Abstract
RNA transcription by all the three RNA polymerases (RNAPs) is tightly controlled, and loss of regulation can lead to, for example, cellular transformation and cancer. While most transcription factors act specifically with one polymerase, a small number have been shown to affect more than one polymerase to coordinate overall levels of transcription in cells. Here we show that TLS (translocated in liposarcoma), a protein originally identified as the product of a chromosomal translocation and which associates with both RNAP II and the spliceosome, also represses transcription by RNAP III. TLS was found to repress transcription from all three classes of RNAP III promoters in vitro and to associate with RNAP III genes in vivo, perhaps via a direct interaction with the pan-specific transcription factor TATA-binding protein (TBP). Depletion of TLS by small interfering RNA (siRNA) in HeLa cells resulted in increased steady-state levels of RNAP III transcripts as well as increased RNAP III and TBP occupancy at RNAP III-transcribed genes. Conversely, overexpression of TLS decreased accumulation of RNAP III transcripts. These unexpected findings indicate that TLS regulates both RNAPs II and III and supports the possibility that cross-regulation between RNA polymerases is important in maintaining normal cell growth.
Proteins regulate each step in gene expression to ensure accuracy and increase efficiency, and they do so by a variety of mechanisms. The TET family of RNA-binding proteins, which includes TLS, EWS, and TAF15 (for a review, see reference 66), appear to function in at least two distinct nuclear processes, transcription and splicing. TLS (translocated in liposarcoma), EWS (Ewing sarcoma), and TAF15 (TATA-binding protein [TBP]-associated factor 15) all contain a glutamine-, serine-, and glycine-rich amino terminus; a conserved RNP-type RNA-binding domain (RBD); RGG repeats that are thought to contribute to RNA binding; and a putative Cys2-Cys2 zinc finger (6, 18). TLS (also called FUS) was originally discovered as part of a fusion protein with the transcription factor CHOP, deriving from a characteristic chromosomal translocation in human liposarcomas (18, 56). Likewise, EWS is encoded by a gene that is translocated in Ewing's sarcoma and fused to the DNA-binding domain of the transcription factor Fli-1 (66). TAF15 was originally identified through its association with a specific subpopulation of the general transcription factor (GTF) TFIID (5, 67).
A role for TET proteins in transcription by RNA polymerase (RNAP) II is suggested by several observations. Early on, the glutamine-rich amino termini of TET proteins were shown to be capable of substituting for the activation domains of certain DNA-binding transcription factors, suggesting a possible role in enhancement of RNAP II transcription (4, 80). TLS, EWS, and TAF15 were each found to copurify with separate and distinct TFIID complexes (6). The multisubunit TFIID, which includes TBP, recognizes and binds to the promoter regions of many genes transcribed by RNAP II. It has been postulated that association of different TET proteins with specific populations of TFIID may influence transcription initiation and promoter choice (6). TET proteins share similarity with Tat-SF1, a cellular factor that was originally found to stimulate the HIV-1 Tat protein but was later shown to be a general transcription elongation factor (44, 78). Finally, TET proteins have been found to associate with RNAP II itself (5, 6).
Considerable evidence also links TET proteins to splicing of mRNA precursors. While TET proteins lack the arginine-serine (RS) domain found in many known splicing factors, they do contain an RBD and were found in systematic analyses of splicing complexes (57, 79). Other studies, using transient transfection and overexpression, have shown that TLS can bring about changes in the splicing pattern of alternatively spliced transcripts produced from cotransfected reporter plasmids (73). Additionally, the carboxy terminus of TLS interacts with various splicing factors, including the SR proteins TASR (also known as SRp38) and SC35 (74). TLS was also found as a component of in vitro transcription and splicing complexes, together with RNAP II and snRNPs (35).
In addition to their roles in transcription and splicing, TET proteins may also function in other cellular processes. For example, TLS was recently implicated in a form of familial amyotrophic lateral sclerosis (ALS) (39, 68) and has been found localized to dendrites and may stabilize RNA transcripts (3, 20, 21). TLS may have a role in DNA damage response and genome stability, since it is a downstream target of the protein kinase ATM, which senses and coordinates DNA repair (22). TLS can promote D-loop formation, which is a step in the homologous recombination pathway for DNA repair (2, 7), and mice lacking TLS have high genomic instability and enhanced sensitivity to radiation (25, 38). Furthermore, TLS is activated by a noncoding RNA after DNA damage and specifically represses transcription of the cyclin D1 gene by inhibiting the histone acetyltransferase activity of CREB-binding protein (CBP) and p300 at the cyclin D1 promoter (69).
While it is unusual for transcription factors to regulate so many cellular processes, other cancer-related transcription factors have been shown to regulate transcription by multiple RNAPs. The tumor suppressor p53 can inhibit transcription of snRNA genes transcribed by both RNAPs II and III (13, 17, 24), while the Rb tumor suppressor protein inhibits RNAP III transcription to control cell growth (14, 27, 72), and the protein Maf1 represses transcription by all three RNAPs (33). Levels of RNAP III transcripts are often elevated in cancer cells (71), and overexpression of an RNAP III transcript, initiator tRNAMet, alone is sufficient to induce cellular growth and oncogenic transformation (48).
Here we show that TLS, in addition to its roles in RNAP II transcription and splicing, also represses transcription by RNAP III. Repression was first detected in vitro during a study of the possible role of TLS in coupling RNAP II transcription and splicing. This unexpected activity was confirmed by in vitro transcription assays with all three types of RNAP III genes, which were all repressed by TLS. Confirming the physiological significance of the in vitro results, TLS is associated with RNAP III genes in vivo, as demonstrated by chromatin immunoprecipitation (ChIP) assays. This may reflect a direct interaction with TBP, as shown by immunoprecipitation from cell extracts and “pulldown” assays with purified proteins. Furthermore, we detected an increase in the levels of RNAP III transcripts after small interfering RNA (siRNA)-mediated knockdown of TLS in HeLa cells and a decrease following TLS overexpression. Additionally, ChIP assays with cells where TLS was depleted showed an increase in occupancy of TBP and RNAP III at RNAP III genes. TLS thus joins a short list of important regulators capable of controlling multiple polymerases.
MATERIALS AND METHODS
In vitro transcription and splicing reactions.
Final concentrations for 25-μl in vitro transcription and splicing reaction mixtures were as follows: 3 mM MgCl2; 50 μM each CTP, GTP, and UTP; 300 μM ATP; 4 mM creatine phosphate; 1.5% polyvinyl alcohol; 100 ng DNA template; 10 μCi [α-32P]GTP; and 15 μl nuclear extract in buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol DTT, 100 mM KCl). Where indicated, α-amanitin (Calbiochem) was added at appropriate concentrations to inhibit RNAP II and RNAP III, DNA was omitted from the reaction mixture, or purified recombinant glutathione S-transferase (GST) or GST-TLS was added.
Reaction mixtures were incubated for 120 min at 30°C. Reactions were stopped by the addition of 500 μl proteinase K buffer (10 mM Tris [pH 7.8], 10 mM EDTA [pH 8.0], 0.5% SDS, 8 M urea), 50 μg proteinase K, and 10 μg tRNA, and then mixtures were incubated at 37°C for 30 min before phenol-chloroform extraction and ethanol precipitation. RNA was analyzed on 7 M urea-4% acrylamide gels.
In vitro RNAP III transcription reactions.
Final concentrations for 20-μl in vitro transcription reaction mixtures were as follows: 500 μM each ATP, CTP, and UTP; 50 μM GTP; 10 μCi [α-32P]GTP; 250 μM DTT; 2.25 mM MgCl2; 75 mM KCl; 20 mM HEPES (pH 7.9); and 8 μl S100 extract in buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 100 mM KCl). Where indicated, α-amanitin (Calbiochem), tagetitoxin (Tagetin; Epicentre Biotechnologies), actinomycin D (Calbiochem), 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (Calbiochem), GST, or GST-TLS was added to reaction mixtures at the indicated concentrations. Reaction mixtures were incubated for 60 min at 30°C, and then reactions were stopped by the addition of proteinase K before extraction and precipitation as described for in vitro transcription and splicing reactions. RNA was run on an 8.3 M urea-6% acrylamide gel and then dried and analyzed using a Storm 860 PhosphorImager (Molecular Dynamics).
Purification of recombinant proteins.
GST-TLS was expressed in baculovirus-infected Hi-5 cells for 48 h; harvested and lysed in 50 mM Tris (pH 8.0), 400 mM NaCl, 1% NP-40, 1 mM DTT, 50 μg/ml bovine serum albumin (BSA), and protease inhibitor cocktail (Sigma) for 2 h at 4°C; and then centrifuged at 15,000 rpm in a Sorvall SS-34 rotor for 30 min. The soluble fraction was bound to prepared glutathione-Sepharose beads (Amersham Pharmacia) for 4 h at 4°C, washed extensively with the same buffer, and then eluted with the same buffer containing 50 mM reduced glutathione and subsequently dialyzed against buffer D. GST was purified from Escherichia coli as previously described (26). GST-hnRNP K protein was a kind gift from C. David, and GST-SRp38 protein was a kind gift from Y. Feng. His-ASF and His-SC35 proteins were expressed in baculovirus-infected Hi-5 cells for 48 h and purified under denaturing conditions (50). pET11d-hTBP plasmid DNA was a gift from R. Prywes, and His-TBP was expressed in E. coli and purified as previously described (12).
Coimmunoprecipitation experiments.
HeLa cells were harvested, pelleted by centrifugation, and washed twice with 10 ml cold 1× phosphate-buffered saline (PBS). Cells were resuspended in cold lysis buffer (0.5% Triton X-100, 0.5% sodium deoxycholate, 50 mM Tris-Cl [pH 7.5], 250 mM NaCl, 5 mM EDTA), rocked on a nutator at 4°C for 20 min, and then centrifuged at 14,000 rpm for 15 min at 4°C. The supernatant was used immediately for immunoprecipitation or stored at −80°C. Where indicated, 50 μg/ml ethidium bromide (Sigma) was added to the supernatant to disrupt any DNA-dependent protein interactions (40). The mixture was incubated on ice for 20 min before centrifugation for 5 min at 4°C. This supernatant was then used, and the ethidium bromide concentration was maintained in all subsequent binding and wash steps.
For each IP, 25 μl of protein A-agarose (Roche Diagnostics) beads was washed twice with 1× PBS and then twice with IP buffer (50 mM Tris pH 7.5, 250 mM NaCl, 0.1% NP-40). Antibodies used for immunoprecipitation were anti-TLS antibody (BL1355) from Bethyl Laboratories, anti-TBP antibody (N-12, sc-204) from Santa Cruz Biotechnology, anti-RNAPIII Ab1900 against subunit RPC155 from R. White (University of Glasgow), and anti-BRF1 from Bethyl Laboratories. Beads were incubated with 1 to 2 μg of appropriate antibodies in 200 μl of IP buffer for 3 to 4 h at 4°C on a nutator. Tubes were centrifuged for 2 min at 4,000 rpm at 4°C and washed once with 1× PBS to remove unbound antibodies.
The HeLa whole-cell lysate supernatant was added to the beads in cold IP buffer to a total of 300 μl and incubated for at least 4 h on a nutator at 4°C. Tubes were centrifuged for 2 min at 4,000 rpm at 4°C and washed three times with 1 ml cold IP buffer and then once with 1 ml IP-50 (50 mM Tris [pH 7.5], 50 mM NaCl). Beads were resuspended in 2× SDS-PAGE loading dye and boiled for 10 min before 20% of IP and 5% of input lysate was loaded onto 6% or 10% SDS-polyacrylamide gels for Western blotting analysis. Proteins were transferred to a nitrocellulose membrane, probed with the specified antibodies, and visualized using enhanced chemiluminescence (ECL) (Amersham Pharmacia).
Protein interaction assays.
One microgram of His-tagged TBP was bound to Ni-nitrilotriacetic acid (NTA) agarose for 2 h at 4°C in 250 μl IP buffer, and then tubes were centrifuged and unbound protein was removed. Five hundred nanograms of GST-TLS, 1 μg of GST, or 500 ng of GST-hnRNP K was added in 500 μl IP buffer with ethidium bromide to a final concentration of 50 μg/ml, and tubes were incubated for 2 h at 4°C. For GST pulldown assays, 500 ng of GST, GST-TLS, GST-hnRNP K, or beads alone (mock pulldown) was bound to glutathione-Sepharose beads for 2 h at 4°C in 250 μl IP buffer before tubes were centrifuged and unbound protein was removed. Five hundred nanograms of His-TBP was added to each 500-μl reaction mixture, and tubes were incubated for 4 h at 4°C.
Beads were washed four times with 1 ml cold IP buffer with 50 μg/ml ethidium bromide and then once with 1 ml cold IP-50 buffer. Beads were resuspended in SDS-PAGE loading buffer, and 25% or 33% of pulldown product and 10% or 20% of input protein was loaded on 8% SDS-polyacrylamide gels for separation and subsequently transferred to nitrocellulose membrane. Western blotting was done using anti-GST (Molecular Probes) or anti-TBP (N-12, sc-204) from Santa Cruz Biotechnology and visualized using enhanced chemiluminescence (ECL) (Amersham Pharmacia).
Cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Gibco) with 10% fetal bovine serum (HyClone). For knockdown experiments, siRNA targeting either TLS (Dharmacon D-009497-06) or luciferase (Dharmacon D-001219-02) as a negative control was used. For overexpression experiments, the TLS gene was cloned into the p3XFLAG-CMV-14 vector (Sigma), and this construct or empty vector alone was transfected into HeLa cells. All transfections were done using Lipofectamine 2000 (Invitrogen) according to the manufacturer's guidelines.
For Western blots, cells were harvested, washed twice with cold PBS, resuspended in 2× SDS-PAGE buffer, sonicated, boiled, and loaded on 8% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes and probed with BL1355 anti-TLS (Bethyl Laboratories) and antiactin (Sigma) antibodies. Other cells from the same transfection were harvested, washed twice with cold PBS, and lysed with TRIzol (Invitrogen). RNA was extracted using TRIzol-chloroform, precipitated using isopropyl alcohol, washed with 70% ethanol, resuspended, and used for reverse transcription with random hexamer primers and avian myeloblastosis virus (AMV) reverse transcriptase (RT) (Promega). This cDNA was subsequently used for real-time PCR using Sybr master mix (Applied Biosystems or Fermentas) on an ABI Prism 7300 real-time PCR machine. Primer sequences are available upon request. Experiments were repeated at least three independent times.
Chromatin immunoprecipitation.
The ChIP protocol was adapted from the fast protocol (53) as previously described (8). Briefly, cells were cross-linked at a final concentration of 1.4% formaldehyde for 15 min at room temperature, and the reaction was stopped by the addition of glycine to a 125 mM final concentration for 5 min at room temperature. Cells were harvested from the plates, washed twice with cold PBS, and then lysed with IP buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.5% NP-40, 1% Triton X-100). Samples were sonicated (Branson Sonifier 250) to generate fragments of ∼500 bp.
Antibodies used for immunoprecipitation were as described above for immunoprecipitation and TFIIIC antibody from R. Roeder (Rockefeller University). Sheared chromatin was incubated with the appropriate antibody for 25 min in an ultrasonic water bath at 4°C and then centrifuged for 10 min at 12,000 × g at 4°C. The supernatant was incubated with protein A beads (Roche) overnight at 4°C. Beads were collected by centrifugation and washed three times with IP buffer, once with wash buffer (50 mM Tris pH 8.8, 1% NP-40, 1% sodium deoxycholate, 0.5 M LiCl), and once more with IP buffer. Complexes were separated from beads with elution buffer (1% SDS and 100 mM NaHCO3), and then RNase A (1 μg) and NaCl (300 mM final concentration) were added and cross-links were reversed by incubating overnight at 65°C. Samples were treated with proteinase K for 30 min at 42°C. DNA was extracted using phenol-chloroform and ethanol precipitation and was subsequently used in PCRs.
Radioactive PCR amplification mixtures contained 0.5 units of Taq polymerase (Invitrogen), 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 10% dimethyl sulfoxide (DMSO), [α-32P]dCTP, and primers corresponding to the indicated genes for 30 cycles. Five percent of input and 20% of ChIP reaction products were analyzed on 8.3 M urea-10% acrylamide gels and using a Storm 860 PhosphorImager (Molecular Dynamics). Real-time PCR was performed using Sybr master mix (Applied Biosystems or Fermentas) on an ABI Prism 7300 real-time PCR machine. Quantitative PCRs were performed in triplicate, and primer sequences are available upon request. Experiments were repeated at least three independent times, and the fold change for each gene and antibody was normalized to the 18S RNA gene signal for each antibody.
RESULTS
TLS inhibits RNA polymerase III transcription.
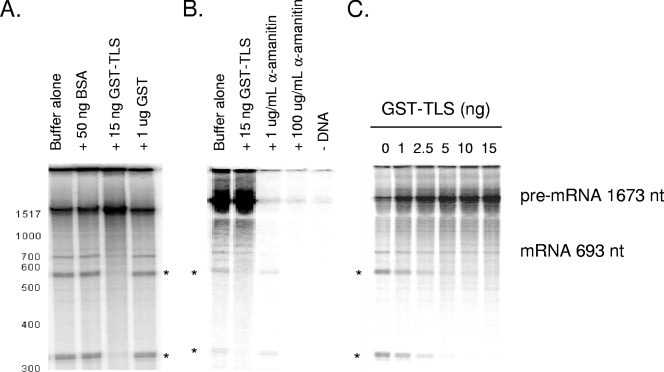
The protein domains within TLS and its association with both RNAP II transcription and splicing factors suggested that TLS may function to link these processes, perhaps by recruiting splicing factors to the RNAP II transcription machinery, and we therefore set out to test this idea directly. Specifically, we utilized a cell-free reaction with HeLa nuclear extract that allows transcription by RNAP II and splicing of the newly synthesized transcript to occur simultaneously, under conditions similar to those that have been described previously (23, 42). Unlike in vitro splicing assays, in which a presynthesized pre-mRNA substrate is added to reaction mixtures, the transcript in the transcription-splicing system is produced by endogenous RNAP II in HeLa nuclear extract from a recombinant DNA template. The newly synthesized transcript is then subject to processing by factors present in the nuclear extract. For these experiments, we used as the DNA template a linearized plasmid expressing the full-length β-globin gene from a cytomegalovirus (CMV) promoter. The pre-mRNA produced in these reactions contains three exons and two introns, and mature spliced mRNA was in fact detected (Fig. 1A and unpublished data). Significantly, addition of only 30 ng recombinant GST-TLS purified from baculovirus-infected insect cells increased transcription (Fig. 1A), while addition of 50 ng BSA or 1 μg GST was without effect. Quantitation of radiolabeled RNAs suggests that addition of GST-TLS increased total transcription by approximately 30% (data not shown).
FIG. 1.
TLS increases RNAP II transcription but not splicing in HeLa nuclear extract. In vitro transcription and splicing of the β-globin gene from the CMV promoter in the presence of [α-32P]GTP are shown. RNA was analyzed by 7 M urea-4% PAGE. Asterisks denote RNAP III products. (A) Buffer alone, BSA (50 ng), GST-TLS (15 ng), or GST (1 μg) was added as indicated. (B) Reactions with buffer alone, GST-TLS (15 ng), α-amanitin (1 μg/ml or 100 μg/ml), or no DNA. (C) Reactions with 0, 1 ng, 2.5 ng, 5 ng, 10 ng, or 15 ng GST-TLS. nt, nucleotides.
In addition to bands reflecting β-globin-derived RNAs, we also detected two prominent transcripts that could not be identified by size. Significantly, the appearance of these species was also repressed by GST-TLS but not by GST (Fig. 1A, bands identified by asterisks). We considered the possibility that these RNAs were RNAP III transcripts, since such products have been described previously as products of the β-globin locus produced during in vitro transcription (10, 46). Indeed, characterization of the transcripts' sensitivity to the inhibitor α-amanitin indicated that they were RNAP III products. Synthesis of both RNAs was resistant to a low concentration of the drug that inhibits RNAP II (1 μg/ml) (Fig. 1B) but was inhibited by a high concentration that inhibits both RNAPs II and III (100 μg/ml) (Fig. 1B). No transcripts were detected in reactions lacking a DNA template, indicating that the apparent RNAP III products were transcribed from the exogenous plasmid (Fig. 1B). GST-TLS is a remarkably potent inhibitor of these transcripts, as inhibition was detected with as little as 2.5 ng and repression was nearly complete with 10 ng GST-TLS (Fig. 1C).
TLS inhibits transcription of all classes of RNAP III genes.
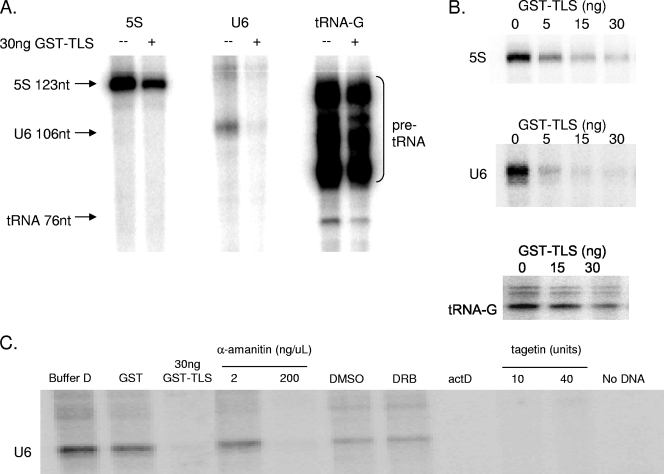
The above data provided evidence that TLS can inhibit transcription by RNAP III in vitro. Rather than investigate this further with the uncharacterized transcripts described above, we decided to examine the effect of GST-TLS on transcription of authentic RNAP III products. RNAP III transcribes small, untranslated RNAs from three general types of promoter elements that are recognized by specific transcription factors (for a review, see reference 60). For this analysis, we utilized HeLa S100 extract, which in our experiments was more efficient than nuclear extract, as has been described previously (37). For templates, we used PCR-amplified DNA fragments containing a representative from each of the three classes of promoter elements: the 5S RNA, U6 snRNA, and tRNAGly genes.
Strikingly, addition of 30 ng of GST-TLS resulted in a significant decrease in transcription of each of these genes (Fig. 2A). GST-TLS strongly inhibited transcription of the 5S and U6 genes, while the tRNAGly gene was also inhibited but to a somewhat lesser extent (note the synthesis of pre-tRNA as well as processed mature tRNAGly). Inhibition was again concentration dependent (Fig. 2B), as increasing amounts of GST-TLS inhibited transcription of the 5S, U6, and tRNAGly genes. Repression was again observed with nanogram levels of GST-TLS in a similar range as observed with the cryptic RNAP III products synthesized in the in vitro transcription and splicing reactions (see above). The observed repression was not a nonspecific property of RNA-binding proteins generally, as addition of several recombinant SR proteins did not affect U6 transcription and an hnRNP protein, hnRNP K, actually enhanced transcription (data not shown). The effect of hnRNP K is intriguing, as previous studies have implicated the protein in RNAP II transcriptional control (see, e.g., reference 52).
FIG. 2.
TLS represses RNAP III transcription in vitro. In vitro transcription by RNAP III from S100 in the presence [α-32P]GTP is shown. (A) In vitro transcription of three classes RNAP III promoter, represented by the 5S, U6 snRNA, and tRNAGly genes. Thirty nanograms of GST-TLS was added to reaction mixtures as indicated. (B) Addition of GST-TLS to in vitro transcription by RNAP III of the 5S (0, 5, 15, or 30 ng), U6 snRNA (0, 5, 15, or 30 ng), and tRNAGly (0, 15, or 30 ng) genes. (C) Buffer alone, 1 μg GST, 30 ng GST-TLS, and various inhibitors were added to reaction mixtures. Specifically, 2 μg/ml α-amanitin, 200 μg/ml α-amanitin, 50 μM DRB (in DMSO) or DMSO alone, 50 ng/μl actinomycin D, and the RNAP III-specific inhibitor tagetin (10 and 40 units) were added. A reaction without DNA template is also shown.
The three RNAP III transcripts analyzed as described above were identified on the basis of their size, which in each case was exactly as expected. However, given the extremely potent and unanticipated inhibition induced by GST-TLS, we wished to verify that the transcription observed was indeed due to RNAP III. To this end, we examined the response of transcription of the U6 gene to several inhibitors (Fig. 2C). Buffer D alone and 1 μg of recombinant GST did not affect U6 transcription, whereas 30 ng GST-TLS again strongly inhibited transcription. As with the cryptic β-globin transcripts, 2 μg/ml of α-amanitin did not affect the amount of U6 RNA produced, while 200 μg/ml of α-amanitin inhibited U6 transcription completely. The RNAP II inhibitor DRB (5,6-dichlorobenzimidazole) had no effect beyond that of DMSO (the solvent in which it is dissolved). Actinomycin D also repressed U6 transcription, as did the RNAP III-specific inhibitor tagetitoxin (64). As expected, transcription was DNA dependent. Thus, GST-TLS specifically and strongly represses transcription of genes transcribed by RNAP III.
Although we are unaware of instances in which accumulation of newly synthesized transcripts in in vitro reactions such as described above is influenced by RNA stability, we wished to ascertain that GST-TLS did not function by in some way facilitating turnover of the RNAP III products. To do so, we examined the effect of adding GST-TLS to transcription reaction mixtures containing the U6 template after 20 min simultaneously with α-amanitin (100 μg/ml) to block transcription and then incubating the reaction mixtures for an additional 30 min. Products were analyzed as described above and the results indicated that GST-TLS had no effect on U6 RNA accumulation under these conditions and thus does not decrease U6 RNA levels by decreasing stability (data not shown).
TLS interacts directly with TBP.
We next wished to gain insight into the possible mechanism by which TLS represses RNAP III transcription. We first examined whether the inhibition might reflect direct interaction of TLS with target DNA. Previous studies had shown that TLS can bind RNA (29, 43) and may bind both single- and double-stranded DNA (2), but evidence of sequence specificity of TLS binding to DNA has not been shown. We found that GST-TLS bound to two putative target genes (U6 snRNA and 5S) by electrophoretic mobility shift assays (data not shown), with some specificity based on competition, but subsequent experiments with DNA templates lacking RNAP III promoter sequences suggested that TLS could bind a variety of DNAs. This correlates with the ability of TLS to bind ends of double-stranded DNA and to assist D-loop formation during homologous recombination (2, 7). It is thus unclear whether TLS specifically and directly binds RNAP III genes.
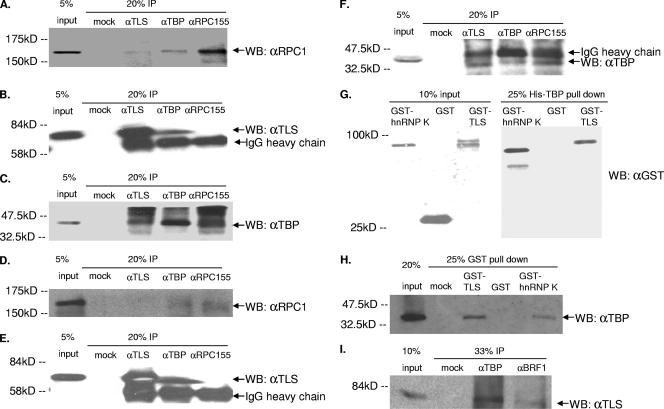
We next examined whether TLS might interact with RNAP III or initiation factors using coimmunoprecipitation (co-IP) experiments. Using anti-TLS specific antibodies, we found that TLS coimmunoprecipitates with RPC155, the largest subunit of RNAP III (Fig. 3A), and with the pan-specific GTF TBP (Fig. 3C). We also found that anti-TBP antibodies coimmunoprecipitate TLS (Fig. 3B) and RPC155 (Fig. 3A). Anti-RPC155 antibody did not appear to immunoprecipitate TLS (Fig. 3B), which may be due to immunoprecipitation of RPC155 being relatively inefficient and/or to weak interactions between TLS and RPC155. These data suggest that TLS may be involved in promoter and polymerase choice by associating with TBP and/or RNAP III, possibly by preventing TBP from associating with TFIIIB subunits or the RNAP III holoenzyme (70) or by inhibiting transcription when bound to TBP and RNAP III at RNAP III promoters (see Discussion).
FIG. 3.
TLS binds to TBP and the RNAP III transcription machinery. (A to C) Coimmunoprecipitation from HeLa cells and subsequent Western blot (WB) analysis of TLS, TBP, and RPC155. (A) Western blotting using an antibody to the RPC155 subunit of RNAP III after immunoprecipitation by TLS, TBP, or RPC155 antibodies. IgG, immunoglobulin G. (B) Western blotting using TLS antibody after immunoprecipitation by TLS, TBP, or RPC155 antibodies. (C) Western blotting using TBP antibody after immunoprecipitation by TLS, TBP, or RPC155 antibodies. (D to F) Western blots in which immunoprecipitation was carried out in the presence of 50 μg/ml ethidium bromide and probed with antibodies to RPC155, TLS, and TBP, respectively. (G) Western blotting using GST antibody for 10% of input proteins GST-hnRNP K, GST, and GST-TLS and 25% of bound proteins after His-TBP pulldown assay with GST-hnRNP K, GST, and GST-TLS. (H) Western blotting using TBP antibody after GST pulldown. Twenty percent of input His-TBP and 25% of protein from mock (no protein), GST-TLS, GST, and GST-hnRNP K pulldowns are shown. (I) Western blotting using TLS antibody after IP by mock, TBP, or BRF1 antibodies.
Since TLS, RNAP III, and TBP bind DNA, one possibility is that this protein interaction may be indirect or bridged by DNA. To test this, we repeated the immunoprecipitation experiments in the presence of ethidium bromide, which disrupts DNA-mediated protein-protein interactions (40). The interaction between TLS and RPC155 was abrogated in the presence of ethidium bromide (Fig. 3D), suggesting that this interaction is at least partly mediated by DNA. However, despite the presence of ethidium bromide, TBP coimmunoprecipitated with TLS (Fig. 3F), indicating that these proteins interact independently of DNA. Furthermore, anti-TBP antibodies also immunoprecipitated TLS in the presence of ethidium bromide (Fig. 3E).
To determine whether TLS and TBP interact directly, we used in vitro “pulldown” assays. His-tagged human TBP protein was expressed in bacteria, and the resulting recombinant protein was purified and bound to nickel-agarose beads. Purified GST-tagged TLS, GST alone, or GST-hnRNP K, which has previously been shown to interact directly with TBP (49), was added to the beads, and after washing the bound fraction was analyzed by SDS-PAGE and Western blotting using an antibody to GST (Fig. 3G). Ethidium bromide was present during the binding and wash steps to prevent DNA-mediated protein-protein interactions. Both GST-hnRNP K and GST-TLS (Fig. 3G), but not GST alone, bound His-TBP. To confirm these results, we also performed the converse experiment, in which GST or GST-TLS and-hnRNP K were bound to glutathione-Sepharose beads and incubated with His-TBP. Complexes were washed extensively, and proteins were separated by SDS-PAGE and analyzed by Western blotting using an antibody to TBP (Fig. 3H). Similarly, we found that TBP interacts with TLS and hnRNP K (Fig. 3H) but not GST alone.
If the interaction of TLS with TBP reflects the role of TBP in RNAP III transcription, i.e., as a component of TFIIIB, then antibodies against other components of TFIIIB should immunoprecipitate TLS from cell extracts. We therefore tested whether anti-Brf1 antibodies can coimmunoprecipitate TLS from HeLa extracts. The results indicate that TLS does associate, directly or indirectly, with Brf1 (Fig. 3I). These data provide evidence that TLS does interact with the TFIIIB complex, likely via the interaction with TBP. Additionally, the DNA-dependent association of TLS with RNAP III supports the idea of a direct role of TLS on RNAP III genes. Indeed, the data below suggest that TLS interacts with TBP and components of the general RNAP III transcription machinery at RNAP III promoters to repress RNAP III transcription.
TLS associates with RNAP III-transcribed genes in vivo.
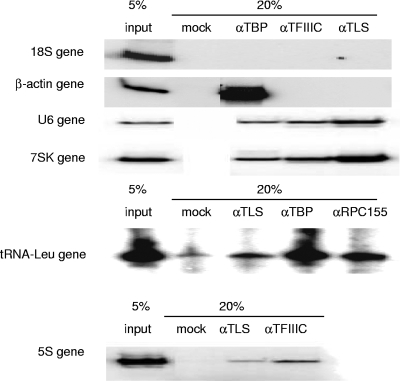
We next wished to investigate whether TLS affects RNAP III transcription in vivo. We first asked whether TLS associates with RNAP III-transcribed genes, using chromatin immunoprecipitation (ChIP). HeLa cells were cross-linked with formaldehyde and sonicated, and DNA-protein complexes were immunoprecipitated with appropriate antibodies. After washing and reversing the cross-links, DNA was purified and analyzed by PCR with [α-32P]dCTP and primers for a variety of genes.
We analyzed six different genes or gene regions; four RNAP III-transcribed genes (U6, 7SK, tRNALeu, and 5S), the promoter region of the β-actin gene, and an internal region of an 18S rRNA gene. Antibodies directed against TBP, TFIIIC, RPC155, or TLS were used. The results (Fig. 4) showed that, as expected, none of the proteins examined associated with the 18S rRNA gene fragment, indicating that the antibodies are specific and background is undetectable, and only TBP localized to the β-actin promoter. Additionally, as expected, TBP and TFIIIC or RPC155 were present at all four RNAP III genes examined. Most importantly, while TLS was not present at the β-actin promoter or 18S rRNA gene, it was found associated with all of the RNAP III-transcribed genes, albeit weakly with the tRNALeu gene (Fig. 4) and other tRNA genes tested (data not shown). Although the ChIP analyses were only semiquantitative (we did analyze the presence of TLS at the 5S gene by real-time PCR, and the results [not shown] were consistent with the radioactive PCR data), they suggest that TLS occupancy varied somewhat at the RNAP III genes, with 7SK the highest and tRNAs the lowest. This apparent difference in RNAP III gene occupancy is in fact consistent with the difference in the extent of transcriptional repression observed in vitro (Fig. 2) and suggests that TLS has a stronger inhibitory effect on RNAP III genes with an upstream promoter (i.e., U6 and 7SK). In any event, these results provide strong evidence that TLS associates with RNAP III-transcribed genes in vivo.
FIG. 4.
TLS localizes to RNAP III genes in vivo. ChIP assays were performed using antibodies against TLS, TFIIIC or RPC155, and TBP. DNA fragments were then amplified using primers specific to genes representing each of the RNA polymerases I (18S), II (β-actin), and III (U6, 7SK, tRNALeu, and 5S).
Altering TLS levels in vivo affects RNA polymerase III transcript levels.
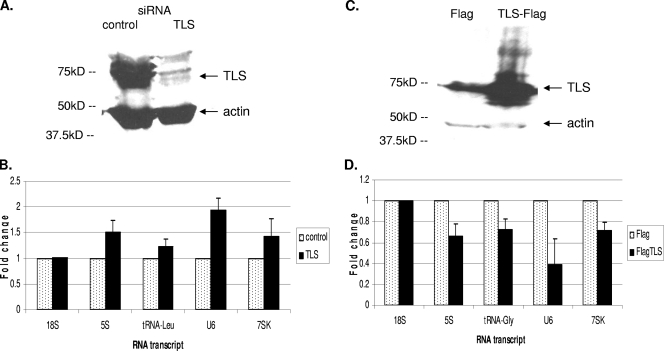
The above results have shown that TLS represses RNAP III transcription in vitro, interacts with components of the general RNAP III transcription machinery, and associates with RNAP III genes in vivo. We next wished to obtain evidence that these findings are physiologically significant, i.e., that TLS levels can influence expression of RNAP III-transcribed genes in vivo. To this end, we first performed a series of experiments in which TLS levels were decreased by siRNA and levels of RNAP III transcripts were analyzed. HeLa cells were transfected with siRNAs targeting either TLS or luciferase (as a control). Cells were lysed at 48 h after transfection, and protein levels were first analyzed by Western blotting. TLS levels were decreased as a result of siRNA knockdown, while actin levels were unaffected (Fig. 5A). RNA from the knockdown cells was then analyzed by real-time RT-PCR with primers designed to detect 5S, tRNALeu, U6, 7SK, and 18S RNAs (see Materials and Methods). Importantly, levels of all four RNAP III products were increased in the TLS knockdown cells, whereas 18S RNA levels were unaffected (Fig. 5B). It should be noted that our analysis was of steady-state RNA. As RNAP III transcripts are quite stable (55), the observed changes in their accumulation due to loss of TLS are thus especially notable. Similar results were obtained with a second, unrelated TLS siRNA (results not shown). These results suggest that TLS naturally represses or controls transcription of these genes in vivo, as it does in vitro.
FIG. 5.
Levels of TLS protein affect levels of RNAP III transcripts in vivo. (A) HeLa cells were transfected with siRNA targeting TLS or luciferase (control), and protein levels were analyzed by Western blotting using antibodies to TLS and actin. (B) Steady-state RNA levels after siRNA knockdown. RNA was extracted at 48 h posttransfection for reverse transcription using random hexamer primers, followed by real-time PCR. Results are depicted as fold increase. Error bars indicate standard deviations. (C) HeLa cells were transfected with plasmids encoding either Flag or TLS-Flag, and protein levels were analyzed by Western blotting using antibodies to TLS and actin. (D) Steady-state RNA levels after overexpression of Flag or TLS-Flag. RNA was extracted at 48 h posttransfection for reverse transcription using random hexamer primers, followed by real-time PCR.
We also tested the effect of overexpressing TLS in HeLa cells on RNAP III transcript levels. Cells were transfected with plasmids containing the CMV promoter driving expression of either the Flag epitope tag alone or TLS-Flag. As with the siRNA knockdown cells, these cells were harvested and lysed at 48 h after transfection, and protein levels were analyzed by Western blotting using antibodies to TLS and actin (Fig. 5C). RNA from these cells was then analyzed by real-time RT-PCR with primers designed to detect 5S, tRNAGly, U6, 7SK, and 18S RNA. Levels of all four RNAP III transcripts were significantly decreased in cells overexpressing TLS-Flag, while 18S RNA levels were unaffected (Fig. 5D). These results confirm that levels of TLS affect expression of RNAP III genes in vivo and provide additional evidence that TLS is a repressor of RNAP III transcription.
Decreasing TLS levels increases TBP and RNAP III occupancy at RNAP III genes.
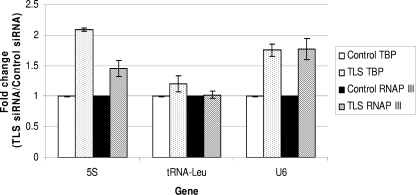
TLS could affect levels of RNAP III transcripts through a variety of mechanisms, including by blocking access of the RNAP III transcription machinery to target genes. To determine whether TLS acts directly on RNAP III transcription, we examined TBP and RNAP III occupancy at the 5S, U6, and tRNALeu genes, one from each class of RNAP III promoter, in cells where TLS protein levels were decreased by siRNA. To this end, HeLa cells were transfected with TLS siRNA or a control siRNA and then 72 h later were cross-linked with formaldehyde, harvested, processed, and analyzed by ChIP with appropriate antibodies, as described above. We used real-time PCR to quantitate association of TBP and RNAP III at the three representative RNAP III genes and again at the internal region of an 18S RNA gene as a control. Significantly, as depicted in Fig. 6, both TBP and RNAP III levels were increased in the TLS-depleted cells. We also examined the occupancy of TBP at the 7SK gene after TLS knockdown and, as expected, found that it was elevated (data not shown). We also used ChIP to examine TFIIIC occupancy at RNAP III genes, which reflects an earlier step in RNAP III transcription complex formation (60), in TLS-depleted cells, but we did not see a significant change compared to control cells (data not shown). Taken together, our results indicate that TLS affects levels of RNAP III transcription and does so by controlling access of TBP and the RNAP III transcription machinery to target genes.
FIG. 6.
Knockdown of TLS protein increases occupancy of TBP and RNAP III at RNAP III genes. TLS protein was depleted by siRNA followed by ChIP using antibodies against TBP and the RPC155 subunit of RNAP III. DNA fragments were amplified by real-time PCR, and results are depicted as a fold change in TLS-depleted cells compared to control cells for each gene and antibody. The graph shown represents data from three independent experiments. Error bars indicate standard deviations.
DISCUSSION
TLS interacts with a number of proteins that affect gene expression at various steps, including those involved in RNAP II transcription and splicing of mRNA precursors. We discovered that TLS inhibits RNAP III transcription while examining the possible role of TLS in linking transcription and splicing. Repression was demonstrated in vitro by inhibition of RNAP III transcription and reflects an association of TLS with TBP. TLS was found to associate with and repress all three classes of RNAP III promoters, and increases and decreases in TLS levels in vivo were found to affect expression of endogenous RNAP III-transcribed genes accordingly. Taken together, the in vitro and in vivo data correlate well and indicate that TLS indeed regulates RNAP III transcription. Below we discuss how this novel function of TLS might work, how it relates to other proteins that regulate more than one RNAP, and why a small group of important regulatory proteins function in cell growth control by regulating both RNAP II and RNAP III.
Our data indicate that TLS modulates transcription of all three classes of RNAP III-transcribed genes, albeit with somewhat different efficiencies. TLS had the strongest effect on transcription of the U6 gene, which, like 7SK, contains an external RNAP III promoter. Although we did not directly measure effects on 7SK transcription in vitro, TLS displayed a very strong association with this gene, and decreasing TLS protein resulted in increased levels of 7SK transcript in vivo. TLS had an intermediate effect on 5S transcription in vitro, but the greatest apparent increase in TBP occupancy after TLS knockdown was seen at this gene. The basis for this is not known, but it may be due to limitations in accurately quantifying and comparing protein levels at different genes by ChIP assays. The weakest effects were observed with tRNA genes, where TLS had a modest repressive effect in vitro and altered steady-state levels of mature tRNAs only modestly, and decreased TLS levels did not significantly alter the TBP and RNAP III binding at the tRNA genes tested.
Why does TLS have a greater effect on transcription of the U6 gene than on that of 5S or tRNA genes? RNAP III genes that contain an internal promoter are recognized by TFIIIC and subsequently bound by TFIIIB containing Brf1, whereas RNAP III genes with an upstream promoter utilize the Brf2-containing form of TFIIIB (61). Recruitment of TFIIIB to genes with an upstream promoter is mediated by direct interaction between TBP and the TATA box and, unlike transcription of RNAP III genes with an internal promoter, is enhanced by the nonconserved amino terminus of TBP, which binds to the snRNA transcription factor SNAPc (51). Given the interaction of TLS with TBP, the greater effect of TLS on U6 transcription could be attributed to TLS affecting the DNA-binding ability of TBP, to TLS binding the amino terminus of TBP, or to masking of other regions of TBP that interact with Brf2. TBP interacts more weakly with Brf2 than with Brf1 (59, 60), and thus alterations in TBP protein-protein interactions could have a greater effect on transcription of U6 than on that of the 5S or tRNA genes. It is noteworthy that levels of U6 RNA are tightly regulated in normal cells, and increased expression in cancer cell lines as a result of increased expression of Brf2 (9) and/or TBP (32) may contribute to oncogenesis.
Our results suggest that TLS functions through interaction with TBP. TBP levels in cells are usually limiting for regulation of growth, and stimulating cells with growth factors results in a protein kinase cascade that increases levels of TBP (77). The level of TBP alters gene expression and has a greater effect on RNAP I and III genes, which are important for protein translation capacity, and a selective effect on RNAP II genes (see, for example, references 8, 15, and 58). The structure of TBP has been well characterized, and surfaces involved in contacting RNAP II- and III-specific transcription initiation factors have been defined (see, e.g., references 16 and 34). The surface of TBP that Brf1 and Brf2 binds is adjacent to and may overlap the regions bound by RNAP II-specific factors (62, 76). Polymerase specificity may thus occur through factors competing for binding to a limited pool of TBP (16), and factors that modulate this process, such as TLS, could in this way affect both RNAP II and III transcription. The direct interaction between TLS and TBP, coupled with the DNA-mediated interaction between TLS and RNAP III (RPC155) and the presence of TLS at RNAP III genes, suggests that whatever interactions TLS interferes with to prevent transcription occur at the gene itself.
The oncogenic transformation in human liposarcoma has been attributed to uncontrolled transcriptional activation by the TLS-CHOP fusion protein (80). However, the loss of RNAP III regulation by TLS may also be a factor. In normal cells, TLS could play an important role in preventing cellular transformation by restraining RNAP III transcription. Transcription by RNAP III is important for transformation, and levels of RNAP III transcripts are elevated and deregulated in tumor cells (30, 31). Increases in RNAP III products alone can cause cellular transformation, and proto-oncoproteins such as c-Myc upregulate RNAP III transcription to induce cell growth and proliferation (31, 48). Myc increases RNAP III transcription by recruiting a histone acetyltransferase to increase chromatin accessibility and increase TFIIIB and RNAP III occupancy at 5S and tRNA genes (36). Loss of TLS regulation of RNAP III genes may result in increased transcription of the 5S RNA ribosome component and tRNAs for protein translation, which in turn may lead to increased protein synthesis and contribute to uncontrolled cell growth. The higher levels of RNAP III transcripts and increased occupancy of TBP and RNAP III at RNAP III genes in TLS-depleted cells support this idea.
The tumor suppressor proteins p53 and Rb have been shown to regulate both RNAP II and III transcription, and loss of such regulation is an important step toward cellular transformation and cancer. p53 is, of course, a well-studied sequence-specific regulator of numerous protein-coding genes (for a review, see reference 41). In addition, p53 inhibits transcription of both U6 snRNA by RNAP III and U1 snRNA by RNAP II in response to cellular stress (13). While ChIP experiments suggest that p53 is associated with these genes in vivo, protein truncation experiments suggest that transcription repression is not mediated by sequence-specific DNA binding but instead occurs through p53 binding to and repressing the snRNA transcription factors SNAPc and TBP (24). While p53 binding to TBP does not prevent TBP associating with the Brf1 subunit of the TFIIIB complex, it does inhibit TFIIIB occupancy at tRNA genes and prevents association of Brf1 with TFIIIC and RNAP III (17).
The tumor suppressor Rb also represses transcription of RNAP III genes with an external promoter such as U6 by binding to subunits of SNAPc and TBP, as suggested by co-IP experiments (27). ChIP experiments demonstrate that like TLS, Rb occupies the promoter region of the U6 snRNA gene (28). For RNAP III genes that contain internal promoters, Rb mediates repression by binding to TFIIIB and disrupting TFIIIB interactions with RNAP III (14, 27, 65, 72). Although it is a general RNAP III repressor, Rb also regulates specific RNAP II genes by recruiting histone deacetylases (65). Furthermore, Rb regulates RNAP I transcription of rRNA genes by binding to and repressing upstream binding factor (UBF), an RNAP I GTF, under conditions of restrictive growth, so inactivation of Rb is a key step in the pathway to uncontrolled cell growth and proliferation (11, 71).
As RNAP III transcribes 5S rRNA and U6 snRNA, the ribosome and spliceosome may also be affected by levels of RNAP III transcription. Other protein factors have previously been found to affect more than one RNAP to regulate levels of the RNA components of these complexes. In the case of the ribosome, the mitogen-activated protein (MAP) kinase extracellular signal-regulated kinase (ERK) coordinates transcription through phosphorylating UBF to enhance RNAP I transcription of 5.8S, 18S, and 28S rRNAs and phosphorylating TFIIIC to enhance RNAP III transcription of 5S rRNA and tRNAs (19, 63). Such coregulation is particularly important since rRNAs are highly transcribed and ribosome assembly affects both protein synthesis and cell growth (71). For the spliceosome, the coordinate regulation of RNAP II and III spliceosome components was examined using ChIP, and RNAP II was found upstream of U6 genes and enhanced RNAP III transcription (45). Not only is TLS a component of the spliceosome, but our results also indicate that it controls levels of a key component, U6 snRNA. Since U6 snRNA is an integral component of the spliceosome, it is important that the levels of this snRNA are approximately equal to those transcribed by RNAP II.
Coordinated regulation of RNA polymerases is an emerging theme, and cross-regulation does not occur solely through protein factors. There are increasing data on the importance of small, untranslated RNAP III transcripts in regulating gene expression by RNAP II. For example, 7SK RNA inhibits RNAP II transcription by binding to elongation factor P-TEFb and repressing the kinase activity of its CDK9 subunit (54, 75). Similarly, in murine cells, the B2 noncoding RNA transcribed by RNAP III binds to and represses RNAP II after heat shock (1), and more recently it was shown that Alu RNA may have a similar role in human cells (47). TLS may regulate RNAP III production of these noncoding RNAs and thus affect RNAP II transcription indirectly. Such examples not only suggest a function for many untranslated RNAs present in cells but also add another layer of complexity to regulation of gene expression.
In summary, our data indicate that TLS is a general repressor of RNAP III transcription. TLS thus provides an additional example of an important regulatory factor that can affect more than one RNAP. In normal cells, TLS could play an important role in preventing cellular transformation by regulating RNAP III transcription. TLS is also involved not only in RNAP II transcription and splicing of mRNA precursors but also in RNAP III transcription, and it thus appears to control gene expression at multiple levels. The many functions of TLS thus allow it, and likely other TET family proteins, to affect and possibly connect many cellular processes.
Acknowledgments
We thank N. Rao for HeLa nuclear extract; P. Richard and E. Rosonina for assistance with ChIP; C. David and Y. Feng for purified proteins; V. Vethantham, S. Bush, C. Wachtel, and other members of the Manley lab for helpful discussions; R. Beckerman and L. Biderman for assistance with quantitative PCR; and R. Prywes for pET11d-hTBP. We are grateful for antibodies from R. Roeder (Rockefeller University) and R. White (University of Glasgow).
A.Y.T. was partially funded by a Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada, and this work was supported by grants from the National Institutes of Health to J.L.M.
Footnotes
Published ahead of print on 19 October 2009.
REFERENCES
- 1.Allen, T. A., S. Von Kaenel, J. A. Goodrich, and J. F. Kugel. 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 11:816-821. [DOI] [PubMed] [Google Scholar]
- 2.Baechtold, H., M. Kuroda, J. Sok, D. Ron, B. S. Lopez, and A. T. Akhmedov. 1999. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 274:34337-34342. [DOI] [PubMed] [Google Scholar]
- 3.Belly, A., F. Moreau-Gachelin, R. Sadoul, and Y. Goldberg. 2005. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci. Lett. 379:152-157. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti, A., B. Bell, and L. Tora. 1999. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene 18:8000-8010. [DOI] [PubMed] [Google Scholar]
- 5.Bertolotti, A., Y. Lutz, D. J. Heard, P. Chambon, and L. Tora. 1996. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15:5022-5031. [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti, A., T. Melot, J. Acker, M. Vigneron, O. Delattre, and L. Tora. 1998. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 18:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand, P., A. T. Akhmedov, F. Delacote, A. Durrbach, and B. S. Lopez. 1999. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene 18:4515-4521. [DOI] [PubMed] [Google Scholar]
- 8.Bush, S. D., P. Richard, and J. L. Manley. 2008. Variations in intracellular levels of TATA binding protein can affect specific genes by different mechanisms. Mol. Cell. Biol. 28:83-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabarcas, S., J. Jacob, I. Veras, and L. Schramm. 2008. Differential expression of the TFIIIB subunits Brf1 and Brf2 in cancer cells. BMC Mol. Biol. 9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson, D. P., and J. Ross. 1983. Human beta-globin promoter and coding sequences transcribed by RNA polymerase III. Cell 34:857-864. [DOI] [PubMed] [Google Scholar]
- 11.Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X., G. Farmer, H. Zhu, R. Prywes, and C. Prives. 1993. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 7:1837-1849. [DOI] [PubMed] [Google Scholar]
- 13.Chesnokov, I., W. M. Chu, M. R. Botchan, and C. W. Schmid. 1996. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol. Cell. Biol. 16:7084-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu, W. M., Z. Wang, R. G. Roeder, and C. W. Schmid. 1997. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J. Biol. Chem. 272:14755-14761. [DOI] [PubMed] [Google Scholar]
- 15.Colgan, J., and J. L. Manley. 1992. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 6:304-315. [DOI] [PubMed] [Google Scholar]
- 16.Cormack, B. P., and K. Struhl. 1993. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science 262:244-248. [DOI] [PubMed] [Google Scholar]
- 17.Crighton, D., A. Woiwode, C. Zhang, N. Mandavia, J. P. Morton, L. J. Warnock, J. Milner, R. J. White, and D. L. Johnson. 2003. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 22:2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crozat, A., P. Aman, N. Mandahl, and D. Ron. 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363:640-644. [DOI] [PubMed] [Google Scholar]
- 19.Felton-Edkins, Z. A., N. S. Kenneth, T. R. Brown, N. L. Daly, N. Gomez-Roman, C. Grandori, R. N. Eisenman, and R. J. White. 2003. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2:181-184. [PubMed] [Google Scholar]
- 20.Fujii, R., S. Okabe, T. Urushido, K. Inoue, A. Yoshimura, T. Tachibana, T. Nishikawa, G. G. Hicks, and T. Takumi. 2005. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 15:587-593. [DOI] [PubMed] [Google Scholar]
- 21.Fujii, R., and T. Takumi. 2005. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 118:5755-5765. [DOI] [PubMed] [Google Scholar]
- 22.Gardiner, M., R. Toth, F. Vandermoere, N. A. Morrice, and J. Rouse. 2008. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 415:297-307. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh, S., and M. A. Garcia-Blanco. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 6:1325-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gridasova, A. A., and R. W. Henry. 2005. The p53 tumor suppressor protein represses human snRNA gene transcription by RNA polymerases II and III independently of sequence-specific DNA binding. Mol. Cell. Biol. 25:3247-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks, G. G., N. Singh, A. Nashabi, S. Mai, G. Bozek, L. Klewes, D. Arapovic, E. K. White, M. J. Koury, E. M. Oltz, L. Van Kaer, and H. E. Ruley. 2000. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 24:175-179. [DOI] [PubMed] [Google Scholar]
- 26.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch, H. A., L. Gu, and R. W. Henry. 2000. The retinoblastoma tumor suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell. Biol. 20:9182-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch, H. A., G. W. Jawdekar, K. A. Lee, L. Gu, and R. W. Henry. 2004. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell. Biol. 24:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iko, Y., T. S. Kodama, N. Kasai, T. Oyama, E. H. Morita, T. Muto, M. Okumura, R. Fujii, T. Takumi, S. Tate, and K. Morikawa. 2004. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 279:44834-44840. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, D. L., and S. A. Johnson. 2008. RNA metabolism and oncogenesis. Science 320:461-462. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, S. A., L. Dubeau, and D. L. Johnson. 2008. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J. Biol. Chem. 283:19184-19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, S. A., L. Dubeau, M. Kawalek, A. Dervan, A. H. Schonthal, C. V. Dang, and D. L. Johnson. 2003. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol. Cell. Biol. 23:3043-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, S. S., C. Zhang, J. Fromm, I. M. Willis, and D. L. Johnson. 2007. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol. Cell 26:367-379. [DOI] [PubMed] [Google Scholar]
- 34.Juo, Z. S., G. A. Kassavetis, J. Wang, E. P. Geiduschek, and P. B. Sigler. 2003. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422:534-539. [DOI] [PubMed] [Google Scholar]
- 35.Kameoka, S., P. Duque, and M. M. Konarska. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenneth, N. S., B. A. Ramsbottom, N. Gomez-Roman, L. Marshall, P. A. Cole, and R. J. White. 2007. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. U.S.A. 104:14917-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel, G. R., R. L. Maser, J. P. Calvet, and T. Pederson. 1986. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl. Acad. Sci. U.S.A. 83:8575-8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuroda, M., J. Sok, L. Webb, H. Baechtold, F. Urano, Y. Yin, P. Chung, D. G. de Rooij, A. Akhmedov, T. Ashley, and D. Ron. 2000. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. EMBO J. 19:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwiatkowski, T. J., Jr., D. A. Bosco, A. L. Leclerc, E. Tamrazian, C. R. Vanderburg, C. Russ, A. Davis, J. Gilchrist, E. J. Kasarskis, T. Munsat, P. Valdmanis, G. A. Rouleau, B. A. Hosler, P. Cortelli, P. J. de Jong, Y. Yoshinaga, J. L. Haines, M. A. Pericak-Vance, J. Yan, N. Ticozzi, T. Siddique, D. McKenna-Yasek, P. C. Sapp, H. R. Horvitz, J. E. Landers, and R. H. Brown, Jr. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323:1205-1208. [DOI] [PubMed] [Google Scholar]
- 40.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laptenko, O., and C. Prives. 2006. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 13:951-961. [DOI] [PubMed] [Google Scholar]
- 42.Lazarev, D., and J. L. Manley. 2007. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. RNA 13:1546-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lerga, A., M. Hallier, L. Delva, C. Orvain, I. Gallais, J. Marie, and F. Moreau-Gachelin. 2001. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 276:6807-6816. [DOI] [PubMed] [Google Scholar]
- 44.Li, X. Y., and M. R. Green. 1998. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 12:2992-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Listerman, I., A. S. Bledau, I. Grishina, and K. M. Neugebauer. 2007. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 3:e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manley, J. L., and M. T. Colozzo. 1982. Synthesis in vitro of an exceptionally long RNA transcript promoted by an AluI sequence. Nature 300:376-379. [DOI] [PubMed] [Google Scholar]
- 47.Mariner, P. D., R. D. Walters, C. A. Espinoza, L. F. Drullinger, S. D. Wagner, J. F. Kugel, and J. A. Goodrich. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 29:499-509. [DOI] [PubMed] [Google Scholar]
- 48.Marshall, L., N. S. Kenneth, and R. J. White. 2008. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133:78-89. [DOI] [PubMed] [Google Scholar]
- 49.Michelotti, E. F., G. A. Michelotti, A. I. Aronsohn, and D. Levens. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16:2350-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millhouse, S., and J. L. Manley. 2005. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol. Cell. Biol. 25:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mittal, V., and N. Hernandez. 1997. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science 275:1136-1140. [DOI] [PubMed] [Google Scholar]
- 52.Moumen, A., P. Masterson, M. J. O'Connor, and S. P. Jackson. 2005. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123:1065-1078. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1:179-185. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 55.Paule, M. R., and R. J. White. 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabbitts, T. H., A. Forster, R. Larson, and P. Nathan. 1993. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 4:175-180. [DOI] [PubMed] [Google Scholar]
- 57.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadovsky, Y., P. Webb, G. Lopez, J. D. Baxter, P. M. Fitzpatrick, E. Gizang-Ginsberg, V. Cavailles, M. G. Parker, and P. J. Kushner. 1995. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 15:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saxena, A., B. Ma, L. Schramm, and N. Hernandez. 2005. Structure-function analysis of the human TFIIB-related factor II protein reveals an essential role for the C-terminal domain in RNA polymerase III transcription. Mol. Cell. Biol. 25:9406-9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 61.Schramm, L., P. S. Pendergrast, Y. Sun, and N. Hernandez. 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 14:2650-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen, Y., G. A. Kassavetis, G. O. Bryant, and A. J. Berk. 1998. Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol. 18:1692-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 64.Steinberg, T. H., D. E. Mathews, R. D. Durbin, and R. R. Burgess. 1990. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 265:499-505. [PubMed] [Google Scholar]
- 65.Sutcliffe, J. E., T. R. Brown, S. J. Allison, P. H. Scott, and R. J. White. 2000. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 20:9192-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan, A. Y., and J. L. Manley. 24 September 2009. The TET family of proteins: functions and roles in disease. J. Mol. Cell Biol. [Epub ahead of print.] doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed]
- 67.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 68.Vance, C., B. Rogelj, T. Hortobagyi, K. J. De Vos, A. L. Nishimura, J. Sreedharan, X. Hu, B. Smith, D. Ruddy, P. Wright, J. Ganesalingam, K. L. Williams, V. Tripathi, S. Al-Saraj, A. Al-Chalabi, P. N. Leigh, I. P. Blair, G. Nicholson, J. de Belleroche, J. M. Gallo, C. C. Miller, and C. E. Shaw. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323:1208-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, X., S. Arai, X. Song, D. Reichart, K. Du, G. Pascual, P. Tempst, M. G. Rosenfeld, C. K. Glass, and R. Kurokawa. 2008. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454:126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Z., T. Luo, and R. G. Roeder. 1997. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 11:2371-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White, R. J. 2005. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6:69-78. [DOI] [PubMed] [Google Scholar]
- 72.White, R. J., D. Trouche, K. Martin, S. P. Jackson, and T. Kouzarides. 1996. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 382:88-90. [DOI] [PubMed] [Google Scholar]
- 73.Yang, L., L. J. Embree, and D. D. Hickstein. 2000. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol. Cell. Biol. 20:3345-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273:27761-27764. [DOI] [PubMed] [Google Scholar]
- 75.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 76.Zhao, X., L. Schramm, N. Hernandez, and W. Herr. 2003. A shared surface of TBP directs RNA polymerase II and III transcription via association with different TFIIB family members. Mol. Cell 11:151-161. [DOI] [PubMed] [Google Scholar]
- 77.Zhong, S., C. Zhang, and D. L. Johnson. 2004. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 24:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 79.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 80.Zinszner, H., R. Albalat, and D. Ron. 1994. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 8:2513-2526. [DOI] [PubMed] [Google Scholar]