our single-copy BAC transgenic line, we additionally rescued the mislocalization defects and returned glycogen staining to within the normal range (Fig. 4A and B), formally excluding a major role for Slc22a18 in these phenotypes.
FIG. 4.
Slc22a18 does not regulate glycogen storage. (A) In situ hybridization of Tpbpa to control (wild type [WT]), Phlda2-deficient (Phlda2−/+), single-copy BAC transgenic (5D3), and double-transgenic (Phlda2−/+ plus 5D3) placentas at E14.5 illustrating restoration of the junctional zone. Tpbpa staining within the labyrinth occurs in the 5D3 transgenic placentas but is infrequent in wild-type and double-transgenic placentas, indicating a rescue of this defect. Scale bar, 500 μm. (B) PAS staining of the four genotypes at E14.5 and E18.5 demonstrates restoration of glycogen levels. Mislocalized cells in the 5D3 labyrinth, indicated by the black arrow, are absent in double transgenics. Scale bar, 200 μm.
Phlda2 alters the ratio between spongiotrophoblast and glycogen cells in the junctional zone.
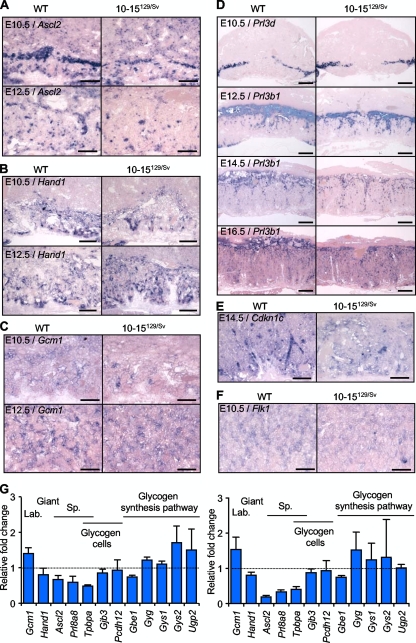
Marker gene analysis was performed for Ascl2 to reveal characteristic expression in the ectoplacental cone, spongiotrophoblast, and labyrinthine trophoblast similarly distributed in both control and transgenic mouse placentas, but at a lower intensity in the transgenics (Fig. 5A). Expression of the gene for heart and neural crest derivatives expressed transcript 1 (Hand1), which is essential for giant trophoblast cell differentiation (44), was similar in control and transgenic placentas (Fig. 5B), as was that of the gene for glial cell missing 1 (Gcm1) (Fig. 5C), which marks branching of the labyrinth (3). The relative abundance of giant cells was examined with the genes for giant-cell-specific probes placental lactogen I (Prl3d) and placental lactogen II (Prl3b1) (Fig. 5D). Prl3d is expressed in the giant cell border, and Prl3b1 is exclusively expressed in the trophoblast giant cell layer beginning around E9.5 and then later in all layers of the placenta (28). These markers showed similar distributions and intensities in the control and experimental placentas, suggesting no overt defect effect in giant trophoblast cells. Cdkn1c (Fig. 5E), which is expressed in the ectoplacental cone, in giant trophoblast cells (44), and transiently in glycogen cells (23), was distributed normally, as was the gene for the kinase insert domain protein receptor (Flk1/Kdr), which marks the fetal vasculature and plays a key role in vascular development (48) (Fig. 5F). These data indicate that the placental defect was confined to cells contributing to the junctional zone and not labyrinthine trophoblast cells or giant cells. Quantitation of mRNAs performed at E12.5 and E16.5 revealed a significant reduction in Ascl2 and Tpbpa at both time points, consistent with a specific loss of the junctional zone (Fig. 5G; Table 1). The subtle differences in the expression levels of Flk1 and the gene for FMS-like tyrosine kinase 1 (Flt1) likely reflected their differential expression in the labyrinth and junctional zone, respectively (6). Gcm1 was also increased reflecting the change in ratio between the labyrinth and the junctional zone. Igf2, insulin-like growth factor 1 receptor (Igf1r), and insulin-like growth factor 2 receptor (Igf2r) gene expression levels were unchanged, suggesting a balanced Igf2 signaling pathway (Table 1).
FIG. 5.
In situ hybridization of placental marker genes. (A) Ascl2 expression at E10.5 and E12.5. Spatially appropriate but reduced intensity of Ascl2 signal in transgenic placentas. Scale bar, 200 μm. (B) Hand1 and (C) Gcm1 at E10.5 and E12.5 showing spatially appropriate localization with similar intensity in transgenic placentas. Scale bar, 200 μm. (D) Relative abundance of giant cells examined by hybridization of midline placental sections with giant-cell-specific probes. Prl3d at E10.5 and Prl3b1 at E12.5, E14.5, and E16.5. Scale bar, 500 μm. (E) Spatially appropriate Cdkn1c expression at E12.5 and (F) Flk1 expression at E10.5. Scale bar, 100 μm. (G) Relative expression levels of critical developmental genes in 10-15129/Sv and wild-type (WT) placentas at E12.5 and E16.5 expressed as relative fold change.
TABLE 1.
Placental marker gene expression analysisa
| Gene | Mean fold change ± SD (P value) |
|
|---|---|---|
| E12.5 | E16.5 | |
| Gcm1 | 1.40 ± 0.16 (0.014) | 1.54 ± 0.35 (0.014) |
| Hand1 | −1.25 ± 0.37 (0.014) | −1.25 ± 0.13 (0.014) |
| Ascl2 | −1.55 ± 0.25 (3.27 × 10−4) | −5.51 ± 1.02 (3.23 × 10−4) |
| Prl8a8 | −1.70 ± 0.52 (4.40 × 10−4) | −3.02 ± 0.65 (4.40 × 10−4) |
| Tpbpa | −2.07 ± 0.17 (0.014) | −2.50 ± 0.51 (0.014) |
| Gjb3 | −1.17 ± 0.15 (3.27 × 10−4) | −1.14 ± 0.16 (1.18 × 10−3) |
| Pcdh12 | −1.07 ± 0.35 (0.369) | −1.07 ± 0.39 (1.0) |
| Gbe1 | −1.35 ± 0.09 (0.014) | −1.34 ± 0.10 (0.014) |
| Gyg | 1.21 ± 0.09 (0.013) | 1.52 ± 0.50 (0.014) |
| Gys1 | 1.10 ± 0.08 (0.219) | 1.23 ± 0.48 (1.0) |
| Gys2 | 1.71 ± 0.47 (0.014) | 1.23 ± 0.48 (1.0) |
| Ugp2 | 1.50 ± 0.30 (0.219) | 1.01 ± 0.10 (1.0) |
| Flk1 | 1.16 ± 0.22 (0.219) | 1.43 ± 0.16 (0.014) |
| Flt1 | −2.06 ± 0.14 (0.014) | −3.01 ± 0.84 (0.014) |
| Igf1 | −1.03 ± 0.11 (0.487) | 1.29 ± 0.30 (0.219) |
| Igf1r | 1.32 ± 0.19 (0.027) | 1.19 ± 0.40 (1.0) |
| Igf2 | −1.07 ± 0.09 (0.219) | −1.13 ± 0.16 (0.219) |
| Igf2r | 1.16 ± 0.12 (0.047) | −1.06 ± 0.33 (1.0) |
Relative changes in expression in transgenic placentas were determined by QRT-PCR at E12.5 and E16.5. Statistical significance was determined by Mann-Whitney test; n = 4 + 4.
Two major cell types originate from the ectoplacental cone, the spongiotrophoblast and the glycogen cells. The glycogen cell population undergoes a dramatic 80-fold expansion from approximately 2,000 cells at E12.5 to 1.4 million cells at E14.5, whereas spongiotrophoblast cells undergo their major proliferative event prior to E12.5, at which time point nearly 2 million cells are present occupying the bulk of the junctional zone (9). The marked reduction in Tpbpa and Ascl2 staining demonstrated that the phenotype was already well established at E10.5. Both proliferation (BrdU incorporation) and apoptosis (TUNEL) were relatively unchanged at E10.5 and E12.5 (see Fig. S1 in the supplemental material), also suggestive of an earlier defect. Markers specific to the spongiotrophoblast and glycogen cell populations were quantified at E12.5 and E16.5. The gene for prolactin family 8 subfamily A member 8 (Prl8a8), which is expressed specifically in spongiotrophoblast cells (50), was markedly reduced at both time points (Fig. 5G; Table 1). In contrast, there was only a small reduction in the expression of the gene for gap junction protein beta 3 (Gjb3/Cx31) and no difference in the expression of the gene for protocadherin 12 (Pcdh12). Both of these genes are expressed exclusively in the glycogen cells of the mouse placenta (5, 60). The relatively normal expression of Gjb3 and Pcdh12 suggested that Phlda2 did not limit expansion of the glycogen cell population. The expression of the genes for several enzymes critical for glycogen metabolism, including those for glycogenin (Gyg), glycogen synthases 1 and 2 (Gys1 and -2), UDP-glucose pyrophosphorylase (Ugp2), and 1,4-α-glucan branching enzyme (Gbe1), was also comparable to wild-type expression (Fig. 5G; Table 1). The marked reduction in stored glycogen appeared to originate not from a loss of glycogen cells but from a change in the microenvironment provided by the spongiotrophoblast cells.
Altered expression of placental nutrient transporters.
A reduction in stored glycogen could occur as a result of reduced glucose transport, but expression of the genes for two placental glucose transporters, solute carrier family 2 (facilitated glucose transporter) members 1 (Slc2a1) and 3 (Slc2a3), was in fact higher in the transgenic placentas (Table 2). Upregulation of Slc2a3 has previously been reported in a mouse model in which the placenta-specific Igf2 promoter has been deleted, causing early placental growth restriction and late embryonic growth restriction (12). In contrast to the Igf2 P0+/− model, no significant effect on the expression level of Slc38a4 was detected although six other nutrient transporters were significantly upregulated in Phlda2 transgenic placentas (Table 2), which may suggest that compensatory mechanisms are triggered in order to overcome the junctional zone deficit.
TABLE 2.
Relative expression levels of genes encoding placental transporters in transgenic placentas of line 10-15 micea
| Gene | Fold change(s) (microarray) | Fold change (QRT-PCR) (P value) |
|---|---|---|
| Slc2a1 | 1.7b | |
| Slc2a3 (2 probes) | 1.7,b 2.9b | 1.7 ± 0.08 (0.014) |
| Slc3a2 | 1.5b | 1.5 ± 0.24 (0.014) |
| Slc19a2 | 2.0b | |
| Slc22a3 | 2.5b | |
| Scl25a3 | 1.7b | |
| Slc38a2 | 2.0b | 1.7 ± 0.07 (0.037) |
| Slc38a4 | 1.3c | 1.0 ± 0.15 (1.00) |
| Slc40a1 | 2.1b | |
| Slc41a1 | 1.7b |
Fold changes in transporter expression at E16.5 were determined by microarray analysis. Transporters altered in a Igf2 P0+/− model (12) were confirmed by QRT-PCR on biological replicates at E16.5. Statistical significance was determined by Mann-Whitney test.
P < 0.05.
No statistically significant difference.
No evidence of embryonic lethality.
A complete loss of the junctional zone is embryonic lethal (26). For line 10-15129/Sv, the number of wild-type and transgenic conceptuses observed at each developmental stage was compared to the number that would be expected according to Mendelian ratios using the chi-square (χ2) test. P values for each analysis are shown in Table 3. Wild-type and transgenic conceptuses were observed at the anticipated Mendelian ratios up to E18.5, indicating that a 60% loss of the junctional zone was compatible with survival.
TABLE 3.
No embryonic lethality associated with the severe placental defecta
| Stage | No. of progeny |
P value (χ2) | |||
|---|---|---|---|---|---|
| Wild type |
Transgenic |
||||
| Observed | Expected | Observed | Expected | ||
| E12.5 | 39 | 37 | 35 | 37 | 0.216 |
| E13.5 | 18 | 19.5 | 23 | 19.5 | 0.610 |
| E14.5 | 31 | 26.5 | 22 | 26.5 | 1.528 |
| E16.5 | 30 | 29 | 28 | 29 | 0.069 |
| E18.5 | 47 | 41.5 | 36 | 41.5 | 1.458 |
The chi-square test was performed to determine if the observed frequencies of wild-type and transgenic progeny at each stage during gestation differed significantly from the expected frequency predicted by Mendelian inheritance.
Progressive slowdown of embryonic growth.
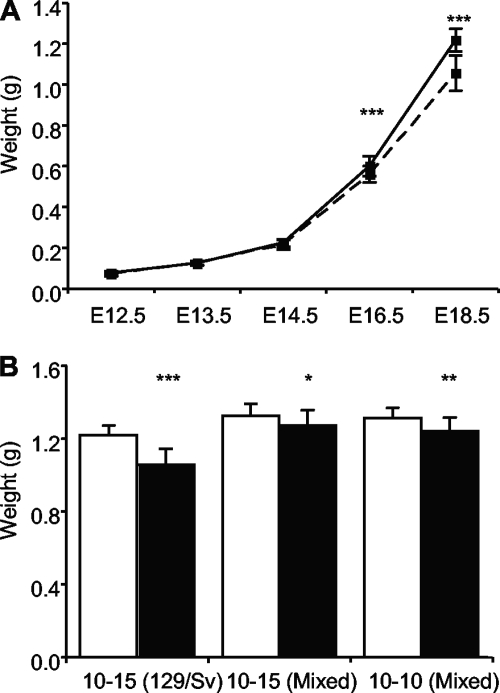
In our previous study transgenic embryos from the three-copy line 10-15 were indistinguishable from control littermates at E14.5 but 10% lighter at E16.5 with marginal statistical significance. Similarly, transgenic embryos in the pure 129/Sv genetic background weighed the same as wild-type embryos at E14.5 but weighed 6.6% less at E16.5 (P = 1.39 × 10−3) and 13.2% less at E18.5 (P = 8.66 × 10−15) (Fig. 6A). Growth restriction was less apparent in the mixed genetic background. At E18.5, 10-15Mixed transgenic embryos weighed 4.4% less than wild-type embryos (P = 0.0278) and 10-10Mixed embryos weighed 5.6% less (P = 0.0089) (Fig. 6B). As observed for placental weights, wild-type embryos were significantly heavier in the mixed genetic background in comparison to pure 129/Sv embryos at both gestational stages examined, being 11.9% heavier at E14.5 (222.5 ± 35.4 versus 249.0 ± 19.4 mg; wild-type weights from 19 litters; n = 73; P = 1.13 × 10−4) and 8.5% heavier at E18.5 (1,215.7 ± 54.9 versus 1,318.6 ± 60.7 mg; wild-type data from 22 litters; n = 82; P = 6.57 × 10−12). We were unable to determine the relationship between the placental phenotype and embryonic weight in the single-copy line due to the transgenic expression of Cdkn1c in the embryo in this line, which carries the unmodified version of the BAC (2). However, increasing Phlda2 and Slc22a18 from three to four doses did not further decrease embryonic weight.
FIG. 6.
Slowdown of embryonic growth. (A) Weights of wild-type and transgenic embryos of line 10-15129/Sv in a pure 129/Sv background at the indicated gestational stages. Transgenic embryos show a 6.6% reduction in wet weight at E16.5 (600.0 ± 48.3 versus 560.5 ± 40.3 mg; 8 litters; n = 58; P = 1.39 × 10−3) and a 13.2% reduction at E18.5 (1,215.7 ± 54.9 versus 1,055.6 ± 88.1 mg; 11 litters; n = 75; P = 8.66 × 10−15). Error bars show standard deviations. (B) Weights of wild-type and transgenic embryos of lines 10-15Mixed and 10-10Mixed in a mixed (129/Sv × C57BL/6) genetic background at E18.5. 10-15Mixed transgenic embryos show a 4.4% reduction in wet weight (1,327.6 ± 64.2 versus 1,269.1 ± 86.9 mg; 6 litters; n = 37; P = 0.0278). 10-10Mixed transgenic embryos are 5.6% lighter (1,322.9 ± 57.7 versus 1,246.2 ± 79.9 mg; 4 litters; n = 26; P = 0.0089). Error bars show standard deviations. Significance levels according to the Student t test are indicated by asterisks as follows: *, P < 0.05; *, P < 0.01; ***, P < 0.001.
In summary, a single extra dose of Phlda2 had serious consequences for placental development, driving the loss of the junctional zone and a progressive mislocalization of cells from the junctional zone into the labyrinth. Excess Phlda2 also dramatically reduced the amount of stored glycogen, and the glycogen cells additionally failed to mobilize into the maternal decidua late in gestation. Despite this severely disorganized placenta, embryos were viable and showed a loss of growth potential only very late in gestation.
DISCUSSION
Previous data have suggested a pivotal role for the imprinted gene Phlda2 as a growth rheostat in the murine placenta which negatively regulates the expansion of the junctional zone in the murine placenta (20, 46). Here we have shown that a single extra dose of Phlda2 leads to a 60% loss of the junctional zone with a dramatic 25 to 35% reduction in stored glycogen, changes that are the reciprocal of those seen with Phlda2 deficiency. Uniquely, excess Phlda2 also causes a failure of glycogen cells to migrate into the maternal decidua in a timely manner. Instead, Tpbpa-positive cells progressively mislocalize into the labyrinth, resulting in a complete collapse of the junctional zone by E18.5. Interestingly, despite these severe placental defects, there is no loss of fetal viability. Instead, there is a progressive slowing of embryonic growth culminating in a 13% decrease in embryonic weight by E18.5. This finding modifies our understanding of the biological function of the junctional zone. Based on the clear observation of lethality in Ascl2-deficient embryos (26), this zone is thought to be essential for the viability of the conceptus, but our data shown here indicate that the junctional zone can be severely stunted and yet remain fully able to support gestation. Thus, there is a considerable reserve capacity in this zone making it a potential drain on maternal resources and thus, according to the genomic conflict model, a prime anatomical and genetic substrate for imprinting.
Our BAC transgene spans two imprinted genes, Phlda2 and Slc22a18, which we have shown were expressed in incrementally increasing doses from 2 to 4 times the endogenous level in the placenta and with the correct spatial and temporal pattern in three transgenic lines. The maximal placental phenotype was achieved by a single extra dose of the transgene, and genetically rescuing the dosage of Phlda2 excluded a significant role for Slc22a18 in the identified placental phenotypes, assigning them to Phlda2.
In mice there are four distinctive layers in the mature placenta, the inner labyrinth layer, an intermediate junctional zone, a layer of trophoblast giant cells, and the maternal decidua. The Phlda2-induced phenotype was primarily apparent at the junctional zone with a 60% loss of volume by E14.5 and a complete breakdown of this zone by E18.5, while the labyrinthine and giant cell components remained relatively unaltered. A second major defect was the decrease in stored glycogen evident as a reduction in the relative intensity of glycogen staining and in the absolute quantity of stored glycogen but not in the level of expression of Gjb3 and Pcdh12, markers specific to the glycogen cell population, or the mRNAs of several enzymes involved in the synthesis of glycogen, suggesting that the proliferation and differentiation of this component of the junctional zone were not affected by the altered Phlda2 dosage. The marked reduction in Tpbpa and Ascl2 staining was already well established at E10.5, prior to the 80-fold expansion of the glycogen cell population that occurs after E12.5, also suggesting the involvement of the second major component of the junctional zone, the spongiotrophoblast cells, which form the bulk of the junctional zone at E12.5 (9). Expression of Prl8a8, an mRNA specific to the spongiotrophoblast cell type, was markedly reduced in transgenic placenta at E12.5 and E16.5, indicative of the loss of this cell type. These data demonstrate that Phlda2 negatively regulates the pool of spongiotrophoblast cells in the placenta. Thus, the ability of glycogen cells to store glycogen is determined by their microenvironment. The simplest explanation is that spongiotrophoblast cells are involved in the hormonal regulation of glycogenesis. Indeed, the spongiotrophoblast layer is already known to have a hormonal role providing pregnancy-related hormones (9, 35). The failure of glycogen cells to migrate into the maternal decidua and the progressive mislocalization of Tpbpa-positive cells into the labyrinth suggest a cell adhesion defect. This provides experimental evidence that spongiotrophoblast cells are also critical in maintaining the integrity of the placental layers. Further studies are required to determine whether these two roles are in some way coupled.
Glycogen cells are thought to provide a readily accessible depot of glycogen that can be rapidly converted into glucose (9). Depletion of these stores might have a consequence for late embryonic growth. Consistent with this theory, we observed a subtle, late intrauterine growth restriction in our model. Several key placental transporters were upregulated in the transgenic placenta. This expression profile is similar to a confirmed model of placental insufficiency, the Igf2 P0+/− mouse, suggesting an imbalance in the growth potential of the embryo and the ability of the defective placenta to support late embryonic growth in our model which is only partly overcome by compensatory pathways (12). A role for Phlda2 in regulating embryonic growth is also suggested by the finding that this gene is overexpressed in placentas from human infants with IUGR (36) and in term placentas from non-IUGR infants with low birth weight (4). Although these data are consistent with a role for increased dosage of Phlda2 in extrinsically limiting embryonic growth, it is important to determine the contribution of excess embryonic expression of Phlda2 and Slc22a18 to any embryonic phenotype.
Phlda2 deficiency causes placentomegaly with an expansion of the junctional zone and increased glycogen (20). Imprinting Phlda2 thus may have acted to increase glycogen stores, providing a rapidly mobilizable energy depot for the final stages of gestation precisely when the fetus is placing the highest demands on maternal resources for growth. The function of Phlda2 protein at the cellular level is still under investigation, but it is already known that this pleckstrin homology domain-containing protein can bind phosphatidylinositide lipids, so it may be acting in cell signaling or subcellular trafficking (47). As has been suggested previously (20, 55), while the biochemical functions of imprinted genes are diverse, these genes increasingly appear to act in concert to precisely regulate fetal and placental growth. A rationale for imprinting the adjacent Cdkn1c locus based on the parental conflict theory is provided by its role in negatively regulating embryonic growth. Excess expression of this maternally expressed gene restricts embryonic growth, while loss of expression enhances embryonic growth, at least at E13.5 (2). Embryos are not overgrown at birth (52, 57, 59), suggesting that their growth potential is not maintained later in gestation. These data could indicate that coimprinting of Cdkn1c and Phlda2 is functionally significant with a reduced dosage of Cdkn1c intrinsically enhancing embryonic growth and a reduced dosage of Phlda2 supplying the necessary increase in extraembryonic resources required to support this growth potential late in gestation.
In the eutherian mammals studied so far the Kvdmr1 locus controls the imprinting of both Cdkn1c and Phlda2 (17). The physical linking of these genes predates the imprinting of this locus (40). It is interesting, therefore, that Cdkn1c is biallelically expressed in marsupials (51), as is Phlda2 (S. Suzuki, M. Renfree, and F. Ishino, personal communication), suggesting that the IC2 domain acquired its imprinted status only after eutherians diverged from marsupials. Little is known about extraembryonic glycogen stores in marsupials, but there is some evidence that a population of glycogenic cells may exist within a chorioallantois-like structure of the Tasmanian bandicoot (18). Imprinting of the IC2 domain may have been instrumental in the transition from a relatively primitive internal development in the common ancestor of marsupials and eutherians to the more complex placentation and more mature internal development found in modern eutherians (see Fig. S2 in the supplemental material).
In summary, we have genetically demonstrated that Phlda2 regulates the storage of glycogen in the eutherian placenta. Our data suggest that imprinting of this locus gave eutherians an advantage by providing a depot of stored energy to support the enhanced embryonic growth potential driven by imprinting Cdkn1c.
Supplementary Material
Acknowledgments
This work was supported by Wellcome Trust grant 074851/Z/04/Z (R.M.J.), BBSRC grant BB/G015465 (R.M.J.), and a grant from the March of Dimes (B.T.). S.J.T. was supported by an MRC Ph.D. studentship.
We thank Martha Salas for technical assistance.
Footnotes
Published ahead of print on 2 November 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ager, E. I., A. J. Pask, H. M. Gehring, G. Shaw, and M. B. Renfree. 2008. Evolution of the CDKN1C-KCNQ1 imprinted domain. BMC Evol. Biol. 8:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., M. D. Wood, S. J. Tunster, S. C. Barton, M. A. Surani, and R. M. John. 2007. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev. Biol. 7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25:311-314. [DOI] [PubMed] [Google Scholar]
- 4.Apostolidou, S., S. Abu-Amero, K. O'Donoghue, J. Frost, O. Olafsdottir, K. M. Chavele, J. C. Whittaker, P. Loughna, P. Stanier, and G. E. Moore. 2007. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J. Mol. Med. 85:379-387. [DOI] [PubMed] [Google Scholar]
- 5.Bouillot, S., C. Rampon, E. Tillet, and P. Huber. 2006. Tracing the glycogen cells with protocadherin 12 during mouse placenta development. Placenta 27:882-888. [DOI] [PubMed] [Google Scholar]
- 6.Breier, G., M. Clauss, and W. Risau. 1995. Coordinate expression of vascular endothelial growth factor receptor-1 (flt-1) and its ligand suggests a paracrine regulation of murine vascular development. Dev. Dyn. 204:228-239. [DOI] [PubMed] [Google Scholar]
- 7.Cerrato, F., A. Sparago, I. Di Matteo, X. Zou, W. Dean, H. Sasaki, P. Smith, R. Genesio, M. Bruggemann, W. Reik, and A. Riccio. 2005. The two-domain hypothesis in Beckwith-Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum. Mol. Genet. 14:503-511. [DOI] [PubMed] [Google Scholar]
- 8.Christie, G. A. 1967. Comparative histochemical distribution of glycogen and alkaline phosphatases in the placenta. Histochemie 9:93-116. [DOI] [PubMed] [Google Scholar]
- 9.Coan, P. M., N. Conroy, G. J. Burton, and A. C. Ferguson-Smith. 2006. Origin and characteristics of glycogen cells in the developing murine placenta. Dev. Dyn. 235:3280-3294. [DOI] [PubMed] [Google Scholar]
- 10.Coan, P. M., A. C. Ferguson-Smith, and G. J. Burton. 2004. Developmental dynamics of the definitive mouse placenta assessed by stereology. Biol. Reprod. 70:1806-1813. [DOI] [PubMed] [Google Scholar]
- 11.Coan, P. M., A. C. Ferguson-Smith, and G. J. Burton. 2005. Ultrastructural changes in the interhaemal membrane and junctional zone of the murine chorioallantoic placenta across gestation. J. Anat. 207:783-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constância, M., E. Angiolini, I. Sandovici, P. Smith, R. Smith, G. Kelsey, W. Dean, A. Ferguson-Smith, C. P. Sibley, W. Reik, and A. Fowden. 2005. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc. Natl. Acad. Sci. U.S.A. 102:19219-19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeChiara, T. M., A. Efstratiadis, and E. J. Robertson. 1990. A growth deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345:78-80. [DOI] [PubMed] [Google Scholar]
- 14.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849-859. [DOI] [PubMed] [Google Scholar]
- 15.Dunwoodie, S. L., and R. S. Beddington. 2002. The expression of the imprinted gene Ipl is restricted to extra-embryonic tissues and embryonic lateral mesoderm during early mouse development. Int. J. Dev. Biol. 46:459-466. [PubMed] [Google Scholar]
- 16.Ferguson-Smith, A. C., B. M. Cattanach, S. C. Barton, C. V. Beechey, and M. A. Surani. 1991. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351:667-670. [DOI] [PubMed] [Google Scholar]
- 17.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32:426-431. [DOI] [PubMed] [Google Scholar]
- 18.Flynn, T. T. 1923. The yolk-sac and allantoic placenta in perameles. Q. J. Microsc. Sci. 76:124-182. [Google Scholar]
- 19.Fowden, A. L., C. Sibley, W. Reik, and M. Constância. 2006. Imprinted genes, placental development and fetal growth. Horm. Res. 65(Suppl. 3):50-58. [DOI] [PubMed] [Google Scholar]
- 20.Frank, D., W. Fortino, L. Clark, R. Musalo, W. Wang, A. Saxena, C. M. Li, W. Reik, T. Ludwig, and B. Tycko. 2002. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc. Natl. Acad. Sci. U.S.A. 99:7490-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank, D., C. L. Mendelsohn, E. Ciccone, K. Svensson, R. Ohlsson, and B. Tycko. 1999. A novel pleckstrin homology-related gene family defined by Ipl/Tssc3, TDAG51, and Tih1: tissue-specific expression, chromosomal location, and parental imprinting. Mamm. Genome 10:1150-1159. [DOI] [PubMed] [Google Scholar]
- 22.Freyer, C., and M. B. Renfree. 2009. The mammalian yolk sac placenta. J. Exp. Zool. B Mol. Dev. Evol. 312:545-554. [DOI] [PubMed] [Google Scholar]
- 23.Georgiades, P., A. C. Ferguson-Smith, and G. J. Burton. 2002. Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23:3-19. [DOI] [PubMed] [Google Scholar]
- 24.Guillemot, F., T. Caspary, S. M. Tilghman, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, D. J. Anderson, A. L. Joyner, J. Rossant, and A. Nagy. 1995. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9:235-242. [DOI] [PubMed] [Google Scholar]
- 25.Guillemot, F., L.-C. Lo, J. E. Johnson, A. Auerbach, D. J. Anderson, and A. L. Joyner. 1993. Mammalian acaete-scute homolog 1 is required for early development of olfactory and autonomic neurons. Cell 75:463-476. [DOI] [PubMed] [Google Scholar]
- 26.Guillemot, F., A. Nagy, A. Auerbach, J. Rossant, and A. L. Joyner. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371:333-336. [DOI] [PubMed] [Google Scholar]
- 27.Hemberger, M. 2007. Epigenetic landscape required for placental development. Cell. Mol. Life Sci. 64:2422-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, L. L., P. Colosi, F. Talamantes, and D. I. Linzer. 1986. Molecular cloning of mouse placental lactogen cDNA. Proc. Natl. Acad. Sci. U. S. A. 83:8496-8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John, R. M., J. F. Ainscough, S. C. Barton, and M. A. Surani. 2001. Distant cis-elements regulate imprinted expression of the mouse p57(Kip2) (Cdkn1c) gene: implications for the human disorder, Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 10:1601-1609. [DOI] [PubMed] [Google Scholar]
- 30.John, R. M., and M. A. Surani. 2000. Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell 101:585-588. [DOI] [PubMed] [Google Scholar]
- 31.Killian, J. K., C. M. Nolan, N. Stewart, B. L. Munday, N. A. Andersen, S. Nicol, and R. L. Jirtle. 2001. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J. Exp. Zool. 291:205-212. [DOI] [PubMed] [Google Scholar]
- 32.Killian, J. K., C. M. Nolan, A. A. Wylie, T. Li, T. H. Vu, A. R. Hoffman, and R. L. Jirtle. 2001. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 10:1721-1728. [DOI] [PubMed] [Google Scholar]
- 33.Lescisin, K. R., S. Varmuza, and J. Rossant. 1988. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 2:1639-1646. [DOI] [PubMed] [Google Scholar]
- 34.Lo, S., J. C. Russell, and A. W. Taylor. 1970. Determination of glycogen in small tissue samples. J. Appl. Physiol. 28:234-236. [DOI] [PubMed] [Google Scholar]
- 35.Malassiné, A., J. L. Frendo, and D. Evain-Brion. 2003. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 9:531-539. [DOI] [PubMed] [Google Scholar]
- 36.McMinn, J., M. Wei, N. Schupf, J. Cusmai, E. B. Johnson, A. C. Smith, R. Weksberg, H. M. Thaker, and B. Tycko. 2006. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27:540-549. [DOI] [PubMed] [Google Scholar]
- 37.Nolan, C. M., J. K. Killian, J. N. Petitte, and R. L. Jirtle. 2001. Imprint status of M6P/IGF2R and IGF2 in chickens. Dev. Genes Evol. 211:179-183. [DOI] [PubMed] [Google Scholar]
- 38.Oh, R., R. Ho, L. Mar, M. Gertsenstein, J. Paderova, J. Hsien, J. A. Squire, M. J. Higgins, A. Nagy, and L. Lefebvre. 2008. Epigenetic and phenotypic consequences of a truncation disrupting the imprinted domain on distal mouse chromosome 7. Mol. Cell. Biol. 28:1092-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill, M. J., R. S. Ingram, P. B. Vrana, and S. M. Tilghman. 2000. Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 210:18-20. [DOI] [PubMed] [Google Scholar]
- 40.Paulsen, M., T. Khare, C. Burgard, S. Tierling, and J. Walter. 2005. Evolution of the Beckwith-Wiedemann syndrome region in vertebrates. Genome Res. 15:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian, N., D. Frank, D. O'Keefe, D. Dao, L. Zhao, L. Yuan, Q. Wang, M. Keating, C. Walsh, and B. Tycko. 1997. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum. Mol. Genet. 6:2021-2029. [DOI] [PubMed] [Google Scholar]
- 42.Rawn, S. M., and J. C. Cross. 2008. The evolution, regulation, and function of placenta-specific genes. Annu. Rev. Cell Dev. Biol. 24:159-181. [DOI] [PubMed] [Google Scholar]
- 43.Renfree, M. B., E. I. Ager, G. Shaw, and A. J. Pask. 2008. Genomic imprinting in marsupial placentation. Reproduction 136:523-531. [DOI] [PubMed] [Google Scholar]
- 44.Riley, P., L. Anson-Cartwright, and J. C. Cross. 1998. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 18:271-275. [DOI] [PubMed] [Google Scholar]
- 45.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 46.Salas, M., R. John, A. Saxena, S. Barton, D. Frank, G. Fitzpatrick, M. J. Higgins, and B. Tycko. 2004. Placental growth retardation due to loss of imprinting of Phlda2. Mech. Dev. 121:1199-1210. [DOI] [PubMed] [Google Scholar]
- 47.Saxena, A., P. Morozov, D. Frank, R. Musalo, M. A. Lemmon, E. Y. Skolnik, and B. Tycko. 2002. Phosphoinositide binding by the pleckstrin homology domains of Ipl and Tih1. J. Biol. Chem. 277:49935-49944. [DOI] [PubMed] [Google Scholar]
- 48.Shalaby, F., J. Rossant, T. P. Yamaguchi, M. Gertsenstein, X. F. Wu, M. L. Breitman, and A. C. Schuh. 1995. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376:62-66. [DOI] [PubMed] [Google Scholar]
- 49.Simmons, D. G., and J. C. Cross. 2005. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284:12-24. [DOI] [PubMed] [Google Scholar]
- 50.Simmons, D. G., S. Rawn, A. Davies, M. Hughes, and J. C. Cross. 2008. Spatial and temporal expression of the 23 murine prolactin/placental lactogen-related genes is not associated with their position in the locus. BMC Genomics 9:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, S., M. B. Renfree, A. J. Pask, G. Shaw, S. Kobayashi, T. Kohda, T. Kaneko-Ishino, and F. Ishino. 2005. Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 122:213-222. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, K., K. Nakayama, and K. Nakayama. 2000. Mice lacking a CDK inhibitor, p57Kip2, exhibit skeletal abnormalities and growth retardation. J. Biochem. 127:73-83. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, M., M. Gertsenstein, J. Rossant, and A. Nagy. 1997. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev. Biol. 190:55-65. [DOI] [PubMed] [Google Scholar]
- 54.Toder, R., S. A. Wilcox, M. Smithwick, and J. A. Graves. 1996. The human/mouse imprinted genes IGF2, H19, SNRPN and ZNF127 map to two conserved autosomal clusters in a marsupial. Chromosome Res. 4:295-300. [DOI] [PubMed] [Google Scholar]
- 55.Tycko, B., and I. M. Morison. 2002. Physiological functions of imprinted genes. J. Cell. Physiol. 192:245-258. [DOI] [PubMed] [Google Scholar]
- 56.Weidman, J. R., K. A. Maloney, and R. L. Jirtle. 2006. Comparative phylogenetic analysis reveals multiple non-imprinted isoforms of opossum Dlk1. Mamm. Genome 17:157-167. [DOI] [PubMed] [Google Scholar]
- 57.Yan, Y., J. Frisen, M. H. Lee, J. Massague, and M. Barbacid. 1997. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 11:973-983. [DOI] [PubMed] [Google Scholar]
- 58.Yokomine, T., H. Shirohzu, W. Purbowasito, A. Toyoda, H. Iwama, K. Ikeo, T. Hori, S. Mizuno, M. Tsudzuki, Y. Matsuda, M. Hattori, Y. Sakaki, and H. Sasaki. 2005. Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2-H19 region. Genome Res. 15:154-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, P., N. J. Liegeois, C. Wong, M. Finegold, H. Hou, J. C. Thompson, A. Silverman, J. W. Harper, R. A. DePinho, and S. J. Elledge. 1997. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387:151-158. [DOI] [PubMed] [Google Scholar]
- 60.Zheng-Fischhöfer, Q., M. Kibschull, M. Schnichels, M. Kretz, E. Petrasch-Parwez, J. Strotmann, H. Reucher, B. D. Lynn, J. I. Nagy, S. J. Lye, E. Winterhager, and K. Willecke. 2007. Characterization of connexin31.1-deficient mice reveals impaired placental development. Dev. Biol. 312:258-271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.