Abstract
mRNAs encoding mitochondrial proteins are enriched in the vicinity of mitochondria, presumably to facilitate protein transport. A possible mechanism for enrichment may involve interaction of the translocase of the mitochondrial outer membrane (TOM) complex with the precursor protein while it is translated, thereby leading to association of polysomal mRNAs with mitochondria. To test this hypothesis, we isolated mitochondrial fractions from yeast cells lacking the major import receptor, Tom20, and compared their mRNA repertoire to that of wild-type cells by DNA microarrays. Most mRNAs encoding mitochondrial proteins were less associated with mitochondria, yet the extent of decrease varied among genes. Analysis of several mRNAs revealed that optimal association of Tom20 target mRNAs requires both translating ribosomes and features within the encoded mitochondrial targeting signal. Recently, Puf3p was implicated in the association of mRNAs with mitochondria through interaction with untranslated regions. We therefore constructed a tom20Δ puf3Δ double-knockout strain, which demonstrated growth defects under conditions where fully functional mitochondria are required. Mislocalization effects for few tested mRNAs appeared stronger in the double knockout than in the tom20Δ strain. Taken together, our data reveal a large-scale mRNA association mode that involves interaction of Tom20p with the translated mitochondrial targeting sequence and may be assisted by Puf3p.
mRNA localization to distinct cellular compartments is important for the efficiency and specificity of the translation process. Synthesis of proteins at their sites of action may decrease the likelihood of ectopic protein expression and facilitate assembly of large multiprotein complexes. Two general modes for mRNA localization are known. The first, which is common for endoplasmic reticulum (ER)-associated mRNAs, necessitates translation of a short region of the protein (the signal peptide). The signal is recognized by the signal recognition particle as it emerges from the ribosome exit tunnel, and the complex that includes the mRNA, ribosome, and signal recognition particle is targeted to the ER (18). As an outcome of this process, mRNAs that encode proteins destined for the ER and the secretory pathway are associated with this compartment (7). The second mode for mRNA localization occurs prior to translation and in many cases prevents initiation of protein synthesis. Sequences or structural elements of the mRNA are bound by RNA-binding proteins, and these interact with transport factors, which direct the mRNA to its destination (5, 35, 42). Genome-wide studies indicate that localization by either mode is a broad phenomenon that encompasses many mRNAs and various cellular destinations (6, 21, 32, 38). Interestingly, we along with others have recently shown that noncoding regions may also be involved in localization of ER-associated mRNAs (1, 26, 38), demonstrating that these two modes are not mutually exclusive.
Most of the mitochondrial proteins are encoded in the nucleus and need to be imported into the organelle. Various in vitro and in vivo assays led to the widely accepted notion that import may occur posttranslationally, i.e., after the protein is fully synthesized in the cytosol (33). However, mounting evidence also supports a cotranslational import of proteins into the mitochondria. Specifically, polysomes were shown to be associated with the mitochondrial surface, and these translated a distinct set of proteins (12, 19, 20). Moreover, isolated mitochondria are associated with many different mRNAs that encode mitochondrial proteins (28, 46). Elements from both the coding region (the mitochondrial targeting signal [MTS]) and the 3′ untranslated region (UTR) were shown to be important for targeting of some of these mRNAs (4, 29). One model for localization suggests association of the nascent peptide chain (specifically, the N-terminal MTS) with receptors on the mitochondria, coupled to cotranslational insertion of the protein (24). As an outcome of this cotranslational mechanism, polysomal mRNAs become associated with the mitochondria, analogously to what is observed in the ER. However, experimental support for this hypothesis is currently lacking.
Recently, Saint-Georges et al. (41) have shown a role for Puf3p in localization of many mRNAs to the mitochondria of Saccharomyces cerevisiae. Puf3p is associated with the mitochondria outer membrane (11), and its role is mediated through interaction with UTRs. This may suggest a translation-independent mode of action. Intriguingly, however, most Puf3 targets appeared to be mislocalized also after treatment with the translation inhibitor cycloheximide (CHX), suggesting that an active translation process is required for their asymmetric localization (41). Moreover, a large number of mRNAs that are not Puf3 targets appeared to be affected from treatments with the translation inhibitors puromycin and cycloheximide (41), further supporting the existence of an additional, translation-dependent mode of mRNA targeting to the mitochondria.
The translocase of the mitochondrial outer membrane (TOM complex) is a multiprotein machinery which mediates the import of the vast majority of proteins into the mitochondria (36, 39). Its core protein (Tom40) forms a β-barrel structure and serves as the main component of the import pore. Tom20 is a peripheral component of the TOM complex that functions as a primary receptor for mitochondrial precursor proteins (15). It was hypothesized that protein receptors interact with the incoming polypeptide while it is translated, and this leads to a local increase in mRNA concentration (24). An open question is whether the TOM complex, through Tom20, interacts with polypeptides while they are translated and thereby leads to higher local concentrations of mRNAs near the mitochondria. To address this issue, we analyzed the effects of TOM20 deletion on mRNA association with mitochondrial fractions and the role of the MTS on mRNA localization. We also tested the interactions between Tom20 and Puf3. We found that Tom20 is involved in mitochondrial association of many mRNAs by a process that requires the MTS. Tom20 deletion affects the localization of Puf3p, and a strain with deletions of both Tom20 and Puf3 exhibits a growth defect under conditions that require mitochondrial optimal function.
MATERIALS AND METHODS
Yeast growth.
For mitochondrial fractionation cells were grown in YP-galactose medium (1% yeast extract, 1% peptone, and 2% galactose) at 30°C. Cells carrying expression plasmids were grown in a defined medium (0.17% yeast nitrogen base and 0.5% ammonium sulfate) with the necessary amino acids and a carbon source (2% galactose unless otherwise indicated). For P-body analysis, strains were grown to mid-log phase, and P-body assembly was induced by washing cells in distilled water and further incubation in water for 10 min before fixation. Cells either before or after induction were fixed with 4% formaldehyde, washed twice with phosphate-buffered saline (PBS), and suspended with 10% antifade solution (2% propyl-gallate, 90% glycerol, 10% PBS) in PBS.
Yeast strains and plasmids.
The following strains were used in this study: yA1 (BY4741; MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Euroscarf), yA163 (MATα ura3 leu2 his4 lys2 tom20::LEU2) and its isogenic parental strain (yA162) (25), yA329 (MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 puf3::PUF3-TAP-URA3) (Euroscarf), yA330 (BY4741; puf3::kanMX4), yA332 (MATα leu2 tom20::LEU2 puf3::kanMX4) (this study), yA333 (MATα his4 leu2 ura3 tom20::LEU2 puf3::PUF3-TAP-URA3) (this study), and yA409 (BY4741; tom7::kanMX4).
The following plasmids were used in this study: pA381 (TOM20 promoter-TOM20 ORF-Flag tag-TOM20 3′ UTR in pRS416 vector [this study], where ORF is open reading frame), pA429 (PUF3 promoter-PUF3 ORF-ADH1 3′ UTR in pRS416 vector [this study]), pA520 (ADH1 promoter-Su9 MTS-GFP-CYC 3′ UTR [48], where GFP is green fluorescent protein), pA534 (same as pA520 except that the Su9 MTS was deleted by HindIII and KpnI restriction, fill-in, and ligation), pA559 (same as pA534 but with addition of 448 bp downstream of the BCS1 ORF, cloned into the XhoI and XbaI sites), pA560 (same as pA559 except that the Su9 MTS was deleted as in pA534), and pA554 (pDHH1-GFP [47]). pA552 (Dcp2-RFP [44], where RFP is red fluorescent protein) Constructs were created using a PCR amplification reaction and specific primers, with either genomic DNA or other plasmids as a template. PCR products were first cloned into pGEM-T easy vector (Promega) and then cut with the appropriate restriction enzymes and ligated into the multicloning site of pRS416. All clones were verified by sequencing.
Cell fractionation and RNA extraction.
Cells (100 ml) were grown at 30°C in YP-galactose medium to an optical density at 600 nm of 0.8. Spheroplasts were prepared by a 20-min incubation with zymolyase 20T (6 mg per gram of dry cells; ICN Pharmaceuticals) in sorbitol buffer (1.2 M sorbitol and 20 mM KPO4, pH 7.4). Cells were then recovered for 2 h in galactose medium supplemented with 1 M sorbitol. Cycloheximide was added to the medium (100 μg/ml), and cells were harvested immediately into 4 ml of mannitol buffer (0.6 M mannitol, 30 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM Mg acetate, 0.5 mM phenylmethylsulfonyl fluoride, 1 mg/ml heparin and 100 μg/ml CHX) and homogenized with 15 strokes in a Dounce homogenizer. The lysate was cleared from cell debris and nuclei by centrifugation (1,300 × g for 6 min at 4°C), and the supernatant was centrifuged again (10,000 × g for 10 min at 4°C). The resulting supernatant was designated the cytosolic fraction, whereas the pellet was washed with mannitol buffer and resuspended in 1 ml of mannitol buffer to form the mitochondrial fraction. Puromycin treatment was imposed by the addition of puromycin (final concentration, 0.1 mg/ml) for 30 min to the spheroplast recovery medium. For RNA extraction each fraction was mixed with an equal volume of 8 M guanidinium HCl and two volumes of ethanol. Samples were spun down, resuspended in water, and precipitated again with ethanol and sodium acetate. Half of the pellet was removed for Northern analysis (31).
Western blot analysis.
Proteins were precipitated by addition of trichloroacetic acid (15% final concentration), followed by incubation at 4°C for 1 h and centrifugation for 20 min at 13,000 × g. Precipitates were dissolved in Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were blotted onto a nitrocellulose membrane and stained with 0.2% Ponceau S to evaluate protein loading in each lane. The following antibodies were used: anti-Tom20 (dilution, 1:10,000), anti-Aco1 (1:10,000), anti-Bip1 (1:10,000), anti-Sec61 (1:500; a gift from R. P. Jensen), anti-Gas1 (1:6,000; a gift from L. Popolo), anti-GFP (1:5,000; ENCO M048-3), horseradish peroxidase-conjugated anti-Hxk1 (1:50,000; Acris R1093HRP), anti-nuclear pore complex proteins (1:5,000; Covance A488-120L), and anti-TAP (1:1,500; Sigma P1291). Enhanced chemiluminescent reactions were performed with an EZ-ECL kit (Biological Industries) according to the manufacturer's instructions.
Microarray procedures.
RNA samples (half of the amounts resulting from the cell fractionation) were purified as previously described (30). Amino-allyl cDNA synthesis was performed by standard procedures utilizing Improm II Reverse Transcriptase (Promega) in the presence of 2 mM amino-allyl dUTP and oligo(dT) primer, followed by fluorescent dye (Amersham) coupling to the amino-allyl group (30). In two experimental repeats, the two fractions were labeled with the same fluorescent dye and hybridized separately, each with a green-labeled unrelated reference RNA (type II experiment). In a third repeat the fractions were labeled with different dyes and hybridized to the same DNA microarray (type I experiment). DNA microarrays included either the entire coding region which was PCR amplified by specific primers (experiments 1 and 2) or 70-mer oligonucleotides (Operon AROS for yeast) (experiment 3). DNAs were spotted on glass slides and hybridized as described above.
Data analysis.
The DNA microarrays were scanned with a GenePix 4000B apparatus (Molecular Devices) at two wavelengths to detect emissions from both Cy5 and Cy3. The images were acquired with GenePix Pro, version 5.1, software and analyzed with Acuity, version 4.0, software (Molecular Devices). To minimize artifacts that can arise from low expression values, genes were filtered out if less than 80% of their Cy3 or Cy5 raw intensity values exceeded 2 standard deviations above background levels. Genes with inconsistent Cy5/Cy3 raw intensity ratios (regression R2 of <0.6) or gene features that were smaller than 55 μm were also removed. The microarray data were normalized using Bacillus subtilis RNA spike-in mix derived from lys (ATCC 87482), trp (ATCC 87485), dap (ATCC 87486), thr (ATCC 87484), and phe (ATCC 87483) clones. The spike-in mix (80 pg/μl lys, 160 pg/μl trp, 200 pg/μl dap, 240 pg/μl thr, and 320 pg/μl phe) was added in equal amounts (40 μl) to either cytosolic or mitochondrial fractions immediately after RNA extraction and to the reference RNA samples (5 μl). About 20 spots per spike were spotted across the complete grid of the microarray in a spatially even manner. Changes in unfractionated (total) mRNA abundances were normalized to the median of the signal intensity ratios of all data points in the microarray.
RESULTS
Mislocalized mRNAs in a TOM20 deletion strain.
Recent studies revealed that many mRNAs that encode mitochondrial proteins are associated with mitochondria (10, 28). An attractive working model for this association suggests that mRNAs are anchored to the mitochondria through their translated peptide in a manner that is analogous to cotranslational association of polysomes with the ER (24). The TOM complex, as the major protein translocase in the outer mitochondria membrane, is likely to be involved in such cotranslational targeting of mRNAs to the organelle. To test this possibility, we analyzed the effects on mRNA localization to the mitochondria following deletion of TOM20, which is a central protein receptor in the TOM complex.
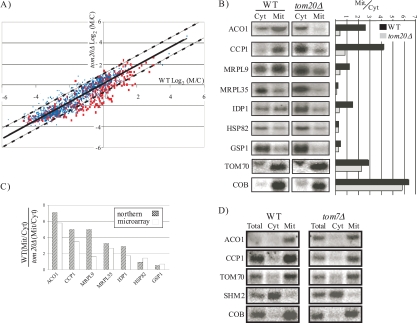
A tom20Δ strain and its parental strain (25) were grown to mid-log phase, cell walls were lysed with zymolase, and spheroplasts were recovered in high-osmolarity medium. Spheroplasts were then lysed in a buffer that lacks detergent, and cell debris or nuclei were removed by centrifugation. The mitochondria fraction was separated from the cytosol by differential centrifugation at 10,000 × g, and RNA or proteins were extracted from each fraction. Northern analyses were performed for several marker RNAs (Fig. 1A). A one-third equivalent of unfractionated RNA (Total) was also analyzed by Northern blotting, and for every gene the sum of signals in the mitochondrial and cytosolic fractions appeared about three times higher than that of the Total RNA. This indicates that no significant RNA losses occurred during fractionation. Analysis of mitochondrially encoded RNAs (15S rRNA and COB) revealed the presence of minor amounts of mitochondria in the cytosolic fraction, which appeared more significant in the more abundant 15S rRNA. This may indicate that during preparation a small fraction of the mitochondria is lysed, and some of its soluble content is released into the cytosolic fraction. Mitochondrion protein markers (Aco1p and Tom20p), however, appeared exclusively in the mitochondrial fraction (Fig. 1B) and were absent from the cytosolic fraction. The mitochondrial fraction appears to have low levels of mRNA encoding a soluble cytosolic protein (Fig. 1A, SHM2) and of a cytosolic protein marker like hexokinase (Fig. 1B, Hxk1p). These signals may indicate a limited association of these markers with mitochondria. Minor amounts of Hxk1 were indeed reported to be functionally associated with mitochondria (2). Alternatively, the signals may suggest the sedimentation of nonmitochondrial compartments in the mitochondrial fraction. We reason that further purification steps are unwanted herein as they strip the mitochondria from important complexes that are associated with them (e.g., polyribosomes) and therefore may limit conclusions regarding the in vivo roles of Tom20. Importantly, the markers' levels appeared to be the same in the wild-type and tom20Δ strains (Fig. 1B; see also Fig. 2A and B and 5; also data not shown). This suggests that similar levels of nonmitochondrial elements are present in the mitochondrial fraction of both strains. Thus, it is conceivable that effects that will be observed in the tom20Δ cells are due to the deletion itself and not the extraction procedure.
FIG. 1.
Analysis of mitochondrial and cytosolic fractions. Yeast cells were treated with zymolase and lysed with a Dounce homogenizer. Cellular compartments were fractionated to crude mitochondrial (Mit) or cytosolic (Cyt) fractions by sequential centrifugation. (A) RNA from the cytosolic or mitochondrial fractions was subjected to Northern analyses with probes recognizing mitochondrially encoded RNAs (15S rRNA and COB) or nuclear-encoded mRNAs (SEC61 and SHM2). The Total lane includes RNA that was collected before fractionation, and a one-third equivalent of the fractionated RNA was loaded. The signal in the Total lane was therefore usually one-third of the sum of mitochondrial and cytosolic signals. (B) Subcellular fractions of wild-type (WT) and tom20Δ cells were analyzed by Western blotting. Antibodies recognizing markers for the mitochondria (Aco1 and Tom20), nuclear-membrane (Npc), cytosol (Hxk1), ER (Sec61 and Bip1), and plasma membrane (Gas1) were used. Equal amounts of unfractionated protein sample (Total) were included as controls to demonstrate that there are no significant losses during fractionation. Note that the two bands that appear in the cytosolic fraction of Tom20p are nonspecific signals because they appear also in the tom20Δ strain upon longer exposure (not shown).
FIG. 2.
TOM20 deletion specifically affects mitochondrial association of hundreds of mRNAs. (A) Scatter plot where each spot indicates the relative amount of each mRNA in the mitochondrial fraction in the parental strain (wild type [WT]) and the tom20Δ cells. The solid line indicates the best-fit linear trend line, and the two dashed lines represent 2 SDs above and below the trend line. Red spots indicate genes that are designated mitochondrial in the SGD, and blue spots represent all other genes. (B) Northern analyses of the change in distribution of the indicated mRNAs in the wild-type and tom20Δ cells. Bars represent the ratio of the M/C signal in the WT and tom20Δ cells. (C) Comparison of the change in mitochondrial association obtained by the microarray analysis and the Northern analysis. (D) Northern analyses of the distribution of the indicated mRNAs in the wild-type and tom7Δ cells. Lanes are as described in the legend to Fig. 1A.
FIG. 5.
Deletion of TOM20 results in overexpression of Puf3 and its partial cytosolic location. (A) Wild-type and tom20Δ cells expressing TAP-tagged Puf3p were grown in glucose-containing medium, and a fraction of these was shifted to a medium containing either galactose (for 2 h) or ethanol (for 30 min). The level of PUF3 mRNA was determined by Northern analysis. Values were normalized to the levels of ACT1 mRNA and are relative to the value in glucose-containing medium. (B) Wild-type and tom20Δ cells were grown in a medium containing galactose and subjected to fractionation. Proteins were analyzed by Western blotting using antibodies that recognize the indicated proteins. For the analysis of the P-body marker, cells were transformed with a plasmid expressing Dhh1-GFP and grown in the appropriate selective medium. Representative fractionation markers (Aco1p and Hxk1p) are shown. (C) Wild-type and tom20Δ cells were transformed with a P-body marker (Dcp2-RFP) and grown in galactose (upper panels) or glucose, washed, and incubated for 10 min in water (lower panels). Images were acquired on an Olympus BX61TRF microscope, equipped with a DP70 digital camera under the same exposure conditions. Arrows point to P bodies. DIC, differential interference contrast.
Mitochondria were shown to be extensively associated with the ER, in part in order to facilitate transport of lipids and Ca2+ between these organelles (reviewed by references 14 and 37). Indeed, our mitochondrial preparations contain ER markers (Sec61 mRNA and protein and Bip1 protein) (Fig. 1). These markers appear at similar levels in the mitochondrial fraction from the parental and tom20Δ strain (Fig. 1B), suggesting that TOM20 deletion has no impact on localization to the ER. Consistent with that, the association of mRNAs encoding ER proteins with this fraction was not affected in the deletion strain (Fig. 2).
RNA from either the mitochondrial or cytosolic fractions was isolated, fluorescently labeled, and hybridized to DNA microarrays that contain probes for all known and predicted ORFs in the genome of S. cerevisiae. The entire experimental scheme, from cell growth to DNA microarray analysis, was repeated three times, and the results appear in Table S1 in the supplemental material (data from the third repeat is shown in Fig. 2A). A lack of signal in one fraction (mitochondrial or cytosolic) may represent exclusive localization in the other fraction, and indeed we found that the group of genes that were absent from the cytosolic fraction was enriched in genes encoding mitochondrial proteins. Yet, it is also possible that a lack of signal in a fraction is due to technical problems. Because we could not distinguish between these two possibilities, we excluded such cases from the analysis and restricted the analysis to those genes that passed the quality filtration criteria in both fractions. More than 1,000 genes passed the quality selection criteria in both fractions (mitochondria and cytosol) of both the wild type and the deletion strain in each repeat (1,025, 1,139, and 1,437 genes in repeats 1, 2, and 3, respectively). For the wild-type strain, the 10% of mRNAs with the highest mitochondria-to-cytosol signal (M/C) in experimental repeat 3 many mRNAs that encode mitochondrial proteins (P value of <10−10). We also found a high correlation between the M/C values in our experiments with values from the previous analysis of ribosomal association (46) (Spearman correlation, 0.62 ± 0.1) although the exact value per each mRNA may differ because of the different experimental and analysis methods.
Comparison of the M/C values of the wild type to those of the tom20Δ strain revealed an overall positive correlation (Fig. 2A), indicating that most mRNAs did not change their relative mitochondrial enrichment. The majority of the mRNAs that fell within ± 1 standard deviations (SD) of the trend line encoded ER, nuclear, or cytosolic proteins (P value of <10−5; based on Gene Ontology [GO] database analysis), consistent with the notion that deletion of TOM20 does not have a global effect on mRNA association with these compartments. On the other hand, the group of mRNAs with decreased mitochondrial association in tom20Δ cells (i.e., deviating by >1 SD from the trend line) was highly enriched in mRNAs encoding mitochondrial proteins (P value of <10−5 in all three experimental repeats) (see Table S1 in the supplemental material). Northern analysis was performed for several mRNAs in order to verify the microarray data by an alternative method (Fig. 2B). mRNA encoding the mitochondrial proteins Aco1, Ccp1, Mrpl9, Mrpl35, and Idp1 had reduced association with mitochondria in the tom20 mutant. This was not the case for mRNAs encoding nonmitochondrial proteins like Hsp82 and Gsp1. For all tested mRNAs, there is good agreement between the two methods regarding changes in the M/C values (Fig. 2C). We performed a genome-wide analysis for changes in steady-state, unfractionated mRNA levels in two of the experimental repeats (see Table S1 in the supplemental material). Comparison of changes in steady-state levels to changes in the M/C value revealed low, if any, correlation between them (Pearson values of 0.2 and 0.01 in experiments 1 and 3, respectively). This excludes the possibility that the changes in M/C are an outcome of an indirect effect on transcript levels.
To investigate whether any mutation in the TOM complex would lead to alteration in mRNA association, we analyzed mRNA distribution in cells lacking the small structural subunit Tom7 (24). Tom7p is not involved in recognition of precursor proteins with presequence and was shown to have an import role only after the preprotein interacts with one of the receptors in the TOM complex (e.g., Tom20 or Tom70) (17). None of the tested mRNAs appeared to be affected by Tom7 deletion (Fig. 2D), indicating that not all TOM complex subunits mediate mitochondrial association. The TOM receptors are probably the primary mediators of association, presumably with distinct targets to each; future genome-wide analyses may support this hypothesis.
The protein products from about 1,000 genes are designated as mitochondrial (GO term, mitochondrion) in the S. cerevisiae Genome Database (SGD). Of these, 155, 180, and 370 genes passed the quality selection criteria in each of the three experimental repeats (see Table S1 in the supplemental material). In each repeat, most of these genes (>80%) appeared to have lower association with mitochondria in the tom20Δ cells. Among them, about one-third showed a decrease larger than 1 SD from the general trend line of all genes (for a list of these genes, see Table S1 in the supplemental material). Thus, Tom20 is involved in mitochondrial association of most mitochondrial mRNAs. Yet the extent of decrease in tom20Δ (i.e., the dependence on Tom20 for mitochondrial association) varies between mRNAs. We aimed to find common properties in those mitochondrial mRNAs whose association with mitochondria was affected by the loss of Tom20. GO term analysis did not reveal enrichment in any function or process category that was specific to the group of affected genes. Comparison of chemical properties (8) between the mitochondrial mRNAs that were affected by TOM20 deletion and those that were not revealed a difference in the number of positive amino acids in the N terminus and the hydrophobic moment (μH) (Table 1). Moreover, on average, the mRNAs that are affected by deletion of Tom20 have a slightly higher hydrophobic moment (Table 1). However, we did not find a simple correlation between the extent of change in association and the number of positive charges in the MTS (Pearson correlation, −0.01 to 0.01). This indicates that multiple factors are involved in the Tom20-dependent association.
TABLE 1.
Comparison between affected and nonaffected mitochondrial mRNAs
| Featurea | Avg valueb |
|
|---|---|---|
| Decrease | No effect | |
| mRNA abundance (mol/cell) | 1.6 ± 0.2 | 2.0 ± 0.4 |
| ORF length (nt) | 1419 ± 140 | 1268 ± 254 |
| MTS featuresc | ||
| Hmax | 4.4 ± 0.17 | 4.5 ± 0.2 |
| μHδ | 7.6 ± 0.15 | 6.9 ± 0.3 |
| Positive amino acids (no.) | 6.9 ± 0.65 | 4.2 ± 0.7 |
| Net charge of the proteinh | 6.3 ± 2.3 | 3.1 ± 2.8 |
| Proteins with predicted cleavable MTS (% of group)d | 59.3 ± 11.2 | 36.3 ± 7.3 |
| Genes of prokaryotic origin (% of group)e | 86 ± 4.0 | 71% ± 5.6 |
| Puf3 targets | ||
| mRNAs with Puf3 binding site (% of group)f | 40.5 ± 6.7 | 35 ± 7.9 |
| mRNAs bound by Puf3 (% of group)f | 28.3 ± 1.3 | 17.3 ± 8.7 |
| Decreased association in puf3Δ cells (% of group)g | 28.6 ± 8.7 | 20.8 ± 8.6 |
| mRNAs affected by puromycin (% of group)g | 72.7 ± 14.4 | 49.5 ± 13 |
Values for each feature per experiment were averaged. Presented is the average of the three values ± SEM. Feature analysis was performed only for those genes that encode mitochondrial proteins (as listed in the SGD).
“Decrease” indicates mRNAs that decreased by more than 1 SD from the trend line. “No effect” indicates genes that were within ±1 SD from the trend line. For the complete list of genes in each group see Table S2 in the supplemental material.
Physical features of the encoded N terminal sequence were taken from reference 8. Hmax, maximal hydrophobicity of the hydrophobic face of the helical structure; μHδ, hydrophobic moment of the N-terminal sequence. Positive amino acids (K or R) were counted in the N terminus until the first negative amino acid or within the first 50 amino acids.
Assignment of prokaryotic origin was done according to data from reference 46.
Data were taken from reference 13.
Genes were considered as having decreased association if their mitochondrial localization rate decreased by >8% according to reference 41.
Sum of the charges along the protein (8).
mRNA association with mitochondria requires translating ribosomes.
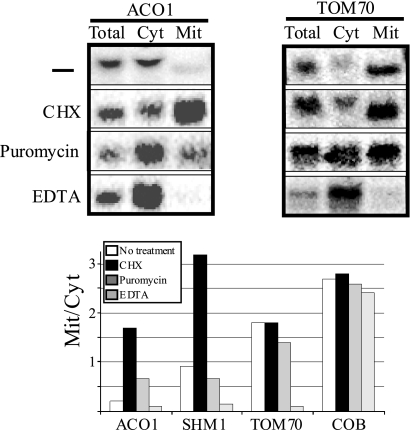
The experiments described above were performed in the presence of CHX, a drug that is known to inhibit ribosomal translocation yet maintains ribosomes on the mRNA. To pinpoint the involvement of ribosomes in the mitochondrial association, mitochondrial extracts from the wild-type strain were prepared either in the presence or absence of CHX. Samples were then fractionated, and RNAs were analyzed by Northern blotting together with a sample of unfractionated RNA (Fig. 3). Absence of CHX led to a significant reduction in the mitochondrial association of ACO1, SHM1, and FUM1, mRNAs that are affected by Tom20 deletion (Fig. 3 and data not shown). No change in association was observed for TOM70, an mRNA that was not affected by Tom20 deletion (Fig. 2B). It should be noted that previous genome-wide study has shown that CHX imposes differential effects on mRNA association (41). However, this is unlikely to be the basis for the tom20Δ effects that we observed in the experiment shown in Fig. 2 because both tom20Δ and its parental strain were treated with CHX.
FIG. 3.
Association of Tom20 target mRNAs is translation dependent. Cells were grown in a medium containing galactose and subjected to fractionation in the absence or presence of polyribosome stabilizer (CHX) or destabilizers (puromycin) or with a divalent ion chelator (EDTA). RNA samples were subjected to Northern analysis using probes that recognize Tom20 target mRNAs (ACO1 and SHM1), non-Tom20 targets (TOM70), and mitochondrially transcribed mRNA (COB). Lane loading is as described in the legend of Fig. 1. Northern blots for two representative mRNAs and a bar chart of quantitation of all mRNAs are shown.
We also tested the impact of puromycin, a drug that leads to dissociation of active ribosomes from mRNAs. The high level of association of TOM70 mRNA was not reduced by this treatment, further supporting the notion that its association is not through a translational process. These results may have a global implication because the vast majority of genes with decreased mitochondrial association in tom20Δ were shown previously to be affected by puromycin (Table 1). A treatment with the general chelator EDTA led to a significant decrease in TOM70 association (as well as all other mRNAs). This indicates that complexes that are critical for association necessitate divalent ions and that the observed association of TOM70 under all other conditions can be affected. All effects appear to be specific to nuclear-encoded mRNAs since the fractionation of a mitochondrially encoded transcript (COB) is not changed. Taken together, our data imply that an active translational process is necessary for association of Tom20 target mRNAs with mitochondria.
Role of the MTS in mRNA localization.
A possible mechanism for Tom20-mediated mRNA localization may involve the translated MTS. It can be envisaged that during translation the MTS becomes associated with an import receptor like Tom20, and this association, in turn, captures the translating mRNA near the mitochondria (24). To test this, the localization of a reporter mRNA with the MTS from subunit 9 of the Neurospora crassa Fo ATPase fused to GFP (Su9-GFP) (48) was determined (Fig. 4). This MTS interacts with Tom20p and is commonly used to study protein translocation into mitochondria (16, 48). In wild-type cells, about 50% of MTS-GFP mRNAs were localized to the mitochondria (Fig. 4). The association is due to the MTS moiety because mRNA encoding only GFP (without Su9 MTS) appeared exclusively in the cytosolic fraction. Importantly, tom20Δ cells had decreased levels of MTS-GFP mRNAs in the mitochondrial fraction (Fig. 4), thus indicating a role for Tom20 in MTS-dependent mRNA localization. It should be noted that the deletion of Tom20 did not lead to complete depletion of MTS-GFP mRNA from the mitochondria. This suggests that additional protein receptors are involved in mRNA localization and partially compensate for the absence of Tom20p.
FIG. 4.
mRNA association with mitochondria is MTS-dependent. (A) Wild-type (WT) or tom20Δ cells were transformed with a plasmid expressing either a GFP reporter alone (GFP) or GFP fused to the Su9 MTS (MTS-GFP), to the BCS1 3′ UTR (GFP-3′ UTR), or to the MTS and the 3′ UTR (MTS-GFP-3′ UTR). Cells were fractionated, and RNA samples were subjected to Northern analysis with probe recognizing GFP. The bar chart presents the quantitation of the data from the wild-type or tom20Δ cells. Error bars indicate the standard errors of the means where at least three experiments were performed. (B) Plasmids expressing either the normal ACO1 coding region (WT) or ACO1 variants (H, J, and L) carrying point mutations in the MTS (indicated in bold) were introduced into aco1Δ cells. Cells were fractionated, and the M/C signal ratio is shown (average of three experiments). The number of positive amino acids until the first negatively charged (at position 25) and the hydrophobic moment for this MTS are also indicated (40).
The 3′ UTR of some mRNAs was implicated in mitochondrial association. We therefore tested the impact of the 3′ UTR taken from a mitochondrion-associated mRNA (BCS1) on GFP localization. A putative Puf3 binding site within this 3′ UTR was shown previously to have a contribution to the mitochondrial association of BCS1 mRNA (41). In agreement with this, the GFP-3′ UTR reporter mRNA appeared to have a slightly higher mitochondrial association than GFP alone in both the wild-type and tom20Δ strains (Fig. 4A). Addition of the 3′ UTR to the MTS-GFP construct resulted in an even higher contribution to the mitochondrial association. Thus, in the context of a heterologous ORF, the BCS1 3′ UTR role in mitochondrial association appears to be more significant when an MTS and Tom20p are present. This is in agreement with the notion that a translation process is involved in targeting Puf3 target mRNAs (41). Importantly, however, the MTS contribution is much more significant for Tom20-mediated localization than the role of this 3′ UTR.
Our genome-wide analysis suggested that features that are related to the charge of the MTS may contribute to association through Tom20 (Table 1). To test this experimentally, we analyzed Tom20 target mRNAs (ACO1 variants) that carry replacements of positive amino acids in the MTS region (40) (Fig. 4B). Plasmids expressing these transcripts were inserted into aco1Δ, and the mitochondrial association of each was determined (Fig. 4B). Interestingly, the normal ACO1 mRNA (wild-type ACO) appeared to be less associated with mitochondria than the endogenously expressed transcript (M/C ratio of 1.1 for the plasmid-expressed versus 2.8 for the endogenous transcript) (Fig. 4B versus 2B). This may suggest that features outside the coding region, such as the type of promoter, genomic context, expression level, etc., may also be involved in mRNA localization. Comparison of the M/C values of ACO1 transcripts that contain mutations within the MTS (and are expressed from identical plasmids at the same level) revealed that replacements of one or two positive amino acids (Fig. 4B, mutants H and J, respectively) led to a small decrease in the M/C ratio. Replacement of three positive amino acids (mutant L) led to a more significant reduction. There is no simple linear correlation between the number of replaced amino acids and the extent of change, suggesting that other features may also contribute to the association. The changes in amino acid composition also affect other features in the MTS, such as the hydrophobic moment (μH). Indeed, mutations with higher impact on the hydrophobic moment led to a stronger decrease in mitochondrial association (Fig. 4B). This is consistent with the observation that Tom20 target genes have a higher μH than genes that were not affected by Tom deletion (Table 1). Thus, features that are related to the charge of the MTS have important roles in mRNA association with the mitochondria.
Interactions between TOM20 and PUF3.
The RNA-binding protein Puf3 was recently implicated in mRNA association with the mitochondria. To explore the relation between TOM20 and PUF3, we examined the expression levels of PUF3 in a tom20Δ strain (Fig. 5A). To that end, we constructed a strain that has a Tom20p deletion and expresses TAP-tagged Puf3 protein. Northern analysis revealed that PUF3 levels are at least three times higher in tom20Δ than in the wild-type strain in all tested media (YP supplemented with glucose, galactose, or ethanol). A similar increase occurs also at the protein level (Fig. 5B, Total lanes; also data not shown). Fractionation analysis revealed that in the TOM20 deletion strain substantial amounts (>40%) of Puf3p appear in the cytosolic fraction (Fig. 5B). This is in clear contrast to the wild-type strain, in which Puf3p is exclusively associated with the mitochondrial fraction (Fig. 5B) (11). Thus, in the absence of Tom20, Puf3 is less associated with mitochondria and is expressed at elevated levels. The increase is probably in order to maintain sufficient levels of this protein near the mitochondria.
Puf3 protein is known to be involved in mRNA degradation and was shown recently to accumulate in specific cellular loci (P bodies) under certain stress conditions (i.e., incubation in water for 10 min) (22). Analysis of the sedimentation of a P-body marker (Dhh1-GFP) revealed that it appears mostly in the cytosolic fraction and is not affected by Tom20 deletion (Fig. 5B). This excludes the possibility that the mitochondrial association of Puf3 is due to cosedimentation of P bodies in this fraction. Microscopic analysis did not reveal any accumulation of P bodies in the tom20Δ strain when it is grown in standard galactose medium (Fig. 5C, upper panels). This is not due to a problem in the P-body assembly machinery because after 10 min in water this strain accumulates P-bodies at levels similar to those of the wild-type strain (Fig. 5C, lower panels). Because there is no accumulation of P bodies in a tom20Δ strain grown in standard medium, we suggest that the higher levels of cytoplasmic Puf3p in this strain are not in order to direct its target mRNAs to these loci. Consistent with this, we did not observe any decrease in the polysomal association of a Puf3 target mRNA (MRPL9) in the tom20Δ strain (E. Eliyahu et al., unpublished data), a decrease that is characteristic for mRNAs that are targeted to P bodies (3).
We also generated a tom20Δ puf3Δ double-deletion strain. The cells of this strain appear normal in size and express COB mRNA to the same level as the parental strains (Fig. 6A), thus indicating that it contains mitochondrial genome (i.e., it is not [rho0]). The import of a mitochondrial marker also appears normal in this strain (Fig. 6B). Growth assays on various media revealed that in a glucose-containing medium (YP-glucose), this strain grows similarly to its parental strain. In contrast, a severe growth defect of the double-deletion strain was observed with media that necessitate higher aerobic functions (YP-glycerol and YP-ethanol) (Fig. 6C and data not shown). The single-deletion strains appear to grow almost normally in these media (Fig. 6D) (13), with the exception of tom20Δ in glycerol medium (Fig. 6D); this growth defect, however, appeared to be temporary as longer incubation of the plates resulted in growth similar to that of the parental strain ((25; also data not shown). Complementation of the double-deletion strain with a plasmid that expresses either Puf3p or Tom20p from its endogenous promoter resulted in rescue of the growth defect (Fig. 6D). These results reveal that both TOM20 and PUF3 are necessary for cell viability under conditions with higher dependence on mitochondrial functions. They coincide with the notion that both translation-dependent mechanisms and untranslated domains are involved in mitochondrial association of mRNAs.
FIG. 6.
Genetic interaction between TOM20 and PUF3. (A) Strains with deletions of puf3, tom20, or both were fractionated, and the RNA levels of a mitochondrially transcribed gene (COB) were tested and appeared to be the same in all strains. (B) Western analysis of the mitochondria marker Aco1p in the different fractions of tom20Δ puf3Δ and tom20Δ strains. (C) Strains with deletions of puf3, tom20, or both and their isogenic wild types were replica plated on YP plates supplemented with glucose, galactose, or glycerol and incubated for 3 days at 30°C. (D) Rescue of the double deletion phenotype by transformation of a plasmid expressing either Puf3 (pPUF3) or TOM20 (pTOM20). The indicated strains were plated in serial dilutions (10−1, 10−2, 10−3, 10−4, and 10−5) on YP plates supplemented with glucose, galactose, or glycerol.
We tested whether the double deletion affects the mitochondrial association of a representative group of mRNAs, two that appeared to be highly associated with Puf3p (MRPL9 and TIM44) and two that had low association with this protein (ACO1 and FUM1) (13). Cells were grown for several hours on galactose medium and treated with CHX, and Northern analysis of mRNA association with mitochondrial and cytosolic fractions was performed. All four mRNAs appeared less associated with mitochondria in the double-deletion strain than in the wild-type strains (Fig. 7). Such a reduction was not observed for mRNAs encoding a cytosolic control protein (SHM2) and for the mitochondrially encoded COB mRNA (data not shown). The association (M/C ratio) of the mRNAs appeared to be lower in the double-deletion strain than that observed in the tom20Δ strain. Yet deletion of Puf3 alone did not lower the mitochondrial association of these mRNAs. Thus, under these experimental conditions the impact of the Puf3 deletion on mRNA localization becomes apparent only upon deletion of Tom20. This further indicates that Puf3 is involved in the Tom20-dependent localization mechanism. Its role may not necessarily be imposed by direct interaction with the 3′ UTR because a decrease in association occurred also for mRNAs that are not associated with it (ACO1 and FUM1).
FIG. 7.
Effect of TOM20 and PUF3 deletion on mRNA distribution. Strains with deletions of puf3, tom20, or both and their isogenic parental strains were fractionated. The distribution of the indicated mRNAs between the mitochondrial (Mit) and cytosolic (Cyt) fractions was determined for each of the strains by Northern blotting and quantification of the corresponding bands.
DISCUSSION
Mitochondrial association of mRNAs through Tom20.
In our genome-wide analysis of mRNAs, we observed that many mRNAs that are normally associated with mitochondria were dissociated from the organelle in a tom20Δ strain. This finding reveals a role for Tom20 in mRNA localization to the vicinity of mitochondria. In addition, the presence of the MTS and, specifically, positively charged amino acids within it, appears to be important for such localization. Based on these observations, we propose a working model where the N-terminal signal peptide emerges from the ribosome and interacts with the TOM complex. This engagement by the translocase leads to anchoring of the mRNA, while it is being translated, to the mitochondria. This mechanism appears to be important for many mRNAs that encode mitochondrial proteins. We cannot assign a complete list of these genes due to the technical limitation of our experimental system. Yet we note that the majority (>80%) of mitochondrial mRNAs for which we had reliable data appeared to have lower association in the tom20Δ strain. The observation that MTS-dependent localization of GFP occurs even in the absence of Tom20p suggests that additional protein receptors (presumably of the TOM complex) are involved in mRNA association to the mitochondria. Moreover, our data do not exclude the possibility that other translated features in addition to the MTS are involved in mitochondrial association of mRNAs. One such feature might be the rate of protein synthesis; proteins which are synthesized at a relatively slow rate can better interact with the TOM complex and therefore display a higher dependence on its receptors.
What is the physiological significance of mRNA association with the mitochondria? It is well established that some mitochondrial proteins can be transported posttranslationally into the mitochondria. Thus, under standard experimental procedures, and by common analysis tools, cytosolic localization of mRNA does not necessarily imply mislocalization of the protein. This is also established for mRNA targeting to the bud, where in various deletion strains mRNAs appeared mislocalized to the mother cell, yet the encoded proteins were properly localized (43). Herein, we observed that Aco1p is localized to the mitochondria in tom20Δ cells even though its mRNA is mainly cytosolic (Fig. 1). Therefore, mRNA localization might become important under conditions which require high efficiency of mRNA translation, for example, under limiting growth conditions. This was observed for the ER, which was shown to be a privileged site for translation under conditions that inhibit global cellular translation upon coxsackie B virus infection (23). Alternatively, since all mitochondria proteins need to be inserted in an unfolded state, cotranslational insertion minimizes the chances of premature folding or the necessity of specific chaperones. This process might be especially relevant for proteins (like Fum1, Sod2, and Aky2) where a premature cytosolic folding may alter their subcellular distributions (27, 45, 49). Identifying the contribution of mRNA localization to protein import will probably necessitate experimental tools with high temporal and spatial resolution.
Interaction between TOM20 and PUF3.
Puf3p was implicated in the mitochondrial localization of about 150 to 200 mRNAs by a mechanism that is mediated through elements in the 3′ UTR (41). Comparison of the mRNAs that were affected in tom20Δ to those affected in puf3Δ cells revealed a rather partial overlap (Table 1). In addition, about half of the mRNAs affected by TOM20 deletion do not have a predicted Puf3 binding site and are not associated with it (as revealed by pull-down assay) (13). This suggests that at least two alternative mechanisms of mRNA targeting to the mitochondria exist: one that is mediated by Tom20 and involves cotranslational targeting and a one that is mediated by Puf3p and utilizes sequence elements from the 3′ UTR. The observed synthetic lethality of TOM20 and PUF3 on a nonfermentable carbon source suggests that both pathways are necessary for viability when fully functional mitochondria are crucial. These two mechanisms are unlikely to be mutually exclusive, and a functional overlap of both pathways is very likely. Such cross talk is supported by the higher expression levels of Puf3 and its cytosolic location in the absence of Tom20. Moreover, many mRNAs that appeared to be mislocalized in puf3Δ were also affected by CHX treatment (41).
Tom20p and Puf3p are associated with the outer membrane of the mitochondria and involved in mRNA localization. However, we did not detect direct interactions between them, either by two-hybrid analysis or coimmunoprecipitation with TAP-Puf3 (data not shown). This may indicate that the interaction between Tom20p and Puf3p is indirect. Our current working model is that Tom20p induces association of many mRNAs with the mitochondria while they are being translated, and Puf3p serves as a secondary anchor for some of these mRNAs through interaction with their 3′ UTRs.
We conclude that yeast cells possess an mRNA mitochondrial association mode that involves Tom20 and the translated MTS. The RNA binding protein Puf3 assists in the association of some mRNAs, presumably through interaction with elements in their 3′ UTRs. Both Puf3p and Tom20p are necessary for viability under conditions that depend on optimal mitochondrial function. Future studies will concentrate on the cross talk between Puf3 and Tom20 and will address the potential involvement of additional members of the TOM complex in mRNA localization.
Supplementary Material
Acknowledgments
We thank M. Choder, M. Glickman, A. Harel, R. P. Jensen, and L. Popolo for strains and reagents.
This work was supported by grants from the ISF and the Niedersachsen ministry (Y.A.), the Swiss National Science Foundation (A.P.G.), and the Deutsche Forschungsgemeinschaft (D.R.).
Footnotes
Published ahead of print on 26 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aragon, T., E. van Anken, D. Pincus, I. M. Serafimova, A. V. Korennykh, C. A. Rubio, and P. Walter. 2009. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandina, I., J. Graham, C. Lemaitre-Guillier, N. Entelis, I. Krasheninnikov, L. Sweetlove, I. Tarassov, and R. P. Martin. 2006. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta 1757:1217-1228. [DOI] [PubMed] [Google Scholar]
- 3.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corral-Debrinski, M., C. Blugeon, and C. Jacq. 2000. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20:7881-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czaplinski, K., and R. H. Singer. 2006. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 31:687-693. [DOI] [PubMed] [Google Scholar]
- 6.Diehn, M., R. Bhattacharya, D. Botstein, and P. O. Brown. 2006. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehn, M., M. B. Eisen, D. Botstein, and P. O. Brown. 2000. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 25:58-62. [DOI] [PubMed] [Google Scholar]
- 8.Dinur-Mills, M., M. Tal, and O. Pines. 2008. Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS ONE 3:e2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, M., X. Darzacq, T. Delaveau, L. Jourdren, R. H. Singer, and C. Jacq. 2007. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell 18:362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Rodriguez, L. J., A. C. Gay, and L. A. Pon. 2007. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 176:197-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George, R., P. Walsh, T. Beddoe, and T. Lithgow. 2002. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 516:213-216. [DOI] [PubMed] [Google Scholar]
- 13.Gerber, A. P., D. Herschlag, and P. O. Brown. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2:E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz, J. G., and I. R. Nabi. 2006. Interaction of the smooth endoplasmic reticulum and mitochondria. Biochem. Soc. Trans. 34:370-373. [DOI] [PubMed] [Google Scholar]
- 15.Hachiya, N., K. Mihara, K. Suda, M. Horst, G. Schatz, and T. Lithgow. 1995. Reconstitution of the initial steps of mitochondrial protein import. Nature 376:705-709. [DOI] [PubMed] [Google Scholar]
- 16.Haucke, V., T. Lithgow, S. Rospert, K. Hahne, and G. Schatz. 1995. The yeast mitochondrial protein import receptor Mas20p binds precursor proteins through electrostatic interaction with the positively charged presequence. J. Biol. Chem. 270:5565-5570. [DOI] [PubMed] [Google Scholar]
- 17.Honlinger, A., U. Bomer, A. Alconada, C. Eckerskorn, F. Lottspeich, K. Dietmeier, and N. Pfanner. 1996. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 15:2125-2137. [PMC free article] [PubMed] [Google Scholar]
- 18.Keenan, R. J., D. M. Freymann, R. M. Stroud, and P. Walter. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755-775. [DOI] [PubMed] [Google Scholar]
- 19.Kellems, R. E., V. F. Allison, and R. A. Butow. 1974. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem. 249:3297-3303. [PubMed] [Google Scholar]
- 20.Kellems, R. E., V. F. Allison, and R. A. Butow. 1975. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J. Cell Biol. 65:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecuyer, E., H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak, and H. M. Krause. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131:174-187. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. I., A. M. Dudley, D. Drubin, P. A. Silver, N. J. Krogan, D. Pe'er, and D. Koller. 2009. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 5:e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerner, R. S., and C. V. Nicchitta. 2006. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA 12:775-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lithgow, T. 2000. Targeting of proteins to mitochondria. FEBS Lett. 476:22-26. [DOI] [PubMed] [Google Scholar]
- 25.Lithgow, T., T. Junne, C. Wachter, and G. Schatz. 1994. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J. Biol. Chem. 269:15325-15330. [PubMed] [Google Scholar]
- 26.Loya, A., L. Pnueli, Y. Yosefzon, Y. Wexler, M. Ziv-Ukelson, and Y. Arava. 2008. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA 14:1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luk, E., M. Yang, L. T. Jensen, Y. Bourbonnais, and V. C. Culotta. 2005. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 280:22715-22720. [DOI] [PubMed] [Google Scholar]
- 28.Marc, P., A. Margeot, F. Devaux, C. Blugeon, M. Corral-Debrinski, and C. Jacq. 2002. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margeot, A., C. Blugeon, J. Sylvestre, S. Vialette, C. Jacq, and M. Corral-Debrinski. 2002. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 21:6893-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melamed, D., and Y. Arava. 2007. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol. 431:177-201. [DOI] [PubMed] [Google Scholar]
- 31.Melamed, D., L. Pnueli, and Y. Arava. 2008. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 14:1337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mili, S., K. Moissoglu, and I. G. Macara. 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Paquin, N., and P. Chartrand. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105-111. [DOI] [PubMed] [Google Scholar]
- 36.Perry, A. J., K. A. Rimmer, H. D. Mertens, R. F. Waller, T. D. Mulhern, T. Lithgow, and P. R. Gooley. 2008. Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol. Biochem. 46:265-274. [DOI] [PubMed] [Google Scholar]
- 37.Pinton, P., C. Giorgi, R. Siviero, E. Zecchini, and R. Rizzuto. 2008. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyhtila, B., T. Zheng, P. J. Lager, J. D. Keene, M. C. Reedy, and C. V. Nicchitta. 2008. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 14:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapaport, D. 2005. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 171:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regev-Rudzki, N., O. Yogev, and O. Pines. 2008. The mitochondrial targeting sequence tilts the balance between mitochondrial and cytosolic dual localization. J. Cell Sci. 121:2423-2431. [DOI] [PubMed] [Google Scholar]
- 41.Saint-Georges, Y., M. Garcia, T. Delaveau, L. Jourdren, S. Le Crom, S. Lemoine, V. Tanty, F. Devaux, and C. Jacq. 2008. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 3:e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shav-Tal, Y., and R. H. Singer. 2005. RNA localization. J. Cell Sci. 118:4077-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shepard, K. A., A. P. Gerber, A. Jambhekar, P. A. Takizawa, P. O. Brown, D. Herschlag, J. L. DeRisi, and R. D. Vale. 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 100:11429-11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheth, U., and R. Parker. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strobel, G., A. Zollner, M. Angermayr, and W. Bandlow. 2002. Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell 13:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sylvestre, J., S. Vialette, M. Corral Debrinski, and C. Jacq. 2003. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol. 4:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng-Rogenski, S. S., J. L. Chong, C. B. Thomas, S. Enomoto, J. Berman, and T. H. Chang. 2003. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 31:4995-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westermann, B., and W. Neupert. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421-1427. [DOI] [PubMed] [Google Scholar]
- 49.Yogev, O., S. Karniely, and O. Pines. 2007. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 282:29222-29229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.