Abstract
Inhibition of cell growth and transformation can be achieved in transformed glial cells by disabling erbB receptor signaling. However, recent evidence indicates that the induction of apoptosis may underlie successful therapy of human cancers. In these studies, we examined whether disabling oncoproteins of the erbB receptor family would sensitize transformed human glial cells to the induction of genomic damage by γ-irradiation. Radioresistant human glioblastoma cells in which erbB receptor signaling was inhibited exhibited increased growth arrest and apoptosis in response to DNA damage. Apoptosis was observed after radiation in human glioma cells containing either a wild-type or mutated p53 gene product and suggested that both p53-dependent and -independent mechanisms may be responsible for the more radiosensitive phenotype. Because cells exhibiting increased radiation-induced apoptosis were also capable of growth arrest in serum-deprived conditions and in response to DNA damage, apoptotic cell death was not induced simply as a result of impaired growth arrest pathways. Notably, inhibition of erbB signaling was a more potent stimulus for the induction of apoptosis than prolonged serum deprivation. Proximal receptor interactions between erbB receptor members thus influence cell cycle checkpoint pathways activated in response to DNA damage. Disabling erbB receptors may improve the response to γ-irradiation and other cytotoxic therapies, and this approach suggests that present anticancer strategies could be optimized.
Keywords: apoptosis/epidermal growth factor receptor/glioblastoma/p185neu/trans-receptor inhibition
The molecular parameters that determine how a cell becomes more or less sensitive to DNA damage induced by radiation or chemotherapeutic agents are poorly understood. Status of cell cycle checkpoint-signaling pathways has been argued to be an important determinant of the response to DNA damage, and mutations in checkpoint components are prevalent in human cancers (reviewed in refs. 1 and 2). A recently introduced paradigm suggests that tumor cells exhibit growth arrest or apoptosis in response to cytotoxic therapies depending on the functional state of checkpoint pathways and that radiation-induced apoptosis may result from impaired growth arrest pathways (3). Similarly, in other systems using nontransformed cells, incomplete mechanisms of DNA repair, occurring during checkpoint phase delay, increase the tendency to apoptosis (4).
Human glioblastomas exhibit many genetic alterations, including amplification and/or mutation of the gene encoding the epidermal growth factor receptor (EGFR) (reviewed in refs. 5 and 6) in some cases resulting in expression of a constitutively activated EGF receptor kinase (7–9). We have shown that expression of a trans-receptor inhibitor of the EGFR, derived from the ectodomain of the p185neu oncogene (T691stop neu), forms heterodimers with both full-length EGFR and a constitutively activated extracellular-deleted mutant EGFR form (ΔEGFR) commonly observed in human glial tumors, particularly those of higher pathologic grade (9, 10). Cell growth and transformation of EGFR-positive or EGFR/ΔEGFR-coexpressing human glioma cells is inhibited by kinase-deficient deletion mutants of p185neu (9, 10). The surface-localized T691stop neu mutant/EGFR heterodimeric receptor complex has decreased affinity for the EGF ligand, impaired internalization kinetics, reduced phosphotyrosine content, and diminished enzymatic kinase activity relative to full-length EGFR and ΔEGFR homodimeric complexes (9, 10).
The specific pathways mediating oncogenic transformation in EGFR positive-transformed human cells have not been characterized completely. Naturally occurring ΔEGFR oncoproteins may increase constitutive activity of a Grb2/Shc/Ras pathway (11) and signaling through phosphatidyl inositol-3 (PI-3) kinases (12), presumably by binding to distinct adaptor proteins (13). Particular mitogen-activated protein kinases, such as those of the c-jun amino terminal kinase family, may be constitutively activated by ligand-independent oncogenic ΔEGF receptors (14). Although holo-EGFRs have been found to be weakly transforming only in a ligand-dependent manner at high levels of receptor expression in fibroblasts, many human tumors exhibit elevated levels of EGFR and this may contribute to unregulated kinase activity in transformed cells (15, 16).
We sought to address whether specific inhibition of signaling through the overexpressed EGFR in radioresistant human glioma cells would alter the physiologic response of these cells to the induction of genomic damage. γ-irradiation combined with erbB receptor inhibition resulted in a greater degree of radiation-induced growth arrest and apoptosis in cancer cells normally resistant to ionizing radiation. These results have implication for the design of receptor-specific agents capable of sensitizing cells to cytotoxic therapies and suggest that erbB receptor-specific inhibition combined with cytotoxic treatments may improve the response to anticancer regimens.
MATERIALS AND METHODS
Vector Construction.
The derivation of the T691stop neu mutant receptor construct has been detailed previously (10).
Maintenance of Cells and Development of Stably Transfected Cell Lines.
The U87MG human glioblastoma cell line was obtained from Webster Cavenee (Ludwig Cancer Institute, San Diego). U373MG human glioma cells, originally isolated from a human anaplastic astrocytoma, were obtained through the American Type Tissue Collection (ATCC; Rockville, MD). Maintenance of cells lines, methods for deriving subclones expressing p185neu-derived proteins and transfection procedures have been described previously (9, 10).
Flow Cytometric Analysis of Cell Cycle Distribution.
Cells were stained for flow cytometry by sequential treatment with 0.003% trypsin solution, followed by 0.05% trypsin inhibitor, 0.01% RNase A solution, and then 0.0416% propidium iodide and 5 mM spermine tetrachloride solution. Each treatment was performed for 10 min with continuous shaking at room temperature. All reagents were ordered from Sigma. Cell cycle analysis was performed within 2 h of staining on a Becton Dickinson FACScan flow cytometer. Ten thousand events were collected for each sample and the data analyzed by using the modfit cell cycle analysis program (Becton Dickinson, version 2.0).
Nuclei Staining and Morphologic Analysis of Apoptosis.
Cells were plated onto coverslips for at least 12 h before irradiation. Irradiation was performed in conditions identical to the colony formation assays. Coverslips were then washed twice with PBS at the indicated times and fixed in 50:50 mix of ice-cold methanol/acetone for 1 min. Coverslips were subsequently stained with 4′,6-diamidino-2-phenylindole dihydrochloride hydrate (DAPI) (Sigma) at a concentration of 0.1–.25 μg/ml in PBS. Inter-observer consistency in apoptosis counts was confirmed with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling-staining and by three independent observers.
Cell counts were performed within 30 min of staining, and photographs were taken on a Zeiss Axioplan epifluorescence microscope. At least three independent fields of 100 cells were counted for each sample.
Colony Formation Assay.
Cell survival after irradiation was assessed by the colony formation assay. The number of cells to be plated was calculated to form 20–200 colonies per dish at each radiation dose and plated into 10-cm culture dishes (Fisher Scientific). By using a J. L. Shepherd (San Fernando, CA) model 30 Mark I Cesium-137 irradiator, 12.8 Gy/min of irradiation was delivered to the cells on a rotating platform to ensure uniform dosing. Cells were incubated after irradiation at 37°C with 5% CO2 for 7–10 days and then stained with crystal violet. Colonies containing >50 cells were counted under a dissecting microscope. The surviving fraction is the ratio of the number of colonies formed to the number of cells plated and was corrected for plating efficiency. At least three cell concentrations were used for each radiation dose.
Western Blotting.
For each time point, 105 cells per 6-cm plate were harvested by lysis in 400 μl of sample buffer (10% glycerol/2% SDS/100 mM DTT/50 mM Tris, pH 6.8). Thirty microliters of each lysate was loaded per lane and separated by electrophoresis on a 15% SDS-polyacrylamide gel before overnight transfer to a nitrocellulose membrane (Bio-Rad). Membranes were probed with mouse anti-human p53 mAb (NeoMarkers, Fremont, CA), followed by goat anti-mouse Ig secondary antibody coupled to horseradish peroxidase (Amersham). To reduce background antibody binding, incubation with secondary antibody in 2.5% powdered milk in PBS was performed. Detection was performed by enhanced chemiluminescence (ECL, Amersham). Relative levels of p53 expression were determined by scanning the blots using a scanning densitometer (Molecular Dynamics).
Antibodies.
The mAb 7.16.4 reactive against the p185neu ectodomain has been described (10). Polyclonal antibodies reactive with p53 and p21 were obtained from NeoMarkers. Antibodies reactive with bcl-2, bax, and bcl-xL were obtained from Oncogene Science.
RESULTS
Cell Cycle Distribution of Cycling Human Glioblastoma Cells Treated with γ-Irradiation: Effects of Disabling erbB Signaling on Growth Arrest.
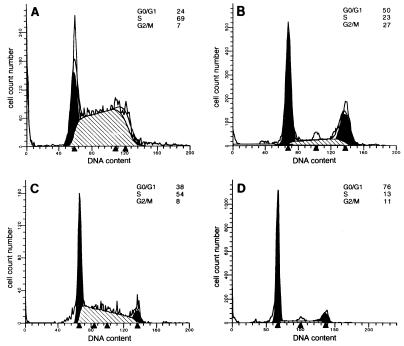
For both U87MG and U87/T691 cells, prolonged serum starvation alone (72–100 h) led to increased accumulation of cells in G0/G1, with modest reductions in both the S and G2/M populations. U87/T691 cells exhibited a higher G0/G1 fraction than parental U87MG cells either in the presence of serum (Fig. 1 A and C) or after prolonged serum deprivation (data not shown). The relative increase in growth arrest induced by expression of the T691stop neu mutant receptor in U87MG cells was thus not overcome by growth in full serum.
Figure 1.
Cell cycle distribution of human glioblastoma cells with or without radiation treatment in 10% serum. Parental U87MG cells (A and B) and U87/T691 transfectants (C and D) were studied. Cells were plated in 60-mm dishes and allowed to attach before either being γ-irradiated (10 Gy) (B and D) or mock-irradiated (A and C). After 72 h, cells were than analyzed by flow cytometry after propidium iodide staining. The distributions of cells according to DNA content are indicated in each panel. The data shown in this experiment are representative of four independent experiments.
Induction of growth arrest by exposure of asynchronously cycling transformed human glial cell populations to γ-irradiation was greater than that induced by prolonged serum deprivation alone. In both U87MG and U87/T691 cells, irradiation of cells grown under full serum growth conditions caused robust increases in G0/G1 and G2/M, and a decrease in the percentage of cells in S phase, as determined by flow cytometric staining for DNA content (Fig. 1 B and D). Reduction of the S phase fraction and accumulation of cells in G2 is characteristic of cells sustaining DNA damage (4, 17). The data in Fig. 1 depict a representative experiment of cells analyzed 72 h after γ-irradiation. Earlier time points indicated similar trends, but analysis 72 h after irradiation was chosen to be consistent with subsequent experiments (see below). An analysis of three independent experiments revealed the following changes in cell cycle distribution [mean percent of cells ± SEM; ± radiation treatment (RT)]: (i) U87MG parental cells: G0/G1: 26 ± 2.8, +RT 51.5 ± 2.1; S: 66 ± 4.2, +RT 21 ± 2.8; G2/M: 8 ± 1.4, +RT 28.5 ± 0.7; (ii) U87/T691 cells: G0/G1: 34.5 ± 4.9, +RT 71 ± 7.1; S: 57.5 ± 4.9, +RT 16 ± 4.2; G2/M: 7.5 ± 0.7, +RT 12.5 ± 3.5. U87/T691 cells exhibited a higher G0/G1 fraction and reduced S and G2/M populations when compared with parental glioblastoma cells when grown asynchronously in culture either with or without radiation treatment, and the largest difference was in the G0/G1 population. Radiation-induced increases in the G2/M fraction were seen in both U87MG and U87/T691 cells, although to a greater degree in parental U87MG cells. Serum deprivation and radiation treatment in these cell populations was not additive and did not appreciably alter the cell cycle distributions in either cell line from that observed with radiation treatment in full serum (data not shown). Thus, disabling EGFR-mediated signaling appears to induce a growth arrest by a mechanism distinct from that observed with prolonged serum deprivation.
Trans-Receptor Inhibition Sensitizes Human Glioblastoma Cells to Radiation-Induced Apoptosis.
Human glioblastoma cells have been shown to be especially resistant to radiation treatment both experimentally and clinically. EGFR overexpression and/or mutation has been correlated with particularly aggressive human glial tumors and oncogenicity was suggested to be caused by reduced apoptosis in vitro and in vivo (18). We examined whether inhibition of EGFR-mediated signaling in human glioblastoma cells by the T691stop neu mutant receptor could sensitize cells to apoptotic cell death.
With prolonged serum deprivation, we observed only 0–1% apoptosis in U87MG parental cells by either 4′-6-diamidino-2-phenylindole (DAPI) staining or terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling staining, which was less than that observed in other studies (18). We found that U87MG-derived cells do not exhibit a sub-G0 peak by flow cytometric analysis after propidium iodide staining under conditions causing apoptosis, which is in agreement with others (19). Expression of the T691stop neu inhibitor in U87MG cells resulted in only 0–2% apoptosis with prolonged serum deprivation as determined by immunohistochemical identification of apoptotic nuclei with DAPI.
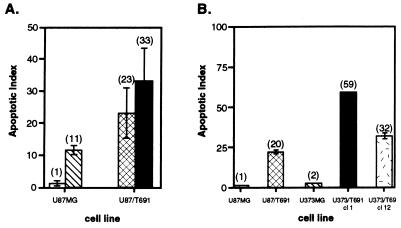
Apoptosis was maximal in repeated studies at 72 h, and this time point was selected for all additional experiments. Expression of the T691stop neu protein in the U87MG cell background increased the level of radiation-induced apoptosis to 23 ± 7.9% (mean ± SEM) at 72 h in four independent experiments in full growth media (Fig. 2A). Prolonged serum deprivation combined with radiation resulted in 33 ± 10.6% apoptosis in U87/T691 cells and in 11 ± 1.5% apoptosis in parental U87MG cells, a comparable increase in both populations above that observed with radiation of cells in full growth media. These data indicate that T691stop neu expression induced greater apoptosis than prolonged serum deprivation in U87MG cells. The morphological changes of nuclear blebbing and fragmentation characteristic of apoptosis are shown by immunohistochemical analysis of U87MG-derived cultured cells stained with DAPI (Fig. 3A–D). The apoptotic indices represent an underrepresentation of total cell death after radiation in U87/T691 cells because we were unable to examine floating cells immunohistochemically.
Figure 2.
Determination of apoptosis and clonogenic survival after γ-irradiation of human glioblastoma cells. (A) Cells were plated and allowed to attach before being exposed to γ-irradiation (10 Gy) in 10% serum or serum-free media. After 72 h, quantitation of apoptosis was conducted by two independent observers. The apoptotic index is the percentage of apoptotic cells with morphologic evidence of apoptosis as determined by staining of nuclei with DAPI. Results presented are mean ± SEM of four independent experiments, and the mean is indicated in parentheses. U87MG cells were grown in 10% serum (□) or serum-free media (▧) and U87/T691 cells were grown in 10% serum (▩) or serum-free media (■). (B) U87MG and U373MG human glioma cells and derivatives were stained with DAPI and analyzed for apoptotic morphology 72 h after γ-irradiation. The mean is indicated in parentheses, and the index shown in this representative experiment is mean ± SD. These results are representative of two additional experiments. Apoptotic indices were felt to be an underestimate because floating cells could not be assayed by this technique.
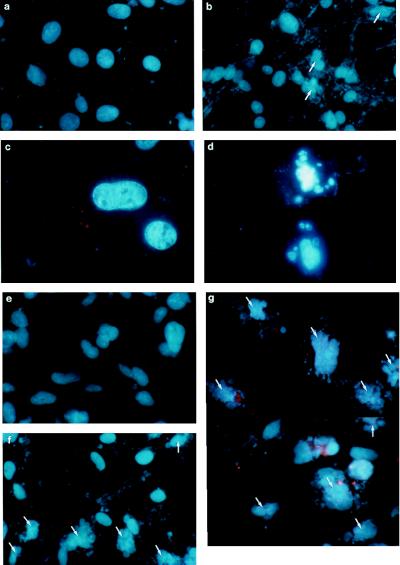
Figure 3.
Morphologic assessment of apoptosis in human glioma cells after γ-irradiation. All cells were stained with DAPI 72 h after being exposed to γ-irradiation. Parental U87MG cells (a and c) and U87/T691 clonal derivatives (b and d) are depicted at two magnifications. Nuclei exhibiting apoptotic morphology are indicated by the arrows. Parental U373MG cells (e) and U373/T691 subclones 1 (g) and 12 (f) are shown after staining with DAPI.
Clonogenic Survival of Irradiated Human Glioblastoma Cells.
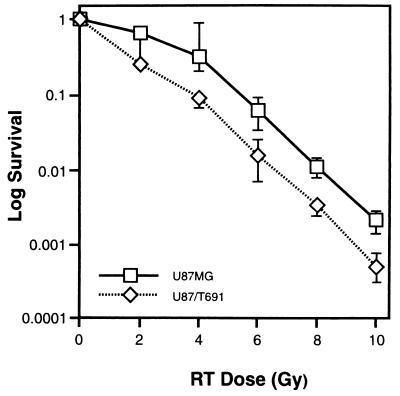
We measured the number of cells that escape growth arrest or death and are able to go on to form a colony, an assay commonly used to determine radiosensitivity. In certain cases, clonogenic growth assays have not correlated with sensitivity to radiation or chemotherapy (3), presumably because the fate of the dead or stably arrested cells is not determined in this assay (20). As shown in Fig. 4, U87/T691 cells exhibited increased sensitivity to radiation across a range of radiation concentrations (2–10 Gy). U87/T691 cells were approximately one-half log more sensitive to radiation than their untransfected parental counterparts at all radiation doses tested. These data suggest a correlation between increases in growth arrest and apoptosis and increased radiosensitivity after inhibition of erbB signaling in irradiated human glioma cells. We confirmed these results by analysis of additional T691stop neu-expressing subclones.
Figure 4.
Clonogenic survival after irradiation. Cells were plated and γ-irradiated with varying doses of radiation followed by incubation for 7–10 days at 37°C with 5% CO2. Colonies were then stained and those with >50 cells were counted under a dissecting microscope. The log survival was then determined by calculating the ratio of the number of colonies formed to the number of cells plated, after correcting for plating efficiency. Similar experiments were performed three times.
Relationship of Radiation Sensitivity of Human Glioblastoma Cells to p53 Status.
U87MG cells and their derivatives contain wild-type p53 and p21 proteins. p53 status has been shown to influence the response to ionizing radiation in a number of transformed and nontransformed cell types (21–23). Western analysis of cell lysates obtained at distinct time points after radiation treatment indicated persistent increases in p53 protein levels detected at all times between 6 and 72 h after radiation in both U87MG and their T691stop neu-transfected derivatives (Fig. 5). The zero time point indicates cells which were γ-irradiated and immediately lysed for analysis. p53 densities were comparable at this time point to mock-irradiated, cycling cells (data not shown). We observed a 10-fold increase in p53 density 12 h after radiation in U87/T691 cells, as compared with only 1.5- to 3-fold increases in both U87MG cells and U87/T691 cells at all other time points examined. This trend was consistently observed (four experiments), and was seen in U87/T691 cells as early as 6 h after radiation in some experiments, and suggests that p53-dependent signaling pathways may be more efficiently activated by disabling the EGFR in the presence of genomic damage. Alterations in p53-regulated checkpoint proteins have been observed 12 h after the induction of genomic damage by γ-irradiation (24) Growth inhibition and differentiation of human breast cancer cells after ligation of erbB receptors has been associated with activation of a p53-dependent pathway (25).
Figure 5.
Analysis of p53 induction in human glioblastoma cells after γ-irradiation. 105 U87MG and U87/T691 cells containing a wild-type p53 gene product were plated and γ-irradiated (10 Gy) after attachment overnight. Lysates were then taken at the indicated times after radiation, subjected to SDS/PAGE and immunoblotted with an antibody reactive with p53. Control (C) cells were MCF-7 breast cancer cells containing immunoreactive p53 protein. We consistently demonstrated more robust induction of the p53 protein at 12 h after γ-irradiation in U87/T691 subclones on four independent occasions.
p21 was induced in both U87MG and U87/T691 cells after radiation treatment, with highest levels seen 24 h after radiation exposure in both cell lines. In both U87MG cells and U87/T691 cells, p21 protein density 6–24 h after radiation was comparable (data not shown). Although others (18) have suggested that up-regulation of bcl-xL is associated with reduced apoptosis in human glioma cells, we detected no changes in bcl-xL protein expression after radiation in either U87MG or U87/T691 cells. Both constitutive and radiation-induced bcl-xL levels were comparable in U87MG and U87/T691 cells (data not shown). Examination of bax and bcl-2 protein levels did not reveal differences between glioblastoma cells and their inhibited subclones.
Apoptosis in p53-Mutated Human Glioblastoma Cells.
U373MG human glioma cells contain a mutated p53 gene product, have undetectable levels of the p21 protein (26, 27), and display a comparable elevation of surface EGFR to U87MG cells by flow cytometric analysis. These cells were used to determine whether the observed apoptosis after inhibition of EGFR-mediated signaling and γ-irradiation was dependent on a wild-type p53 protein. U373MG cells exhibited increases in levels of a mutated p53 protein after γ-irradiation but do not express p21 constitutively or after radiation treatment (data not shown, ref. 27).
We expressed the truncated T691stop neu protein in U373MG glioma cells and confirmed expression at levels comparable with U87/T691 cells in four U373/T691 subclones by metabolic labeling and flow cytometic analysis (data not shown). Surface levels for the T691stop neu mutant receptor was equivalent in U87/T691, U373/T691 cl 1 and U373/T691 cl 12 subclones, and two additional T691stop neu-expressing U373MG derivatives. U373/T691 subclones were capable of growth arrest in low serum and displayed a lawn of confluent cells without the development of morphologically transformed foci in vitro (data not shown), indicating that wild-type p53 and p21 proteins were not required to arrest growth or inhibit transformation of glioma cells in which erbB signaling was disabled.
U373/T691 cl 1 and U373/T691 cl 12 subclones exhibited increased levels of apoptosis over parental U373MG cells after radiation (Figs. 2B and 3E-G). In the representative experiment shown, two U373/T691 subclones exhibited 32% and 59% apoptosis, respectively, 72 h after γ-irradiation, compared with 2% apoptosis in parental U373MG cells and 20% apoptosis in U87/T691 cells. Disabling EGFR signaling by expression of T691stop neu in two distinct human glioma cell lines containing differences in p53 and p21 status resulted in increased radiation-induced apoptosis in each case. Sensitization of human glioblastoma cells to genomic damage can thus occur in the absence of wild-type p53 and p21 proteins. Taken together, these data suggest that both p53-dependent (Fig. 5) and p53-independent pathways may mediate sensitization to cell death induced by a combination of trans-receptor inhibition and genomic damage. Of note, human glioblastoma cells in which EGFR signaling is disabled do not appear to be more sensitive to either prolonged serum deprivation or tumor necrosis factor α-mediated cell death than parental cells (data not shown).
DISCUSSION
Specific inhibition of EGFR signaling inhibits cell growth and transformation and also sensitizes radioresistant human glioma cells to radiation-induced genomic damage. Glioblastoma cells expressing a trans-dominant p185neu-derived mutant receptor exhibit a greater G1 phase arrest and higher levels of apoptosis after radiation than their parental counterparts. In mammalian fibroblasts (28) and in specialized neuronal cells (29, 30), serum or growth factor deprivation can lead to apoptosis under particular conditions. Prolonged serum deprivation alone did not induce apoptosis in human glioblastoma cells in these studies. DNA damage combined with either disabling of erbB receptor signaling or serum deprivation was required to induce apoptosis. Apoptosis was induced by radiation in 23% of U87MG derivatives and in 32–59% of U373MG-derived subclones in which EGFR was disabled (compared with only 1–2% in parental cells) in full growth media, indicating that inhibition of EGFR signaling by trans-receptor inhibition could not be overcome by growth in serum. Serum deprivation combined with radiation damage increased observed levels of apoptosis in both parental U87MG cells and T691stop neu-expressing human glioblastoma derivatives to the same degree. Notably, after DNA damage, the apoptosis observed by disabling erbB receptor signaling at the cell surface was greater than that seen with serum deprivation. Counterintuitively, these data also reveal that growth-inhibited glioma cells are more sensitive to radiation-induced cell death. Induction of an inhibitory pathway occurring as a result of EGFR inhibition may thus sensitize cancer cells to radiation-induced growth arrest and/or cell death.
Resistance of γ-irradiated cells is affected by the functional state of distinct oncogenes. Expression of oncogenic Ras or Raf diminishes radiosensitivity in NIH 3T3 cells (31–34) and expression of the RasH plus either c- or v-myc oncogenes conferred resistance to rat embryo fibroblasts exposed to γ-irradiation (35). It is also true that expression of various oncogenes can sensitize cells to apoptosis, upon exposure to low serum (28) or to anticancer agents (21, 36). Division delay occurring in both the G1 and G2 phases of the cell cycle is influenced by the expression of dominant oncoproteins such as H-ras (17). Expression of a wild-type p53 protein has been associated with decreased survival after γ-irradiation, due to the induction of a higher fraction of apoptosis over cells containing a mutated p53 protein (21, 22). However, tumor cells containing a mutated p53 protein (37) and proliferating lymphoid cells derived from p53−/− mice (38) have been shown to undergo apoptosis after radiation, suggesting p53-independent mechanisms of cell death following genomic damage.
We have shown that p53-dependent mechanisms may influence the response of inhibited glioma cells to undergo relative growth arrest and/or apoptosis. Our results in U373MG-derived cells also indicate that apoptotic cell death occurring after genomic damage in transformed human cells in which EGFR signaling is inhibited involves mechanisms that do not require wild-type p53 and p21 proteins. p21−/− mice develop normally and do not appear to have defects in programmed cell death required for normal organ development, indicating that p21 is not likely to be required for apoptosis (39). p53−/− mice display genetic instability and contain elevated c-myc levels (40). These mice undergo significant levels of apoptosis in vivo, indicating that p53-independent mechanisms of apoptosis are functional in both normal tissues (40) and transformed cells (37).
Interestingly, recent work demonstrates that the absence of p21 in isogenically matched colorectal carcinoma cells resulted in reduced growth arrest when compared with p21-positive derivatives of the same cell line and this was correlated to more inhibited tumor growth in vivo (3). These observations were ascribed to increased apoptosis due to defects in p21-mediated checkpoint growth arrest, though the increased tendency to apoptose by p21−/− cells was not directly shown in this work. Induction of apoptosis was suggested to be preferable to growth arrest as a response to anticancer therapy in vivo (3). In our studies, unlike those of Waldmann et al. (3), there was a correlation between apoptosis, increased growth arrest, and reduction in clonogenic survival after radiation. Pathways distal to the specific inhibitory interaction between the T691stop mutant neu protein and the EGF receptor determine tumor responsiveness to genomic damage and these pathways can be modulated by proximal erbB receptor associations. Specific inhibitory pathways initiated at the level of the cell membrane and associated with growth arrest and/or apoptosis may modulate subsequent checkpoint outcomes in response to DNA damage.
Under certain circumstances, particularly in cancer cells, apoptosis may be favored after genomic damage if defects in pathways mediating growth arrest are present (3). Additionally, when cells are capable of undergoing both growth arrest and apoptosis, as in the case of p21-containing and -deficient human glioma cells in which EGFR signaling was disabled in these studies, apoptosis may be induced after certain signals, such as radiation. The ability or inability to induce growth arrest per se does not appear to be a major determinant of radiosensitivity because both radioresistant parental human glioblastoma cell lines and more radiosensitive derivatives exhibited growth arrest with prolonged serum deprivation or exposure to radiation, and radiosensitive subclones displayed a greater degree of growth arrest.
Our data indicate that the relative proportion of growth arrest or apoptosis induced by genomic damage is influenced by both the integrity of specific checkpoints and alterations in erbB-signaling pathways. Notably, modulating receptor tyrosine kinase signaling pathways may influence checkpoint outcomes after DNA damage in transformed cells. Others (41) have shown that activation of erbB-signaling pathways in breast cancer cells contributes to radioresistance, suggesting that erbB family-signaling pathways influence the response to DNA damage in many tumor types. By combining biologic inhibition of signaling with agents capable of specifically inhibiting receptor oncoproteins of the tyrosine kinase family, we may be able to influence the kinetics of tumor cell response to standard cytotoxic agents. The timing of administration of cytotoxic therapies may be optimized in such combination therapies ,and these data suggest that selective antitumor effects of presently available anticancer regimens could be improved, even in the treatment of advanced human malignancies containing alterations in multiple checkpoint signal transduction pathways.
Acknowledgments
This work was supported by a Merit Review Grant from the Veterans Administration (to D.M.O.), by grants from the Lucille P. Markey Trust and the American Association of Neurological Surgeons Research Foundation (to D.M.O.), and by grants from the National Cancer Institute, the National Institutes of Health, the American Cancer Society, the U.S. Army, and the Abramson Institute to M.I.G.
ABBREVIATIONS
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride hydrate
- EGFR
epidermal growth factor receptor
- RT
radiation treatment
References
- 1.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 2.Nurse P. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 3.Waldman T, Zhang Y, Dillehay L, Yu J, Kinzler K, Vogelstein B, Williams J. Nat Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 4.Orren D K, Petersen L N, Bohr V A. Mol Biol Cell. 1997;8:1129–1142. doi: 10.1091/mbc.8.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis D N, Gusella J F. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- 6.Westermark B, Nister M. Curr Opin Oncol. 1995;7:220–225. doi: 10.1097/00001622-199505000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa R, Ji X D, Harmon R C, Lazar C S, Gill G N, Cavenee W K, Huang H J. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moscatello D K, Montgomery R B, Sundareshan P, McDanel H, Wong M Y, Wong A J. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- 9.O’Rourke D M, Nute E J L, Davis J G, Wu C, Lee A, Murali R, Zhang H-T, Qian X, Kao C-C, Greene M I. Oncogene. 1998;16:1197–1207. doi: 10.1038/sj.onc.1201635. [DOI] [PubMed] [Google Scholar]
- 10.O’Rourke D M, Qian X, Zhang H-T, Davis J G, Nute E, Meinkoth J, Greene M I. Proc Natl Acad Sci USA. 1997;94:3250–3255. doi: 10.1073/pnas.94.7.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prigent S A, Nagane M, Lin H, Huvar I, Boss G R, Feramisco J R, Cavenee W K, Huang H-J S. J Biol Chem. 1996;271:25639–25645. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- 12.Moscatello D K, Holgado M M, Emlet D R, Montgomery R B, Wong A J. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 13.Holgado-Madruga M, Moscatello D K, Emlet D R, Dieterich R, Wong A J. Proc Natl Acad Sci USA. 1997;94:12419–12424. doi: 10.1073/pnas.94.23.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonyak M A, Moscatello D K, Wong A J. J Biol Chem. 1998;273:2817–2822. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- 15.Samanta A, LeVea C M, Dougall W C, Qian X, Greene M I. Proc Natl Acad Sci USA. 1994;91:1711–1715. doi: 10.1073/pnas.91.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiyokawa N, Yan D H, Brown M E, Hung M C. Proc Natl Acad Sci USA. 1995;92:1092–1096. doi: 10.1073/pnas.92.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna W G, Bernhard E J, Markiewicz D A, Rudoltz M S, Maity A, Muschel R J. Oncogene. 1996;12:237–245. [PubMed] [Google Scholar]
- 18.Nagane M, Coufal F, Lin H, Bogler O, Cavenee W K, Huang H-J S. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 19.Haas K D, Yount G, Haas M, Levi D, Kogan S S, Hu L, Vidair C, Deen D F, Dewey W C, Israel M A. Int J Radiat Oncol Biol Phys. 1996;36:95–103. doi: 10.1016/s0360-3016(96)00244-1. [DOI] [PubMed] [Google Scholar]
- 20.Lamb J R, Friend S H. Nat Med. 1997;3:962–963. doi: 10.1038/nm0997-962. [DOI] [PubMed] [Google Scholar]
- 21.Lowe S W, Ruley H E, Jacks T, Housman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 22.Lowe S W, Bodis S, McClatchey A, Remington L, Ruley H E, Fisher D E, Housman D E, Jacks T. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 23.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 24.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 25.Bacus S S, Yarden Y, Oren M, Chin D M, Lyass L, Zelnick C R, Kazarov A, Toyofuku W, Gray B J, Beerli R R, Hynes N E, Nikiforov M, Haffner R, Gudkov A, Keyomarsi K. Oncogene. 1996;12:2535–2547. [PubMed] [Google Scholar]
- 26.Russell S J, Ye Y W, Waber P G, Shuford M, Schold S J, Nisen P D. Cancer. 1995;75:1339–1342. doi: 10.1002/1097-0142(19950315)75:6<1339::aid-cncr2820750616>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Willingham T, Shuford M, Bruce D, Rushing E, Smith Y, Nisen P D. Oncogene. 1996;13:1395–1403. [PubMed] [Google Scholar]
- 28.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 29.Greene L A. J Cell Biol. 1978;78:747–755. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batistatou A, Greene L A. J Cell Biol. 1993;122:523–532. doi: 10.1083/jcb.122.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang E H, Pirollo K F, Zou Z Q, Cheung H Y, Lawler E L, Garner R, White E, Bernstein W B, Fraumeni J J, Blattner W A. Science. 1987;237:1036–1039. doi: 10.1126/science.3616624. [DOI] [PubMed] [Google Scholar]
- 32.Kasid U, Pfeifer A, Weichselbaum R R, Dritschilo A, Mark G E. Science. 1987;237:1039–1041. doi: 10.1126/science.3616625. [DOI] [PubMed] [Google Scholar]
- 33.Kasid U, Pfeifer A, Brennan T, Beckett M, Weichselbaum R R, Dritschilo A, Mark G E. Science. 1989;243:1354–1356. doi: 10.1126/science.2466340. [DOI] [PubMed] [Google Scholar]
- 34.Sklar M D. Science. 1988;239:645–647. doi: 10.1126/science.3277276. [DOI] [PubMed] [Google Scholar]
- 35.McKenna W G, Weiss M C, Endlich B, Ling C C, Bakanauskas V J, Kelsten M L, Muschel R J. Cancer Res. 1990;50:97–102. [PubMed] [Google Scholar]
- 36.Harrington E A, Fanidi A, Evan G I. Curr Opin Genet Dev. 1994;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 37.Bracey T S, Miller J C, Preece A, Paraskeva C. Oncogene. 1995;10:2391–2396. [PubMed] [Google Scholar]
- 38.Strasser A, Harris A W, Jacks T, Cory S. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 39.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 40.Fukasawa K, Wiener F, Vande W G, Mai S. Oncogene. 1997;15:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 41.Wollman R, Yahalom J, Maxy R, Pinto J, Fuks Z. Int J Radiat Oncol Biol Phys. 1994;30:91–98. doi: 10.1016/0360-3016(94)90523-1. [DOI] [PubMed] [Google Scholar]