Abstract
Intrinsically disordered (ID) regions are disproportionately higher in cell signaling proteins and are predicted to have much larger frequency of phosphorylation sites than ordered regions, suggesting an important role in their regulatory capacity. In this study, we show that AF1, an ID activation domain of the glucocorticoid receptor (GR), adopts a functionally folded conformation due to its site-specific phosphorylation by p38 mitogen-activated protein kinase, which is involved in apoptotic and gene-inductive events initiated by the GR. Further, we show that site-specific phosphorylation-induced secondary and tertiary structure formation specifically facilitates AF1's interaction with critical coregulatory proteins and subsequently its transcriptional activity. These data demonstrate a mechanism through which ID activation domain of the steroid receptors and other similar transcription factors may adopt a functionally active conformation under physiological conditions.
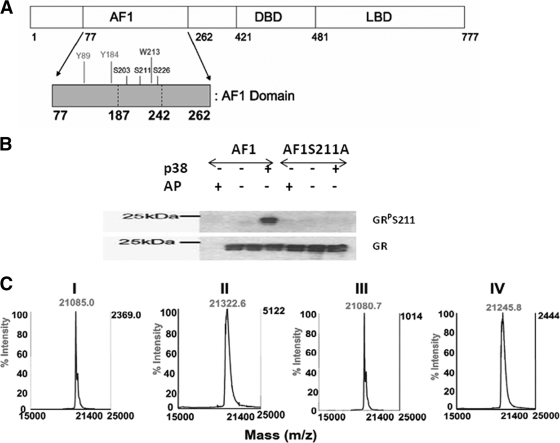
Protein phosphorylation is generally an important phenomenon in the regulation of protein function in eukaryotic cells and is often concerned with switching of a cellular activity from one state to another (29). For transcription factors (TFs), three main mechanisms of regulation by phosphorylation can be identified: (i) the DNA-binding affinity of TFs can be modulated negatively or positively, (ii) the interaction of transactivation domains of TFs with components of the transcription initiation complex can be regulated, and (iii) the shuttling of TFs between the cytoplasmic compartments can be influenced (16, 35, 43). Like many other TFs, the glucocorticoid receptor (GR) is a phosphoprotein, and it has been suggested that phosphorylation plays an important role in the regulation of GR activity (5, 9, 19, 40, 41). There are also reports suggesting that phosphorylation may affect GR stability and thus alter receptor activity (4, 41). All seven phosphorylation sites identified in the mouse GR are found in the N-terminal domain (NTD), in or near the AF1 domain (20, 31, 41). Except for one threonine, all of these sites are serine residues (41). All of the GR phosphorylation sites that are conserved among human, mouse, and rat are located within the AF1 domain (4, 31, 40, 41). In the human GR (hGR) AF1, major functionally important known phosphorylated residues are S203, S211, and S226 (Fig. 1A). At least two of these (S211 and S226) are thought to be important for transcriptional activity of the GR (4). Further, GR S211 is reported to be a substrate for p38 mitogen-activated protein kinase (MAPK), suggesting a role for p38 MAPK signaling in glucocorticoid-induced apoptosis of lymphoid cells (11, 14, 21, 31), and AF1 appears to be a main player in this process (31).
FIG. 1.
p38 phosphorylates conserved Ser residues in GR's AF1, as shown by S211-phospho-specific antibody and MALDI-TOF MS analysis. (A) Topological diagram of the hGR with expended AF1 domain containing conserved phosphorylation sites. (B) AF1 but not AF1-S211A (AF1-A) is phosphorylated at site S211. In vitro phosphorylation of AF1 and AF1-S211A was performed using p38 MAPK. AF1 or AF1-S211A and active p38 MAPK or calf alkaline phosphatase (AP) were incubated, followed by SDS-PAGE and by immunoblot analysis with GR-P S211- and GR-specific antibodies. (C) Recombinant AF1 and AF1-S211A were phosphorylated in vitro as described above, and the phosphorylation status was confirmed by MALDI MS. Subpanels: I, AF1 without p38 MAPK; II, AF1 phosphorylated with p38 MAPK; III, AF1-S211A without p38 MAPK; IV, AF1-S211A with p38 MAPK. The difference in mass between panels I and II and between panels III and IV is equivalent to three and two phospho groups, respectively.
However, it is not yet known exactly how phosphorylation influences the structure and functions of the GR AF1. Although, the importance of the AF1 domain as a major activation region was established long ago (14, 27), we are only beginning to understand its structure-function relationship. To understand how the GR transmits the transcriptional signal from ligand to specific gene(s), it is essential to gain this information. However, the structure of AF1 has been difficult to determine because in solution it exists as an intrinsically disordered (ID) domain, frequently found in TFs (2, 3, 15, 33). It is generally believed that ID sequences usually achieve structure to carry out their functions. These ID regions or domains promote molecular recognition primarily by creating propensity to form large interaction surfaces suitable for interactions with their specific binding partners (2, 17, 18, 42). It is generally accepted that the structural uniqueness of most proteins determines their biological function. This raises the question: what is the structural basis of the functional activity of such ID proteins or domains? Recent studies have suggested that signaling via phosphorylation-regulated protein-protein interaction often involves ID regions, and ID regions have a much higher frequency of known phosphorylation sites than ordered regions, suggesting a strong preference for locating phosphorylation sites in the ID regions (10, 17, 18, 22, 42, 36).
Site-specific phosphorylation represents an important regulatory mechanism in the activities of signaling proteins including steroid receptors (23, 24, 32). Since all of the known phosphorylation sites in the GR are located in the ID AF1/NTD, we hypothesize that site-specific phosphorylation of GR AF1 leads to changes in its conformations that are important for AF1's interaction with other critical coregulatory proteins and subsequent transcriptional activity. We report that GR's ID AF1 domain adopts a functionally active folded conformation due to site-specific (S211) phosphorylation by p38 MAPK that we have earlier shown to be involved in the apoptotic and gene-inductive events initiated by the GR (31).
MATERIALS AND METHODS
Protein expression and purification.
Recombinant AF1 protein was expressed in Escherichia coli BL21(DE3) using recombinant vector pGEX-4T1-AF1 (Amersham Biosciences, Piscataway, NJ). AF1-S211A and -S211E mutants were generated by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and appropriate primers were designed. Recombinant protein expressed in E. coli were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h, lysed, and extracted. The bacterial extracts were loaded onto a glutathione-Sepharose column at 4°C as described previously (21). Final protein purity of each protein was >98% as verified by presence of a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Plasmids.
The pGRE_SEAP vector (BD Biosciences, Palo Alto, CA) contains three copies of a GRE consensus sequence in tandem, fused to a TATA-like promoter (pTAL) upstream from the reporter gene for secreted alkaline phosphatase (SEAP). GR500 encodes amino acids 1 to 500 of the hGR, plus a five-residue nonspecific extension (21). The GR500 variants (GR500-S211A and -S211E) were generated via PCR by using phCMV2-GR500 as the starting template and inserting the PCR fragments into pECFP-C1 (BD Biosciences) using XhoI/SmaI cloning sites. TATA box binding protein (TBP) was cloned into the pcDNA3.1(+) expression vector (Invitrogen, Carlsbad, CA) and into pEYFP-C1 (BD Biosciences). DNA sequencing was performed on all clones to confirm correct sequence.
In vitro phosphorylation assays.
In vitro phosphorylation of recombinant AF1 at S211 was carried out by using a p38α/SAPKa assay kit (Upstate, Cell Signaling solutions, Lake Placid, NY). In each case, 10 μg of AF1 or AF1-S211A and 10 ng of active p38 MAPK was incubated for 2 h at 37°C in a shaking water bath. The level of S211 phosphorylation was determined by using immunoblot analysis with specific antibodies against GR-S211-P (Cell Signaling Technology, Beverly, MA) and GR. We further analyzed the level of phosphorylation by using mass spectroscopy (MS). Samples were prepared for matrix-assisted laser desorption ionization time-of-flight MS (MALDI-TOF MS). A 1.0-μl portion of the sample was deposited onto the MALDI plate and allowed to dry. Then, 1.0 μl of matrix (sinapinic acid or 3,5-dimethoxy-4-hydroxycinnamic acid; Aldrich Chemical Co.) was applied to the sample spot and allowed to dry. External calibration was performed on each sample spot using a nearby spot consisting of cytochrome c (Sigma-Aldrich) mixed with matrix as described previously. MALDI-TOF MS was performed by using an Applied Biosystems 4800 MALDI TOF/TOF proteomics analyzer in positive-ion and linear modes. For MS data, 2,000 to 4,000 laser shots were acquired and averaged from each sample spot.
Fluorescence emission spectroscopy.
Fluorescence emission spectra of purified recombinant AF1 in solution were recorded by using a Spex FluoroMax spectrometer at excitation wavelengths of 278 or 295 nm as described previously (3). Measurements were taken in a 1.0-cm rectangular cuvette at 22°C, and all data were corrected for the contribution of the buffer.
CD spectroscopy.
Circular dichroism (CD) spectra of proteins (at 200 μg/ml with or without p38 MAPK in 5 mM Tris [pH 7.9] and 25 mM NaCl) were recorded on an Aviv 62 spectropolarimeter using a 0.1-cm quartz cell, with a bandwidth of 1.0 nm and a scan step of 0.5 nm, as described previously (3). Each spectrum is representative of at least three independent experiments, corrected for the contribution of the buffer and smoothed.
Limited proteolytic digestion.
Digestion of 10 μg of purified AF1, AF1-S211A, or AF1-S211E was carried out using sequencing-grade trypsin (Sigma-Aldrich), chymotrypsin (Sigma-Aldrich), or Endopeptidase-Glu-C (Sigma-Aldrich) at 4°C for 15 min in 20 mM Tris-150 mM NaCl (pH 8.3) at a protein/enzyme mass ratio of 100:1. Reactions were terminated by adding SDS loading buffer and boiling the mixture for 5 min. Digested samples were run on an SDS-PAGE gel and stained with Coomassie blue R-250.
Immunoprecipitation.
HeLa nuclear extract containing 1 mg of total protein, 5 μl of antibody (TBP, CREB binding protein [CBP], or steroid receptor coactivator 1 [SRC-1]), and 50 μl of protein A-agarose conjugate was incubated for 4 h at 4°C. Then, 10 μg of purified AF1 with or without p38 MAPK or AF1-S211A with or without p38 MAPK was added, followed by incubation for another 2 h at 4°C. The beads were centrifuged, washed thoroughly, resuspended in SDS loading buffer, and boiled for 5 min to release bound proteins. The released proteins were resolved by SDS-PAGE and immunoblotted with a GR antibody after transfer onto a polyvinylidene difluoride membrane as described previously (21). The results are expressed as means ± the standard error. Levels of significance were evaluated by a two-tailed paired Student t test, and a P value of <0.05 was considered significant.
Cell culture and transient transfection.
CV-1 cells (American Type Culture Collection) were grown at 370 C in minimal essential medium with Earle's salts (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (Atlanta Biologicals, Norcross, GA). Cells were subcultured every 2 to 3 days. CV-1 cells were plated on a 24-well plate (1000 μl/well) 1 day before the transfection and transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Transfected cells were maintained at 37°C in 5% CO2 and 95% air for the duration of the experiment (27 h). The level of transfection was estimated to be >60% by using fluorescence microscopy of the cells receiving cyan fluorescent protein (CFP). Transfection efficiency was normalized by using immunoblot analysis with specific antibodies against AF1, green fluorescent protein (GFP), TBP, CBP, and/or SRC-1. Protein extracts from whole-cell lysates were loaded on SDS-PAGE gels, transferred to polyvinylidene difluoride membranes, blocked with 5% milk at room temperature, and incubated with 5% bovine serum albumin containing appropriate antibody at 4°C overnight. Membranes were washed with TBS-Tween, incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature, and visualized by enhanced chemiluminescence (Amersham Biosciences).
Fluorescence microscopy and FRET analysis.
CV-1 cells grown on a tissue culture dish with integrated slide (Matec) 1 day before transfection were cotransfected with 1 μg of pGRE-SEAP reporter and 3 μg of pYFP-ECFP (positive control), 1.5 μg of pECFP-C1, and/or 1.5 μg of pEYFP-C1 (negative control). To test the dependence of fluorescence resonance energy transfer (FRET) on AF1 in the GR, cells were cotransfected with 1.5 μg of pEYFP-TBP, -CBP, or -SRC-1 and 1 μg of pGRE-SEAP. Pairs of pGRE-SEAP received 1.5 μg of either pECFP-GR500, pECFP-GR500S211A, or pECFP-GR500S211E. Cells were washed 24 h later twice with isotonic phosphate-buffered saline (PBS; pH 7.4), fixed with 4% formaldehyde-PBS for 10 min, and washed twice with PBS. The cells were visualized by using a Zeiss LSM-510 META confocal microscope (Carl Zeiss, Thornwood, NY) with a Plan-Apochromat 63× 1.4 oil-immersion objective lens and 6.1-Å argon laser. Pre- and postbleach (PB) images were collected at 12-bit resolution on two channels: 458 nm for CFP and 514 nm for yellow fluorescent protein (YFP). Five images were taken, two before and three after bleaching, with 20-s intervals. To assure more than 90% postbleach (PB), an arbitrarily selected region of interest containing examples of both nuclear and cytoplasmic compartments was irradiated with a 100% intensity laser line at 514 nm at 200 to 2,000 iterations. Increased CFP (donor) fluorescence intensity upon YFP (acceptor) was indicative of positive FRET, and its efficiencies (FE) were calculated by the following equation: FE% = (I DA − I DB)/(I DA) × 100, where IDA is the donor intensity after bleaching (extracted from image 2 of time series) corrected for background and fractional PB result and IDB is donor intensity before PB background correction (estimated from image 3 of the PB time series). Images that showed any focal plane drift were eliminated. In addition, we tested CFP, CFP-GR500, YFP, and YFP-TBP alone each time to account for any bleedthrough and background FRET as recommended (data not shown).
Reporter gene assays.
We used the SEAP reporter system due to its high signal-to-noise ratio and quantifiable transcriptional activity without the need for cell disruption. In the experiments with holo-GR, CV-1 cells were cotransfected as described above with 0.13 μg of pGRE_SEAP reporter vector; 0.13 μg of pECFP-GR500, -S211A, or -S211E; and 0.5 μg of pcDNA3.1-TBP, pRSLV-CBP, or SRC-1. The total amount of DNA added was kept fixed at 0.8 μg by the addition of empty pECFP vector. Medium (25 μl) was collected 27 h later and tested for the presence of SEAP (Great EscAPe SEAP detection kit; BD Biosciences) according to the manufacturer's protocol. Experiments were performed at least three times, in triplicate. The data from different experiments were normalized to GR500 activity.
Inhibition of p38 MAPK.
CV-1 cells were cotransfected with CFP-GR500 and TBP, CBP, or SRC-1 as described above. After 24 h of transfection the cell cultures were treated with 1 μg of SB203580 or SB202190 (Sigma-Aldrich)/ml, respectively, for an additional 24 h, and the reporter gene was detected as described above. In the experiments with holo-GR, CV-1 cells were cotransfected as described above with 0.13 μg of pGRE_SEAP reporter vector, 0.13 μg of pEGFP-GR, and 0.5 μg of pcDNA3.1_TBP. Cells were allowed 24 h to recover and then were treated with 1 μM dexamethasone (Dex). Medium was collected 24 h later and tested for the presence of SEAP (Great EscAPe SEAP detection kit) according to the manufacturer's protocol.
RESULTS
Site-specific phosphorylation induces secondary or tertiary structure in otherwise ID GR AF1 domain.
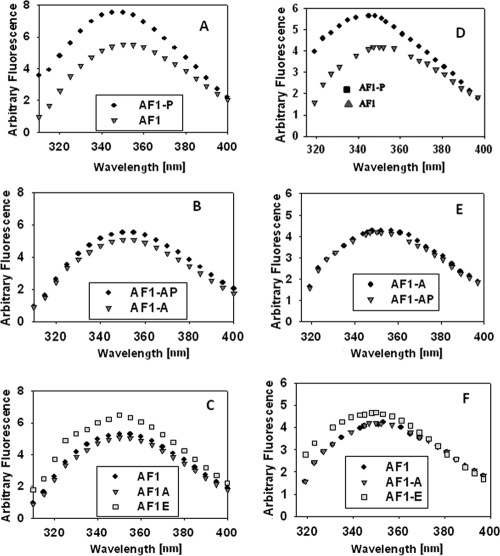
Similar to other steroid receptors, the GR contains several major functional domains. These are shown diagrammatically for the hGR in Fig. 1A. The AF1 transactivation region, amino acids 77 to 262 is highlighted, with vertical lines above the bar indicating the location of the two Tyr and one Trp residues, and three potential phosphorylation sites within AF1. Recombinant wild-type and two AF1 mutants S211A or S211E were expressed in a bacterial system and purified to homogeneity. To test the effects of phosphorylation on the conformational changes in AF1, we first established conditions that can phosphorylate AF1 in vitro. Using GR S211-phospho site-specific antibody (Fig. 1B) and MALDI-TOF MS (Fig. 1C), we confirmed p38 MAPK-mediated in vitro phosphorylation of AF1. We recorded the far-UV CD spectra of unphosphorylated and phosphorylated AF1. As expected, unphosphorylated AF1 shows characteristics of an ID protein, whereas phosphorylated AF1 adopts significantly higher secondary structural elements in it with helical content increased by >50% at the expense of random coil compared to unphosphorylated AF1 (Fig. 2A and Table 1). Under similar conditions, mutation of S211A does not show any significant secondary structural changes in AF1 (Fig. 2B and Table 1), suggesting that p38 MAPK-mediated phosphorylation of S211 is important for inducing secondary structure in an otherwise ID AF1 domain. We also compared the far-UV CD spectra of unphosphorylated AF1, AF1-S211A, and AF1-S211E to determine whether addition of negative charge by way of substituting Ser to Glu results in similar effects as seen due to phosphorylation of S211. Our results show that S211A mutant does not alter AF1 conformation, whereas S211E mutation showed only a modest increase in helical content compared to wild-type AF1 (Fig. 2C), suggesting that site-specific phosphorylation of ID AF1 leads to the induction of secondary structural elements in it and negative charge per se is not sufficient to mimic these effects.
FIG. 2.
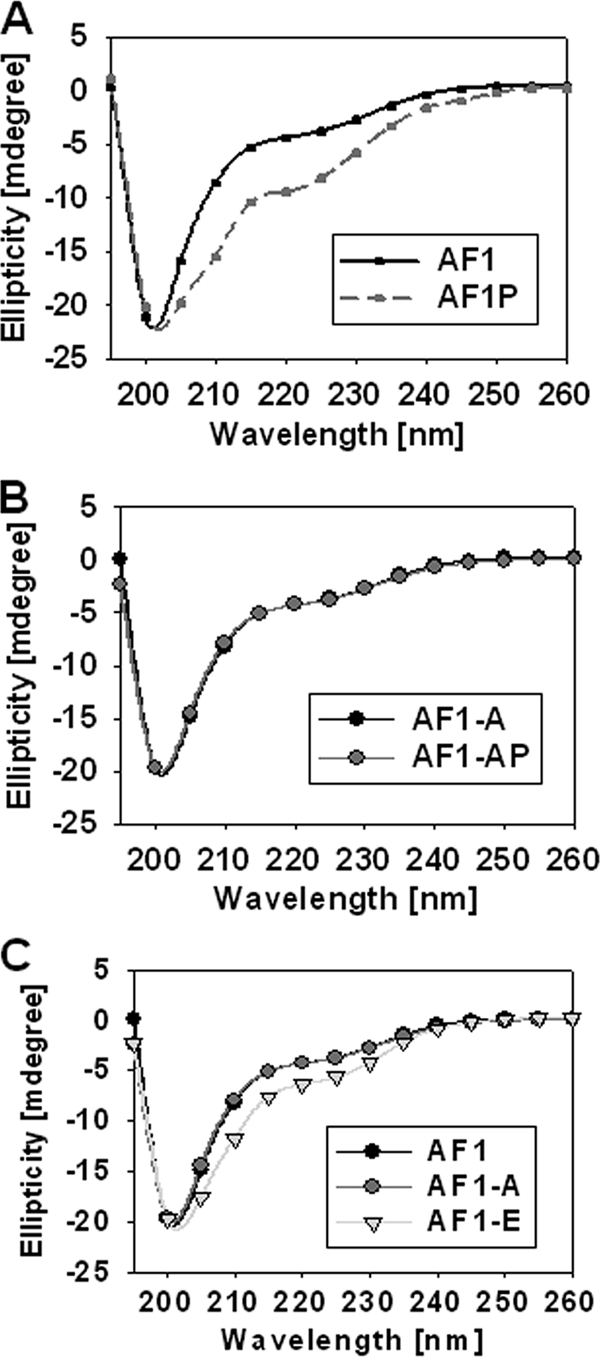
Site-specific phosphorylation induces secondary structure in ID AF1 domain. (A to C) Far-UV CD spectra of recombinant AF1 and AF1-S211A with or without p38 MAPK and AF1-S211E. AF1, unphosphorylated AF1; AF1-P, phosphorylated; AF1-A, AF1S211A mutant; AF1-E, AF1S211E mutant. Each spectrum represents an average of five spectra recorded, corrected for the contribution of the buffer, and smoothed.
TABLE 1.
Summary of changes in secondary structural elements in AF1 after p38-mediated phosphorylationa
| Element | % Change |
|||
|---|---|---|---|---|
| AF1 | AF1-P | AF1S211A | AF1S211A-P | |
| α-Helix | 26 | 45 | 25 | 26 |
| β-Sheet | 13 | 19 | 15 | 14 |
| Random coil | 61 | 36 | 60 | 60 |
We further carried out partial proteolytic digestion experiments with three different proteases (trypsin, chymotrypsin, and Endo Gluc-C) to determine tertiary structural changes in AF1 with or without p38 MAPK-mediated phosphorylation. The results of one such partial proteolysis experiment are shown in Fig. 3. The upper panel shows the products of proteolysis of AF1 (as resolved by a Coomassie blue-stained SDS-PAGE gel) after digestion with chymotrypsin. Partial proteolysis data indicate protection of peptides in phosphorylated AF1 (a major protected band indicated by an arrow) compared to unphosphorylated AF1 (compare lanes 3 and 5). Under similar conditions, digestion of AF1S211A did not show any significant difference in the cleavage pattern (compare lanes 7 and 9), suggesting that blocking of S211 phosphorylation site fails to produce any tertiary structural changes in AF1 even after treatment with p38. However, mutation of AF1S211E results in similar cleavage pattern (at least for a major band) as seen in case of phosphorylated AF1. Similar results were obtained with Endo Gluc-C (middle panel) and trypsin (lower panel) proteases.
FIG. 3.
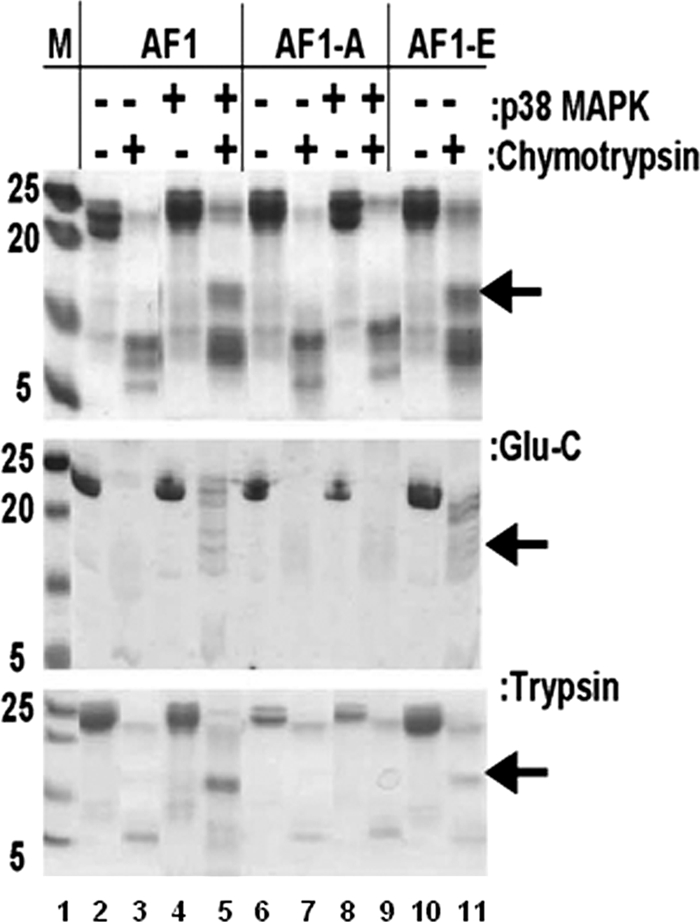
Phosphorylated AF1 resists partial proteolysis. A Coomassie blue-stained SDS-PAGE gel showing limited proteolytic digestion of purified AF1 and AF1-S211A with (+) or without (−) p38 MAPK and AF1-S211E is shown. AF1-A, AF1S211A mutant; AF1-E, AF1S211E mutant. Experiments were performed with sequencing-grade trypsin, chymotrypsin, or Endo Glu-C. Experiments were repeated at least five times with similar results.
We also recorded fluorescence emission spectra of AF1 (Fig. 3) after excitation at 278 or 295 nm, which reflect the changes coming from Tyr (Y89 and Y184) and Trp (W213) residues or from of the single Trp (W213), respectively. In both sets of spectra, the quantum yield of the fluorescence is significantly increased in the phosphorylated AF1 compared to unphosphorylated AF1, and corresponding blue shifts in the emission maxima (from 351 to 346 nm and from 355 to 341 nm at 278- and 295-nm excitation wavelengths, respectively; Fig. 4A and D and Table 2) . Under similar conditions, AF1-S211A failed to show any significant changes both in terms of fluorescence intensity and shift in wavelength maxima (Fig. 4B and E and Table 2). A comparison of spectra from AF1 and S211E showed only moderate changes in case of S211E (Fig. 4C and F). These fluorescence changes due to site-specific phosphorylation of AF1 are typical of those accompanying the removal of aromatic residues from polar, aqueous solution into a more hydrophobic environment within the protein. Because the three amino acids excited are located well apart in AF1 (Fig. 1A), the conformational alterations reflected in the fluorescence emission changes should be happening throughout the AF1 peptide. Both partial proteolysis and fluorescence emission data clearly indicate the formation of tertiary structure in the GR AF1 following p38-mediated phosphorylation of S211. Taken together, these results suggest that site-specific (S211) phosphorylation of otherwise ID AF1 results in a compact structure formation in it. Some modest structural changes observed in case of S211E mutants (Fig. 2 and 3) may be simply due to effects of phosphorylation of other sites (S203 and S226), which can be phosphorylated by treating AF1 with p38 MAPK (Fig. 1C). In the case of proteolytic digestion experiments, observation of some similar protected bands both with phosphorylated AF1 and AF1S211E mutant could be due to local structural rearrangements arising from the mutation of S211E, which could have buried the available site(s) for these proteases. However, our data with S211A mutant do not support this notion unless phosphorylation of other two sites is dependent upon S211 phosphorylation.
FIG. 4.
Site-specific phosphorylation induces tertiary structure in ID AF1 domain. (A to F) Fluorescence emission spectra of AF1 and AF1-S211A with or without p38 MAPK and AF1-S211E recorded at a 278-nm (A to C) or 295-nm (D to F) excitation wavelength. AF1, unphosphorylated AF1; AF1-P, phosphorylated; AF1-A, AF1S211A mutant; AF1-E, AF1S211E mutant. Each spectrum was recorded in the emission wavelength range of 300 to 400 nm, and the result of one representative spectrum of at least three independent experiments is presented, corrected for the contribution of the buffer, and smoothed.
TABLE 2.
Summary of changes in the relative fluorescence intensity and wavelength maximum after AF1 phosphorylationa
| Parameter | Change in relative fluorescence intensity |
|||
|---|---|---|---|---|
| AF1 | AF1-P | AF1S211A | AF1S211A-P | |
| F/F0 max (278 nm) | 5.31 | 7.55 | 5.08 | 5.50 |
| λmax (278 nm) | 351 | 346 | 352 | 352 |
| F/F0 max (295 nm) | 4.22 | 5.71 | 4.28 | 4.17 |
| λmax (295 nm) | 355 | 341 | 357 | 356 |
Values were calculated from fluorescence data obtained at a 278- or 295-nm excitation wavelength (Fig. 4). F/F0, relative fluorescence intensity at λmax; λmax, the wavelength at the maximum fluorescence intensity. AF1, unphosphorylated AF1; AF1-P, phosphorylated AF1.
Phosphorylation-induced structure formation in the GR AF1 facilitates its interaction with specific coregulatory proteins.
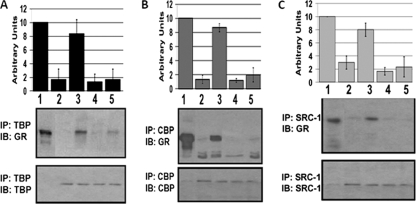
It is known that AF1 makes physical interactions with other cofactors in order to transactivate gene(s) and that conditional folding is presumed to be important for these interactions (12, 21, 28, 30). It has recently been hypothesized that one of the reasons why ID proteins particularly in TFs are rich in phosphorylation sites is that kinase-mediated phosphorylation can induce conformation(s) in ID region, which can facilitate its interaction with other binding partners. We therefore evaluated whether the conformation induced in ID AF1 domain by site-specific phosphorylation is important for its specific protein-protein interactions. Using immunoprecipitation assay, we tested these AF1 interactions with specific coregulatory proteins (TBP, CBP, and SRC-1) from HeLa nuclear extracts. Separate HeLa nuclear extracts supplemented with purified AF1/AF1-S211A protein with or without p38 MAPK treatment were prepared. The extracts were then incubated with antibody-linked beads specific to each of the partner proteins. The antibody-linked beads were recovered and washed extensively, and the bound proteins were released and resolved by SDS-PAGE. An antiserum to amino acids 150 to 175 of the GR was then used to identify AF1 on the gels. Consistent with previous reports (11, 20, 21), in the case of unphosphorylated AF1, we detected a very weak interaction with each of these coregulatory proteins. These interactions were increased significantly when AF1 was phosphorylated, suggesting that phosphorylation of AF1 facilitates its interaction with all of the three coregulatory proteins tested (Fig. 5, compare lanes 2 and 3). Mutation of S211A does not show any significant differences in AF1's interaction with these coregulatory proteins under similar conditions (Fig. 5, compare lanes 4 and 5).
FIG. 5.
Phosphorylation-induced conformational changes facilitate interactions of AF1 with critical coregulatory proteins from HeLa nuclear extracts. Immunoprecipitation (IP) with indicated antibodies was carried out with HeLa cell nuclear extracts with AF1. Shown are representative immunoblots (IB) of at least three independent experiments. Graphs show the results of densitometric analysis of three experiments. The results are expressed as means ± the standard deviation. Lanes: 1, control AF1; 2, unphosphorylated AF1; 3, phosphorylated AF1; 4, unphosphorylated AF1-S211A; 5, phosphorylated AF1-S211A.
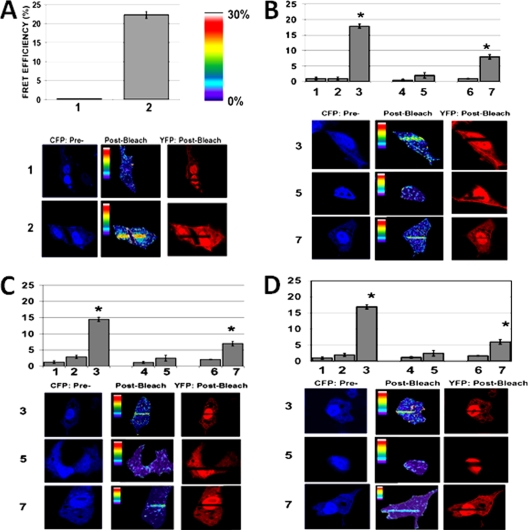
We further determined whether these interactions are taking place within a cellular environment. To test this, we used the FRET method. Plasmids expressing CFP and YFP were obtained, and from that we generated constructs that express the fluorophores linked to GR500 (CFP-GR500), (CFP-GR500-S211A), or (CFP-GR500-S211E) and to TBP (YFP-TBP), CBP (YFP-CBP), or SRC-1 (YFP-SRC-1). The GR500 construct is constitutively active as a TF, while avoiding the possibility of any contribution from AF2 (31). The constructs were cotransfected into GR-deficient CV-1 cells with cotransfection of a promoter-reporter construct containing GRE sites (GRE-SEAP). Several control experiments were included. Independent CFP- and YFP-expressing constructs were cotransfected with GRE-SEAP and tested for FRET as a negative control (data not shown). As a positive control, a CFP-YFP construct that linked CFP-YFP by eight amino acids was coexpressed with GRE-SEAP. Figure 6A shows examples of results from such controls. Further, we tested the FRET efficiency of GR500 interaction with each of the coregulators in a cotransfection assay. Our results show that GR500 interacts directly with TBP (Fig. 6B), CBP (Fig. 5C), and SRC-1 (Fig. 6D) in the nuclei of GR-deficient CV-1 cells cotransfected with GR500 with or without each coregulator (compare lanes 2 and 3). Interaction of the GR500-S211A mutant with each binding partner was greatly diminished (Fig. 6B to D, compare lanes 3 and 5), whereas the S211E mutant indicates only a modest interaction (Fig. 6B to D, compare lanes 3 and 7). Since the interactions of these coregulators with GR500 is known to take place only with the AF1 domain, these results suggest that phosphorylation-induced conformational changes in ID AF1 allow its protein surfaces to facilitate AF1's interactions with specific coregulatory proteins, an essential requirement for activation domains of TFs to regulate transcriptional activity of the target gene (21, 34). To confirm the localization of the CFP-GR500 plasmids and its mutants, CFP-GR500S211A and CFP-GR500S211E, we cotransfected them with YFP-TBP into CV-1 cells and evaluated their localization via confocal microscopy (Fig. 7). These results suggest that all GR500 constructs, including the mutant forms, are mainly located in the nuclei of the cells and that the coregulator is dispersed throughout the cells. This reestablishes that the S211A or -E mutation does not affect the translocation of the GR500 and that the interactions reported in the present study are specific to phosphorylation-induced changes and not due to alterations in the nuclear translocation.
FIG. 6.
Phosphorylation-induced conformational changes facilitate interactions of AF1 with critical coregulatory proteins in CV-1 cell as assessed by FRET analyses. Representative same-cell images in the donor (CFP) channel before and after PB and the YFP channel are shown postbleaching to demonstrate the bleach efficiency. The upper panel graphs show the average FRET efficiency in each case, whereas the lower panel represents one such image in each case. (A) Controls receiving fluorescent proteins (CFP-YFP) without TBP or GR to establish basal and maximal FRET efficiency under the experimental conditions. Graph lanes: 1, CFP-empty + YFP-empty negative control; 2, CFP-YFP fusion constructs positive control. (B to D) Interactions between GR500 and TBP (B), CBP (C), or SRC-1 (D). 1, CFP-empty + YFP-TBP; 2, CFP-GR500 + YFP-empty; 3, CFP-GR500 + YFP-TBP, CBP, or SRC-1; 4, CFP-GR500-S211A + YFP-empty; 5, CFP-GR500-S211A + YFP-TBP, CBP, or SRC-1; 6, CFP-GR500-S211E + YFP-empty; 7, CFP-GR500-S211E + YFP-TBP, CBP, or SRC-1. Experiments were carried out three independent times and analyzed, and the calculated average FRET efficiencies ± the standard deviation of 15 cells were graphed for each of the conditions.
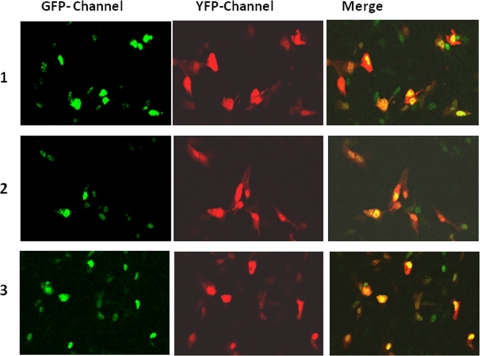
FIG. 7.
Confocal microscopic images showing that mutations (S211A or -E) within AF1 do not affect GR500's translocation to the nucleus in CV-1 cells. CV-1 cells constitutively expressing GFP-GR500, YFP-TBP, or GFP-GR500 plus YFP-TBP (row 1), GFP-GR500S211A, YFP-TBP, or GFP-GR500S211A plus YFP-TBP (row 2), or GFP-GR500S211E, YFP-TBP, or GFP-GR500S211E plus YFP-TBP (row 3) (left panels) are shown.
Effect of phosphorylation-induced interactions of TBP, SRC-1, or CBP on AF1-driven transcription.
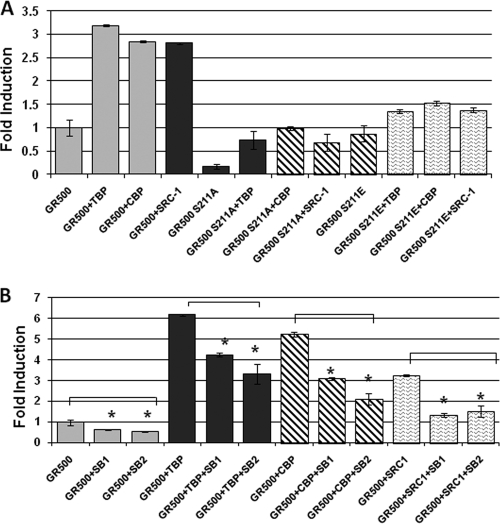
We tested the effects of phosphorylation-induced folding or binding events on AF1-driven transcription using GR-responsive promoters in transient transfection-based reporter assays in GR-deficient CV-1 cells. The promoter-reporter plasmid (GRE-SEAP) contains three GREs upstream from a TATA box and a reporter gene that encodes alkaline phosphatase secreted into the medium. To test the effects of these coregulators on transcription driven by hGR AF1, we cotransfected CV-1 cells with a GRE-dependent reporter gene and a constant amount of GR500 expression vector alone or with added vectors expressing TBP, SRC-1 or CBP. Lacking the ligand binding domain (LBD), GR500 is transcriptionally active without steroid and can induce genes and/or apoptosis in cells to nearly the same extent as steroid-bound holo-GR (20, 31). GR500 alone significantly increased reporter activity compared to empty CFP vector alone (Fig. 8A, upper panel, lane 1), and input of the plasmids expressing TBP, SRC-1, or CBP gene enhanced the GR500 induction of the GRE-SEAP reporter severalfold (Fig. 8A, upper panel, lanes 2 to 4). These reporter activities were significantly reduced when the GR500-S211A mutant was used (Fig. 8A, upper panel, lanes 5 to 8), whereas GR500-S211E mutant showed only a modest activity compared to GR500 (Fig. 8A, upper panel, lanes 9 to 12). The level of expression of GR500 was assessed by using GR antibody. The level of GR500 expression was also assessed using antibody for GFP and that of each coregulatory protein using the respective antibody. The results show the expected levels of expression of each protein (data not shown). These results strongly suggest that the enhancement of GR-induced transcription by TBP, SRC-1, or CBP is achieved through the AF1 region and that p38-mediated site-specific phosphorylation plays an important role in it by inducing more helical structure in the otherwise ID AF1 region, confirming that phosphorylation-induced structure formation in ID regions aids in facilitating protein-protein interactions and, subsequently, GRE-mediated AF1 transcriptional activity. To further confirm that these changes in transcriptional activity are in fact caused by phosphorylation-dependent changes and mediated by p38, we cotransfected GR500 with TBP, CBP, or SRC-1 and used two pharmacological inhibitors of p38 (SB203580 or SB202190) to see the effect on GRE-mediated transcriptional activity (Fig. 8B). Treatment of SB203580 or SB202190 significantly reduces GRE-driven AF1 activity in the GR500 construct alone (Fig. 8B, lanes 1 to 3), as well as with respect to cotransfection of TBP (lanes 4 to 6), CBP (lanes 7 to 9), or SRC-1 (lanes 10 to 12). The results show that p38 is indeed responsible for the increase of transcriptional activity.
FIG. 8.
Phosphorylation-dependent cofactor-binding increases AF1-mediated transcriptional activity of a promoter containing 3×GRE. (A) S211A mutant inhibits GRE-mediated AF1 activity, as assessed by SEAP-based promoter-reporter assay in CV-1 cells. (B) Treatment of pharmacological inhibitors of p38 (SB203580 or SB202190) inhibits GRE-mediated GR AF1 activity in a promoter-reporter assay. SB1, SB203580; SB2, SB202190. The results are expressed as means ± the standard error. Experiments were repeated five times. The levels of significance were evaluated by a two-tailed paired Student t test, and a P value of <0.05 was considered significant. Graphs were normalized to the transfection efficiency of each construct assayed by immunoblotting with specific antibodies for GR, TBP, CBP, SRC-1, and GFP (data not shown).
Effect of S211 site-specific phosphorylation on the transcriptional activity of holo-GR.
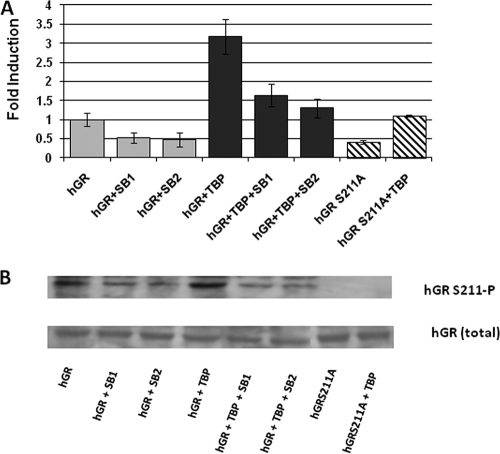
To better understand the effect of phosphorylation on the GR-dependent transcriptional activity, we measured the effects of p38-mediated S211 phosphorylation in the full-length GR. To test these effects, we cotransfected CV-1 cells with a GRE-dependent reporter gene and a constant amount of hGR expression vector alone or with added vector expressing TBP. hGR alone significantly increased reporter activity compared to empty vector alone (Fig. 9A), and input of the plasmid expressing TBP gene enhanced the hGR induction of the GRE-SEAP reporter severalfold (Fig. 9A). These reporter activities were significantly reduced when the hGR-S211A mutant was used (Fig. 9A). To further confirm that these changes in transcriptional activity are in fact mediated by p38, we cotransfected hGR with or without TBP and used two pharmacological inhibitors of p38 (SB203580 or SB202190) to see the effect on GRE-mediated transcriptional activity (Fig. 9A). Treatment of SB203580 or SB202190 significantly reduces GRE-driven activity in the hGR construct alone (Fig. 9A), as well as with respect to cotransfection of TBP. The results show that p38 is indeed responsible for the increase of GRE-mediated transcriptional activity of the holo-GR. The level of expression of hGR was assessed using GR antibody (Fig. 9B). The level of S211 phosphorylation was also assessed using antibody for GRS211-phospho-specific antibody. The results show that treatment of p38 inhibitors significantly lowered the level of hGRS211 phosphorylation, whereas total GR level of similar in each case. It is evident from the immunoblots that the level of GRS211 phosphorylation was not completely abolished by the treatment of these inhibitors, which could be the reason for only a partial blocking of receptor activity after treatment with these inhibitors. However, the role of other kinases and/or phosphorylation of other sites (mainly, S203 and S226) cannot be ruled out. Nevertheless, these results suggest that the enhancement of GR-induced transcription is achieved through p38-mediated site-specific phosphorylation and that AF1 appear to be the major player in the process.
FIG. 9.
Phosphorylation-dependent cofactor-binding increases GRE-mediated transcriptional activity of full-length GR. (A) S211A mutation in hGR or treatment of pharmacological inhibitors of p38 (SB203580 or SB202190) inhibits GRE-mediated hGR activity in a promoter-reporter assay. SB1, SB203580; SB2, SB202190. The results are expressed as means ± the standard error. The levels of significance were evaluated by a two-tailed paired Student t test, and a P value of <0.05 was considered significant. (B) Immunoblots showing the levels of hGR expression in transfected CV-1 cells. The upper panel shows the levels of S211 phosphorylation in hGR, and the lower panel shows the levels of total hGR in each case.
DISCUSSION
Compared to prokaryotes, eukaryotic genomes contain a much higher number of ID proteins, indicating toward greater need for signaling and regulation in nucleated cells (10, 17, 38). ID regions commonly exist within TFs and are often located in their transactivation domain (15). The mechanisms by which TFs control gene expression poses a central problem in molecular biology and the role of their transcriptional activation domains in this complex process is of immense importance. TFs remodel chromatin structure in an extremely dynamic situation such that they have the capacity to rapidly form and reform multiprotein complexes involving coactivators/corepressors and/or proteins from the fundamental initiation complex. In this context the role of ID domain(s) with flexible conformations becomes extremely important by providing platforms for inclusion and/or exclusion of specific protein complexes; thus, the ultimate composition of the complex assembly may dictate final outcome responsible for specific gene regulation. Therefore, the notion that ID domains of TFs must have significantly ordered conformation in their normal cellular milieu poses a paradox that must be resolved before we can fully understand their role in gene regulation (17). It has been predicted that ID regions have a much higher frequency of known phosphorylation sites than ordered regions and that signaling via phosphorylation-regulated protein-protein interactions often involves ID regions (9, 26, 37, 43).
In recent years it has become clear that the cross talk exists between GR signaling and other receptor cascades including inflammatory kinases (MAPKs, ERK, p38, and JNK), as well as the cyclic AMP-driven protein kinase A pathways (1a, 6, 13, 24, 25, 39). There is evidence that that differential phosphorylation is a potential regulator of species-specific actions of the GR (6, 25, 31, 32). It has been shown that p38 MAPK is a component of the glucocorticoid-evoked pathway to lymphoid cell apoptosis in human and mouse cell systems and that the p38-specific upstream kinase kinase MKK3 appears to be involved. It is clear that GR-mediated glucocorticoid signaling is a multifaceted process involving cross talk with various regulatory kinase pathways (6, 31). Thus, signaling cascades that induce phosphorylation of the GR and its coactivator proteins are critical factors in determining the physiological actions of the GR. However, the underlying mechanism that governs this important and yet complex phenomenon is not well understood. Our results indicate a mechanism for how site-specific phosphorylation within its activation domain leads to GR-mediated signaling. Further, phosphorylation-induced functional conformation in AF1 may also shed light on why most if not all of the major known phosphorylation sites are located in the ID activation domain of the GR and several other TFs. Based upon our findings here, we propose that p38 MAPK-mediated site-specific phosphorylation of the GR results in the recruitment of critical coregulatory proteins (by providing AF1 surfaces available for these interactions) that subsequently leads to transcription of target gene(s).
Site-specific phosphorylation represents an important regulatory mechanism in the activities of signaling proteins including TFs and regulates the binding affinity of TFs to their coactivators and DNA thereby altering gene expression, cell growth, and differentiation (43). It has been hypothesized that protein phosphorylation of Ser residue predominantly occurs within ID regions of signaling molecules rather than merely on surface residues. Our data clearly support this idea and provide a physical proof of it. Relatively few ID regions have been structurally characterized, and yet a significant fraction of them contain phosphorylation sites (17, 18, 42). One of the main reasons for such propensity is to facilitate extensive formation of hydrogen binding between the backbones and/or side chains that can occur through disorder-order transition within the ID region. The formation of these hydrogen bonds would be difficult if the sites of phosphorylation were located within ordered regions. In fact, using a computer modeling approach, it has recently been proposed that the phosphorylation of S211 may result in the formation of a more compact structure in the surrounding peptide and can facilitate specific hydrogen binding, which exposes novel surfaces potentially facilitating cofactor interaction (7). In that study, the kinetics of GR phosphorylation at S226, S203, and S211 was also studied (7), and the authors found that Dex-dependent phosphorylation of both S211 and S226 followed a similar pattern, whereas S203 had a higher basal level of phosphorylation and a less Dex-induced effect (7). Further, they showed that common surfaces within GR affected by phosphorylation may be responsible for influencing the regulation of selective genes (7). Phosphorylation-induced conformational changes have also been proposed in the ID NTD of the estrogen receptor (8).
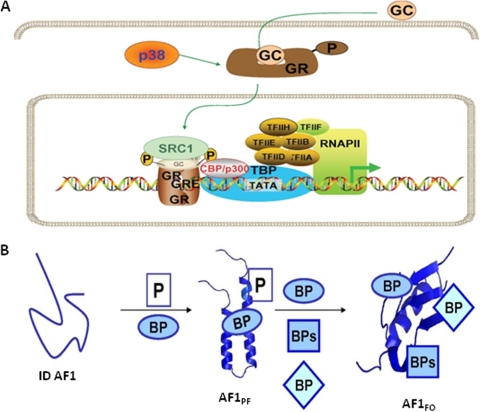
ID activating regions of many TFs have been shown to function by recruiting the transcriptional apparatus to the promoter (24), and ID regions do indeed fold into a more ordered helical structure under physiological conditions (43). The characterization of phosphorylation-induced structure formation in facilitating protein-protein interactions should be of particular importance in understanding the mechanism by which kinase(s) regulate the transcriptional regulation of the target genes in the nucleus. In sum, we propose that under physiological conditions, site-specific phosphorylation plays a crucial role in allowing the ID activation domain of the GR to adopt functionally active conformation(s) in vivo (Fig. 10). The resulting structurally modified forms of ID region suit it for its varied interactions with other critical coregulatory proteins and possibly additional modulations in its structure essential for gene regulation by the signaling molecules. These interactions give a set of functionally active folded structure to ID AF1 region and form the basis for the multiprotein assemblies involved in GR-mediated regulation of transcription. This information will lead to an understanding of the role of this important phenomenon in the transcription process and provide a biological mechanism, information essential to understanding how GR affects gene regulation. Since several protein kinase pathways are involved in cell signaling leading to diverse cellular responses through TFs, phosphorylation-induced conformational changes observed in the present study should provide a potential mechanism thorough which ID activation domains of signaling molecules exert their effects in gene regulation.
FIG. 10.
(A) Model showing the proposed effect of phosphorylation by p38 on GR-mediated transcription (based on our data and the available literature). (B) Working model of the ID AF1 domain undergoing conformational changes due to phosphorylation and/or interaction with binding partners that lead to interaction with one or more binding partners adapting functionally active ordered conformation. AF1PF, partially folded; AF1FO, fully folded AF1.
Acknowledgments
This study was supported by National Institutes of Health grant NIDDK DK058829 (to R.K.).
We thank A. Mishra for technical help in creating some of the AF1 mutants, E. B. Thompson (University of Texas Medical Branch [UTMB]) for providing GR plasmids, D. W. Bolen (UTMB) for CD analysis, M. Mancini (Baylor College of Medicine) for the pYFP-CBP and pYFP-SRC-1 constructs, and L. Vergara and the Optical Imaging Core staff (UTMB) for technical support and help with data analysis.
Footnotes
Published ahead of print on 19 October 2009.
REFERENCES
- 1.Andrade, M.A., P. Chacón, J. J. Merelo, and F. Morán. 1993. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 1a.Bai, Y., and V. Giguére. 2003. Isoform-selective interactions between estrogen receptors and steroid receptor coactivators promoted by estradiol and ErbB-2 signaling in living cells. Mol. Endocrinol. 17:589-599. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, R. L., and G. D. Rose. 1999. Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem. Sci. 24:77-83. [DOI] [PubMed] [Google Scholar]
- 3.Baskakov, I. V., R. Kumar, G. Srinivasan, Y. Ji, D. W. Bolen, and E. B. Thompson. 1999. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J. Biol. Chem. 274:10693-10696. [DOI] [PubMed] [Google Scholar]
- 4.Blind, R. D., and M. J. Garabedian. 2008. Differential recruitment of glucocorticoid receptor phospho-isoforms to glucocorticoid-induced genes. J. Steroid Biochem. Mol. Biol. 109:150-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodwell, J. E., L. M. Hu, J. M. Hu, E. Ortí, and A. Munck. 1993. Glucocorticoid receptors: ATP-dependent cycling and hormone-dependent hyper-phosphorylation. J. Steroid Biochem. Mol. Biol. 47:31-38. [DOI] [PubMed] [Google Scholar]
- 6.Bodwell, J. E., J. C. Webster, C. M. Jewell, J. A. Cidlowski, J. Hu, and A. Munck. 1998. Glucocorticoid receptor phosphorylation: overview, function and cell cycle dependence. J. Steroid Biochem. Mol. Biol. 65:91-99. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., T. Dang, R. D. Blind, Z. Wang, C. N. Cavasotto, A. B. Hittelman, I. Rogatsky, S. K. Logan, and M. J. Garabedian. 2008. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol. Endocrinol. 22:1754-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. X., J. T. Du, L. X. Zhou, X. H. Liu, Y. F. Zhao, H. Nakanishi, and Y. M. Li. 2006. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational Disturbances to the N terminus of murine estrogen receptor β. Chem. Biol. 13:937-944. [DOI] [PubMed] [Google Scholar]
- 9.Dalman, F. C., E. R. Sanchez, A. L. Lin, F. Perini, and W. B. Pratt. 1998. Localization of phosphorylation sites with respect to the functional domains of the mouse L cell glucocorticoid receptor. J. Biol. Chem. 263:12259-12267. [PubMed] [Google Scholar]
- 10.DeFranco, D. B., M. Qi, K. C. Borror, M. J. Garabedian, and D. L. Brautigan. 1991. Protein phosphatase types 1 and/or 2A regulate nucleocytoplasmic shuttling of glucocorticoid receptors. Mol. Endocrinol. 5:1215-1228. [DOI] [PubMed] [Google Scholar]
- 11.Dieken, E. S., and R. L. Miesfeld. 1992. Transcriptional transactivation functions localized to the glucocorticoid receptor N terminus are necessary for steroid induction of lymphocyte apoptosis. Mol. Cell. Biol. 12:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunker, A. K., C. J. Brown, J. D. Lawson, L. M. Iakoucheva, and Z. Obradović. 2002. Intrinsic disorder and protein function. Biochemistry 41:6573-6582. [DOI] [PubMed] [Google Scholar]
- 13.Dunker, A. K., and V. N. Uversky. 2008. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 4:229-230. [DOI] [PubMed] [Google Scholar]
- 14.Godowski, P. J., S. Rusconi, R. L. Miesfeld, and K. R. Yamamoto. 1987. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature 325:365-368. [DOI] [PubMed] [Google Scholar]
- 15.Härd, T., and D. R. Kearns. 1990. Reduced DNA flexibility in complexes with a type II DNA binding protein. Biochemistry 29:959-965. [DOI] [PubMed] [Google Scholar]
- 16.Hay, T. J., and D. W. Meek. 2000. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 478:183-186. [DOI] [PubMed] [Google Scholar]
- 17.Iakoucheva, L. M., P. Radivojac, C. J. Brown, T. R. O'Connor, J. G. Sikes, Z. Obradovic, and A. K. Dunker. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32:1037-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, C. C., E. M. Gardiner, F. T. Zenke, B. P. Bohl, A. C. Newton, B. A. Hemmings, and G. M. Bokoch. 2000. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1). J. Biol. Chem. 275:41201-41209. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, R., and E. B. Thompson. 2005. Gene regulation by the glucocorticoid receptor: structure:function relationship. J. Steroid Biochem. Mol. Biol. 94:383-394. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, R., J. M. Serrette, S. H. Khan, A. L. Miller, and E. B. Thompson. 2007. Effects of different osmolytes on the induced folding of the N-terminal activation domain (AF1) of the glucocorticoid receptor. Arch. Biochem. Biophys. 465:452-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, R., J. C. Lee, D. W. Bolen, and E. B. Thompson. 2001. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J. Biol. Chem. 276:18146-18152. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, R., R. Betney, J. Li, E. B. Thompson, and I. J. McEwan. 2004. Induced α-helix structure in AF1 of androgen receptor upon binding transcription factor TFIIF. Biochemistry 43:3008-3013. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, R., B. H. Johnson, and E. B. Thompson. 2004. Overview of the structural basis for transcription regulation by nuclear hormone receptors. Essays Biochem. 40:27-39. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, R., D. E. Volk, J. Li, J. C. Lee, D. W. Gorenstein, and E. B. Thompson. 2004. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc. Natl. Acad. Sci. USA 101:16425-16430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurl, R. N., and S. T. Jacob. 1984. Phosphorylation of purified glucocorticoid receptor from rat liver by an endogenous protein kinase. Biochem. Biophys. Res. Commun. 119:700-705. [DOI] [PubMed] [Google Scholar]
- 26.Kyriakis, J. M. 2000. MAP kinases and the regulation of nuclear receptors. Sci. STKE PE1. [DOI] [PubMed]
- 27.Lavery, D. N., and I. J. McEwan. 2005. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem. J. 391:449-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J., N. B. Perumal, C. J. Oldfield, E. W. Su, V. N. Uversky, and A. K. Dunker. 2006. Intrinsic disorder in transcription factors. Biochemistry 45:6873-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marks, F. 1996. Protein phosphorylation. VCH Weinheim, Basel, Switzerland.
- 30.McEwan, I. J., K. Dahlman-Wright, J. Ford, and A. P. Wright. 1996. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: evidence for an induced fit model of transactivation domain folding. Biochemistry 35:9584-9593. [DOI] [PubMed] [Google Scholar]
- 30a.Merelo, J. J., M. A. Andrade, A. Prieto, and F. Morán. 1994. Proteinotopic feature maps. Neurocomputing 6:443-454. [Google Scholar]
- 31.Miller, A. L., M. S. Webb, A. J. Copik, Y. Wang, B. H. Johnson, R. Kumar, and E. B. Thompson. 2005. p38 mitogen-activated protein kinase (MAPK) is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol. Endocrinol. 19:1569-1583. [DOI] [PubMed] [Google Scholar]
- 32.Miller, A. L., A. S. Garza, B. H. Johnson, and E. B. Thompson. 2007. Pathway interactions between MAPKs, mTOR, PKA, and the glucocorticoid receptor in lymphoid cells. Cancer Cell Int. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minezaki, Y., K. Homma, A. R. Kinjo, and K. Nishikawa. 2006. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 359:1137-1149. [DOI] [PubMed] [Google Scholar]
- 34.Parker, D., U. S. Jhala, I. Radhakrishnan, M. B. Yaffe, C. Reyes, A. I. Shulman, L. C. Cantley, P. E. Wright, and M. Montminy. 1998. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell 2:353-359. [DOI] [PubMed] [Google Scholar]
- 35.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 36.Rogatsky, I., S. K. Logan, and M. J. Garabedian. 1998. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 95:2050-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogatsky, I., C. L. Waase, and J. M. Garabedian. 1998. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J. Biol. Chem. 273:14315-14321. [DOI] [PubMed] [Google Scholar]
- 38.Vetter, S. W., and E. Leclerc. 2001. Phosphorylation of serine residues affects the conformation of the calmodulin binding domain of human protein 4.1. Eur. J. Biochem. 268:4292-4299. [DOI] [PubMed] [Google Scholar]
- 39.Wada, T., and J. M. Penninger. 2004. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23:2838-2849. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Z., J. Frederick, and M. J. Garabedian. 2002. Deciphering the phosphorylation “code” of the glucocorticoid receptor in vivo. J. Biol. Chem. 277:26573-26580. [DOI] [PubMed] [Google Scholar]
- 41.Webster, J. C., C. M. Jewell, M. Sar, J. E. Bodwell, A. Munck, and J. A. Cidlowski. 1997. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J. Biol. Chem. 272:9287-9293. [DOI] [PubMed] [Google Scholar]
- 42.Zenke, F. T., C. C. King, B. P. Bohl, and G. M. Bokoch. 1999. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J. Biol. Chem. 274:32565-32573. [DOI] [PubMed] [Google Scholar]
- 43.Zor, T., B. M. Mayr, H. J. Dyson, M. R. Montminy, and P. E. Wright. 2002. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J. Biol. Chem. 277:42241-42248. [DOI] [PubMed] [Google Scholar]