Abstract
The mammalian circadian rhythm is observed not only at the suprachiasmatic nucleus, a master pacemaker, but also throughout the peripheral tissues. Its conserved molecular basis has been thought to consist of intracellular transcriptional feedback loops of key clock genes. However, little is known about posttranscriptional regulation of these genes. In the present study, we investigated the role of the 3′-untranslated region (3′UTR) of the mouse cryptochrome 1 (mcry1) gene at the posttranscriptional level. Mature mcry1 mRNA has a 610-nucleotide 3′UTR and mediates its own degradation. The middle part of the 3′UTR contains a destabilizing cis-acting element. The deletion of this element led to a dramatic increase in mRNA stability, and heterogeneous nuclear ribonucleoprotein D (hnRNP D) was identified as an RNA binding protein responsible for this effect. Cytoplasmic hnRNP D levels displayed a pattern that was reciprocal to the mcry1 oscillation. Knockdown of hnRNP D stabilized mcry1 mRNA and resulted in enhancement of the oscillation amplitude and a slight delay of the phase. Our results suggest that hnRNP D plays a role as a fine regulator contributing to the mcry1 mRNA turnover rate and the modulation of circadian rhythm.
Circadian rhythms in living organisms are regulated by an endogenous clock. In mammals, it is well known that the circadian clock is located at the suprachiasmatic nucleus (SCN) in the brain. The SCN orchestrates circadian oscillations in various peripheral tissues (21, 36, 56). Because the circadian rhythm is conserved from the entire organism to the level of the single cell, many efforts have been made to understand the molecular basis of the regulation of circadian timing (15, 20).
Enormous progress has been made toward understanding the secrets of the rhythm generator, providing a view of the circadian clock mechanism. Each cell of the body contains a circadian core oscillator composed of essential components for normal circadian behavior. The RNA and protein levels of the components, called clock genes, are known to oscillate in a circadian rhythm, based on studies of the expression of genes, including Clock (49); Bmal1 (5); mPer1 and mPer2 (55); mCry1 and mCry12 (22); Rev-erbα (34); and Rorα (44). These key clock genes are involved in interacting positive and negative transcriptional feedback loops (9, 34, 37, 44).
In mammals, cryptochromes are expressed in every organ, and there are two homologues, mcry1 and mcry2. In general, mcry1 is expressed at a high level in the SCN, whereas mcry2 expression in this region is almost negligible (41). The oscillation of mcry1 transcripts persists when mice are kept in a “free-running” state, such as constant darkness (27), as has been observed for other clock genes and as would be expected of a true circadian regulator. mcry1 expression also shows circadian oscillation in other organs, most notably in the liver; however, the phase of its expression in internal organs is delayed or advanced compared to the phase of its expression in the SCN. Both mCRY1 and mCRY2 bind to mPER1, mPER2, and mPER3. CRY-PER interaction results in nuclear transport and subsequent inhibition of the transcription of clock genes and clock-controlled genes mediated by the CLOCK/BMAL1 complex (41).
Whereas many lines of evidence have accumulated regarding the transcriptional feedback loops of clock proteins, these feedback loops would not be sufficient to shape the amplitude and phase of the key clock genes. Basically, to sustain the oscillation of mRNA and proteins, the degradation of these products of expression should harmonize with their synthesis. Although degradation of the clock proteins has been reported as a result of posttranslational modifications (24, 26, 43), much less is known about their posttranscriptional regulation, including mRNA stability. In Drosophila melanogaster, several pieces of evidence indicate that posttranscriptional regulation involves mRNA oscillation (10, 46). However, these studies did not reveal which events of posttranscriptional regulation are responsible for the clock-gene oscillation. In mammals, the 3′-untranslated region (3′UTR) of mouse Period 1 (mper1) repressed its expression in a posttranscriptional manner, which explains the time lag of the mRNA and the protein products of mper1 (18).
Individual mRNAs have widely varying turnover rates. The regulation of the turnover rate of mRNAs is critical in determining the abundance and dynamics of transcripts. The stability of mRNA is determined by the interaction of specific cis-acting elements (CAEs) within the 3′UTR regions and the trans-acting factors that bind to them (51). The AU-rich RNA destabilizing element (ARE) in the 3′UTR is well characterized and is the most commonly occurring CAE involved in posttranscriptional regulation (3, 4, 7).
ARE binding factor 1 (Auf-1), also known as heterogeneous nuclear ribonucleoprotein (hnRNP) D, is a shuttling RNA binding protein that is abundant in many tissues. hnRNP D binds to putative AREs in the UTRs of target mRNAs and regulates mRNA stability or the promotion of translation (2, 25). Much research has focused on the action of hnRNP D in various mRNAs involved in the immune system, cell cycle regulation (29, 47), and related areas, including studies using knockout animals (28, 39).
In contrast to the translational modulation associated with the 3′UTR of mper1 (18), we show here that the mRNA stability of the circadian core clock gene mcry1 is modulated by the 3′UTR and cytoplasmic hnRNP D; in addition, mRNA instability is mediated by cytoplasmic increases in hnRNP D protein. Our results suggest that cytoplasmic hnRNP D-mediated mRNA decay serves to modulate peak amplitude and phase or to damp down the mcry1 mRNA circadian oscillation.
MATERIALS AND METHODS
Cell culture, transient transfection, and circadian synchronization.
HEK 293T and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (Welgene/HyClone) supplemented with 10% fetal bovine serum (HyClone) and 1% antibiotics (Welgene) in a humidified 95% air, 5% CO2 incubator at 37°C. Circadian oscillation of NIH 3T3 cells was synchronized by treatment with 100 nM dexamethasone for 2 h. Instead of the usual single dexamethasone treatment, dexamethasone shock was applied twice at −24 h and 0 h before the onset of the circadian period, and cells were monitored throughout the following day.
For expression of the reporter constructs, cells were counted and plated in a 24-well plate at a density of 105 cells/well. After 24 h, 0.7 μg of DNA was introduced into each well by lipid-mediated transfection using Welfect transfection reagent (Welgene) following the manufacturer's instructions. After transfection, cells were treated with the transcription blocker actinomycin D (5 μg/ml) at 0 h and harvested thereafter at the times indicated in the figures.
Rapid amplification of cDNA 3′ ends.
Single-stranded cDNA was synthesized from total RNA of mouse SCN by using oligo(dT) and Moloney murine leukemia virus reverse transcriptase (Roche). A first PCR was performed with the cDNA, using an mcry1-specific primer (5′-GGAATTCAGCAGTAACTGATACGAA) and the (dT)17-adaptor primer (5′-GCTCTAGAGGCCTCGAGTTTTTTTTTTTTTTTTT). In a second PCR, 1/50 of the product of this reaction was reamplified with a nested mcry1-specific primer (5′-GGAATTCTACGAAAGCGTGTGGGAGGAG) and just adaptor primer (5′-GCTCTAGAGGCCTCGAG). Sequencing was performed on the resulting product, representing the mcry1 3′UTR.
Plasmid construction and AANAT assay.
To generate pcNAT-wt610 (with the wild-type, full-length, 610-nucleotide mcry1 3′UTR inserted into the 3′UTR of AANAT) and pSK′-wt610, PCR products were digested with EcoRI/XbaI and inserted into the EcoRI/XbaI site of the control vector pcNAT, which expresses rat Aanat without the 3′UTR (32), and into modified pSK′ (16), respectively. To dissect the regions involved in posttranscriptional regulation, serially deleted 3′UTR fragments were cloned into the 3′UTR region of the reporter using the EcoRI/XbaI digestion site. The Aanat assay for reporter activity was performed as previously described (6).
Northern blot analysis and real-time reverse transcription-PCR (RT-PCR).
Northern blot analysis was carried out as described previously (16). Briefly, total RNA was extracted from HEK 293T cells using TRI Reagent (Molecular Research Center). The total RNA (5 μg) was size separated by 1% formaldehyde-agarose gel electrophoresis containing 0.66 M formaldehyde, transferred to nylon membranes (ICN), and hybridized with a specific probe labeled with [α-32P]dCTP (NEN). mRNA levels of reporters were also detected by quantitative real-time PCR using a MyIQ single-color real-time detector system (Bio-Rad) with SYBR green PCR mixture (Bio-Rad, Takara), as described previously (23). Specific primer pairs for Aanat, endogenous mcry1, per2, mgapdh, mrpl32, and mtbp were used (primer sequences are provided in Table S1 in the supplemental material).
In vitro transcription and UV cross-linking.
To identify proteins specifically bound to mcry1 3′UTR, in vitro binding and UV cross-linking assays were performed as described previously (37). pSK′ constructs with serially deleted 3′UTR were constructed for T7 RNA polymerase-based in vitro transcription (Promega, Roche). RNAs labeled with [α-32P]UTP (NEN) and unlabeled competitor RNAs were transcribed in vitro using T7 RNA polymerase and XbaI-linearized pSK′-3′UTR constructs. Equal molar amounts of labeled RNA were incubated with 15 μg of nuclear extract or 30 μg of cytoplasmic extract of either HEK 293T cells or NIH 3T3 cells for 20 min at 30°C. The RNA-protein binding reaction was carried out in a 30-μl reaction mixture containing 0.5 mM dithiothreitol, 5 mM HEPES (pH 7.6), 75 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 20 U RNasin (Promega), and 4 μg of yeast tRNA. Where indicated, homopolymeric ribonucleotides [poly(A), poly(C), and poly(U)] or unlabeled competitor transcripts were included in the reaction mixture. After incubation, the samples were UV irradiated on ice for 10 min with a CL-1000 UV cross-linker (UVP). Unbound RNAs were digested with 5 μl of RNase cocktail (2.5 μl of RNase A [10 mg/ml] [Roche] and 2.5 μl of RNase T1 [100 U/ml] [Roche]) at 37°C for 1 h. The reaction mixtures were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
For immunoprecipitation, RNase-digested lysates were incubated with specific antibodies or immunoglobulin for the negative control. After overnight incubation, protein G agarose beads (Amersham Bioscience) were added to the sample, which was further incubated for 3 h. Washed beads were analyzed by SDS-PAGE and autoradiography.
RNA interference (RNAi).
Small interfering RNAs (siRNAs) for endogenous hnRNP D knockdown were purchased from Dharmacon (siGENOME SMARTpool HNRPD M-042940-00). For siRNA transfection into NIH 3T3 cells, a microporator (Digital-Bio/Invitrogen) was used as recommended by the manufacturer (two 20-ms pulses at 1,300 V) 12 h prior to dexamethasone shock.
Cytoplasmic/nuclear protein preparation and immunoblot analysis.
Fractionation of HEK 293T cells and NIH 3T3 cells into cytoplasmic and nuclear extracts was performed as described previously (12, 17). Immunoblot analyses were performed with monoclonal anti-hnRNP D (Upstate), monoclonal anti-14-3-3ζ (Santa Cruz), and monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ICN) primary antibodies. Horseradish peroxidase-conjugated species-specific secondary antibodies (KPL) were visualized using a SUPEX ECL solution kit (Neuronex) and a LAS-4000 chemiluminescence detection system (FUJIFILM), and acquired images were analyzed using Image Gauge (FUJIFILM) according to the manufacturer's instructions.
RESULTS
The mcry1 3′UTR contains a destabilizing element.
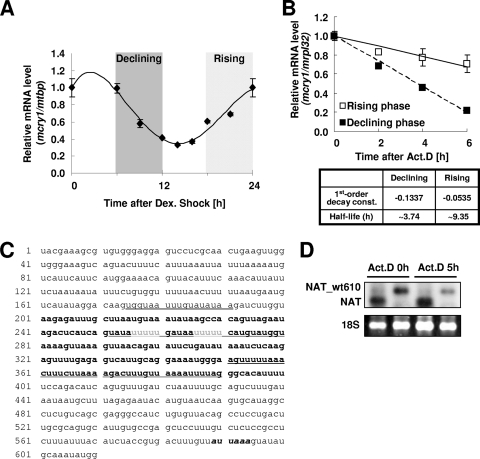
mRNA oscillation of mcry1 in various tissues, including the brain, and even in fibroblast cell lines, is well established (1, 38). To elucidate the mechanism of phase-dependent mRNA decay, we monitored the mcry1 mRNA oscillation pattern in NIH 3T3 fibroblast cells after synchronization by dexamethasone treatment. Robust and reproducible mcry1 mRNA oscillation was observed (Fig. 1A). In this oscillatory condition, mRNA degradation kinetics were compared in the declining and rising phases (Fig. 1A). When actinomycin D was used to inhibit transcription, more-dramatic decay was observed in the declining phase (half-life [t1/2], 3.74 h) than in the rising phase (t1/2, 9.35 h) (Fig. 1B). The role of transcriptional regulation in circadian mRNA oscillation is well established, and such oscillation is generally thought to be mainly governed by the action of transcription factors, such as the BMAL1/CLOCK heterodimer. However, our results indicate that mRNA decay also plays a role in the circadian mRNA regulatory mechanism. Because mRNA decay is typically mediated by the 3′UTR, the mcry1 3′UTR sequence was determined, using rapid amplification of cDNA 3′ ends with total RNA obtained from mouse SCN. Sequence analysis showed that the mcry1 3′UTR was 610 nucleotides long and contained a poly(A) addition signal (AUUAAA) at nucleotide 588 (Fig. 1C). A detailed analysis revealed that several U-rich elements were located throughout the region. We introduced AANAT (serotonin N-acetyltransferase) reporter constructs containing the full-length mcry1 3′UTR (wt610) into HEK 293T cells (16) to determine the UTR's function with regard to the stability of mcry1. In cells treated with the transcription blocker actinomycin D for the times indicated (Fig. 1D), the full-length mcry1 3′UTR triggered dramatic decreases in mRNA levels, whereas the reporter gene lacking a 3′UTR was relatively stable. This difference in stability was also observed at the level of the reporter protein (see Fig. S1A in the supplemental material). These trends in stability were also observed in NIH 3T3 cells (Fig. 2B), which are typically used as a circadian model system. This result implies that the mcry1 3′UTR contributes to the stability of mcry1 mRNA.
FIG. 1.
mcry1 3′UTR contains a destabilizing element. (A) Oscillation of endogenous mcry1 mRNA in NIH 3T3 cells was measured by quantitative real-time PCR. NIH 3T3 cells were treated with 100 nM dexamethasone (Dex.) twice for 2 h, at −24 h and 0 h, to synchronize the circadian oscillation. Total RNA (1 μg) from each time point was subjected to quantitative real-time RT-PCR with specific oligonucleotides for mcry1 and mtbp (TATA box binding protein, used as a control for normalization). Declining and rising phases are indicated. All results are expressed as the mean ± standard error of the mean of the results of three independent experiments. (B) mRNA degradation kinetics were measured during both declining (closed squares) and rising (open squares) phases within a 1-day cycle after dexamethasone treatment. Cells were treated with 5 μM actinomycin D (Act. D) at 6 h during the declining phase and at 18 h during the rising phase and then harvested at 3-h intervals. Total RNA (1 μg) was subjected to quantitative real-time RT-PCR with specific primer pairs against mcry1 or mrpl32, used as a control for normalization. All results are expressed as the mean ± standard error of the mean of the results of three independent experiments. First-order decay constants and mRNA half-lives were calculated from the formula for each fitted line. (C) RNA sequence of the mcry1 3′UTR (immediately after the stop codon). Nucleotides 201 to 400 of the 3′UTR are shown in bold font. Underlining indicates the sequences of competitive oligonucleotides used in later experiments. Gray font shows poly(U) tracts. Italicized bold font shows a putative poly(A) signal. (D) Change of reporter mRNA levels after actinomycin D (Act. D) treatment. HEK 293T cells were transiently transfected with the pcNAT-wt610 and pcNAT reporters and incubated with actinomycin D for the times indicated. The reporter mRNA levels were analyzed by Northern blot analysis, using a specific probe for Aanat mRNAs. Ethidium bromide-stained 18S rRNA was used as a loading control.
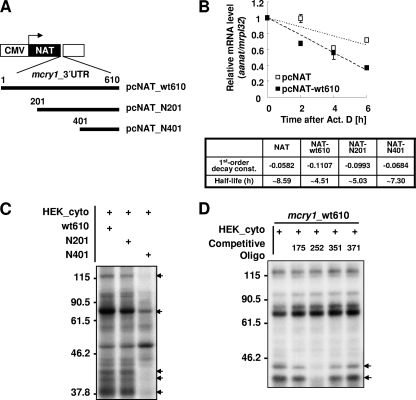
FIG. 2.
U tracts within the mcry1 3′UTR affect mRNA stability. (A) Diagram of reporter constructs containing the full-length or serially deleted mcry1 3′UTR, with numbers indicating the positions of the starting nucleotide within the 3′UTR. CMV, cytomegalovirus promoter; NAT, AANAT open reading frame. (B) NIH 3T3 cells were transiently transfected by electroporation with the NAT reporter or pGL3 control or the NAT reporter fused to serially deleted mcry1 3′UTR constructs (shown in the schematic in panel A) or pGL3 control. Transfected cells were incubated for 12 h before treatment with 5 μM actinomycin D (Act. D). Total RNA (1 μg) from the time points indicated (0 h or 6 h after treatment with 5 μM actinomycin D) was subjected to quantitative real-time RT-PCR with specific oligonucleotides for Aanat, fluc (a control for transfection efficiency), and mrpl32 (a control used for normalization). All results are expressed as the mean ± standard error of the mean of the results of three independent experiments. First-order decay constants (const.) and mRNA half-lives were calculated from the formula for each fitted line. (C) Each 3′UTR, transcribed in vitro with [α-32P]-rUTP, was subjected to an in vitro binding and UV cross-linking assay with HEK 293T cytoplasmic extract (HEK_cyto). The arrows indicate the proteins that show differential binding. Molecular sizes in kDa are shown on the left. +, present. (D) The radiolabeled 3′UTR was subjected to an in vitro binding and UV cross-linking assay with HEK 293T cytoplasmic extract (HEK_cyto) with or without the deoxyoligonucleotide competitors (1 μM), indicated above the gel by the positions of the first nucleotide of the target sequences (target sequences are shown in Fig. 1C). The arrows indicate the proteins that show differential binding. Molecular sizes in kDa are shown on the left. +, present.
U tracts in the mcry1 3′UTR affect mRNA stability.
To identify the functional destabilizing element in the mcry1 3′UTR, we created serially deleted reporter constructs by fusing DNA encoding the mcry1 3′UTR with the AANAT reporter (Fig. 2A). The reporters containing the mcry1 3′UTR were transiently transfected into NIH 3T3 cells, and then the cells were treated with the transcription blocker actinomycin D for 6 h. The full-length (wt610) and 5′-truncated (N201) mcry1 3′UTR triggered dramatic decreases in mRNA levels (t1/2, ∼4.51 h for wt610 and ∼5.03 h for N201) compared to the levels in the same cells immediately after treatment with actinomycin D (0 h), whereas the reporter gene alone (t1/2, ∼8.59 h) or the reporter fused to a 3′ region (N401) (t1/2, ∼7.30 h) were relatively stable (Fig. 2B; also see Fig. S1C in the supplemental material). This difference in stability was also observed at the level of the activity of reporter protein. Whereas full-length mcry1 3′UTR reduced reporter activity to 35% of that obtained with the reporter lacking the 3′UTR, the removal of nucleotides 1 to 200 of the mcry1 3′UTR caused an increase of 59% in reporter activity, and further truncation by removal of nucleotides 201 to 400 resulted in complete recovery of AANAT activity (see Fig. S1B in the supplemental material). These trends in stability suggest that the region between nucleotides 201 and 401 bears some type of destabilizing element. Next, we looked for proteins that might bind in this region to affect mRNA stability.
To identify the cellular protein factors interacting with mcry1 3′UTR, a UV cross-linking experiment was performed with fractionated cytoplasmic and nuclear cell lysates prepared from HEK 293T or NIH 3T3 cells. Although the contribution of indirectly bound proteins could not be excluded, the direct binding profile of many proteins was examined. Several proteins bound specifically to the in vitro-transcribed mcry1 3′UTR were identified by using cytoplasmic lysate from HEK 293T cells (Fig. 2C; also see Fig. S1D in the supplemental material). Interestingly, although the 3′ truncation of nucleotides (N201) had little effect on proteins binding to the 3′UTR, when nucleotides 201 to 400 were also deleted (N401), proteins that were approximately 120, 80, 42, and 40 kDa in size failed to bind to the mcry1 3′UTR. These changes also occurred when the same experiment was performed using the nuclear fraction or NIH 3T3 cytoplasmic lysate (see Fig. S1D and F in the supplemental material), implying that the proteins described bind to the middle region of the mcry1 3′UTR. This finding agrees with the results of the reporter assay using deletion constructs. Although the presence of additional proteins cannot be excluded, proteins of approximately 120, 80, 42, and 40 kDa may mediate mcry1 mRNA degradation by binding between nucleotides 201 to 400.
To define possible CAEs of the mcry1 3′UTR that interact with the RNA binding proteins discussed above, a competition assay was combined with the UV cross-linking experiment. When homopolymeric ribonucleotides were used as competitors, only poly(U) ribonucleotide efficiently blocked the binding of 42- and 40-kDa proteins to the mcry1 3′UTR. In contrast, poly(A) and poly(C) produced no change in binding (see Fig. S1H in the supplemental material). Thereafter, we designed competitors using four oligodeoxyribonucleotides matching the diverse U-rich segments located in the mcry1 3′UTR (Fig. 1C) (45, 53). Surprisingly, a competitive oligodeoxyribonucleotide spanning nucleotides 252 to 280 (c. oligo-252) completely inhibited binding of the proteins of approximately 40 and 42 kDa to the mcry1 3′UTR (Fig. 2D). This oligonucleotide has two U repeats in tandem, each 5 nucleotides in length. Interestingly, other oligodeoxyribonucleotides with similar U-rich sequences did not compete. Thus, it appears that the proteins of approximately 42 and 40 kDa bind to U-rich elements between bases 252 and 280 of the mcry1 3′UTR, whereas the proteins of approximately 120 and 80 kDa bind to other unique sites.
hnRNP D binds to the middle region of the mcry1 3′UTR.
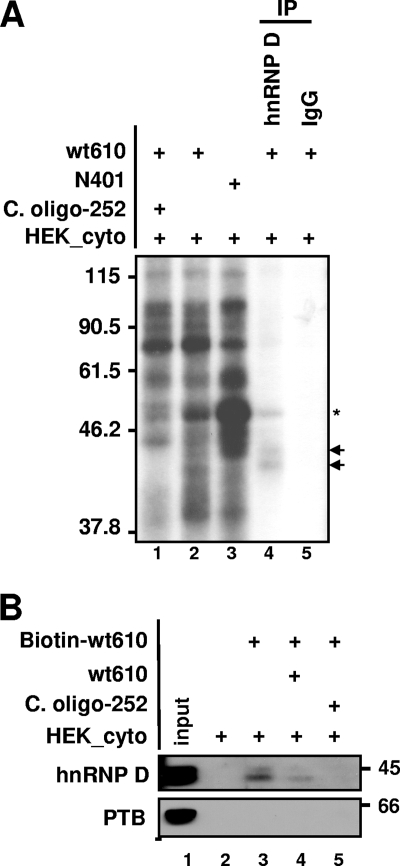
We next investigated the identity of the mRNA binding proteins which bound differentially to the truncated UTRs. The results from the competition assay performed with poly(U) ribonucleotide or the competitive oligodeoxyribonucleotide c. oligo-252 imply that a U-tract binding protein is the responsible factor. hnRNP D has four isoforms (37, 40, 42, and 45 kDa) derived from alternative splicing (42). Because the approximately 40- and 42-kDa proteins showed dramatic changes in binding in the presence of nucleotide competitors, immunoprecipitation was performed on the in vitro binding/UV cross-linked extracts using a monoclonal anti-hnRNP D antibody to confirm the identity of the protein and its binding to the mcry1 3′UTR. As shown in Fig. 3A, hnRNP D was detected by the anti-hnRNP D antibody, indicating that hnRNP D interacts directly with the mcry1 3′UTR (lane 4). We also confirmed binding of hnRNP D to biotin-labeled mcry1 3′UTR by biotin-streptavidin affinity purification followed by immunoblotting with anti-hnRNP D antibody (Fig. 3B). Binding of hnRNP D to the full-length 3′UTR was dramatically inhibited by competition with unlabeled 3′UTR (Fig. 3B, lane 4) and c. oligo-252, which is specific for hnRNP D binding (Fig. 3A, lane 1; 3B, lane 5; 2D, lane 3; and 1C, underlined sequence) (45); however, polypyrimidine tract binding protein (PTB), which is known to bind to mper2 mRNA (53), did not bind to the mcry1 3′UTR. These results suggest that hnRNP D binds directly to the mcry1 3′UTR and that binding occurs preferentially on the U tract between nucleotides 252 and 280.
FIG. 3.
hnRNP D binds to the mcry1 3′UTR. (A) In vitro binding/UV cross-linking/immunoprecipitation were performed with the full-length 3′UTR as described in Materials and Methods. In the experiment whose results are shown in lane 1, the reaction mixture was coincubated with 1 μM c. oligo-252, which binds specifically to the changed bands. Lanes 4 and 5 show immunoprecipitation (IP) with monoclonal anti-hnRNP D antibody or rabbit immunoglobulin G (IgG), as a control, respectively. The arrows indicate hnRNP D, and the asterisk indicates unidentified proteins binding to the hnRNP D complex. Molecular sizes in kDa are shown on the left. +, present. (B) The in vitro-transcribed 3′UTR construct was labeled with biotin-UTP and incubated with HEK 293T cell cytoplasmic extract (HEK_cyto). Streptavidin-affinity-purified samples were separated by SDS-PAGE and subjected to Western blotting with anti-hnRNP D antibody or anti-PTB, as a negative control. hnRNP D was detected with biotin-labeled mRNA (lane 3). hnRNP D binding decreased in the presence of a threefold excess of unlabeled wild-type 3′UTR mRNA (wt610; lane 4) or when 1 μM of the competitor deoxyoligonucleotide c. oligo-252 was added (lane 5), but PTB was not detected. Molecular sizes in kDa are shown on the right. +, present.
Cytoplasmic hnRNP D oscillates and binds to the mcry1 3′UTR with a rhythmic profile.
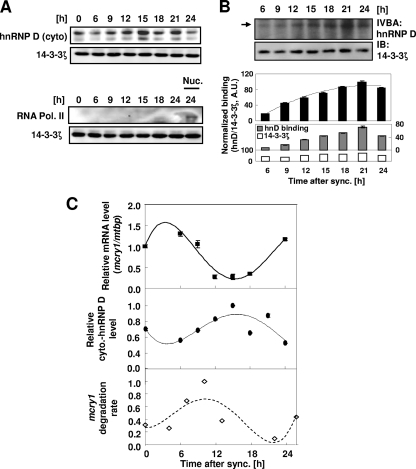
hnRNP D plays many roles in posttranscriptional regulation, and hnRNP D levels in the total cell lysate of various cell lines are very high (data not shown). hnRNP D is well known to shuttle between the nucleus and the cytoplasm; however, because decay of cellular mRNA occurs mainly in the cytoplasm, we examined the cytoplasmic hnRNP D level during circadian synchronization using dexamethasone treatment. Interestingly, the amount of cytoplasmic hnRNP D increased according to the specific period of circadian time following a 2-h exposure of NIH 3T3 cells to dexamethasone (Fig. 4A and C, middle), but the total level of hnRNP D did not change (data not shown). Furthermore, the pattern of cytoplasmic hnRNP D levels was almost reciprocal to that of mcry1 mRNA levels (Fig. 4C, top versus bottom). To examine the amount of cytoplasmic hnRNP D that bound to mRNA, aliquots of the cytoplasmic protein fraction from each time point were coincubated in vitro with radiolabeled mcry1 3′UTR. In comparison with the consistent amount in the input lysate, the amount of cytoplasmic hnRNP D binding to mRNA increased during the phase of mRNA decline (6 to 15 h) and showed a profile similar to the levels of cytoplasmic hnRNP D (Fig. 4B). Interestingly, when the decay kinetics at each time point were compared for 1 day (Fig. 4C, bottom), the declining phase (∼6 to 15 h) had much higher decay kinetics than the rising phase (∼15 to 24 h). Since the decay of mature mRNA occurs mainly in the cytoplasm, our results suggest that the increase in cytoplasmic hnRNP D and its binding to target mRNA are responsible for the rapid decay of mcry1 mRNA during the declining phase.
FIG. 4.
Cytoplasmic hnRNP D oscillates and binds to mcry1 3′UTR with a rhythmic profile. (A) NIH 3T3 cells were treated with 100 nM dexamethasone for 2 h and then harvested at the indicated times, and cytoplasmic extract (cyto) was prepared for Western blot analysis. Cytoplasmic hnRNP D was detected with monoclonal hnRNP D antibody, and 14-3-3ζ was included as an internal control. RNA polymerase II (RNA Pol. II), used for a nuclear marker, was not detected in cytoplasmic extract but appeared in the nuclear fraction (Nuc.). The results shown are representative of three independent experiments. (B) The same cytoplasmic extracts used for the experiments whose results are shown in panel A were subjected to an in vitro binding assay (IVBA) with radiolabeled 3′UTR mRNA. The arrow indicates cytoplasmic hnRNP D binding to 3′UTR mRNA (upper gel). 14-3-3ζ was used as an internal control to demonstrate that the same amount of extract was used for all time points (lower gel). hnRNP D binding intensities were quantified by normalization (black bars) to 14-3-3ζ levels in the same extracts (gray and white bars in lower graph). The error bars represent the standard errors of the means of the results. The results shown are representative of three independent experiments. sync., synchronization. IB, immunoblot; A.U., arbitrary unit. (C) Relative levels of cytoplasmic (cyto.) hnRNP D, normalized to 14-3-3ζ levels, were calculated and plotted (closed ovals/dotted line) for the second day after treatment. Relative levels of endogenous mcry1 mRNA (closed squares/solid line) were also quantified, using mtbp as an internal control for each time point. Relative degradation rates of mcry1 mRNA were calculated for each time point for 4 h after actinomycin D treatment; mgapdh was used as an internal control for each value (open diamonds/dashed line). The results shown are representative of three independent experiments. Error bars represent the standard errors of the means of the results. sync., synchronization.
hnRNP D modulates mcry1 stability and circadian rhythm.
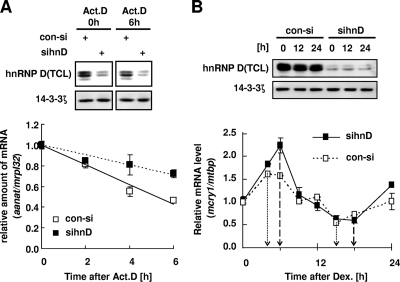
Although there are some reports that hnRNP D acts as a stabilizing regulator (35, 54), studies have generally shown that hnRNP D binds to 3′UTR regions to destabilize target mRNAs. The effect of hnRNP D on mcry1 mRNA stability was examined using transient expression analysis with a 3′UTR-containing reporter and actinomycin D pulse-chase experiments in NIH 3T3 cells without dexamethasone synchronization. Reduction of endogenous hnRNP D was successfully achieved by targeted RNAi (Fig. 5A, gel). When hnRNP D was knocked down by RNAi, the reporter mRNA containing the 3′UTR (wt610) was very stable after treatment with actinomycin D compared with its stability following control siRNA treatment (Fig. 5A, graph).
FIG. 5.
hnRNP D modulates mcry1 stability and circadian rhythm. (A) NIH 3T3 cells were transiently transfected with a pcNAT reporter containing the mcry1 3′UTR, β-galactosidase as a control used for normalization, and siRNA for knockdown of endogenous hnRNP D (sihnD) or control siRNA (con-si) and then incubated for 24 h, followed by treatment with 5 μM actinomycin D (Act. D). Total RNA (1 μg) was reverse transcribed using oligo(dT) primer and then quantified by real-time PCR (graph). Each value was normalized to the level of mrpl32. Open or closed squares indicate control siRNA or siRNA against hnRNP D, respectively. The error bars represent the standard errors of the means of the results of three independent experiments. Reduction of the hnRNP D protein level in total cell lysate (TCL) by siRNA compared to the level of 14-3-3ζ (used as an internal control) was validated by Western blotting (gel). +, present. (B) Effect of hnRNP D depletion on endogenous mcry1 mRNA oscillation. NIH 3T3 cells were transfected with control siRNA (con-si; open squares) or siRNA against hnRNP D (sinhD; closed squares) and grown to confluence prior to treatment with 100 nM dexamethasone (Dex.) to synchronize the circadian oscillation (time zero). Total RNA (1 μg) from each time point was subjected to quantitative real-time RT-PCR with primers specific to mcry1 or to mtbp as a normalizing control (graph). Each value was relative to that for control siRNA at time zero. Results shown are representative of four independent experiments. The error bars represent the standard errors of the means of the results. Reduction of the hnRNP D protein level in total cell lysate (TCL) by siRNA compared to the level of 14-3-3ζ (used as an internal control) was validated by Western blotting (gel).
The influence of hnRNP D on mcry1 mRNA's stability and oscillation was further examined by measuring the mcry1 mRNA oscillation profile in NIH 3T3 cells when hnRNP D was downregulated. Transfection with siRNA efficiently knocked down endogenous hnRNP D proteins throughout the experimental period (Fig. 5B, gel). Interestingly, hnRNP D-knockdown cells showed a significant increase in the peak amplitude of mcry1 mRNA expression (Fig. 5B, graph) but did not exhibit changes in their mcry1 mRNA oscillation pattern other than a slight delay of phase compared to the pattern in cells treated with control siRNA. The mRNA level of bmal1, the transcriptional activator of the mcry1 gene, was not significantly different between hnRNP D-depleted cells and control cells (data not shown), indicating that hnRNP D acts specifically at the posttranscriptional level on the 3′UTR of mcry1 mRNA. Taken together, these results show that the cytoplasmic level of hnRNP D plays a crucial role in regulating mcry1 mRNA stability and damping the oscillation rhythm, critical functions for the maintenance of an mRNA oscillation pattern.
DISCUSSION
Autonomous circadian biorhythms have been studied for many years; however, we still do not fully understand the regulation of such rhythms, and it is generally assumed that other molecular regulatory mechanisms must be involved in addition to transcriptional-posttranslational feedback. A growing body of evidence suggests that posttranscriptional regulation contributes to circadian maintenance or entrainment (8, 16, 17, 19, 23, 46, 53). Although transcriptional activation and repression of circadian genes is an important regulatory mechanism (48), our results indicate that mRNA stability also plays an important role and implicate the mcry1 3′UTR as a major regulatory element in this process. We have previously shown that a reporter gene containing the full-length 3′UTR but lacking a circadian promoter did not oscillate (53), indicating that 3′UTR-mediated regulation of mRNA stability is not sufficient to generate circadian oscillation but is suitable for the function of a fine regulator. The deletion mapping and the competition analysis made it possible to identify a candidate CAE that may interact with the trans-acting factors. Typically, the sequences required for mRNA decay are only loosely conserved (52); however, the CAE identified is located in the middle of the 3′UTR and conserved among species (see Fig. S1H in the supplemental material). This conserved CAE might recruit similar RNP complexes, which may affect mcry1 mRNA stability in a similar manner. The best-studied element, the ARE, has been defined as consisting of repeated AUUUA motifs ranging from 50 to 150 nucleotides in length and divided into five groups according to the pattern of repetition. Among the different ARE groups, the 3′UTRs of some labile mRNAs, such as c-jun, have no AUUUA motif but instead contain a U-rich region (33). The U-repeat element found in the mcry1 3′UTR may fall within this group, because it has no AUUUA motif. One possible explanation for this intriguing phenomenon is that this conserved CAE might be critical for maintaining the secondary structure of mcry1 mRNA among species. Even though the U repeats in the mcry1 3′UTR are shorter than other AREs, hnRNP D binding to this region, similar to that of canonical AREs, was confirmed in the present study.
The level of hnRNP D displayed an oscillating profile in the cytoplasm, and binding to the UTR was concomitantly related to mcry1 oscillation. Even though the binding pattern showed an increase during the rising phase, the beginning of massive transcription may overcome hnRNP D-mediated decay in the rising phase and then cause mRNA increase (Fig. 4B). In this research, we excluded proteins other than hnRNP D because their binding was unchanged in the presence of competitive oligonucleotides (Fig. 2D). Nonetheless, other binding proteins may function in mRNA degradation or at any other steps in mCRY1 expression and its circadian rhythm. Some proteins bind in vitro to several of the clock genes studied, but other proteins show a unique binding pattern. This indicates that both specific and common factors may cooperate to control the turnover of each clock gene. Overall, the physiology of mcry1 oscillation appears to be due to various mRNA binding factors acting in a concerted manner. We speculate that hnRNP D interacts with exosome components, which are crucial for cytoplasmic mRNA decay, and it is related to circadian mRNA regulation (K.-C. Woo, T.-D. Kim, K.-H. Lee, D.-Y. Kim, S.-J. Kim, H.-R. Lee, and K.-T. Kim, unpublished data). Elucidation of other factors and how they function in circadian clock gene regulation will be our next challenge.
In the present study, we determined only whether the binding of hnRNP D promotes the decay of mcry1 mRNA and whether the change in expression of mcry1 results from the dynamic binding of hnRNP D. A previous study (16) demonstrated that trans-acting factors are responsible for the degradation of the Aanat mRNA in the regulation of circadian expression in the rat pineal gland. Therefore, as in other tissues, the circadian expression of the degrading factors might induce the dynamic binding of these factors to the mcry1 3′UTR. Whether or not this is the case, dynamic binding and the timely combination of factors can be expected to control mRNA stability. In addition, as we performed RNAi using siRNA-based knockdown to examine the function of hnRNP D and could not observe that all four isoforms in the cytoplasmic fraction oscillate and bind in the same manner, isotype-specific activity of hnRNP D could not be completely ruled out. Further research is needed.
Complex regulation of gene expression involves various factors acting at the posttranscriptional level, for example, in splicing regulation, mRNA export, mRNA degradation, and surveillance mechanisms, such as nonsense-mediated mRNA decay and nonstop-mediated mRNA decay (14), in addition to translational regulation. These regulatory mechanisms involve many types of RNA binding proteins, including hnRNPs and splicing-related proteins, and various noncoding RNAs, such as microRNA. With respect to the fine regulation of circadian mRNA oscillation, it is important to determine which proteins bind to the mRNA, the function of these partners, how they interact with each other, and how they trigger changes in target mRNA metabolism. This study investigated the significance of mRNA decay and its damping effect in the regulation of circadian oscillation, with particular emphasis on the role of cytoplasmic hnRNP D.
Oscillating mRNAs in all living organisms are expected to have several unique features. First, they should be under the control of rhythmic transcription. Second, they should have relatively short half-lives. If they have long half-lives, they will remain at high levels long after transcription stops. Thus, it is natural to assume that they may be regulated by the mechanism of mRNA decay. Third, they should be sufficiently robust to allow for maintenance of oscillation during environmental change. These characteristics would require diverse coordinated modes of regulation to maintain stable oscillation of the transcripts. A previous study reported a delay of Drosophila per mRNA accumulation compared with the usual transcription profile during the rising phase only (46), suggesting that mRNA in the rising phase may be more stable than that in the declining phase, which might be due to temporal posttranscriptional regulation. A study using transgenic reporter mice showed that mper1 promoter-driven luciferase (luc) mRNA decays more slowly than endogenous mper1 after induction by light exposure, whereas the luc mRNA lacking the 3′UTR displays circadian rhythm in SCN, similar to endogenous mper1 mRNA (50). Many reports have demonstrated that certain physiological treatments or the disruption of certain genes can have an effect on different aspects of circadian oscillation, such as a peak amplitude change in mRNA or protein oscillation, phase shift in locomotor activity, or even elimination of oscillation (30, 31, 40). We found that the oscillation of mcry1 mRNA continued with increased peak amplitude and s slight delay of phase when the level of mcry1 mRNA binding protein hnRNP D was reduced. Because circadian oscillation is the result of complicated regulation and interconnected regulatory circuits, the oscillatory pattern may not be readily disrupted. Under our experimental conditions, the induction of peak amplitude and a small phase delay may be caused by a reduction of hnRNP-mediated mRNA decay regulation (16), which is less dominant than transcriptional regulation by transcription factors. Circadian rhythm is linked to cellular energy metabolism and the cell cycle, and therefore, an organism will face serious physiological problems if the regulation of circadian rhythm is disturbed. As is the case for many other fundamental physiological phenomena, the complete regulatory mechanism controlling circadian rhythm remains to be revealed.
In summary, increasing evidence supports an important role for posttranscriptional regulation in the circadian rhythms of the clock genes (reviewed in reference 13). Our results demonstrated that a key clock gene, mcry1, undergoes posttranscriptional regulation in the form of mRNA degradation. Another result of our study is that mRNA decay is critically dependent on a small element of the mcry1 3′UTR. We are tempted to predict that the short U-rich sequences present in this element function as a determinant of mRNA stability. However, this role requires further verification. Finally, dynamic interactions of hnRNP D with the mcry1 3′UTR may explain the mechanism by which mRNA degradation participates in circadian rhythms. These interactions can be concomitant with changes in the level of transcription of the factors. In fact, the results of several microarray studies have indicated that the expression of RNA binding proteins is controlled by circadian rhythm (11, 32).
In future work, it will be important to identify other trans-acting factors binding to the mcry1 3′UTR and assess their role in mRNA oscillation. This work is in progress now. The functional database of the proteins identified will also allow us to predict other mechanisms of transcriptional regulation, such as splicing or regulation of translational efficiency. On this basis, we hope to be able to reveal hidden aspects of the complex strategy for achieving the 24-h cycle in mammals.
Supplementary Material
Acknowledgments
This work was supported by grants 20090063547 and 20090081464 from the National Research Foundation of Korea (NRF), funded by the South Korean Government (MEST). This work was also supported by grants from the South Korea Ministry of Education, Science and Technology (the Regional Core Research Program/Anti-Aging and Well-Being Research Center/Brain Korea 21 Program).
Footnotes
Published ahead of print on 26 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Balsalobre, A., S. A. Brown, L. Marcacci, F. Tronche, C. Kellendonk, H. M. Reichardt, G. Schutz, and U. Schibler. 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344-2347. [DOI] [PubMed] [Google Scholar]
- 2.Banihashemi, L., G. M. Wilson, N. Das, and G. Brewer. 2006. Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay. Mol. Cell. Biol. 26:8743-8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreau, C., L. Paillard, and H. B. Osborne. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevilacqua, A., M. C. Ceriani, S. Capaccioli, and A. Nicolin. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 195:356-372. [DOI] [PubMed] [Google Scholar]
- 5.Bunger, M. K., L. D. Wilsbacher, S. M. Moran, C. Clendenin, L. A. Radcliffe, J. B. Hogenesch, M. C. Simon, J. S. Takahashi, and C. A. Bradfield. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae, H. D., T. J. Park, Y. K. Lee, T. G. Lee, and K. T. Kim. 1999. Rapid and simple measurement of serotonin N-acetyltransferase activity by liquid biphasic diffusion assay. Neurochem. Int. 35:447-451. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., N. Xu, and A. B. Shyu. 2002. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 22:7268-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, H. Y., J. W. Papp, O. Varlamova, H. Dziema, B. Russell, J. P. Curfman, T. Nakazawa, K. Shimizu, H. Okamura, S. Impey, and K. Obrietan. 2007. microRNA modulation of circadian-clock period and entrainment. Neuron 54:813-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 10.Frisch, B., P. E. Hardin, M. J. Hamblen-Coyle, M. Rosbash, and J. C. Hall. 1994. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12:555-570. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez, R. A., R. M. Ewing, J. M. Cherry, and P. J. Green. 2002. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. U. S. A. 99:11513-11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahm, B., O. H. Cho, J. E. Kim, Y. K. Kim, J. H. Kim, Y. L. Oh, and S. K. Jang. 1998. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 425:401-406. [DOI] [PubMed] [Google Scholar]
- 13.Harms, E., S. Kivimae, M. W. Young, and L. Saez. 2004. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19:361-373. [DOI] [PubMed] [Google Scholar]
- 14.Isken, O., and L. E. Maquat. 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21:1833-1856. [DOI] [PubMed] [Google Scholar]
- 15.Izumo, M., T. R. Sato, M. Straume, and C. H. Johnson. 2006. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput. Biol. 2:1248-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, T. D., J. S. Kim, J. H. Kim, J. Myung, H. D. Chae, K. C. Woo, S. K. Jang, D. S. Koh, and K. T. Kim. 2005. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 25:3232-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, T. D., K. C. Woo, S. Cho, D. C. Ha, S. K. Jang, and K. T. Kim. 2007. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 21:797-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima, S., M. Hirose, K. Tokunaga, Y. Sakaki, and H. Tei. 2003. Structural and functional analysis of 3′ untranslated region of mouse Period1 mRNA. Biochem. Biophys. Res. Commun. 301:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Kojima, S., K. Matsumoto, M. Hirose, M. Shimada, M. Nagano, Y. Shigeyoshi, S. Hoshino, K. Ui-Tei, K. Saigo, C. B. Green, Y. Sakaki, and H. Tei. 2007. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. U. S. A. 104:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornmann, B., O. Schaad, H. Bujard, J. S. Takahashi, and U. Schibler. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornmann, B., O. Schaad, H. Reinke, C. Saini, and U. Schibler. 2007. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb. Symp. Quant. Biol. 72:319-330. [DOI] [PubMed] [Google Scholar]
- 22.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 23.Kwak, E., T. D. Kim, and K. T. Kim. 2006. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281:19100-19106. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 25.Liao, B., Y. Hu, and G. Brewer. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14:511-518. [DOI] [PubMed] [Google Scholar]
- 26.Lin, J. M., V. L. Kilman, K. Keegan, B. Paddock, M. Emery-Le, M. Rosbash, and R. Allada. 2002. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 420:816-820. [DOI] [PubMed] [Google Scholar]
- 27.Liu, W., Y. Rong, M. Baudry, and S. S. Schreiber. 1999. Status epilepticus induces p53 sequence-specific DNA binding in mature rat brain. Brain Res. Mol. Brain Res. 63:248-253. [DOI] [PubMed] [Google Scholar]
- 28.Lu, J. Y., N. Sadri, and R. J. Schneider. 2006. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 20:3174-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazan-Mamczarz, K., Y. Kuwano, M. Zhan, E. J. White, J. L. Martindale, A. Lal, and M. Gorospe. 2009. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 37:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido, T., M. Akiyama, T. Moriya, and S. Shibata. 2001. Sensitized increase of period gene expression in the mouse caudate/putamen caused by repeated injection of methamphetamine. Mol. Pharmacol. 59:894-900. [DOI] [PubMed] [Google Scholar]
- 31.Oster, H., A. Yasui, G. T. J. van der Horst, and U. Albrecht. 2002. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 16:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panda, S., M. P. Antoch, B. H. Miller, A. I. Su, A. B. Schook, M. Straume, P. G. Schultz, S. A. Kay, J. S. Takahashi, and J. B. Hogenesch. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307-320. [DOI] [PubMed] [Google Scholar]
- 33.Peng, S. S., C. Y. Chen, and A. B. Shyu. 1996. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol. 16:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preitner, N., F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht, and U. Schibler. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251-260. [DOI] [PubMed] [Google Scholar]
- 35.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinke, H., C. Saini, F. Fleury-Olela, C. Dibner, I. J. Benjamin, and U. Schibler. 2008. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 22:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reppert, S. M., and D. R. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418:935-941. [DOI] [PubMed] [Google Scholar]
- 38.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63:647-676. [DOI] [PubMed] [Google Scholar]
- 39.Sadri, N., and R. J. Schneider. 2009. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J. Investig. Dermatol. 129:657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto, K., and N. Ishida. 2000. Light-induced phase-shifts in the circadian expression rhythm of mammalian Period genes in the mouse heart. Eur. J. Neurosci. 12:4003-4006. [DOI] [PubMed] [Google Scholar]
- 41.Sancar, A. 2000. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 69:31-67. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar, B., J. Y. Lu, and R. J. Schneider. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 278:20700-20707. [DOI] [PubMed] [Google Scholar]
- 43.Sathyanarayanan, S., X. Zheng, R. Xiao, and A. Sehgal. 2004. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116:603-615. [DOI] [PubMed] [Google Scholar]
- 44.Sato, T. K., S. Panda, L. J. Miraglia, T. M. Reyes, R. D. Rudic, P. McNamara, K. A. Naik, G. A. FitzGerald, S. A. Kay, and J. B. Hogenesch. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527-537. [DOI] [PubMed] [Google Scholar]
- 45.Shih, S. C., and K. P. Claffey. 1999. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 274:1359-1365. [DOI] [PubMed] [Google Scholar]
- 46.So, W. V., and M. Rosbash. 1997. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16:7146-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrisani, J., A. Unterberger, S. R. Tendulkar, K. Shikimi, and M. Szyf. 2007. AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability. Mol. Cell. Biol. 27:395-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ukai-Tadenuma, M., T. Kasukawa, and H. R. Ueda. 2008. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. [DOI] [PubMed]
- 49.Vitaterna, M. H., D. P. King, A. M. Chang, J. M. Kornhauser, P. L. Lowrey, J. D. McDonald, W. F. Dove, L. H. Pinto, F. W. Turek, and J. S. Takahashi. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilsbacher, L. D., S. Yamazaki, E. D. Herzog, E. J. Song, L. A. Radcliffe, M. Abe, G. Block, E. Spitznagel, M. Menaker, and J. S. Takahashi. 2002. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc. Natl. Acad. Sci. U. S. A. 99:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20:491-497. [DOI] [PubMed] [Google Scholar]
- 52.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 53.Woo, K. C., T. D. Kim, K. H. Lee, D. Y. Kim, W. Kim, K. Y. Lee, and K. T. Kim. 2009. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37:26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, N., C. Y. Chen, and A. B. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21:6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, B., U. Albrecht, K. Kaasik, M. Sage, W. Lu, S. Vaishnav, Q. Li, Z. S. Sun, G. Eichele, A. Bradley, and C. C. Lee. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683-694. [DOI] [PubMed] [Google Scholar]
- 56.Zylka, M. J., L. P. Shearman, D. R. Weaver, and S. M. Reppert. 1998. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1103-1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.