Abstract
Replication fork stalling caused by deoxynucleotide depletion triggers Rad53 phosphorylation and subsequent checkpoint activation, which in turn play a crucial role in maintaining functional DNA replication forks. How cells regulate checkpoint deactivation after inhibition of DNA replication is poorly understood. Here, we show that the budding yeast protein phosphatase Glc7/protein phosphatase 1 (PP1) promotes disappearance of phosphorylated Rad53 and recovery from replication fork stalling caused by the deoxynucleoside triphosphate (dNTP) synthesis inhibitor hydroxyurea (HU). Glc7 is also required for recovery from a double-strand break-induced checkpoint, while it is dispensable for checkpoint inactivation during methylmethane sulfonate exposure, which instead requires the protein phosphatases Pph3, Ptc2, and Ptc3. Furthermore, Glc7 counteracts in vivo histone H2A phosphorylation on serine 129 (γH2A) and dephosphorylates γH2A in vitro. Finally, the replication recovery defects of HU-treated glc7 mutants are partially rescued by Rad53 inactivation or lack of γH2A formation, and the latter also counteracts hyperphosphorylated Rad53 accumulation. We therefore propose that Glc7 activity promotes recovery from replication fork stalling caused by dNTP depletion and that γH2A dephosphorylation is a critical Glc7 function in this process.
Eukaryotic cells require specialized surveillance mechanisms called checkpoints to preserve genome integrity in the presence of genotoxic insults. An efficient checkpoint response is also important during S phase, where it inhibits late origin firing, prevents stalled replication fork breakdown, and promotes the restart of replication (6, 22, 23, 33, 34). Checkpoint activation requires protein phosphorylation cascades that in Saccharomyces cerevisiae are initiated by the two protein kinases Mec1 (ATR in humans), which functions in a complex with Ddc2 (27), and Tel1 (ATM in humans) (reviewed in reference 20).
Mec1 and Tel1 phosphorylate the central effector kinases Rad53 and Chk1, which transfer the arrest signal to a myriad of downstream proteins (reviewed in reference 20). Rad53 and Chk1 activation is not governed by their simple interaction with Mec1 or Tel1 but rather requires a stepwise process. Once recruited to the double-strand break (DSB) ends, Mec1 phosphorylates Rad9, which promotes the recruitment of inactive Rad53 in a forkhead-associated domain (FHA)-dependent manner, thus allowing its activating phosphorylation by Mec1 (31), as well as Rad53 in trans autophosphorylation, by increasing the local concentration of Rad53 molecules (14). Active Rad53 kinase molecules are then released from the complex and can phosphorylate downstream targets to arrest mitotic cell cycle progression. Mec1 activation is supported by independent loading onto DNA of the Ddc1-Rad17-Mec3 complex by Rad24-RFC, which enhances Mec1 ability to transmit and amplify the DNA damage signals (24).
Mec1 and Tel1 also phosphorylate histone H2A on serine 129 (γH2A) in response to DNA DSBs (12, 28, 30) and inhibition of DNA replication (7, 41). Formation of γH2A is necessary for recruitment of DNA repair and chromatin remodeling factors to DSB sites and for efficient DSB repair (reviewed in references 1 and 37).
Rad53 hyperphosphorylation is a hallmark of checkpoint activation by DNA damage or inhibition of DNA replication. As DNA damage is repaired, checkpoint-arrested cells resume cell cycle progression upon checkpoint deactivation. This process is known as recovery and is characterized by the disappearance of hyperphosphorylated Rad53 (reviewed in reference 16). Rad53 deactivation is also required for replication restart following transient exposure to methylmethane sulfonate (MMS) during early S phase (26, 32). To date, the S. cerevisiae Ser/Thr phosphatases Ptc2 and Ptc3, as well as the protein phosphatase 4 (PP4)-like protein phosphatase Pph3, have been found to be important for checkpoint recovery after a single DSB (17, 18). Furthermore, Pph3/PP4 is required for γH2A dephosphorylation, which contributes to recovery from a DSB-induced checkpoint (4, 17, 25). Finally, Pph3 and Ptc2 have been implicated in Rad53 dephosphorylation and hence deactivation during recovery from MMS exposure (26, 32).
Notably, none of the above phosphatases is required for Rad53 deactivation after inhibition of DNA replication by deoxynucleoside triphosphate (dNTP) depletion with hydroxyurea (HU), as pph3Δ ptc2Δ ptc3Δ triple mutant cells are not defective in recovering from HU-induced S phase arrest and do not show hypersensitivity to HU treatment (32, 35). Thus, a yet-unidentified phosphatase might be responsible for checkpoint termination under these conditions.
The essential S. cerevisiae gene GLC7 encodes the catalytic subunit of PP1, which is involved in a variety of cellular processes including glucose and glycogen metabolism, establishment of cell polarity, vesicle trafficking, chromatin remodeling, chromosome segregation, transcription, spindle checkpoint inactivation, and meiosis (reviewed in reference 40). Here we show that Glc7 promotes disappearance of phosphorylated Rad53 and recovery from replication fork stalling caused by HU. We also provide evidence that Glc7 regulates γH2A level in vivo and dephosphorylates it in vitro. The lack of γH2A partially suppresses the replication recovery defects and impairs Rad53 phosphorylation in HU-treated glc7 mutants, indicating that the signal keeping Rad53 active in glc7 mutant cells involves γH2A molecules. As Rad53 inactivation suppresses the replication recovery defects of glc7 mutant cells, these findings suggest that Glc7-mediated γH2A dephosphorylation allows replication fork restart by contributing to Rad53 inactivation.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
Yeast strains generated during this study were derivatives of W303. Strain genotypes are listed in Table 1. Strains expressing fully functional tagged versions of Mre11 and Ddc2 were constructed as previously described (5, 27). Strains expressing fully functional hemagglutinin (HA)-tagged versions of Glc7 were constructed by inserting three tandem HA epitopes into a NotI restriction site introduced by PCR at GLC7 codon 1. Strains carrying the glc7-129 (D137A, E138A) or glc7-132 (D165A, E166A, K167A) alleles at the GLC7 locus were constructed by transforming W303 cells with SalI-digested plasmids pRS306-glc7-129 and pRS306-glc7-132, kindly provided by K. Tatchell (North Carolina State University) (3). To generate the glc7-T152K mutant, W303 cells were transformed with SalI-digested plasmid pML582, lacking the first 124 bp of the GLC7 gene. Plasmid excisions were then selected on 5-fluoroorotic acid (FOA) plates, and clones containing the glc7-129, glc7-132, and glc7-T152K alleles were identified by searching for those able to grow on sucrose in the presence of 2-deoxyglucose, which prevents wild-type cells from fermenting sucrose (3). Plasmid pML582 was generated by cloning a HindIII-SacI DNA fragment containing the entire GLC7 open reading frame (ORF) encoding the T152K mutation from plasmid pJTL18T152K, kindly provided by M. Carlson (Columbia University, NY), into the HindIII-SacI sites of plasmid YIplac211. The HindIII-BglII DNA fragment was then excised. To generate strains carrying the GAL-GLC7 fusion, strain W303 was transformed with a centromeric plasmid carrying the entire GLC7 coding region fused to the GAL1 promoter. Strains carrying the hta2-S129A allele at the HTA2 locus were constructed by transforming W303 hta1Δ cells with MfeI-digested plasmid pRS406-hta2-S129A, kindly provided by D. Durocher (University of Toronto, Canada). The strain expressing both the nucleoside transporter hENT and the herpes simplex virus thymidine kinase (HSV TK) was kindly provided by J. Diffley (Clare Hall Laboratories, South Mimms, United Kingdom) (38).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| W303 | MATaade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 rad5-535 | 21 |
| YLL2144.1 | W303 glc7-T152K | This study |
| YLL2163.3 | W303 glc7-129 | This study |
| YLL2064.8 | W303 glc7-132 | This study |
| GLC7 | W303 trp1::TRP1 GAL-dNK leu2::LEU2 GAL-hENTb | 29 |
| glc7-T152K | W303 trp1::TRP1 GAL-dNK leu2::LEU2 GAL-hENT glc7-T152Kb | This study |
| YLL2409 | W303 hta1Δ::KanMX4 hta2-S129A::URA3 | This study |
| DMP5052/15C | W303 hta1Δ::KanMX4 hta2-S129A::URA3 glc7-T152K | This study |
| YLL2228 | W303 GLC7-3HA | This study |
| YLL683.8 | W303 DDC2-3HA::URA3 | 27 |
| YLL2264.5 | W303 glc7-129 DDC2-3HA::URA3 | This study |
| YLL2266.1 | W303 glc7-132 DDC2-3HA::URA3 | This study |
| YLL2268.9 | W303 glc7-T152K DDC2-3HA::URA3 | This study |
| YLL1072.1 | W303 MRE11-3HA::URA3 | 5 |
| YLL2263.8 | W303 glc7-129 MRE11-3HA::URA3 | This study |
| YLL2265.1 | W303 glc7-132 MRE11-3HA::URA3 | This study |
| YLL2267.3 | W303 glc7-T152K MRE11-3HA::URA3 | This study |
| YLL2379 | W303 pph3Δ::KanMX4 | This study |
| YLL2584 | W303 [URA3 CEN] | This study |
| YLL2585 | W303 [URA3 CEN GAL1-GLC7] | This study |
| YLL2586 | W303 glc7-T152K [URA3 CEN] | This study |
| YLL2587 | W303 glc7-T152K [URA3 CEN GAL1-RAD53] | This study |
| YLL2588 | W303 glc7-T152K [URA3 CEN GAL1-rad53-K227A] | This study |
| DMP5095/1A | W303 ptc2Δ::NAT ptc3Δ::HPH | This study |
| DMP5097/15D | W303 glc7-T152K ptc2Δ::NAT ptc3Δ::HPH | This study |
| DMP5097/18C | W303 glc7-T152K pph3Δ::KanMX4 | This study |
| DMP5094/2C | W303 ptc2Δ::NAT ptc3Δ::HPH pph3Δ::KanMX4 | This study |
| DMP5097/37D | W303 glc7-T152K ptc2Δ::NAT ptc3Δ::HPH pph3Δ::KanMX4 | This study |
Plasmids are indicated by brackets.
BrdU labeling.
Strains expressing the GAL-RAD53 and GAL-rad53K227A alleles were constructed by transforming wild-type and glc7-T152K strains with plasmids pNB187, pNB187-RAD53, and pNB187-rad53K227A, kindly provided by D. F. Stern (Yale University School of Medicine, New Haven, CT). Homothallic endonuclease (HO)-expressing W303 strains were obtained by transformation with a centromeric plasmid carrying a GAL-HO fusion.
Cells were grown in synthetic complete medium lacking uracil supplemented with 2% raffinose (SCraf-Ura) or 2% raffinose and 2% galactose (SCgal-Ura) or in YP medium (1% yeast extract, 2% Bacto peptone, 50 mg/liter adenine) supplemented with 2% glucose (YPD), 2% raffinose (YPraf), or 2% raffinose and 2% galactose (YPgal). Unless otherwise stated, all the experiments were carried out at the temperature of 25°C.
Histone purification.
Yeast histones were purified by acid extraction of isolated chromatin. Briefly, 500 ml of YPD medium containing 2 × 107 cells/ml in exponential growth was incubated with 10 μg/ml phleomycin. After 2 h, cells were harvested by centrifugation and resuspended in 50 ml of 20 mM HEPES (pH 7.4) and 1.2 M sorbitol and, after addition of 2.75 mg (per g of cells) of Zymolyase 20T, were incubated for 45 min at 30°C. All the subsequent steps were performed at 4°C. Following the addition of 100 ml of wash buffer [20 mM PIPES {piperazine-N,N′-bis(2-ethanesulfonic acid)}, 1.2 M sorbitol, 1 mM MgCl2, pH 6.8], the spheroplasts were pelleted by centrifugation at 3,500 × g for 5 min and lysed in 50 ml of NIB buffer (0.25 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM MES [morpholineethanesulfonic acid; pH 6.6], 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.8% Triton X-100, protease inhibitor cocktail). After incubation for 20 min, the lysate was centrifuged at 4,000 × g for 5 min and the pellet was resuspended in 50 ml of buffer A (10 mM Tris [pH 8.0], 0.5% NP-40, 75 ml NaCl, 1 mM PMSF). After incubation for 15 min, the pellet was centrifuged at 4,000 × g for 5 min and resuspended in 25 ml of buffer B (10 mM Tris [pH 8.0], 0.4 M NaCl, 1 mM PMSF). After incubation for 5 min, the pellet was centrifuged at 4,000 × g for 5 min and resuspended in 3 volumes of cold water supplemented with 5 N HCl to a final concentration of 0.25 N. After incubation for 30 min, the sample was centrifuged (30,000 × g, 10 min), and the supernatant, containing extracted histones, was supplemented with 100% trichloroacetic acid (TCA) to a final concentration of 20%. The sample was centrifuged at 30,000 × g for 30 min, and the pellet, washed with cold 0.5% HCl-acetone, was resuspended in 150 μl of 10 mM Tris, pH 8.0.
Immunoprecipitation and phosphatase assay.
Protein extracts for Glc7 immunoprecipitation were prepared from exponentially growing cells collected by centrifugation and resuspended in an equal volume (wt/vol) of 50 mM HEPES (pH 7.4), 200 mM NaCl, 0.1 mM EDTA, 0.1% sodium dodecyl sulfate (SDS), 0.3% Tween 20, 1 mM dithiothreitol (DTT), and protease inhibitor cocktail. After addition of an equal volume of acid-washed glass beads, clarified protein extracts were incubated for 1 h 40 min at 4°C with anti-HA affinity matrix (Roche). Immunoprecipitates were then washed four times with 1 ml of 50 mM HEPES (pH 7.4), 200 mM NaCl, 0.1 mM EDTA, 0.1% SDS, 0.3% Tween 20, 1 mM DTT, and protease inhibitor cocktail and resuspended in 50 mM HEPES (pH 7.4), 2 mM MnCl2, and 2 mM DTT to a final volume of 60 μl. Immunoprecipitates were preincubated with or without inhibitor 2 (1 mM) at 30°C for 15 min and then supplemented with 1.5 μg of purified histones. At the 0-, 40-, and 80-min time points after histone addition, 20 μl of the reaction mixtures was supplemented with SDS-gel loading buffer and proteins were resolved by electrophoresis on SDS-polyacrylamide gels and then subjected to Western blot analysis.
Other techniques.
Flow cytometric DNA analysis was determined on a Becton-Dickinson FACScan. Rad53 and Rad9 were detected using polyclonal antibodies kindly provided by J. Diffley and N. Lowndes (University of Galway, Ireland), respectively. γH2A was detected using anti-γH2A polyclonal antibodies kindly provided by W. M. Bonner (NIH, Bethesda, MD). H2A was detected using anti-H2A polyclonal antibodies (active motif). For Western blot analysis, protein extracts were prepared by TCA precipitation. Secondary antibodies were purchased from Amersham, and proteins were visualized by an enhanced chemiluminescence system according to the instructions of the manufacturer. Southern blot analysis at the MAT locus was performed using a 506-bp probe containing part of the MAT locus (201082 to 201588, coordinates of chromosome III). This probe is complementary also to part of the HML locus (13826 to 13918, coordinates of chromosome III). The pulse-chase bromodeoxyuridine (BrdU) experiment and immunodetection of BrdU-labeled DNA were performed as described in reference 38.
RESULTS
Glc7 promotes replication recovery after HU-induced fork stalling.
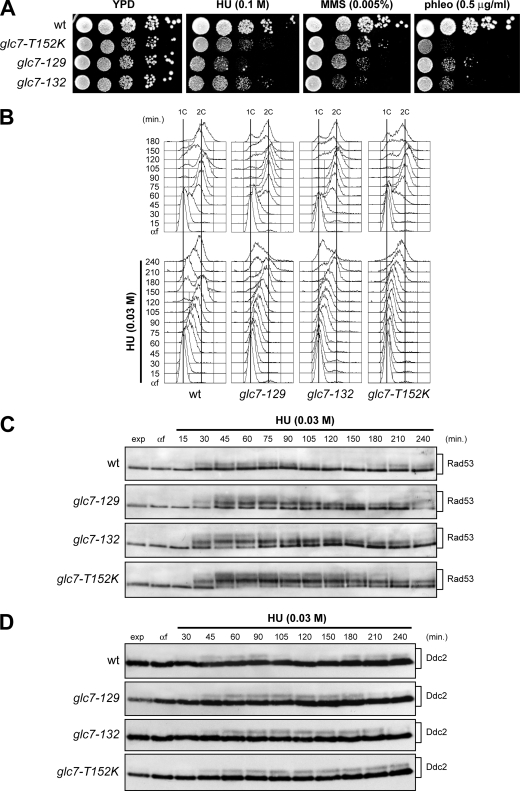
Depletion of dNTPs by HU treatment causes replication fork stalling and activation of the S phase checkpoint, which preserve the integrity of DNA replication forks and promote fork restart (reviewed in reference 42). To examine whether the Glc7 phosphatase could be involved in the inactivation of the checkpoint induced by exposure to HU, we first analyzed the sensitivity to HU of the glc7-129 and glc7-132 mutants generated by alanine scanning (3) and of the glc7-T152K mutant, which was shown to be defective in meiotic prophase (2, 36). Although to different extents, all these glc7 mutants were hypersensitive to HU, indicating that Glc7 contributes to cell viability in conditions of replicative stress (Fig. 1A).
FIG. 1.
DNA replication in HU-treated glc7 mutants. (A) Serial dilutions (1:10) of exponentially growing cultures of cells with the indicated genotypes were spotted onto YPD plates with or without MMS, HU, and phleomycin (phleo) and incubated at 25°C for 3 days. wt, wild type. (B to D) Exponentially growing wild-type, glc7-129, glc7-132, and glc7-T152K cells were arrested in G1 with α-factor (αf) and released from the pheromone block in YPD or YPD containing 0.03 M HU. Aliquots of each culture were harvested at the indicated times after α-factor release to determine DNA content by fluorescence-activated cell sorting (FACS) analysis (B) and to detect Rad53 (C) and Ddc2 (D) by Western blot analysis with anti-Rad53 and anti-HA antibodies, respectively. exp, exponentially growing cells.
Since decreased proliferation in the presence of HU might be due to defects in supporting DNA replication, we monitored progression through S phase of the glc7 mutants during a constant exposure to a low HU dose (Fig. 1B and C). Cells were arrested in G1 with α-factor and then released from the pheromone block either in the absence or in the presence of 0.03 M HU. Although glc7-T152K mutant cells completed DNA replication 15 min later than wild-type cells under unperturbed conditions, they showed a dramatic delay in completing DNA replication in the presence of HU compared to similarly treated wild-type cells (Fig. 1B). A less severe, but still significant delay was observed under the same conditions in glc7-129 and glc7-132 cells, which completed DNA replication about 60 and 90 min later, respectively, than wild-type cells (Fig. 1B). The delay of glc7 mutants in completing DNA replication upon HU-induced stress correlated with increased levels of hyperphosphorylated Rad53, which persisted longer in HU-treated glc7 mutants than in similarly treated wild-type cells, with the glc7-T152K mutant showing the strongest effect (Fig. 1C). Similarly, HU-treated glc7 mutants exhibited a more persistent Ddc2 phosphorylation than wild-type cells, where phosphorylated Ddc2 was detectable concomitantly with S phase progression in the presence of HU (Fig. 1D). Thus, defective Glc7 hyperactivates the S phase checkpoint and delays replication recovery after HU-induced fork stalling.
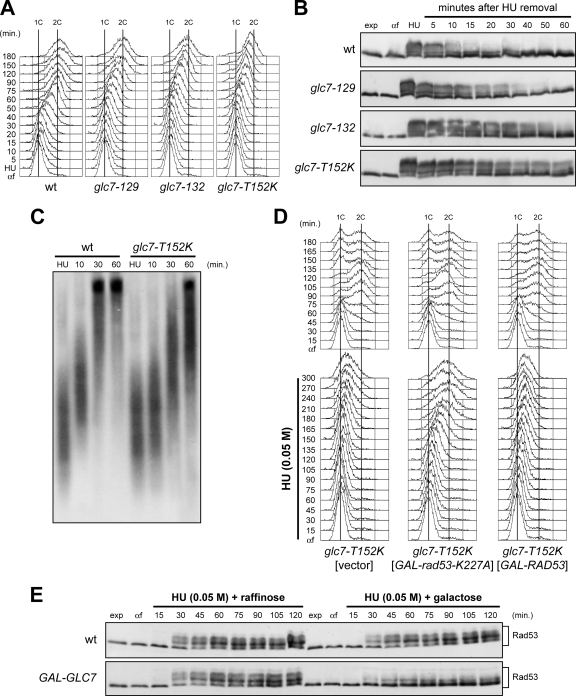
Next, we asked whether Glc7 is required for resumption of DNA replication after transient HU-induced S phase arrest (Fig. 2A and B). Cells were released from G1 arrest in the presence of 0.2 M HU for 210 min and then transferred to HU-free medium that contained nocodazole to prevent passage through mitosis. As shown in Fig. 2A, wild-type cells completed DNA replication within 60 min after release from HU, whereas completion of DNA replication occurred at about 120, 150, and 180 min after release in similarly treated glc7-129, glc7-132, and glc7-T152K cells, respectively. As in the experiment in Fig. 1B and C, these delays correlated with hyperactivation of Rad53, whose phosphorylated forms persisted longer in glc7-T152K, glc7-129, and glc7-132 cells than in similarly treated wild-type cells upon HU removal (Fig. 2B).
FIG. 2.
Restart of a stalled replication fork is defective in glc7 mutants, and this defect is suppressed by antagonizing Rad53 activity. (A and B) Wild-type (wt), glc7-129, glc7-132, and glc7-T152K cells were released from G1 arrest (αf) into YPD medium containing 0.2 M HU and incubated for 210 min (HU). Then, cultures were released into fresh medium, and samples were taken at the indicated times after HU removal to determine DNA content by FACS analysis (A) and to detect Rad53 by Western blot analysis with anti-Rad53 antibodies (B). (C) Immunodetection of BrdU-pulsed DNA. G1-arrested cells were released into YPgal containing 0.2 M HU plus 25 mM BrdU. After 1 h (HU), cells were chased with 2 mM thymidine into fresh medium, and DNA from samples taken at the indicated times after chase was prepared to detect BrdU-labeled DNA with anti-BrdU antibody. (D) glc7-T152K cells containing a URA3 centromeric plasmid, either empty or carrying the GAL-RAD53 or the GAL-rad53-K227A allele, were blocked in G1 with α-factor (αf) in SCraf-Ura and released into YPgal in the absence (top) or presence (bottom) of 0.05 M HU. Cell samples were collected at the indicated times after α-factor release to determine DNA content by FACS analysis. (E) Wild-type and GAL-GLC7 cells were released from G1 arrest (αf) in SCraf-Ura into YPraf or YPgal with 0.05 M HU. Cell samples were collected at the indicated times after α-factor release to detect Rad53 protein by Western blot analysis.
The fate of stalled replication forks after HU removal was followed more directly by labeling nascent strands with BrdU (Fig. 2C). We used isogenic strains that can incorporate BrdU into DNA when present at low concentration in the medium, because they express both the nucleoside transporter hENT and the herpes simplex virus thymidine kinase (HSV TK) (38). Wild-type and glc7-T152K cells were synchronized in G1 with α-factor and released into medium containing 0.2 M HU and BrdU. After the nascent DNA associated with the stalled forks was labeled, the BrdU was chased by transferring cells to fresh medium lacking both HU and BrdU and containing thymidine at high concentration. Consistent with previous similar experiments (29), labeled nascent DNA replication intermediates appeared as a smear in both HU-treated cultures (Fig. 2C). After release, the sizes of DNA fragments increased very quickly in wild-type cells, where almost all the incorporated BrdU was present in the high-molecular-weight fraction by 30 min, as expected for rapid restarting of stalled replication forks (Fig. 2C). In contrast, formation of high-molecular-weight molecules of nascent DNA was delayed in similarly treated glc7-T152K cells (Fig. 2C), indicating defective replication restart after HU-induced fork stalling in cells crippled for Glc7 activity.
Rad53 inactivation suppresses the replication recovery defects of glc7 mutants.
The persistence of hyperphosphorylated Rad53 in HU-treated glc7 cells raises the possibility that Glc7 might promote resumption of DNA replication by inactivating Rad53. We therefore asked whether impairing Rad53 activity could suppress the replication recovery defects of HU-treated glc7 cells. We could not use rad53Δ cells to address this point, as they fail per se to resume DNA replication after HU treatment (11). We therefore generated strains expressing from the GAL1 promoter the dominant-negative, kinase-dead Rad53-K227A variant (13), whose galactose-induced overproduction antagonizes the previously HU-activated endogenous Rad53. Cells were arrested in G1 with α-factor in raffinose-containing medium and then released from the pheromone block in the presence of galactose with or without 0.05 M HU. Analysis of bulk DNA synthesis showed that all cell cultures displayed similar kinetics of DNA replication in the absence of HU (Fig. 2D, top), while HU-treated glc7-T152K cells carrying GAL-rad53-K227A on a centromeric plasmid completed DNA replication faster than similarly treated glc7-T152K cells either overexpressing wild-type RAD53 or carrying the empty vector (Fig. 2D, bottom). Thus, Rad53 inactivation overcomes the requirement of Glc7 for recovery from DNA replication inhibition, indicating that Rad53 inactivation is the critical function of Glc7 during this recovery.
If Glc7 promotes Rad53 dephosphorylation/inactivation, its excess is predicted to counteract HU-induced Rad53 phosphorylation. Indeed, when G1-arrested cell cultures were released from a G1 arrest in the presence of 0.05 M HU, galactose-induced cells expressing an additional GLC7 copy from the GAL1 promoter (GAL-GLC7) on a centromeric plasmid showed a decreased amount of phosphorylated Rad53 compared to either uninduced GAL-GLC7 cells or galactose-induced wild-type cells carrying the empty vector (Fig. 2E). Thus, Glc7 appears to be involved, directly or indirectly, in Rad53 dephosphorylation/inactivation after HU exposure.
Glc7 is required for recovery from a DSB-induced checkpoint, while it is dispensable for checkpoint inactivation during MMS exposure.
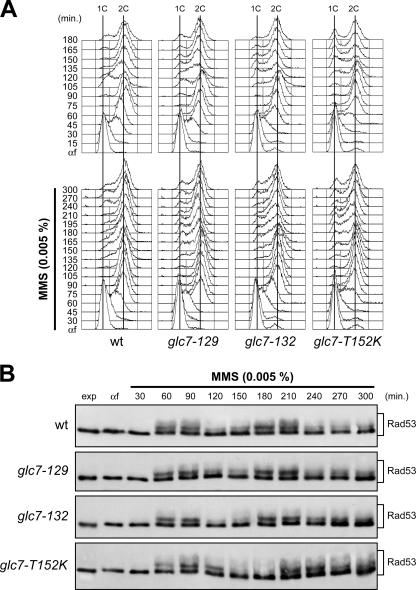
Like HU-induced nucleotide depletion, DNA methylation by MMS interferes with replication fork progression (reviewed in reference 42). Since the glc7-T152K, glc7-129, and glc7-132 mutants were hypersensitive to MMS (Fig. 1A), we asked whether Glc7 might regulate checkpoint termination also during a constant exposure to a low MMS dose (Fig. 3). Cells were arrested in G1 with α-factor and then released from the pheromone block either in the absence or in the presence of 0.005% MMS. MMS-treated wild-type, glc7-129, and glc7-132 cells progressed through S phase with similar kinetics, whereas glc7-T152K cells completed DNA replication about 15 min later than wild-type cells both in the absence and in the presence of MMS (Fig. 3A). Furthermore, both the kinetics and the amounts of Rad53 phosphorylation were similar in MMS-treated wild-type and glc7 mutants (Fig. 3B). Thus, in contrast to what was observed in recovery from HU-induced S phase arrest, Glc7 appears to be dispensable for checkpoint deactivation in the presence of MMS.
FIG. 3.
Glc7 is not involved in the recovery from MMS-induced DNA damage. Exponentially growing wild-type (wt), glc7-129, glc7-132, and glc7-T152K cells were arrested in G1 with α-factor (αf) and released from the pheromone block in YPD or YPD containing 0.005% MMS. Aliquots of each culture were harvested at the indicated times after α-factor release to determine DNA content by FACS analysis (A) and to detect Rad53 by Western blot analysis with anti-Rad53 antibodies (B).
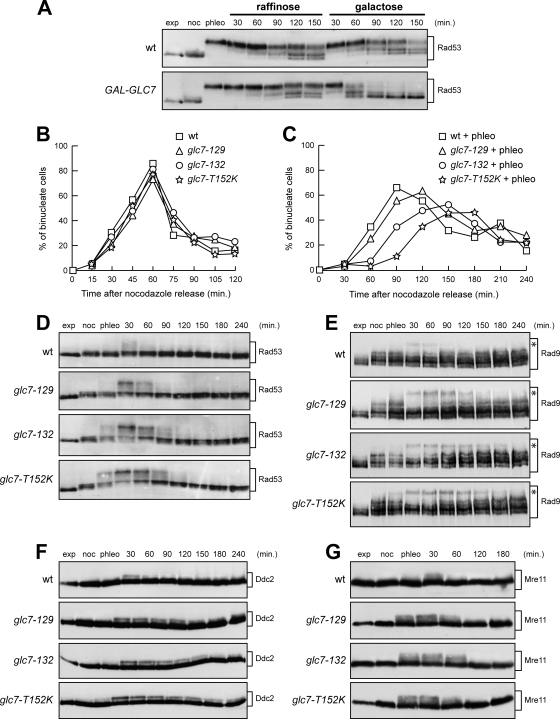
The glc7-T152K, glc7-129, and glc7-132 mutants were also hypersensitive to the radiomimetic agent phleomycin (Fig. 1A). Thus, we investigated whether Glc7 might regulate checkpoint termination not only after inhibition of DNA replication but also after treatment with this DSB-inducing drug. We first asked whether Glc7 overproduction could counteract Rad53 phosphorylation under these conditions by treating G2-arrested cells with phleomycin for 40 min before transferring them to medium lacking phleomycin and containing either raffinose or galactose. As shown in Fig. 4A, Rad53 phosphorylation decreased to a greater extent in galactose-induced GAL-GLC7 cells than in either uninduced GAL-GLC7 cells or galactose-induced wild-type cells carrying the empty vector. This finding suggests that Glc7 participates in checkpoint inactivation after chemically induced DSBs.
FIG. 4.
Recovery from a DSB-induced checkpoint involves Glc7. (A) Nocodazole-arrested (noc) wild-type (wt) cells containing URA3 centromeric plasmids, either empty or carrying the GAL-GLC7 allele, were incubated with 10 μg/ml phleomycin (phleo) in YPraf. After 40 min, two samples of each cell culture were transferred to either YPraf or YPgal medium, both lacking phleomycin but still containing nocodazole. Cell samples were collected at the indicated times after phleomycin removal to detect Rad53 by Western blot analysis. exp, exponentially growing cells. (B and C) Nocodazole-arrested wild-type, glc7-129, glc7-132, and glc7-T152K cells were incubated in YPD or YPD with 5 μg/ml phleomycin (+ phleo). After 15 min, cells were released into YPD lacking both phleomycin and nocodazole (time zero) and the percentages of binucleate cells in untreated (B) and phleomycin-treated (C) cultures were determined at the indicated times. (D to G) Wild-type, glc7-129, glc7-132, and glc7-T152K cells arrested in G2 with nocodazole (noc) were incubated with 5 μg/ml (D to F) or 10 μg/ml (G) phleomycin. After 15 min (phleo), cells were transferred to medium lacking phleomycin but still containing nocodazole. Aliquots of each culture were harvested at the indicated times after phleomycin removal to detect Rad53 (D), Rad9 (E), Ddc2-HA (F), and Mre11-HA (G) by Western blot analysis. Asterisks in panel E indicate hyperphosphorylated Rad9.
If this were the case, defective Glc7 should cause a prolonged checkpoint response after phleomycin treatment. To test this prediction, we exposed nocodazole-arrested cells to a 15-min phleomycin treatment, followed by release into the cell cycle in the absence of both phleomycin and nocodazole. Under these conditions, phleomycin-treated glc7-129, glc7-132, and glc7-T152K cells underwent nuclear division about 15, 30, and 60 min later, respectively, than wild-type cells (Fig. 4C), while nuclear division kinetics after release were similar in all untreated cell cultures (Fig. 4B). In order to assess whether this behavior correlated with abnormal Rad53 activation, nocodazole-arrested cells were treated for 15 min with phleomycin and then transferred to medium lacking phleomycin but containing nocodazole to exclude effects due to different cell cycle phases. As shown in Fig. 4D, glc7-129, glc7-132, and glc7-T152K cells displayed increased and more persistent Rad53 phosphorylation than the wild type, with glc7-T152K cells again showing the strongest effect. Thus, dysfunctional Glc7 causes prolonged checkpoint activation also in response to DSBs.
Rad53 phosphorylation and activation require Rad9, Ddc2, and Mre11 proteins, which are phosphorylated by Mec1 and Tel1 kinases independently of Rad53 (9, 27, 39). Rad9, Ddc2, and Mre11 phosphorylated forms persisted longer in all phleomycin-treated glc7 mutants than in similarly treated wild-type cells (Fig. 4E, F, and G). Thus, Glc7 regulates the phosphorylation state not only of Rad53, but also of upstream checkpoint components.
Glc7 is not required for homologous recombination-mediated DSB repair.
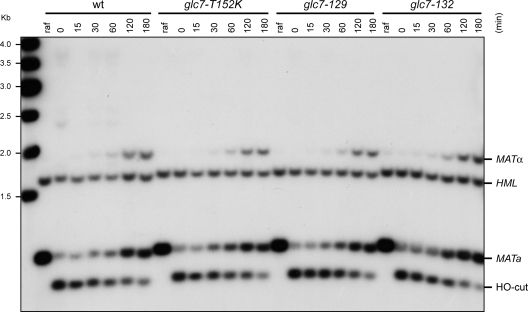
As replication fork stalling after HU treatment might lead to DSB formation, the inability of glc7 mutants to resume DNA replication after HU exposure and/or cell cycle progression after treatment with DSB-inducing agents might be due to defects either in inactivating the checkpoint or in repairing DSBs. To discriminate between these two possibilities, we investigated the ability of glc7 mutants to repair a DSB via homologous recombination (HR). S. cerevisiae cells switch mating type by using the homothallic endonuclease (HO) to create a DSB at the MAT locus, followed by repair of the break by HR with the donor sequence HML or HMR (reviewed in reference 15). Galactose was added to exponentially growing MATa wild-type and glc7 cells expressing HO from the GAL1 promoter. HO induction was allowed for 1 h, and then cells were transferred to glucose-containing medium to allow repair of the HO-induced break. As shown in Fig. 5, efficient HO cleavage at the MATa locus was detectable in 95% of wild-type, glc7-129, glc7-132, and glc7-T152K cells 1 h after galactose addition. Then, MATa and MATα repair products appeared with very similar kinetics in all cell cultures. Similar results were obtained also when the same experiment was performed on GLC7 and glc7 cells lacking the nonhomologous end joining factor Lig4, where only HR repair of the HO-induced break can occur (data not shown). Thus, defective Glc7 does not affect HR DSB repair, suggesting that the prolonged checkpoint activation in glc7 cells after HU exposure or treatment with DSB-inducing agents is likely due to defects in checkpoint inactivation rather than in DNA damage repair.
FIG. 5.
DSB formation and repair in glc7 mutant cells. Exponentially growing wild-type (wt), glc7-129, glc7-132, and glc7-T152K SCraf-Ura cell cultures (raf), all carrying the MATa allele and expressing the HO gene from the GAL1 promoter, were transferred to YPgal to induce HO expression. After 1 h, cells were transferred to medium containing glucose to allow cells to repair the HO-induced break (time zero). StyI-BamHI-digested genomic DNA prepared from cell samples collected at the indicated time points after galactose removal was subjected to Southern blot analysis with a MAT probe that detects 0.9-kb fragments (MATa) in the absence of HO-cut, while HO-induced DSB formation results in generation of HO-cut (0.7-kb fragment), which can be eventually repaired by HR with donor sequence HMR or HML, generating MATa (0.9-kb) and MATα (1.8-kb) repair products, respectively.
Glc7 controls histone H2A phosphorylation in vivo and dephosphorylates γH2A in vitro.
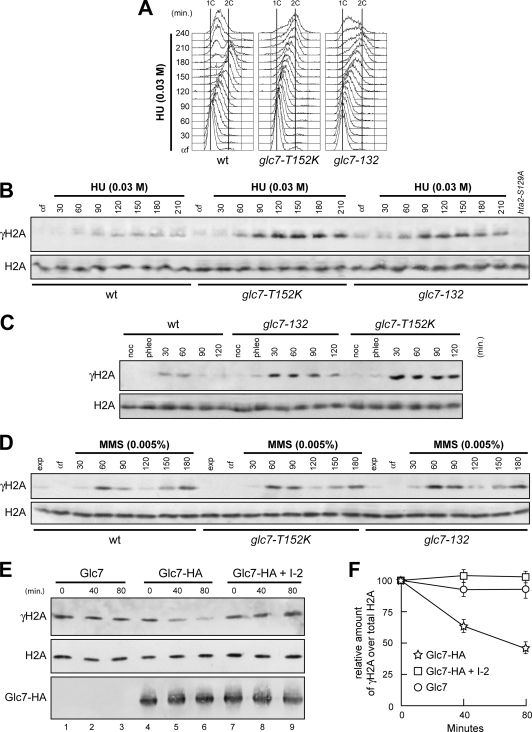
One of the earliest events in the response to DNA damage is the phosphorylation of histone H2A on serine 129 (γH2A) by either Tel1 or Mec1 (12, 28, 30). This event takes place also after inhibition of DNA replication by HU in both mammalian and yeast cells (7, 41). As Glc7 regulates phosphorylation of proteins acting upstream of Rad53, we monitored γH2A formation in glc7-132 and glc7-T152K cells by using phospho-specific antibodies recognizing phosphorylated S129 on histone H2A. When G1-arrested cell cultures were released in the presence of 0.03 M HU (Fig. 6A), γH2A was more abundant in both glc7-132 and glc7-T152K mutant cells than in the wild type (Fig. 6B). Similarly, the appearance of γH2A after phleomycin treatment was enhanced in both glc7-132 and glc7-T152K mutants compared to the wild type (Fig. 6C). Thus, Glc7 appears to regulate γH2A levels in response to both DNA synthesis inhibition and DSB formation. In contrast, and consistent with the finding that Glc7 did not seem to promote Rad53 dephosphorylation during MMS exposure (Fig. 3), both the kinetics and the extent of γH2A phosphorylation in wild-type cells were similar to those in glc7 mutant cells upon release of the cells from G1 arrest in the presence of a sublethal MMS dose (Fig. 6D).
FIG. 6.
Glc7 regulates γH2A formation. (A and B) α-Factor-arrested (αf) wild-type (wt), glc7-T152K, and glc7-132 cells were released in YPD with 0.03 M HU. Aliquots of each culture were harvested at the indicated times after α-factor release to determine the DNA content by FACS (A) and to detect γH2A and H2A by Western blot analysis with anti-γH2A and anti-H2A antibodies, respectively (B). Specificity of the latter was checked with protein extracts from hta1Δ cells expressing the H2A-S129A variant (hta2-S129A), which were treated with 0.03 M HU for 120 min. (C) Cell cultures arrested in G2 with nocodazole (noc) were incubated with 5 μg/ml phleomycin. After 15 min (phleo), cells were transferred to medium lacking phleomycin but still containing nocodazole. (D) α-Factor-arrested (αf) cell cultures were released in YPD with 0.005% MMS. In panels C and D, aliquots of each culture were harvested at the indicated times after α-factor release to detect γH2A and H2A by Western blot analysis with anti-γH2A and anti-H2A antibodies, respectively. (E) Protein extracts from cells carrying either untagged GLC7 (Glc7) or fully functional HA-tagged GLC7 (Glc7-HA) at the GLC7 chromosomal locus were immunoprecipitated with anti-HA antibody. Immunoprecipitates were assayed for phosphatase activity toward purified histones at 30°C (lanes 1 to 6). The Glc7-HA immunoprecipitate was also preincubated for 15 min with inhibitor 2 (I-2) prior to phosphatase assay (lanes 7 to 9). At the indicated time points after histone addition, samples of each reaction mixture were subjected to Western blot analysis with anti-γH2A, anti-H2A, and anti-HA antibodies. (F) Densitometric analysis. Plotted values are the mean values ± standard deviations (SD) from three independent experiments as in panel E. The amount of γH2A was determined as the ratio between γH2A and total H2A band intensities.
If Glc7 were directly involved in γH2A dephosphorylation, then purified Glc7 should be able to dephosphorylate γH2A in vitro. To test this prediction, we immunoprecipitated HA-tagged Glc7 from yeast cell extracts and assayed its ability to dephosphorylate γH2A among histones purified from phleomycin-treated cells. A clear reduction in γH2A amount was detected in the phosphatase reaction mixture containing Glc7-HA immunoprecipitates compared to that containing HA immunoprecipitates from cells expressing untagged Glc7 (Fig. 6E and F). This effect was due to dephosphorylation and not degradation, as similar levels of total H2A were detected in all the phosphatase reaction mixtures. Moreover, this phosphatase activity was further confirmed to be Glc7 dependent, as it was blocked by inhibitor 2 (I-2) (Fig. 6E and F), which specifically inhibits type 1 protein phosphatases (8). Thus, a Glc7-associated phosphatase activity dephosphorylates γH2A in vitro. This finding, together with our previous data, strongly supports the hypothesis that Glc7 directly regulates γH2A levels in vivo.
The lack of γH2A partially rescues the replication recovery defects of glc7 mutants.
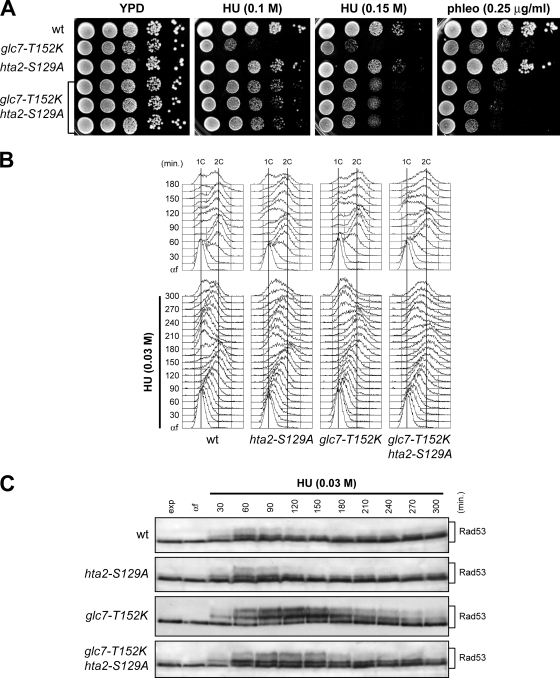
Dephosphorylation of γH2A is necessary for efficient recovery from a DSB-induced checkpoint (17). To elucidate whether the checkpoint recovery defect of glc7 mutants after nucleotide depletion was a direct consequence of the increased γH2A levels, we generated hta2-S129A and glc7-T152K hta2-S129A mutant strains, where the H2A Ser129 was replaced with a nonphosphorylatable alanine residue. Since two genes (HTA1 and HTA2) encode H2A histone, hta2-S129A and glc7-T152K hta2-S129A strains also carried the deletion of HTA1. We found that glc7-T152K hta2-S129A double mutant cells formed colonies on HU-containing plates more efficiently than glc7-T152K single mutant cells (Fig. 7A), indicating that the HU hypersensitivity of glc7-T152K is linked to H2A hyperphosphorylation. In contrast, the same hta2-S129A allele did not significantly increase glc7-T152K cell survival following phleomycin treatment (Fig. 7A), suggesting that hypersensitivity to this drug may be due to defective functions of Glc7 in the DNA damage response other than γH2A dephosphorylation.
FIG. 7.
Lack of γH2A formation alleviates the recovery defects of HU-treated glc7 mutants. (A) Serial dilutions (1:10) of exponentially growing cultures of wild-type (wt), glc7-T152K, hta2-S129A, and glc7-T152K hta2-S129A cells were spotted onto YPD plates with or without HU or phleomycin at the indicated concentrations and incubated at 25°C for 3 days. (B and C) Cells were released from G1 arrest (αf) in YPD medium with 0.03 M HU. Aliquots were harvested at the indicated times after release to determine DNA content by FACS analysis (B) and to detect Rad53 by Western blot analysis (C). The hta2-S129A and glc7-T152K hta2-S129A strains also carry the HTA1 deletion.
The reduced HU sensitivity of glc7-T152K hta2-S129A compared to glc7-T152K cells prompted us to analyze whether the unphosphorylatable H2A-S129A variant also partially restored the ability of glc7-T152K cells to complete DNA replication in the presence of a sublethal concentration of HU. In accordance with the previous finding that the lack of γH2A does not affect S phase checkpoint activation by MMS (12), wild-type and hta2-S129A cells replicated DNA with similar kinetics and contained similar amounts of phosphorylated Rad53 when released from a G1 arrest into fresh medium containing 0.03 M HU (Fig. 7B and C). Interestingly, HU-treated glc7-T152K hta2-S129A cells completed DNA replication faster than similarly treated glc7-T152K cells (Fig. 7B). The increased DNA replication efficiency of HU-treated glc7-T152K hta2-S129A cells correlated with a less active checkpoint, as these cells contained smaller amounts of phosphorylated Rad53 that disappeared faster than similarly treated glc7-T152K cells (Fig. 7C). Thus, the defective recovery from HU of glc7 cells results at least partially from persistent γH2A phosphorylation, indicating that γH2A dephosphorylation is a critical function of Glc7 in the recovery from a HU-induced checkpoint.
Relationships between Glc7, Pph3, Ptc2, and Ptc3 in DNA replication recovery after HU-induced fork stalling.
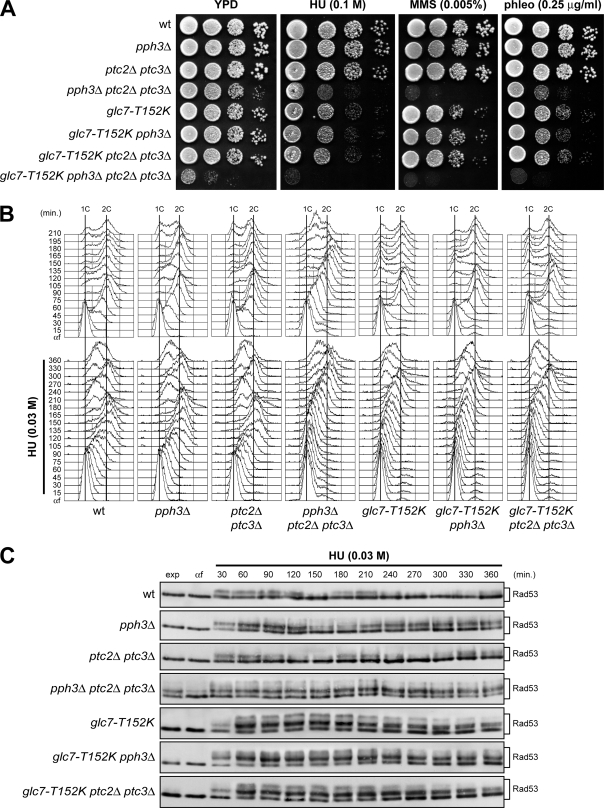
The protein phosphatases Pph3, Ptc2, and Ptc3 appear to be dispensable for Rad53 deactivation following HU-induced S phase arrest, while they allow Rad53 deactivation and replication fork restart after MMS exposure (26, 32, 35). Although Rad53 dephosphorylation is delayed in HU-treated glc7 mutants, it still takes place, raising the possibility that Pph3, Ptc2, and/or Ptc3 might be engaged in cells crippled for Glc7 activity. Thus, we investigated their epistatic relationships in supporting cell survival of HU treatment and in promoting replication recovery after HU-induced fork stalling. According to previous studies (35), we found that pph3Δ and ptc2Δ ptc3Δ cells, which displayed a slight sensitivity to MMS that was enhanced in the triple mutant pph3Δ ptc2Δ ptc3Δ, were not more sensitive to HU than wild-type cells (Fig. 8A). Furthermore, pph3Δ and ptc2Δ ptc3Δ cells did not delay completion of DNA replication compared to wild-type cells when released from a G1 arrest in the presence of low HU doses (Fig. 8B), although pph3Δ cells showed increased amounts of phosphorylated Rad53 (Fig. 8C). On the other hand, pph3Δ ptc2Δ ptc3Δ triple mutant cells formed colonies on HU-containing plates less efficiently than wild-type cells (Fig. 8A). As pph3Δ ptc2Δ ptc3Δ cells were defective in progressing through S phase in the absence of genotoxic agents (Fig. 8B) and showed hyperphosphorylated Rad53 even when exponentially growing (Fig. 8C), their loss of viability in the presence of HU can be likely ascribed to their intrinsic S phase defects.
FIG. 8.
Epistatic relationships between GLC7, PPH3, PTC2, and PTC3. (A) Serial dilutions (1:10) of exponentially growing cells with the indicated genotypes were spotted onto YPD plates with or without MMS, HU, and phleomycin at the indicated concentrations and incubated at 25°C for 3 days. wt, wild type. (B and C) Exponentially growing cells with the indicated genotypes were arrested in G1 with α-factor (αf) and released from the pheromone block in YPD or in YPD containing 0.03 M HU. Aliquots of each culture were harvested at the indicated times after α-factor release to determine DNA content by FACS analysis (B) and to detect Rad53 by Western blot analysis with anti-Rad53 antibodies (C).
Importantly, the absence of Pph3 or both Ptc2 and Ptc3 did not enhance either the sensitivity to HU of glc7-T152K cells (Fig. 8A) or their S phase progression defects during a constant exposure to a low HU dose (Fig. 8B). Altogether, these findings indicate that Pph3, Ptc2, and Ptc3 do not contribute to replication restart after HU exposure in cells where Glc7 activity is compromised. Given that Glc7 is dispensable for DNA replication in the presence of MMS (Fig. 3), these findings suggest that different phosphatases might be specifically engaged in the presence of particular types of DNA perturbations. Interestingly, the glc7-T152K allele greatly enhanced the growth defects of pph3Δ ptc2Δ ptc3Δ cells in the absence of genotoxic agents (Fig. 8A), indicating that Glc7, Pph3, Ptc2, and Ptc3 act redundantly in supporting cell viability.
DISCUSSION
Inactivation of Rad53 allows budding yeast cells to recover from checkpoint-mediated cell cycle arrest. To date, less is known about the mechanisms that silence the checkpoint after inhibition of DNA replication by nucleotide depletion. Here, our results indicate that Glc7/PP1 promotes recovery from inhibition of DNA replication, and this control is linked to γH2A downregulation and Rad53 inactivation. In fact, defective Glc7 impairs resumption of DNA replication after HU-induced fork stalling, and this defect correlates with persistent Rad53 hyperphosphorylation. The latter does not seem to be due to defects in repairing DSBs that may arise as a consequence of replication fork stalling, as glc7 mutants are proficient in DSB repair by HR. Moreover, the artificial inactivation of Rad53 suppresses the replication recovery defects of glc7-T152K mutant cells, indicating that the critical function of Glc7 in replication resumption is likely Rad53 dephosphorylation. This finding also implies that the restart of DNA synthesis at stalled replication forks is coupled to Rad53 deactivation. Thus, although Rad53 activation plays a crucial role in the stabilization of stalled replication forks, possibly by phosphorylating DNA replication proteins (reviewed in reference 42), its subsequent Glc7-mediated inactivation is important for replication fork restart after HU treatment. Similarly, Rad53 deactivation was shown to allow replication fork restart in pph3Δ cells recovering from MMS-induced DNA damage (32).
Interestingly, although MMS treatment interferes with S phase progression similarly to nucleotide depletion, Glc7 does not seem to promote Rad53 dephosphorylation after MMS exposure. On the contrary, the phosphatases Ptc2, Ptc3, and Pph3, which are required to turn off the checkpoint induced by MMS, are dispensable for replication recovery during HU-induced S phase arrest (26, 32, 35). Furthermore, the lack of Pph3 or of both Ptc2 and Ptc3 did not enhance either the sensitivity to HU of glc7-T152K cells or their defects in completing DNA replication when exposed to low HU dose. These findings suggest that different phosphatases might be specifically engaged in response to particular types of signals. Interestingly, cells concomitantly lacking Pph3, Ptc2, and Ptc3 show S phase progression defects and hyperphosphorylated Rad53 accumulation even in the absence of genotoxic treatment, and their growth defects are greatly enhanced by the presence of the glc7-T152K allele. Thus, Glc7, Pph3, Ptc2, and Ptc3 appear to act redundantly in supporting cell viability under unperturbed conditions.
Glc7 function in checkpoint termination is not limited to the checkpoint triggered by nucleotide depletion. In fact, Glc7 overproduction causes premature disappearance of phosphorylated Rad53 after phleomycin-induced DSB formation. Furthermore, the lack of Glc7 impairs resumption of cell cycle progression and Rad53 dephosphorylation under the same conditions, indicating that it is involved also in recovery from a DSB-induced checkpoint. Interestingly, Glc7 seems to act redundantly with Pph3 in checkpoint deactivation in the response to DSBs, as the glc7-T152K allele enhances the sensitivity to phleomycin of pph3Δ cells, but not their sensitivity to HU (Fig. 8A).
How does Glc7 promote termination of the checkpoint response? A checkpoint signal is triggered by the recruitment to stalled replication forks or DSB sites of the Mec1 and Tel1 checkpoint kinases, which phosphorylate H2A, Ddc2, Rad9, and Mre11 proteins, thus propagating the checkpoint signal to the downstream kinase Rad53. Rad53 phosphorylation and activation require Ddc2, Rad9, and Mre11, while γH2A, Rad9, Ddc2, and Mre11 phosphorylation occurs independently of Rad53 (9, 27, 39). We find that Glc7 counteracts phosphorylation not only of Rad53 but also of γH2A, Ddc2, Rad9, and Mre11 after HU exposure or chemically induced DSBs, suggesting that Glc7 can reverse phosphorylation of Rad53 and/or of proteins upstream. However, since there is no evidence that Rad53 regulates γH2A, Ddc2, Mre11, and Rad9 phosphorylation in a positive feedback loop, it seems unlikely that Glc7 promotes checkpoint termination by acting exclusively on Rad53 phosphosites. On the other hand, Glc7 dephosphorylates γH2A in vitro, and the lack of γH2A formation not only counteracts Rad53 phosphorylation in HU-treated glc7-T152K cells but also partially suppresses their defects in replication recovery. Therefore, we favor the hypothesis that Glc7 may promote resumption of DNA replication by reversing γH2A formation. However, we cannot exclude the possibility that Glc7 is involved in checkpoint termination indirectly, by regulating multiple targets in the DNA damage response. Indeed, Glc7 is known to target a wide range of substrates through interactions with specific factors, and the analyzed glc7-132 and glc7-T152K mutations, which map on the protein surface (3, 36), might affect protein-protein interactions. Identification of the factor(s) possibly targeting Glc7 to γH2A will shed light on the mechanistic connections between histone modifications, DNA replication, and checkpoint signaling.
How can γH2A dephosphorylation promote resumption of DNA replication? Formation of γH2A is dispensable for checkpoint activation when the dNTP pool is limiting. However, the lack of γH2A in HU-treated glc7-T152K cells results in reduced Rad53 phosphorylation, indicating that the signal keeping Rad53 active in glc7 cells consists in γH2A molecules. Thus, γH2A formation supports the maintenance of Rad53 activation, possibly by promoting congregation of DNA damage response proteins at stalled replication forks. As artificial Rad53 inactivation suppresses the defects of glc7-T152K cells in replication recovery, removal of γH2A may reduce the checkpoint signal to a level below which it is unable to activate Rad53, thus permitting replication fork restart.
As the lack of γH2A has been shown to contribute to recovery from a DSB-induced checkpoint (17), γH2A persistence in phleomycin-treated glc7 mutants might account also for their defects in recovering from DSBs. However, the lack of γH2A formation does not suppress the hypersensitivity to phleomycin of the glc7-T152K mutant, suggesting that their increased sensitivity may be due to Glc7 functions in the DNA damage response other than γH2A dephosphorylation. Consistent with this hypothesis, it has been shown that Schizosaccharomyces pombe PP1/Dis2 dephosphorylates the checkpoint kinase Chk1 (10) and mammalian PP1a has been shown to dephosphorylate a fragment of BRCA1 in vitro (19).
In summary, we have found that the yeast PP1 phosphatase Glc7 is involved in checkpoint termination after both HU-induced replication block and DSBs and that the critical function of Glc7 in this process is dephosphorylation of γH2A. The emerging view is that different sets of phosphatases participate in checkpoint inactivation after specific types of replication stress. Unraveling how these phosphatases are engaged in response to different stimuli will be a challenging goal for further studies.
Acknowledgments
We thank W. M. Bonner, M. Carlson, J. Diffley, D. Durocher, N. Lowndes, D. F. Stern, and K. Tatchell for providing yeast strains, plasmids, and antibodies. We also thank A. Verrault for advice in H2A purification and Michela Clerici for critical reading of the manuscript.
This work was supported by grants from Associazione Italiana Ricerca sul Cancro to M.P.L.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Altaf, M., A. Auger, M. Covic, and J. Côté. 2009. Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell Biol. 87:35-50. [DOI] [PubMed] [Google Scholar]
- 2.Bailis, J. M., and G. S. Roeder. 2000. Pachytene exit is controlled by reversal of Mek1-dependent phosphorylation. Cell 101:211-221. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. H., D. L. Frederick, A. Bloecher, and K. Tatchell. 1997. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury, D., X. Xu, X. Zhong, F. Ahmed, J. Zhong, J. Liao, D. M. Dykxhoorn, D. M. Weinstock, G. P. Pfeifer, and J. Lieberman. 2008. A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol. Cell 31:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clerici, M., V. Baldo, D. Mantiero, F. Lottersberger, G. Lucchini, and M. P. Longhese. 2004. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 24:10126-10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb, J. A., T. Schleker, V. Rojas, L. Bjergbaek, J. A. Tercero, and S. M. Gasser. 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19:3055-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, P., D. L. Schelling, and M. J. Stark. 1989. Remarkable similarities between yeast and mammalian protein phosphatases. FEBS Lett. 250:601-606. [DOI] [PubMed] [Google Scholar]
- 9.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Elzen, N. R., and M. J. O'Connell. 2004. Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J. 23:908-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 13.Fay, D. S., Z. Sun, and D. F. Stern. 1997. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr. Genet. 31:97-105. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 15.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 16.Heideker, J., E. T. Lis, and F. E. Romesberg. 2007. Phosphatases, DNA damage checkpoints and checkpoint deactivation. Cell Cycle 6:3058-3064. [DOI] [PubMed] [Google Scholar]
- 17.Keogh, M. C., et al. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439:497-501. [DOI] [PubMed] [Google Scholar]
- 18.Leroy, C., S. E. Lee, M. B. Vaze, F. Ochsenbien, R. Guerois, J. E. Haber, and M. C. Marsolier-Kergoat. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11:827-835. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y., D. M. Virshup, R. L. White, and L. C. Hsu. 2002. Regulation of BRCA1 phosphorylation by interaction with protein phosphatase 1a. Cancer Res. 62:6357-6361. [PubMed] [Google Scholar]
- 20.Longhese, M. P., D. Mantiero, and M. Clerici. 2006. The cellular response to chromosome breakage. Mol. Microbiol. 60:1099-1108. [DOI] [PubMed] [Google Scholar]
- 21.Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani, and G. Lucchini. 1997. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16:5216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 23.Lucca, C., F. Vanoli, C. Cotta-Ramusino, A. Pellicioli, G. Liberi, J. Haber, and M. Foiani. 2004. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23:1206-1213. [DOI] [PubMed] [Google Scholar]
- 24.Majka, J., A. Niedziela-Majka, and P. M. Burgers. 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakada, S., G. I. Chen, A. C. Gingras, and D. Durocher. 2008. PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill, B. M., S. J. Szyjka, E. T. Lis, A. O. Bailey, J. R. Yates III, O. M. Aparicio, and F. E. Romesberg. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA 104:9290-9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 28.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 29.Segurado, M., and J. F. Diffley. 2008. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 22:1816-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. B. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweeney, F. D., F. Yang, A. Chi, J. Shabanowitz, D. F. Hunt, and D. Durocher. 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15:1364-1375. [DOI] [PubMed] [Google Scholar]
- 32.Szyjka, S. J., J. G. Aparicio, C. J. Viggiani, S. Knott, W. Xu, S. Tavaré, and O. M. Aparicio. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22:1906-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 34.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 35.Travesa, A., A. Duch, and D. G. Quintana. 2008. Distinct phosphatases mediate the deactivation of the DNA damage checkpoint kinase Rad53. J. Biol. Chem. 283:17123-17130. [DOI] [PubMed] [Google Scholar]
- 36.Tu, J., W. Song, and M. Carlson. 1996. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Attikum, H., and S. M. Gasser. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell. Biol. 19:207-217. [DOI] [PubMed] [Google Scholar]
- 38.Vernis, L., J. Piskur, and J. F. Diffley. 2003. Reconstitution of an efficient thymidine salvage pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 31:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virshup, D. M., and S. Shenolikar. 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33:537-545. [DOI] [PubMed] [Google Scholar]
- 41.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 42.Zegerman, P., and J. F. Diffley. 2009. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst). 8:1077-1088. [DOI] [PubMed] [Google Scholar]