Abstract
Nascent-polypeptide-associated complex and coactivator alpha (αNAC) is a protein shuttling between the nucleus and the cytoplasm. Upon phosphorylation at residue serine 43 by integrin-linked kinase, αNAC is translocated to the nuclei of osteoblasts, where it acts as an AP-1 coactivator to increase osteocalcin gene transcription. To determine the physiological role of nuclear αNAC, we engineered a knock-in mouse model with a serine-to-alanine mutation at position 43 (S43A). The S43A mutation resulted in a decrease in the amount of nuclear αNAC with reduced osteocalcin gene promoter occupancy, leading to a significant decrease in osteocalcin gene transcription. The S43A mutant bones also expressed decreased levels of α1(I) collagen mRNA and as a consequence had significantly less osteoid tissue. Transient transfection assays and chromatin immunoprecipitation confirmed the α1(I) collagen gene as a novel αNAC target. The reduced quantity of bone matrix in S43A mutant bones was mineralized faster, as demonstrated by the significantly reduced mineralization lag time, producing a lower volume of immature, woven-type bone characterized by poor lamellation and an increase in the number of osteocytes. Accordingly, the expression of the osteocyte differentiation marker genes DMP-1 (dentin matrix protein 1), E11, and SOST (sclerostin) was increased. The accelerated mineralization phenotype was cell autonomous, as osteoblasts isolated from the calvaria of S43A mutant mice mineralized their matrix faster than did wild-type cells. Thus, inhibition of αNAC nuclear translocation results in an osteopenic phenotype caused by reduced expression of osteocalcin and type I collagen, accelerated mineralization, and immature woven-bone formation.
Nascent-polypeptide-associated complex and coactivator alpha (αNAC) is a protein shuttling between the cytoplasm and the nucleus (50). In the cytoplasm, αNAC is part of a protein complex termed the nascent-polypeptide-associated complex (NAC), which consists of an α and a β subunit. NAC associates with ribosomes and nascent polypeptide chains in a chaperone-like manner (31). NAC has also been reported to play a role in the endoplasmic reticulum (ER) stress response pathway and to directly interact with the signal recognition particle (SRP) (15). NAC has further been reported to be involved in protein translocation into the ER (26) or mitochondria (13). The importance of NAC is highlighted by the fact that mutations introduced into βNAC (8) or αNAC (O. Akhouayri and R. St-Arnaud, unpublished data) result in embryonic lethal phenotypes in mice.
The α subunit of NAC has specifically been detected in the nuclei of differentiated osteoblasts (50). This translocation from the cytoplasm to the nucleus is regulated by phosphorylation of αNAC by integrin-linked kinase (ILK) at residue serine 43 (S43) (35). The kinase activity of ILK is stimulated upon binding of extracellular matrix components to integrin receptors (47). Phosphorylation of residue S43 by ILK potentiates the coactivating function of αNAC (35). In the nucleus, αNAC acts as a coactivator of AP-1-mediated transcription by specifically potentiating the activity of c-Jun homodimers (1, 2, 27). One of the currently identified targets of the αNAC coactivating activity is the osteocalcin gene promoter (1). αNAC potentiates osteocalcin gene transcription by stabilizing the binding of a c-Jun dimer to its AP-1 binding site in the osteocalcin gene promoter (2) and by acting as a protein bridge between the c-Jun dimer and the basal transcription machinery (TATA-binding protein) (27). It has further been shown that αNAC can specifically bind DNA and that this DNA-binding capacity is required to potentiate AP-1-mediated osteocalcin gene expression in osteoblasts (1).
Osteocalcin is a terminal marker of osteoblastic differentiation and was suggested to play an inhibitory role during bone formation (10). This inhibitory role possibly involves alteration of osteoblast function and was initially thought to affect matrix mineralization (6). However, recent molecular genetic studies do not support a direct role for osteocalcin in matrix mineralization (28). Osteocalcin exists as a noncarboxylated molecule but can also undergo posttranslational modification, during which it gets γ carboxylated at its glutamic acid residues (4, 33). The γ-carboxylated form shows a high affinity for hydroxyapatite (HA) crystals, the main component of mineralized bone tissue (18, 40). The noncarboxylated form of osteocalcin circulates in the bloodstream and has recently been shown to mediate the endocrine function of bone in glucose and fat metabolism (21, 22).
In order to assess the physiological importance of the ILK-αNAC signaling cascade and the role of nuclear αNAC in vivo, we engineered a mouse model carrying a serine-to-alanine single-nucleotide mutation at position S43 within the αNAC gene (S43→S43A). In vitro, this mutation has been shown to hinder the ability of ILK to phosphorylate αNAC at position S43 and subsequently to reduce αNAC translocation into the nucleus (35). The S43A knock-in mouse model also showed reduced αNAC nuclear levels. Consequently, osteocalcin gene transcription was significantly reduced in the S43A mutants, along with transcription of the α1(I) collagen gene, identifying a novel target of the AP-1-coactivating function of αNAC. The S43A mutation resulted in an osteopenic phenotype characterized by increased woven bone containing a larger number of osteocytes. This study is the first demonstration of the importance of nuclear αNAC in osteoblastic gene expression and bone formation in vivo.
MATERIALS AND METHODS
Generation of S43A knock-in mice.
αNAC knock-in mice were generated using a pMC-loxP-NEO-loxP vector (Specialty Media, Phillipsburg, NJ) consisting of a loxP-NEO-loxP selection marker flanked by two fragments of the mouse αNAC gene. The 5′ fragment stretched from exon 1 to intron 3 (4.7 kb) and the other one from intron 3 to exon 9 (3 kb). A single-base-pair mutation (T→G) to replace the serine at residue 43 (TCC) in exon 4 with an alanine (GCC) was introduced by the recombineering technique (7, 43). The resulting targeting vector was used for homologous recombination in 129sv embryonic stem cells and subsequent C57BL/6 blastocyst injection. Highly chimeric mice were crossed with wild-type C57BL/6 mice, and the resulting brown babies were genotyped by single-nucleotide polymorphism (SNP) genotyping (see below). S43/S43A-heterozygous mice were interbred to obtain all three genotypes (S43/S43, wild type [WT]; S43/S43A, heterozygous [het]; S43A/S43A, S43A mutant). The NEO selection cassette was excised by crossing S43A mutant mice with CMV-Cre mice (20). Animals were sacrificed and analyzed at 6 weeks of age. All animal experimentation was approved by the Institutional Animal Care and Use Committee and followed the guidelines of the Canadian Council on Animal Care.
SNP genotyping.
Tail clips of 2- to 4-week-old mice were digested overnight in lysis buffer (100 mM Tris-HCl [pH 8.5], 200 mM NaCl, 5 mM EDTA, 0.2% sodium dodecyl sulfate, 100 μg/ml of proteinase K) and centrifuged at maximum speed for 5 min at 4°C, and the resulting supernatant was transferred to a new tube. The DNA was precipitated using isopropanol (1:1), washed with 70% ethanol, and resuspended in 200 μl of TE-light (10 mM Tris, 0.1 mM EDTA, pH 8.0). The genotype was determined with a custom TaqMan SNP genotyping assay from Applied Biosystems (Foster City, CA) using αNAC primers (5′-TCAGTACCAGAGCTCGAGGAA-3′ and 5′-ACACACCTGGGCTTGCT-3′) flanking the single-point mutation site S43-S43A. Genotyping was carried out according to the custom TaqMan SNP genotyping assay protocol supplied by Applied Biosystems.
Protein extraction and Western blotting.
Calvaria from wild-type and S43A mutant mice were snap-frozen using liquid nitrogen, crushed using a mortar and pestle, and suspended in 600 μl of lysis buffer solution (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.8% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.15 U/ml aprotinin, 10 μg/ml leupeptin, 1 mM NaVO3). Tissue samples were homogenized using a Polytron PT-MR 3000 (Kinematica AG, Littau, Switzerland) at 20,000 rpm for 20 s and then placed on a rotating wheel overnight at 4°C. Samples were then centrifuged at 4°C (10 min at 500 × g), and the supernatants were collected. Pellets were resuspended (600 μl of lysis buffer), rehomogenized, and recentrifuged. The resulting supernatants were pooled with the previously collected protein extracts. The pooled extracts were centrifuged (5,000 rpm for 10 min at 4°C) to remove any remaining bone tissue. Proteins were precipitated by addition of 300 μl of trichloroacetic acid (TCA), mixing, and incubation for 10 min on ice. The samples were centrifuged (5 min at 14,000 rpm), and the precipitated protein pellets were washed twice with 200 μl of cold acetone, dried in a 95°C heat block for 1 to 3 min, and resuspended in 1× Laemmli buffer without dithiothreitol (DTT).
Cytoplasmic and nuclear extracts from primary osteoblast cultures obtained as described below were prepared by scraping primary cells into ice-cold phosphate-buffered saline (PBS). The cells were pelleted, resuspended in 200 μl of lysis buffer A (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF), and allowed to swell on ice for 10 min. After vortexing, the samples were centrifuged and the supernatant was saved as the cytoplasmic fraction at −80°C. The pellet was resuspended in 50 μl of ice-cold lysis buffer B (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF), incubated on ice for 20 min, and then centrifuged. The supernatant was stored as the nuclear fraction at −80°C. Protein concentrations were determined by the Bradford assay, and equal amounts were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
A purified polyclonal anti-αNAC chicken antibody raised against recombinant αNAC was used to probe immunoblots of total bone protein extracts, as well as nuclear and cytoplasmic extracts from wild-type and heterozygous or homozygous mutants. Anti-chicken antibodies conjugated to horseradish peroxidase were used as secondary antibodies and detected using ECL Western blotting detection reagents from GE Healthcare Bio-Sciences (Baie d'Urfé, Quebec, Canada).
Primary cultures.
Primary osteoblast cultures were prepared from 2- to 6-week-old mouse calvaria as previously described (52). Von Kossa staining was performed using 5% silver nitrate solution under strong UV light, followed by treatment with 5% sodium thiosulfate for 5 min. Pictures were taken immediately. The assay was repeated three times with two to six mice of each genotype per experiment. Representative results from one experiment are shown.
Proliferation experiments (n = 6) were performed using the colorimetric cell proliferation enzyme-linked immunosorbent assay (ELISA), bromodeoxyuridine (BrdU) kit from Roche Diagnostics (Mannheim, Germany).
For nuclear localization studies, primary calvarial cells were grown to 70% confluence on coverslips, starved in 0.5% serum overnight, and then incubated with 1 μM proteasome inhibitor III for 5 h at 37°C before fixation in 4% paraformaldehyde (PFA). αNAC localization was visualized using a rabbit anti-αNAC primary antibody (1/200) (51), an Alexa Fluor 594 goat anti-rabbit immunoglobulin G (IgG; 1/2,000) secondary antibody (Invitrogen, Burlington, Ontario, Canada), and 1 drop of Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Four independent fields were counted from cultures of cells from five different mice per genotype.
ChIP.
For quantitative assessment of osteocalcin gene promoter occupancy by wild-type and S43A αNAC in primary osteoblast cultures from wild-type and mutant mice, we used the MAGnify chromatin immunoprecipitation (ChIP) system (Invitrogen Canada, Burlington, Ontario, Canada) by following the manufacturer's instructions. After elution, the amount of ChIP products was measured on a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Equivalent amounts were subjected to quantitative real-time PCR using SYBR green QuantiFast Master Mix (Qiagen, Mississauga, Ontario, Canada) and primers specific for the αNAC binding site within the osteocalcin gene promoter (1), in triplicate. The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s (denaturation), 65°C for 45 s, and 72°C for 45 s (annealing and elongation, respectively). Threshold cycle (CT) numbers were determined with the ABI PRISM 7500 sequence detection system (version 1.1 software). The DNA levels from ChIP-quantitative PCR (qPCR) assays were calculated as follows: fold enrichment = R(CTInput − CTIP), where R is the normalized reporter. After multiple runs with the primers designated for the osteocalcin gene promoter, 95% rates of amplification were achieved with an R value of 1.9, allowing us to solve the equation. As a negative control and to measure the background, IgG antibodies were used in parallel ChIPs and qPCRs to determine the relative enrichment.
ChIP of the α1(I) collagen gene AP-1 intronic response element was performed with the following modification. To first stabilize protein-protein interactions, cells were washed with PBS and then incubated with 5 mM dimethyl-3,3-dithiobispropionimidate-2HCl (DTBP) for 30 min on ice. The DTBP was inactivated by washing with cold inactivation buffer (100 mM Tris-HCl [pH 8.0], 150 mM NaCl), followed by PBS. The ChIP assay was then performed as described previously (1) using the anti-αNAC antibody or control IgG and the following primers: forward, 5′-AGAGTGAGGAAAGCACGTCG-3′ (starting at position +507 relative to the transcription start site); reverse, 5′-GCTGCCCAGCACCCCCA-3′ (position +667). The control coding sequence primers were as follows: forward, 5′-GTCTCTGTTTATGTATATGTA-3′ (position +1907); reverse, 5′-TGACATCATGCACTTCCTG-3′ (position +2385).
Morphological analysis.
One femur from each mouse was collected in PBS-soaked tissues without removing the surrounding muscle tissue and used for three-point bending tests. The other femur was fixed in 4% PFA, transferred to 70% ethanol, and used for microcomputed tomography (μCT) measurements. Bending tests and μCT analysis were performed at the Centre for Bone and Periodontal Research of McGill University. No fewer than six samples of the same gender were analyzed for each genotype. Results for males are reported, but there were no gender differences. Tibias and L2-to-L4 vertebrae were fixed in 4% PFA and embedded in 4.5% methyl methacrylate (MMA) for histological analysis. MMA-embedded tibia and vertebra sections (5 μm) were stained with either Goldner or toluidine blue (9). For dynamic histomorphometric analysis, mice were subcutaneously injected with 25 μg/g body weight of calcein at 8 and 4 days prior to sacrifice. Between four and eight samples from male animals were analyzed for each genotype. All samples were analyzed using a Leica DMR fluorescence microscope with a polarizer (Leica Microsystems, Richmond Hill, Ontario, Canada) connected to a QImaging camera and the Osteo software from BioQuant Image Analysis Corporation (Nashville, TN).
Real-time reverse transcription-qPCR (RT-qPCR).
Calvaria from 6-week-old mice were collected into RNAlater solution (Ambion, Austin, TX) and stored at −20°C until further processed. RNA was extracted with TRIzol (Invitrogen) by following the manufacturer's instructions. One microgram of RNA was reverse transcribed into cDNA using the High Capacity cDNA Archive kit in accordance with the manufacturer's recommendations (Applied Biosystems). Real-time PCR amplification was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) on a 7500 instrument (Applied Biosystems) and specific Assay-On-Demand TaqMan assays for osteocalcin, αNAC, ATF4, Runx2, osterix, bone sialoprotein II, osteopontin, dentin matrix protein 1, E11, sclerostin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Relative quantification of mRNA was performed according to the comparative CT method with GAPDH RNA as an endogenous control (ABI PRISM 7700 sequence detector user bulletin 2; PE Applied Biosystems, 1997). Five samples per genotype were assayed in duplicates.
Transient transfection assays.
MC3T3-E1 osteoblastic cells (41) were cultured in minimum essential medium alpha containing 10% fetal bovine serum and plated at 1.0 × 105/well in a six-well plate. Cells were transfected with 200 ng each of reporter vector (−2300COL-hGH/luc [23], pGL3-DMP1 [42], ECR5-2kb-SOST-luciferase [25], or −1251bp-E11-Luc [38]) and 400 ng of expression vectors for c-Jun (37), αNAC (35), DNA-binding domain (DBD) mutant αNAC (Δ69-80) (1), or the empty vector by using the LipofectAmine transfection reagent (InvitroGen) and following the instructions provided by the manufacturer. The total amount of transfected DNA was adjusted to 2 μg in each well by using pBlueScript (Stratagene, La Jolla, CA). At 48 h posttransfection, cells were lysed and 20 μl of cell lysate was used to measure luciferase activity. Each transfection was repeated at least three times with triplicate samples, and the data shown represent the mean ± the standard error of the mean (SEM) of a representative experiment.
Blood biochemistry.
Alkaline phosphatase (ALP), calcium, phosphate, and glucose serum levels were determined using an automated analyzer by the McGill University Animal Resources Centre Diagnostic and Research Support Service. A minimum of eight male samples were analyzed for each genotype. Fibroblast growth factor 23 (FGF23) serum levels were assessed by using the FGF23 ELISA kit (Cedarlane Laboratories, Burlington, Ontario, Canada) by following the manufacturer's recommendations. The quantification of fragments of type I collagen (CTX) in serum (five samples per genotype) was done using the RatLaps enzyme immunoassay kit (Immunodiagnostic Systems Ltd., Boldon, United Kingdom). HA-bound osteocalcin serum levels were determined as described by Lee et al. (22). Seven samples of the same gender were analyzed for each genotype.
Statistical analysis.
Statistical analysis used Student's t test when two variables were compared or analysis of variance with a post hoc test when more than two samples were included in the analysis. P < 0.05 was accepted as significant.
RESULTS
Engineering of the S43A knock-in strain.
The serine (TCC)-to-alanine (GCC) mutation at position S43 in the αNAC targeting vector (see Fig. 1A posted at http://www.mcgill.ca/files/humangenetics/aTRMSuppl-Fig01-2b.pdf) was confirmed by sequencing (see Fig. 1B at the URL mentioned above). Following homologous recombination in embryonic stem cells, four positive clones were identified and one was used for blastocyst injection. The resulting chimeric mice were back bred to C57BL/6 wild-type mice, and the resulting progeny was screened for germ line transmission by coat color (agouti). Agouti mice were used for further breeding, and the genotype (S43/S43, WT; S43/S43A, het; S43A/S43A, S43A mutant) was determined using a SNP assay (see Fig. 1C at the URL mentioned above). The NEO selection cassette (see Fig. 1A at the URL mentioned above) was excised by breeding S43/S43A heterozygous mice to wild-type mice containing a universal CMV-Cre exciser gene, and complete excision of NEO and segregation of the Cre transgene were confirmed by PCR. Mice analyzed at 6 weeks showed the same phenotype independently of the presence or absence of the NEO selection cassette in their genome (data not shown). There were no gross phenotypic manifestation of the homozygous S43A mutation, and whole-body X-ray images of wild-type and mutant mice were identical (see Fig. 1D at the URL mentioned above).
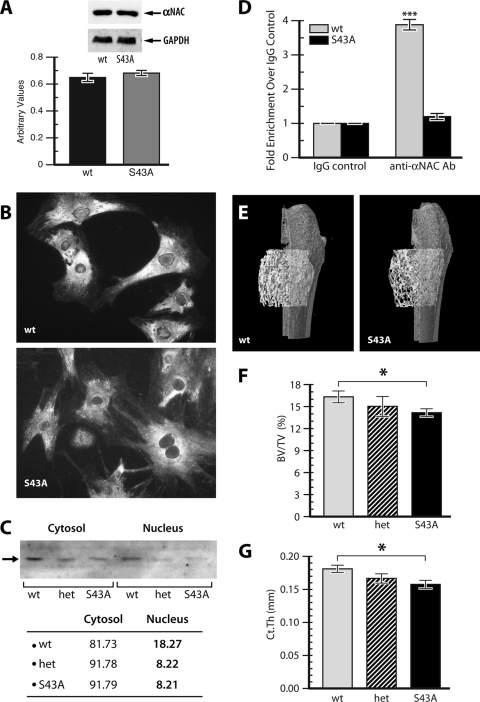
FIG. 1.
Reduced αNAC nuclear translocation and osteopenia in S43A mutant bones. (A) Whole protein extracts from calvaria of wild-type (WT) and S43A mutant littermates were immunoblotted with an anti-αNAC antibody and then stripped and reprobed with an antibody against the ubiquitous marker GAPDH. Relative αNAC protein amounts are graphed below. (B) Primary cultures of osteoblasts from WT and S43A homozygous mutant mice were stained with the anti-αNAC antibody and a fluorescent secondary antibody. Mutant osteoblasts show reduced staining in the nucleus. (C) immunoblotting of cytosol and nuclear fractions for αNAC protein (arrow). The signal intensity was quantified and expressed as the relative percentage of total cellular αNAC present within the cytosol or the nucleus. (D) Quantitative ChIP of the osteocalcin gene promoter fragments using the anti-αNAC antibody (Ab). ***, P < 0.001. (E) Femurs were analyzed by μCT. Data from distal femurs of wild-type animals and S43A mutants was used for three-dimensional reconstruction. Bone volume over tissue volume (BV/TV, panel F) was significantly reduced in S43A mice compared to that of their wild-type littermates. Cortical thickness (Ct.Th, panel G) was also significantly lower. Het, S43/S43A heterozygous mice. *, P < 0.05 (n = 6).
Reduced nuclear αNAC levels in S43A mutated cells.
Immunoblotting of total protein extracts from calvaria of wild-type and S43A homozygote mutant littermates revealed that the mutation did not significantly affect steady-state αNAC protein levels (Fig. 1A). We next determined the impact of the S43A mutation on the subcellular localization of αNAC by immunofluorescence analysis of primary cultures of osteoblastic cells from calvaria of wild-type and S43A mutant littermates. The signals detected in the cytosol of both wild-type and mutant cells were similar in intensity (Fig. 1B). Nuclear staining was clearly reduced in cells from the S43A mutant mice, however (Fig. 1B). When the relative amount of cytosolic and nuclear αNAC was quantified by Western blotting (Fig. 1C), 18% of the cellular αNAC was detected in the nuclei of wild-type cells, while only 8% was found in the nuclei of S43A mutant cells (Fig. 1C). These results confirm that the engineered mutation affected the subcellular localization of αNAC. The lower amount of nuclear S43A-αNAC led to reduced occupancy of the αNAC binding site within the osteocalcin gene proximal promoter, as determined by ChIP with anti-αNAC antibody, followed by quantitative PCR (Fig. 1D).
S43A mutants have less bone, without changes in osteoblast or osteoclast numbers.
To determine the effect of the S43A mutation on bone morphology, we performed μCT analysis and static histomorphometry analyses using Goldner-stained MMA sections. μCT analysis performed at the distal end of femurs from 6-week-old mice (Fig. 1E) showed a significant reduction in bone volume over tissue volume (BV/TV) in the S43A mutants compared to wild-type mice (Fig. 1F). Trabecular thickness (not shown) and cortical thickness (Fig. 1G) were also significantly reduced in the S43A mutants. This finding was confirmed by static histomorphometry analysis of tibia sections from 6-week-old mice: Goldner-stained sections clearly showed decreased bone volume in S43A sections (Fig. 2B) compared to wild-type sections (Fig. 2A), and this was confirmed by a lower BV/TV ratio in the mutants (Fig. 2C). Further analyses of these sections showed a significant reduction in osteoid volume over bone volume (Fig. 2D) in the S43A mutants, as well as significantly reduced bone surface over tissue volume (BS/TV), reduced osteoid surface over bone surface (OS/BS), lower mean osteoid thickness, reduced trabecular thickness, and a lower trabecular number in the S43A mutants compared to those of their wild-type littermates (data not shown). The reduced trabecular thickness and number resulted in significantly increased trabecular separation in the mutants (not shown).
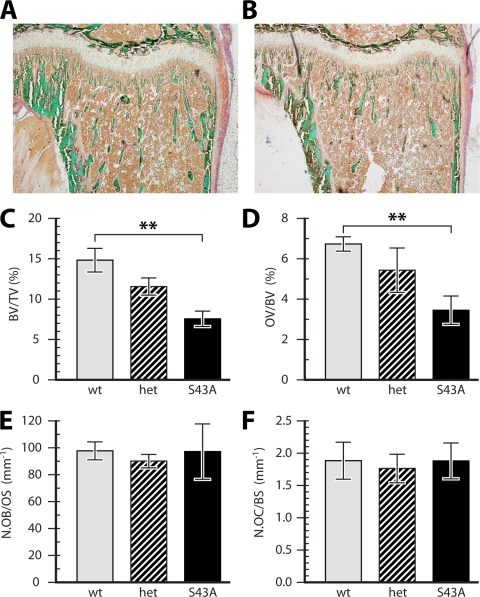
FIG. 2.
Reduced bone volume without changes in osteoblast or osteoclast cell numbers in S43A mutant bones. Proximal tibias were sectioned and analyzed following Goldner staining. Sections from S43A mutant mice (B) showed visibly less trabecular bone than did those of wild-type mice (A). Quantified bone volume (C) and osteoid volume (D) were significantly reduced in S43A mutants compared to those in their wild-type littermates. The numbers of osteoblasts (E) and osteoclasts (F) remained unchanged between mutant and wild-type mice, however. **, P < 0.01 (n = 5).
The reduction in bone volume occurred without changes in the number of bone-forming or bone-resorbing cells, as the number of osteoblasts per osteoid surface (N.Ob/OS) was unchanged (Fig. 2E). Similarly, no difference was measured in osteoclast numbers over bone surface (Fig. 2F) or over erosion surface (not shown), and no change in erosion surface over bone surface (ES/BS) was detected (not shown).
When the mechanical properties of the mutant bones were tested using the three-point bending test, we observed a slight but not significant decrease in stiffness (data not shown).
These results show that S43A mutant mice are osteopenic without changes in the number of osteoblasts or osteoclasts.
S43A mutants have reduced bone formation rates.
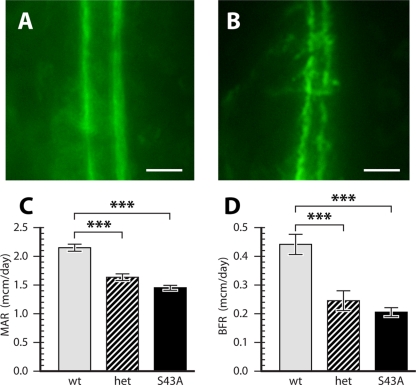
The activity of the S43A mutant osteoblasts was compared to that of wild-type cells using dynamic histomorphometry. Calcein-injected mice were sacrificed at 6 weeks of age and analyzed for calcein incorporation into L3 vertebral trabeculae. While the fluorescent label was incorporated in wild-type bones with sharp boundaries (Fig. 3A), labeling of mutant bone surfaces was irregular and lumpy (Fig. 3B). Measurements of dynamic histomorphometry parameters confirmed the significantly lower BV/TV ratio in the mutants compared to those of their wild-type littermates (not shown). Furthermore, mineral apposition rates (Fig. 3C) and bone formation rates (Fig. 3D) in both heterozygous and homozygous mutant mice (S43A) were significantly decreased compared to those in wild-type animals. These results suggest that there are differences in osteoblast activity between S43A mutants and wild-type mice.
FIG. 3.
Reduced osteoblast activity in S43A mutant bones. Mice were injected with calcein, and dynamic histomorphometry analysis was performed on L3 vertebrae. Calcein labeling boundaries were sharp in wild-type animals (A) but lumpy and irregular in S43A mutants (B). Bars, 10 μm. Quantification confirmed significantly lower mineral apposition rates (MAR) (C) and bone formation rates (BFR) (D) in S43A mutants. ***, P < 0.001 (n = 10).
Osteocalcin and Col1a1 gene expression is downregulated in S43A mutants.
Gene expression was assayed in calvarial mRNA extracted from 6-week-old mice. Gene expression of the known αNAC target osteocalcin was significantly decreased in heterozygous and homozygous mutant mice compared to that in their wild-type littermates (Fig. 4A). There were no significant changes in the expression of the osteoblastic marker genes for Runx2, osterix, ATF4, osteopontin, bone sialoprotein, and αNAC itself (not shown). Also, RANKL expression was unchanged (not shown), which is in accord with the unchanged number of osteoclasts in the mutant mice (Fig. 2F). Since the amount of matrix synthesized by S43A mutant osteoblasts was reduced (Fig. 2D), we measured the expression levels of the α1 chain of the most abundant bone matrix protein, type I collagen [α1(I) collagen gene, Col1a1]. RT-qPCR analysis showed reduced expression of Col1a1 (Fig. 4B), suggesting that the gene for α1(I) collagen could be a target of the αNAC coactivating function. This was tested in transient transfection assays with MC3T3-E1 osteoblastic cells (41) using a Col1a1 promoter reporter vector that included the first intron of the gene, which contains a characterized AP-1-binding site (24). Indeed, c-Jun activated the transcription of the reporter gene (Fig. 4C). This transcriptional activity was potentiated by αNAC coexpression (Fig. 4C). Interestingly, coactivation of c-Jun-mediated transcription from the Col1a1 promoter did not require the DBD of αNAC (residues 69 to 80) since DBD-lacking αNAC, Δ69-80 (reference 1), was as potent as wild-type αNAC in the transfection assay (Fig. 4C). Despite the lack of requirement for DNA binding by αNAC to coactivate Col1a1 transcription, ChIP with the αNAC antibody demonstrated that αNAC is recruited to the intronic α1(I) collagen AP-1 response element in living osteoblasts that express Col1a1 (Fig. 4D).
FIG. 4.
The αNAC S43A mutation results in reduced osteocalcin and α1(I) collagen gene transcription. Osteocalcin (A) and α1(I) collagen (B) gene expression was determined by RT-qPCR on calvarial mRNA from 6-week-old mice. (C) MC3T3-E1 cells were transiently transfected with a Col1a1-luciferase reporter and expression vectors for c-Jun, αNAC, and DBD-lacking αNAC (Δ69-80), alone or in combination. Results are the mean ± SEM of a representative transfection performed in triplicate. RLUs, relative light units. (D) ChIP. Formaldehyde-cross-linked chromatin from osteoblastic MC3T3-E1 cells was immunoprecipitated with IgG or an antibody against αNAC. Ethidium bromide-stained agarose gels of PCR products obtained with primers flanking the AP-1 binding site within the mouse α1(I) collagen gene are shown. Input, amplification of DNA prior to immunoprecipitation; CS, coding sequence control primers; M, molecular size markers. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These data identify Col1a1 as a novel target of the αNAC coactivating activity in osteoblasts and show that inhibition of the translocation of αNAC into the nucleus resulted in decreased Col1a1 and osteocalcin gene transcription.
Blood biochemistry.
Blood analysis revealed no difference between the levels of steady-state, nonfasting glucose, ALP, calcium, phosphorus, FGF23, and type I collagen degradation products (CTX) in the mutant mice and those of their wild-type littermates (Table 1). To determine if a perturbation of the endocrine function of osteocalcin, mediated by the circulating, noncarboxylated form of the protein, might contribute to the phenotype of S43A mutant mice, we determined the level of γ-carboxylation of osteocalcin in serum. γ-carboxylated osteocalcin has a higher affinity for HA than does uncarboxylated osteocalcin (16, 32). Total osteocalcin serum levels (26.3 ± 2.1 ng/ml, 23.6 ± 2.4 ng/ml, and 24.7 ± 1.1 ng/ml [mean ± SEM, n = 7] in WT, het, and S43A mutant littermates, respectively) and relative levels of HA-bound osteocalcin were unchanged in S43A mutants compared to those of their wild-type littermates (Table 1). This supports the notion that the S43A mutation has no effect on the endocrine role of osteocalcin. Taken together, the blood biochemistry results show that the S43A mutant phenotype is not caused by endocrine dysfunction.
TABLE 1.
Blood biochemistry
| Strain | Glucose (mmol/liter) | ALP (U/liter) | Calcium (mmol/liter) | Phosphorus (mmol/liter) | FGF23 (pg/ml) | CTX (ng/ml) | HA-bound OCN (%) |
|---|---|---|---|---|---|---|---|
| Wild type | 13.27 ± 0.95a | 198.4 ± 6.9 | 2.152 ± 0.015 | 3.468 ± 0.169 | 62.19 ± 4.60 | 62.37 ± 3.76 | 70.75 ± 1.17 |
| het | 14.57 ± 0.99 | 187.5 ± 8.9 | 2.167 ± 0.014 | 3.186 ± 0.148 | 60.51 ± 4.60 | 66.38 ± 2.26 | 74.37 ± 1.79 |
| S43A mutant | 13.74 ± 0.81 | 206.1 ± 6.4 | 2.184 ± 0.016 | 3.288 ± 0.168 | 68.78 ± 7.61 | 69.80 ± 5.01 | 74.40 ± 1.09 |
Data are presented as mean ± SEM.
S43A mutants show a decreased mineralization lag time and an increased osteocyte number.
To further characterize the bone phenotype of S43A mutant mice, we analyzed parameters related to matrix mineralization. Analysis of Goldner-stained proximal tibia sections revealed a significant increase in mineralized volume over bone volume (MinV/BV) in the S43A mutants compared to that in wild-type mice (Fig. 5A). This was accompanied by a significant decrease in mineralization lag time (Fig. 5B). We reasoned that this accelerated mineralization could result in the formation of immature bone in which osteoblasts are rapidly encased in the mineralized matrix. Counting of osteocytes revealed a significant increase in osteocyte number per mm2 in the trabeculae of the secondary spongiosa of the proximal tibia (Fig. 5C). Consequently, the expression of the osteocyte differentiation markers DMP-1 (Fig. 5D), E11 (Fig. 5E), and SOST (Fig. 5F) was significantly upregulated in S43A mutants compared to that in the wild type. Analysis of the Goldner-stained bone matrix under polarized light revealed typical lamellar bone in the secondary spongiosa of wild-type proximal tibias (Fig. 5G). However, the S43A mutants showed mainly woven bone with very little lamellar structure (Fig. 5H). Taken together, these results suggest that the low bone volume phenotype of the S43A mutants is due to a mineralization defect in which osteoid tissue gets mineralized too quickly, leading to the formation of immature, woven-type bone rich in osteocytes.
FIG. 5.
Accelerated mineralization associated with an osteocyte-rich, woven-type bone in S43A mutants. Dynamic histomorphometry of proximal tibia sections revealed a significant 3.4% increase in mineralized volume over bone volume (A) in the S43A mutants compared to that in their wild-type littermates. This was due to a significant decrease in mineralization lag time (B). **, P < 0.01; ***, P < 0.001 (n = 10). An elevated number of osteocytes (C), accompanied by increased expression of the osteocytic marker genes DMP-1 (D), E11 (E), and SOST (F), was measured in the S43A mutants compared to that in wild-type mice. Under polarized light, typical lamellar bone was observed in the secondary spongiosa of wild-type proximal tibias (Fig. 5G). However, the S43A mutants showed mainly woven bone with very little lamellar structure (Fig. 5H), a sign of accelerated and immature mineralization. Bars, 50 μm.
S43A primary osteoblast cultures mineralize their matrix faster.
To determine if the phenotype of S43A mutant bone is cell autonomous, primary cells from 2- to 6-week-old mouse calvaria were isolated and expanded. BrdU-based cell proliferation assays showed no significant differences among the three genotypes at 48 h after seeding (not shown). However, 2 weeks after reaching confluence, Von Kossa staining showed more mineralized matrix deposition by primary cells from S43A mutant mice than from wild-type animals (Fig. 6). These results show that there is a cell-autonomous mineralization defect and support an acceleration of osteoblast maturation in S43A mutants.
FIG. 6.
Cell-autonomous, accelerated mineralization in S43A mutant osteoblast cultures. Primary cells isolated from 2- to 4-week-old mouse calvaria mineralized their matrix faster than did cells from their wild-type littermates, as determined by Von Kossa staining. This representative picture shows two wild-type and two S43A mutant cell samples at day 14 postconfluence. The assay was repeated three times with identical results by using two to six mice of each genotype per experiment.
DISCUSSION
Using biochemical and cell culture assays, we have previously shown that αNAC is a substrate for the kinase activity of ILK, the integrin-linked kinase (35). ILK is activated upon adhesion of cells to the extracellular matrix through integrin receptors (47). Activated ILK phosphorylates, among other targets, the αNAC coactivator at residue serine 43 (35). The serine 43-phosphorylated αNAC protein translocates to the nucleus, where it acts as a coactivator of c-Jun-mediated gene transcription (1, 35, 37), and one characterized target is the osteoblast differentiation osteocalcin marker gene (1, 2). In this study, we have engineered a strain of mice in which αNAC serine residue 43 is mutated to a nonphosphorylatable alanine residue. This mutation led to reduced nuclear translocation of the coactivator, decreased osteocalcin gene promoter occupancy and transcription, and a reduced amount of bone characterized by an immature, woven-type-like structure. Our results identified Col1a1 as a novel αNAC coactivating target and are the first demonstration of the in vivo role of αNAC as a coactivator of osteoblastic gene transcription. Interestingly, while the S43A mutation affected all tissues and was not targeted to bone, the major phenotypic manifestation of the mutation was observed in the skeleton. This supports our initial hypothesis that αNAC is a key regulator of osteoblastic gene expression and function (27).
In the cytoplasm, αNAC dimerizes with βNAC to form the NAC heterodimeric complex which appears to be involved in the translation and translocation of proteins (3, 5, 12-15, 44, 45). Conventional targeting and inactivation of the αNAC gene are embryonic lethal before embryonic day 10.5 (Akhouayri and St-Arnaud, unpublished), and we surmise that this phenotype is related to the function(s) of the protein within the cytosol. We therefore aimed at designing a mouse model which would inhibit αNAC's nuclear role while leaving the cytoplasmic functions intact. By altering the serine 43 residue to an alanine (S43A) by knock-in mutagenesis, we were able to inhibit the ILK-mediated translocation of αNAC into the nucleus with no deleterious embryonic lethality phenotype. The S43A mutation resulted in a marked decrease in the amount of nuclear αNAC. However, we were still able to detect residual amounts of the protein within the nucleus, suggesting that mechanisms other than ILK-mediated phosphorylation may control the subcellular shuttling of αNAC. The αNAC protein is extensively posttranslationally modified (34-36), and it will be interesting to determine if phosphorylation at other residues, or another type of processing, can direct the coactivator to the nuclear compartment.
The lower amount of αNAC in the nucleus resulted, as hypothesized, in a significant downregulation of the expression of the osteocalcin gene, thus confirming that the osteocalcin gene is a bona fide αNAC target gene in vivo. Osteocalcin gene transcription was downregulated by approximately 47% in the heterozygous mice and further reduced by 58% in the S43A mutants, suggesting a possible gene dosage effect. The circulating, uncarboxylated form of the osteocalcin protein has recently been shown to mediate the endocrine regulation of energy metabolism by the skeleton (22). Mice lacking osteocalcin (10) have an abnormal amount of visceral fat and exhibit decreased β-cell proliferation, glucose intolerance, and insulin resistance (11, 22). Thus, the phenotype of S43A mutant mice could, in theory, be secondary to a perturbation of the endocrine role of osteocalcin caused by the decrease in osteocalcin gene expression measured in the mutants. We ruled out this possibility by measuring equal amounts of total and uncarboxylated osteocalcin in the serum of S43A mutant mice and their wild-type littermates. It should also be noted that S43A mutant mice are lean. Other biochemical indices such as steady-state glucose levels, calcemia, phosphatemia, and circulating FGF23 levels, which all show no difference between wild-type mice and their mutant littermates, support the conclusion that the phenotype of S43A mutant mice is not caused by endocrine perturbation. In addition, serum concentrations of collagen cross-links, an indirect assessment of resorptive activity, were unchanged in S43A homozygotes, as were the numbers of osteoclasts per bone or erosion surface. This suggests that alterations in osteoclast differentiation and function were not responsible for the mutant phenotype.
Mice homozygous for the S43A allele were osteopenic, with normal amounts of osteoblasts that showed reduced activity. The decreased amount of osteoid (unmineralized) matrix was a combination of reduced type I collagen expression and accelerated mineralization. In examining the mechanism responsible for reduced Col1a1 mRNA levels, we uncovered a role for αNAC in coactivating AP-1-dependent transcription of the α1(I) collagen gene. This activity did not require the DBD of αNAC, and indeed, we did not detect αNAC consensus binding sites in the vicinity of the intronic AP-1 element of the α1(I) collagen gene. Nevertheless, the coactivator was recruited to this AP-1 response element, most likely through protein-protein interactions with c-Jun (27, 37). Thus, Col1a1 was confirmed as a novel target for αNAC coactivation activity in vivo and as the second gene identified that responds to αNAC without the requirement for DNA binding. We have previously shown that the DBD-lacking αNAC protein can coactivate matrix metalloproteinase 9 (MMP-9) gene transcription in transient transfection assays (1). However, MMP-9 expression was not affected in S43A mutant bone (not shown), suggesting that it may not represent a target of the αNAC coactivating function in vivo. Gene expression monitoring and ChIP-chip assays between wild-type and S43A mutant osteoblasts are required to identify all of the relevant αNAC target genes and discriminate between targets that require DNA binding of the coactivator and those that do not.
One striking feature of the phenotype is the observation that the reduced quantity of bone matrix deposited by the mutant osteoblasts was actually mineralized faster, as demonstrated by the significantly reduced mineralization lag time. The reduced amount of matrix and accelerated mineralization translate into a lower BV/TV ratio. The accelerated-mineralization phenotype was cell autonomous, as cells isolated from the calvaria of S43A mutant mice mineralized their matrix faster than did cells from wild-type animals. The faster mineralization also produced an immature, woven type of bone characterized by poor lamellation. Woven bone is mechanically weaker, and it remains puzzling that we did not measure a significant decrease in the biomechanical properties of the S43A mutant bones. We did observe a trend toward reduced stiffness (data not shown). It is possible that this trend could reach statistical significance if we increased the cohort size or assessed biomechanics at a different age. Immature, woven bone often displays high numbers of osteocytes within the matrix (17, 29). When we measured the number of osteocytes in bone matrix, the S43A mutant mice had over 50% more osteocytes in the trabeculae of the proximal tibia than did wild-type animals.
This histomorphometric finding was supported by the detection of increased expression of the genes for the osteocyte differentiation markers DMP-1, E11, and SOST in S43A mutant bones. Since αNAC functions as a coactivator and not as a corepressor, we reasoned that the increase in the expression of these genes in αNAC mutant cells was indirect and that they do not represent coactivating targets of αNAC. This was confirmed for E11 and SOST in transient transfection assays using MLO-Y4 osteocytes and reporter vectors for each gene (see Fig. 2B and C posted at http://www.mcgill.ca/files/humangenetics/aTRMSuppl-Fig02-3bHZ.pdf). Surprisingly, AP-1-dependent transcriptional activity from the DMP-1 promoter was enhanced by cotransfection with wild-type αNAC in osteocytes (see Fig. 2A at the URL mentioned above). This suggests a different mechanism of action of the αNAC protein in regulating DMP-1 expression in mature bone cells that needs to be addressed in future experiments, as it is beyond the scope of the present study.
SOST encodes sclerostin, a protein known to be highly secreted by osteocytes (30, 46). Interestingly, transgenic mice overexpressing SOST show a phenotype similar to that of the S43A αNAC mutants. They are osteopenic, with less trabecular bone, thin cortices, impaired lamellar bone formation, low mineral apposition rates (MAR) and bone formation rates (BFR), a decreased osteoid area, and a decreased osteoblast surface, without changes in bone resorption parameters (46). But in contrast to S43A mutant cells, mesenchymal cells isolated from SOST transgenic mice show a reduced mineralization capacity compared to that of their wild-type littermates, suggesting that impaired osteoblastic differentiation and activity, rather than accelerated osteoblast maturation, could be responsible for the osteopenic phenotype in SOST transgenic mice (46).
A phenotype strikingly similar to that of S43A homozygous mice is that reported for the deletion of the G protein subunit Gsα in early osteoblasts through the action of the osterix-Cre transgene (39) on the floxPed Gsα allele (49). Gsα is a heterotrimeric G protein subunit mediating cyclic-AMP-dependent signaling downstream of G protein-coupled receptors (19). Preliminary results from the analysis of the phenotype of osterix-Cre:Gsα(fl/fl) mice show reduced bone mass, reduced expression of the osteocalcin gene, and abnormal persistence of woven bone (48). Increased expression of E11 and SOST was detected (48). Moreover, similar to S43A mutant osteoblasts, primary calvarial osteoblasts from osterix-Cre:Gsα(fl/fl) mice demonstrated accelerated mineralization when cultured under osteogenic conditions (48). These results suggest the existence of a signaling cascade linking Gsα, αNAC, osteocalcin, and other direct and indirect targets that controls the pace of osteoblast maturation. Whether this putative cascade acts in parallel to ILK-αNAC signal transduction remains to be determined. The characterization of all of the signaling cascades involving αNAC and the identification of additional targets of the αNAC coactivating function will undoubtedly further our understanding of the molecular regulation of osteoblast maturation.
Acknowledgments
Anne George (University of Illinois at Chicago) provided the pGL3-DMP1 promoter reporter plasmid. The ECR5-2kb-SOST-luciferase plasmid was supplied by Gabriela G. Loots (Lawrence Livermore National Laboratory). We obtained the −1251bp-E11-Luc plasmid from Maria I. Ramirez (Boston University) and the −2300COL-hGH/Luc reporter plasmid from Paul Bornstein (University of Washington). The MLO-Y4 osteocytic cell line was generously provided by Lynda Bonewald (University of Missouri). We thank J. Wei and G. Karsenty (Columbia University, New York, NY) for providing the protocol to measure γ-carboxylated osteocalcin levels in serum. We are indebted to Mia Esser, Nathalie Guèvremont, and Nathalie Girard for expert animal care and use. Mark Lepik prepared the figures. Sequencing was conducted by the McGill University and Genome Quebec Innovation Centre. Blastocyst injection was performed by the McGill University core of the Quebec Network for Transgenesis. μCT analysis utilized the services of the McGill Centre for Bone and Periodontal Research Imaging Lab.
T.M. was supported in succession by the Swiss National Fund (Stipendium PBBSA-11158), by a merit scholarship from the Fonds Québécois de la Recherche sur la Nature et les Technologies (file 122244), and by a research fellowship from the Shriners of North America. This research was supported by a grant from the Shriners of North America to R.S.-A.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Akhouayri, O., I. Quélo, and R. St-Arnaud. 2005. Sequence-specific DNA binding by the alphaNAC coactivator is required for potentiation of c-Jun-dependent transcription of the osteocalcin gene. Mol. Cell. Biol. 25:3452-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhouayri, O., and R. St-Arnaud. 2007. Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by jun family members. Calcif. Tissue Int. 80:123-131. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, K. M., C. A. Semple, and R. Hartmann-Petersen. 2007. Characterisation of the nascent polypeptide-associated complex in fission yeast. Mol. Biol. Rep. 34:275-281. [DOI] [PubMed] [Google Scholar]
- 4.Bandyopadhyay, P. K., J. E. Garrett, R. P. Shetty, T. Keate, C. S. Walker, and B. M. Olivera. 2002. Gamma-glutamyl carboxylation: an extracellular posttranslational modification that antedates the divergence of molluscs, arthropods, and chordates. Proc. Natl. Acad. Sci. U. S. A. 99:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatrix, B., H. Sakai, and M. Wiedmann. 2000. The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275:37838-37845. [DOI] [PubMed] [Google Scholar]
- 6.Boskey, A. L., S. Gadaleta, C. Gundberg, S. B. Doty, P. Ducy, and G. Karsenty. 1998. Fourier transform infrared microspectroscopic analysis of bones of osteocalcin-deficient mice provides insight into the function of osteocalcin. Bone 23:187-196. [DOI] [PubMed] [Google Scholar]
- 7.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 8.Deng, J. M., and R. R. Behringer. 1995. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res. 4:264-269. [DOI] [PubMed] [Google Scholar]
- 9.Dickson, G. R. 1984. Methods of calcified tissue preparation. Elsevier, New York, NY.
- 10.Ducy, P., C. Desbois, B. Boyce, G. Pinero, B. Story, C. Dunstan, E. Smith, J. Bonadio, S. Goldstein, C. Gundberg, A. Bradley, and G. Karsenty. 1996. Increased bone formation in osteocalcin-deficient mice. Nature 382:448-452. [DOI] [PubMed] [Google Scholar]
- 11.Ferron, M., E. Hinoi, G. Karsenty, and P. Ducy. 2008. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc. Natl. Acad. Sci. U. S. A. 105:5266-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freire, M. A. 2005. Translation initiation factor (iso) 4E interacts with BTF3, the beta subunit of the nascent polypeptide-associated complex. Gene 345:271-277. [DOI] [PubMed] [Google Scholar]
- 13.Fünfschilling, U., and S. Rospert. 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George, R., P. Walsh, T. Beddoe, and T. Lithgow. 2002. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 516:213-216. [DOI] [PubMed] [Google Scholar]
- 15.Grallath, S., J. P. Schwarz, U. M. Bottcher, A. Bracher, F. U. Hartl, and K. Siegers. 2006. L25 functions as a conserved ribosomal docking site shared by nascent chain-associated complex and signal-recognition particle. EMBO Rep. 7:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Hauschka, P. V., and F. H. Wians, Jr. 1989. Osteocalcin-hydroxyapatite interaction in the extracellular organic matrix of bone. Anat. Rec. 224:180-188. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez, C. J., R. J. Majeska, and M. B. Schaffler. 2004. Osteocyte density in woven bone. Bone 35:1095-1099. [DOI] [PubMed] [Google Scholar]
- 18.Hoang, Q. Q., F. Sicheri, A. J. Howard, and D. S. Yang. 2003. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature 425:977-980. [DOI] [PubMed] [Google Scholar]
- 19.Jüppner, H., A. B. Abou-Samra, M. Freeman, X. F. Kong, E. Schipani, J. Richards, L. F. Kolakowski, Jr., J. Hock, J. T. Potts, Jr., H. M. Kronenberg, et al. 1991. A G protein-linked receptor for parathyroid hormone and parathyroid hormone-related peptide. Science 254:1024-1026. [DOI] [PubMed] [Google Scholar]
- 20.Lakso, M., B. Sauer, B. Mosinger, Jr., E. J. Lee, R. W. Manning, S. H. Yu, K. L. Mulder, and H. Westphal. 1992. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 89:6232-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, N. K., and G. Karsenty. 2008. Reciprocal regulation of bone and energy metabolism. Trends Endocrinol. Metab. 19:161-166. [DOI] [PubMed] [Google Scholar]
- 22.Lee, N. K., H. Sowa, E. Hinoi, M. Ferron, J. D. Ahn, C. Confavreux, R. Dacquin, P. J. Mee, M. D. McKee, D. Y. Jung, Z. Zhang, J. K. Kim, F. Mauvais-Jarvis, P. Ducy, and G. Karsenty. 2007. Endocrine regulation of energy metabolism by the skeleton. Cell 130:456-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liska, D. J., M. J. Reed, E. H. Sage, and P. Bornstein. 1994. Cell-specific expression of alpha 1(I) collagen-hGH minigenes in transgenic mice. J. Cell Biol. 125:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liska, D. J., J. L. Slack, and P. Bornstein. 1990. A highly conserved intronic sequence is involved in transcriptional regulation of the alpha 1(I) collagen gene. Cell Regul. 1:487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loots, G. G., M. Kneissel, H. Keller, M. Baptist, J. Chang, N. M. Collette, D. Ovcharenko, I. Plajzer-Frick, and E. M. Rubin. 2005. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 15:928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Möller, I., M. Jung, B. Beatrix, R. Levy, G. Kreibich, R. Zimmermann, M. Wiedmann, and B. Lauring. 1998. A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. U. S. A. 95:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau, A., W. V. Yotov, F. H. Glorieux, and R. St-Arnaud. 1998. Bone-specific expression of the alpha chain of the nascent polypeptide-associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol. Cell. Biol. 18:1312-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshed, M., T. Schinke, M. D. McKee, and G. Karsenty. 2004. Extracellular matrix mineralization is regulated locally; different roles of two gla-containing proteins. J. Cell Biol. 165:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noble, B. S., and J. Reeve. 2000. Osteocyte function, osteocyte death and bone fracture resistance. Mol. Cell. Endocrinol. 159:7-13. [DOI] [PubMed] [Google Scholar]
- 30.Poole, K. E., R. L. van Bezooijen, N. Loveridge, H. Hamersma, S. E. Papapoulos, C. W. Lowik, and J. Reeve. 2005. Sclerostin is a delayed secreted product of osteocytes that inhibits bone formation. FASEB J. 19:1842-1844. [DOI] [PubMed] [Google Scholar]
- 31.Powers, T., and P. Walter. 1996. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 6:331-338. [DOI] [PubMed] [Google Scholar]
- 32.Price, P. A. 1989. Gla-containing proteins of bone. Connect. Tissue Res. 21:51-60. [DOI] [PubMed] [Google Scholar]
- 33.Pudota, B. N., M. Miyagi, K. W. Hallgren, K. A. West, J. W. Crabb, K. S. Misono, and K. L. Berkner. 2000. Identification of the vitamin K-dependent carboxylase active site: Cys-99 and Cys-450 are required for both epoxidation and carboxylation. Proc. Natl. Acad. Sci. U. S. A. 97:13033-13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quélo, I., O. Akhouayri, J. Prud'homme, and R. St-Arnaud. 2004. GSK3beta-dependent phosphorylation of the alphaNAC coactivator regulates its nuclear translocation and proteasome-mediated degradation. Biochemistry 43:2906-2914. [DOI] [PubMed] [Google Scholar]
- 35.Quélo, I., C. Gauthier, G. E. Hannigan, S. Dedhar, and R. St-Arnaud. 2004. Integrin-linked kinase regulates the nuclear entry of the c-Jun coactivator alpha-NAC and its coactivation potency. J. Biol. Chem. 279:43893-43899. [DOI] [PubMed] [Google Scholar]
- 36.Quélo, I., C. Gauthier, and R. St-Arnaud. 2005. Casein kinase II phosphorylation regulates alphaNAC subcellular localization and transcriptional coactivating activity. Gene Expr. 12:151-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quélo, I., M. Hurtubise, and R. St-Arnaud. 2002. AlphaNAC requires an interaction with c-Jun to exert its transcriptional coactivation. Gene Expr. 10:255-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez, M. I., A. K. Rishi, Y. X. Cao, and M. C. Williams. 1997. TGT3, thyroid transcription factor I, and Sp1 elements regulate transcriptional activity of the 1.3-kilobase pair promoter of T1alpha, a lung alveolar type I cell gene. J. Biol. Chem. 272:26285-26294. [DOI] [PubMed] [Google Scholar]
- 39.Rodda, S. J., and A. P. McMahon. 2006. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133:3231-3244. [DOI] [PubMed] [Google Scholar]
- 40.Romberg, R. W., P. G. Werness, B. L. Riggs, and K. G. Mann. 1986. Inhibition of hydroxyapatite crystal growth by bone-specific and other calcium-binding proteins. Biochemistry 25:1176-1180. [DOI] [PubMed] [Google Scholar]
- 41.Sudo, H., H. A. Kodama, Y. Amagai, S. Yamamoto, and S. Kasai. 1983. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 96:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thotakura, S. R., N. Karthikeyan, T. Smith, K. Liu, and A. George. 2000. Cloning and characterization of rat dentin matrix protein 1 (DMP1) gene and its 5′-upstream region. J. Biol. Chem. 275:10272-10277. [DOI] [PubMed] [Google Scholar]
- 43.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wegrzyn, R. D., D. Hofmann, F. Merz, R. Nikolay, T. Rauch, C. Graf, and E. Deuerling. 2006. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 281:2847-2857. [DOI] [PubMed] [Google Scholar]
- 45.Wiedmann, B., and S. Prehn. 1999. The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett. 458:51-54. [DOI] [PubMed] [Google Scholar]
- 46.Winkler, D. G., M. K. Sutherland, J. C. Geoghegan, C. Yu, T. Hayes, J. E. Skonier, D. Shpektor, M. Jonas, B. R. Kovacevich, K. Staehling-Hampton, M. Appleby, M. E. Brunkow, and J. A. Latham. 2003. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22:6267-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, C., and S. Dedhar. 2001. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J. Cell Biol. 155:505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, J. Y., C. Maes, M. Chen, L. S. Weinstein, and H. M. Kronenberg. 2008. Deletion of the G protein subunit Gs alpha in early osteoblasts leads to accelerated osteoblast maturation and formation of woven bone with abnormal osteocytes, resulting is severe osteoporosis. J. Bone Miner. Res. 23(Suppl. 1):S71. [Google Scholar]
- 49.Wu, J. Y., L. E. Purton, S. J. Rodda, M. Chen, L. S. Weinstein, A. P. McMahon, D. T. Scadden, and H. M. Kronenberg. 2008. Osteoblastic regulation of B lymphopoiesis is mediated by Gsα-dependent signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 105:16976-16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yotov, W. V., A. Moreau, and R. St-Arnaud. 1998. The alpha chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol. Cell. Biol. 18:1303-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yotov, W. V., and R. St-Arnaud. 1996. Differential splicing-in of a proline-rich exon converts alphaNAC into a muscle-specific transcription factor. Genes Dev. 10:1763-1772. [DOI] [PubMed] [Google Scholar]
- 52.Yu, V. W., G. Ambartsoumian, L. Verlinden, J. M. Moir, J. Prud'homme, C. Gauthier, P. J. Roughley, and R. St-Arnaud. 2005. FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J. Cell Biol. 169:591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]