Abstract
Mitochondrial import of cleavable preproteins occurs at translocation contact sites, where the translocase of the outer membrane (TOM) associates with the presequence translocase of the inner membrane (TIM23) in a supercomplex. Different views exist on the mechanism of how TIM23 mediates preprotein sorting to either the matrix or inner membrane. On the one hand, two TIM23 forms were proposed, a matrix transport form containing the presequence translocase-associated motor (PAM; TIM23-PAM) and a sorting form containing Tim21 (TIM23SORT). On the other hand, it was reported that TIM23 and PAM are permanently associated in a single-entity translocase. We have accumulated distinct transport intermediates of preproteins to analyze the translocases in their active, preprotein-carrying state. We identified two different forms of active TOM-TIM23 supercomplexes, TOM-TIM23SORT and TOM-TIM23-PAM. These two supercomplexes do not represent separate pathways but are in dynamic exchange during preprotein translocation and sorting. Depending on the signals of the preproteins, switches between the different forms of supercomplex and TIM23 are required for the completion of preprotein import.
The majority of mitochondrial proteins are nuclear encoded and posttranslationally transported into the organelle. A major class of mitochondrial proteins possess cleavable targeting signals at their amino termini, so-called presequences (5, 9, 12, 19, 30, 32). These α-helical segments are positively charged and direct the proteins across the outer and inner mitochondrial membranes toward the matrix space, where the presequences are proteolytically removed. However, a number of proteins of the inner mitochondrial membrane, among them subunits of the respiratory chain complexes, also utilize presequences as targeting signals. In addition to the presequence, they contain a hydrophobic sorting signal, which arrests precursor translocation across the inner membrane and mediates the lateral release of the polypeptide into the lipid phase (16, 30). In some cases, the membrane-inserted precursors undergo a second processing event by the inner membrane protease that cleaves behind the sorting signal and therefore leads to the release of the protein into the intermembrane space (25, 30, 31). Thus, a large variety of proteins destined for three different intramitochondrial compartments use presequences as the primary signal for transport.
Cleavable preproteins initially enter mitochondria via the TOM complex and are translocated into or across the inner membrane by the TIM23 complex. The TIM23 complex consists of four integral membrane proteins, Tim23, Tim17, Tim50, and Tim21. Tim23 forms the protein-conducting channel of the translocase and is tightly associated with Tim17 (8, 26, 43). Tim50 acts as a regulator for the Tim23 channel and is involved in early steps of precursor transfer from the outer to the inner membranes (23, 29, 41). Tim21 transiently interacts with the TOM complex via binding to the intermembrane space domain of Tom22. This interaction promotes the release of presequences from Tom22 for their further transfer to the Tim23 channel (4). For full matrix translocation of preproteins, the TIM23 complex cooperates with PAM. The central subunit of PAM is mtHsp70, which undergoes ATP-dependent cycles of preprotein binding and release to promote polypeptide movement toward the matrix. The activity of mtHsp70 in the translocation process is regulated by four membrane-bound cochaperones, Tim44, the J complex Pam18/Pam16 (Tim14/Tim16), and Pam17. Tim44 provides a binding site for preproteins and mtHsp70 close to the Tim23 channel (1, 17, 22, 36). The J protein Pam18 stimulates the ATPase activity of mtHsp70 (10, 44), whereas the J-related protein Pam16 controls the activity of Pam18 (11, 13, 20). Pam17 plays an organizing role in the TIM23-PAM cooperation (33, 45).
The following two different views on the organization of the presequence transport machinery are currently discussed. (i) The TIM23 complex and PAM were proposed to exist in different modular states, termed TIM23SORT and TIM23-PAM. The TIM23CORE complex, consisting of Tim23, Tim17 and Tim50, associates with either Tim21 or the subunits of PAM (4, 47, 51). The Tim21-containing form is termed TIM23SORT since this motor-free form was isolated and shown to mediate membrane insertion of sorted preproteins upon reconstitution (46). The TIM23-PAM form (lacking Tim21) is crucial for mtHsp70-driven preprotein translocation into the matrix (4). (ii) On the other hand, it was proposed that presequence translocase and import motor form a single structural and functional entity. Thus, membrane-integrated TIM23 and import motor would always remain in one complex. This model implies that a motor-free form of the TIM23 complex should not exist (27, 33, 42).
To decide between the different views, it is necessary to analyze translocase and motor in their active form, i.e., during their engagement with preproteins. Moreover, the model of modular forms of TIM23 and PAM raises the question whether two strictly separate TIM23 pathways for inner membrane sorting and matrix translocation exist or whether an exchange between the different forms of the presequence translocase occurs. To date, the majority of experimental studies have been performed with the translocases in an inactive, i.e., preprotein-free, state. Studies using preproteins in transit provided only limited information so far and thus did not resolve the controversy, as follows. (i) Mokranjac and Neupert (27) questioned if the in vitro preprotein insertion by purified TIM23SORT in a proteoliposome assay (46) reflected the in organello situation in intact mitochondria. (ii) Popov-Celeketic et al. (33) accumulated a matrix-targeted preprotein in mitochondrial import sites in vivo and performed pulldown experiments. They copurified TIM23, PAM, and Tim21 and thus concluded that the TIM23 and motor subunits formed a single entity. They did not address the possibility that the accumulated preprotein was associated with different pools of translocase complexes. (iii) Wiedemann et al. (51) made use of the observation that TIM23SORT associates with the respiratory chain (47). They reported a copurification of inner membrane-sorted preproteins and matrix-targeted preproteins with respiratory chain complexes. This observation raised the possibility that the pathways for inner membrane sorting and matrix translocation are connected at least at the level of respiratory chain interaction; however, the composition of the TIM23 complexes was not analyzed.
For this study, we used preproteins with variations in the intramitochondrial sorting signal to monitor the active, preprotein-carrying translocases at distinct stages of mitochondrial import. We observed different forms of active translocases on the presequence pathway. The sorting signals of the preproteins are critical for the selection of specific translocase forms. The motor and sorting forms of the TIM23 complex can be isolated as separate entities in support of the modular model. However, the different TIM23 forms are not permanently separated during preprotein import, but a dynamic exchange between the forms takes place for both matrix-targeted preproteins and inner membrane-sorted preproteins.
MATERIALS AND METHODS
Abbreviations.
DHFR, dihydrofolate reductase; Δψ, membrane potential; mtHsp70, mitochondrial heat shock protein 70; PAM, presequence translocase-associated motor; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TIM23, presequence translocase of the inner mitochondrial membrane; TOM, translocase of the outer mitochondrial membrane; TIM23SORT, sorting form containing Tim21; YPG, 1% yeast extract, 2% Bacto peptone, and 3% glycerol; BSA, bovine serum albumin; MOPS, morpholinepropanesulfonic acid; NTA, nitrilotriacetic acid; PMSF, phenylmethylsulfonyl fluoride; i and i*, intermediate forms.
Strains and growth conditions.
The yeast Saccharomyces cerevisiae strains used in this study are listed in Table 1. The strains are derivatives of YPH499, with the exceptions of PRY36 and Tim23His derived from MB29 and Pam18His derived from BY4741. Yeast cells were grown at 19 to 30°C on YPG medium. For overexpression, the TIM21 open reading frame was cloned into plasmid pYeDP10-1 under the control of the PGK1 promoter (7). The resulting plasmid pBG9435 and the corresponding empty vector were transformed into S. cerevisiae YPH499. Cells were grown on selective medium containing 2% galactose at 30°C.
TABLE 1.
Yeast strains
| Strain (type)a | Genotype | Reference |
|---|---|---|
| YPH499 (WT) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | 38 |
| MR103 (Tom22His) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tom22::TOM22His10-HIS3 | 24 |
| NZY2 (mtHsp70His) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 ssc1::SSC1His6-HIS3MX6 | 44 |
| Tim21ProtA | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim21::TIM21ProtA-HIS3MX6 | 4 |
| tim21Δ | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim21::HIS3MX6 | 4 |
| YPH-BG17-9 (tim17-4) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim17-4 [YEp351]b | 4 |
| YPH-BG17-21 (tim17-5) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim17-5 [YEp351]b | 4 |
| yAC58 (tim17-4 Tim21ProtA) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim17-4 tim21::TIM21ProtA-HIS3MX6 [YEp351]b | This study |
| yAC59 (tim17-5 Tim21ProtA) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim17-5 tim21::TIM21ProtA-HIS3MX6 [YEp351]b | This study |
| KNT17-8 (Tim17His) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim17::TIM17His10-HIS3MX6 | This study |
| Tim21↑ | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 [pYeDP10-1-TIM21] | This study |
| MB29 (WT) | MATaade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [YCplac33-TIM23] | 2 |
| Tim23His | MATaade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [YCplac111-TIM23N-His6] | 2 |
| PRY36 (Tim23ProtA) | MATaade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [pRS414-TIM23ProtA] | 14 |
| BY4741 (WT) | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 | 3 |
| Pam18His | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 pam18::PAM18His10-HIS3MX6 | This study |
WT, wild-type strain.
Possible presence of the empty Yep351 vector cotransformed in a procedure to produce chromosomally integrated tim17 mutants.
Import of precursor proteins into mitochondria and generation of import intermediates.
Mitochondria were isolated from yeast cells by differential centrifugation. Precursor proteins b2(220)-DHFR (B18), b2(220)Δ-DHFR (B46), b2(167)Δ-DHFR (B04), b2(167)IC-DHFR (B58), and b2(167)P-DHFR (B62) were synthesized in rabbit reticulocyte lysates or the TNT SP6 coupled transcription/translation kit (Promega) in the presence of [35S]methionine. Import of 35S-labeled precursor proteins into isolated yeast mitochondria was performed in the presence of 2 mM NADH, 2 mM ATP, and an ATP-regenerating system (5 mM creatine phosphate and 0.1 mg/ml creatine kinase) at 25°C for 15 to 30 min unless indicated otherwise. Where indicated, treatment with proteinase K (50 μg/ml) was performed for 15 min on ice. The import buffer contained 3% (wt/vol) BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM KPi, 5 mM methionine, and 10 mM MOPS-KOH, pH 7.2 (34, 39). To generate import intermediates, 5 μM methotrexate was present in the import reaction. To stop the import, the membrane potential was dissipated by addition of 1 μM valinomycin, 8 μM antimycin A, and 20 μM oligomycin. Samples were subjected to SDS-PAGE or blue native electrophoresis, followed by digital autoradiography. Blue native gel electrophoresis was performed essentially as described previously (6, 8, 35). Digitonin-solubilized and clarified mitochondrial extracts were resolved at 4°C on gradient gels. The high-molecular-weight calibration kit for native electrophoresis (GE Healthcare) was used as a molecular weight standard.
Mitoplasts were generated by hypo-osmotic swelling of mitochondria (to rupture the outer membrane and open the intermembrane space) (34, 39), followed by import of b2(220)Δ-DHFR in the presence or absence of methotrexate. When b2(220)Δ-DHFR was imported into mitochondria in the presence of methotrexate, the precursor also accumulated mainly in the inner membrane (TIM23) and not in a TOM-TIM23 supercomplex, indicating that methotrexate retarded only but did not block the translocation of this long matrix-targeted precursor through the TOM complex.
For chase experiments, the b2(220)-DHFR precursor was incubated in the presence of 30 μM NADPH and 30 μM dihydrofolate in import buffer for 10 min at 25°C prior to addition of mitochondria. The import reaction was performed for 15 min at 25°C. Mitochondria were reisolated and resuspended in import buffer. The chase reaction was performed at 25°C, stopped on ice, and followed by a proteinase K treatment (50 μg/ml) for 15 min, where indicated. The samples were analyzed by SDS-PAGE and digital autoradiography.
Import of purified recombinant precursor.
The b2(167)Δ-DHFR precursor was expressed in Escherichia coli and purified as described previously (8). Saturating amounts of the precursor (minimally 3 μg of preprotein/100 μg of mitochondria) were imported into isolated mitochondria at 25°C in the presence of 5 μM methotrexate. After reisolation, mitochondria were washed and lysed in ice-cold digitonin buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [wt/vol] glycerol, and 1 mM PMSF). Clarified mitochondrial extract was subjected to Ni-NTA or IgG affinity chromatography.
Affinity purification procedures.
For isolation of import intermediates via Tom22His, Tim17His, Tim23His, Pam18His, and mtHsp70His, clarified mitochondrial extracts were subjected to Ni-NTA agarose affinity chromatography. After the column was washed, proteins were eluted with elution buffer (20 mM Tris-HCl, 200 mM NaCl, and 400 mM imidazole, pH 7.4). Proteins were separated by SDS-PAGE and analyzed by digital autoradiography or Western blotting.
Purification of import intermediates and complexes via Tim23ProtA or Tim21ProtA was performed essentially as described previously (4). Clarified mitochondrial extracts were subjected to IgG-Sepharose affinity chromatography. Proteins were eluted with SDS sample buffer unless indicated otherwise and analyzed by SDS-PAGE, followed by digital autoradiography or Western blotting.
Mitochondria containing Pam18His were solubilized in buffer A (20 mM HEPES-KOH [pH 8.0], 50 mM NaCl, 10% [vol/vol] glycerol, 10 mM imidazole, 2 mM PMSF, 1% digitonin), and extracts were incubated with Ni-NTA agarose beads for 60 min at 4°C. Beads were washed with buffer B (20 mM HEPES-KOH [pH 8.0], 100 mM NaCl, 10% [vol/vol] glycerol, 40 mM imidazole, 2 mM PMSF, 0.3% [wt/vol] digitonin), and bound proteins were eluted with buffer C (20 mM HEPES-KOH [pH 7.0], 150 mM NaCl, 10% [vol/vol] glycerol, 400 mM imidazole, 2 mM PMSF, 0.3% [wt/vol] digitonin), followed by SDS-PAGE and immunoblotting. Coimmunoprecipitations with antibodies against Tim23 were carried out as described previously (18).
Miscellaneous.
Radiolabeled proteins and protein complexes were detected using digital autoradiography (Storm imaging system, GE Healthcare) and analyzed by ImageQuant software (GE Healthcare). Western blot analysis was performed using the enhanced chemiluminescence system and quantified using the ImageJ program (NIH, Bethesda, MD).
RESULTS
Two forms of mitochondrial TOM-TIM23-preprotein supercomplexes.
In order to analyze the composition of TOM-TIM23 supercomplexes in the presence of substrate (preprotein), we used chemical amounts of a matrix-targeted preprotein and accumulated it in the import sites of isolated mitochondria. The recombinant preprotein, termed b2(167)Δ-DHFR, has been shown to efficiently accumulate in a TOM-TIM23 supercomplex in a two-membrane-spanning fashion in organello (6, 8). b2(167)Δ-DHFR consists of an N-terminal portion of the precursor of the mitochondrial intermembrane space protein cytochrome b2 fused to DHFR. Cytochrome b2 possesses a bipartite presequence, consisting of an N-terminal matrix-targeting sequence and a hydrophobic inner membrane sorting signal (15, 16, 37, 49). In b2(167)Δ-DHFR, the hydrophobic sorting signal is inactivated by a 19-residue deletion, and thus, the preprotein is targeted to the matrix. In the presence of methotrexate, a ligand of DHFR, the C-terminal DHFR domain is stabilized, and thus, the N-terminal b2 part of the b2(167)Δ-DHFR fusion protein becomes arrested in mitochondrial import sites (6, 8).
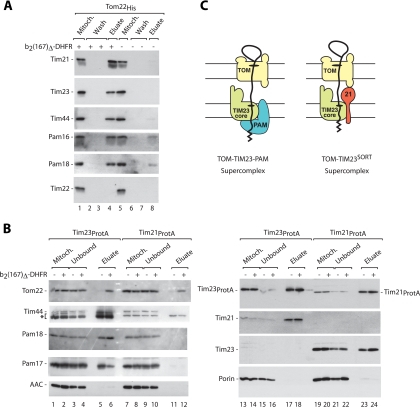
Upon accumulation of b2(167)Δ-DHFR in mitochondria in the presence of methotrexate, the components of the TOM-TIM23 supercomplex, including PAM subunits, can be copurified via a tag at the TOM complex after lysis of the mitochondria with the mild detergent digitonin (the His tag is located at the receptor Tom22) (6, 13, 14, 44, 45). We observed that Tim21 was copurified with the preprotein supercomplex formed in organello (Fig. 1A, lane 4), in agreement with the results obtained in vivo by Popov-Celeketic et al. (33). Two explanations were conceivable for this observation. First, the PAM subunits and Tim21 are permanently associated with the TIM23CORE complex, in line with the suggestion of a single-entity translocase (27, 33). Second, two distinct forms of supercomplexes exist that represent two different pools of the TIM23 translocase, one containing Tim21 and another one bound to PAM.
FIG. 1.
Two forms of preprotein-carrying TOM-TIM23 supercomplexes. (A) Isolated yeast mitochondria harboring Tom22His were incubated with recombinant b2(167)Δ-DHFR in the presence of methotrexate, as indicated. After solubilization in digitonin buffer, mitochondrial extracts were subjected to Ni-NTA affinity chromatography. The samples were analyzed by SDS-PAGE and immunoblotting. Mitoch., 5% mitochondrial extract; wash, 100%; eluate, 100%. (B) Indicated mitochondria containing Tim23ProtA or Tim21ProtA were incubated with recombinant b2(167)Δ-DHFR in the presence of methotrexate and subjected to IgG-Sepharose purification after solubilization. The samples were analyzed by SDS-PAGE and immunoblotting. Asterisk, protein A-containing bands; AAC, ADP/ATP carrier; unbound, 5%; eluate, 100%. (C) Schematic representation of two forms of TOM-TIM23 supercomplexes.
To distinguish between these two possibilities, we performed an affinity purification of the TIM23 translocase from mitochondria containing either tagged Tim23 or tagged Tim21 (protein A tags). The preprotein b2(167)Δ-DHFR was accumulated in the mitochondrial import sites in the presence of methotrexate. Upon lysis with digitonin, an affinity purification of Tim23ProtA led to the copurification of the TOM complex (Tom22), Tim21, and the PAM subunits (Fig. 1B, lanes 6 and 18). As expected, Tim21 and the PAM subunits also copurified with Tim23ProtA in the absence of accumulated preprotein (Fig. 1B, lanes 5 and 17) (4, 13, 33, 44, 46). (The yield of copurification of Pam17 with Tim23ProtA was decreased in the presence of preprotein [Fig. 1B, lane 6 versus lane 5], in agreement with the cross-linking results of Popov-Celeketic et al. [33], supporting the view of a regulatory function of Pam17 in the TIM23-PAM machinery [18, 33, 45].)
In contrast, an affinity purification of Tim21ProtA did not lead to the copurification of motor subunits like Tim44, Pam18, and Pam17 in stoichiometric amounts, in the presence of preprotein (Fig. 1B, lane 12), or in the absence of preprotein (Fig. 1B, lane 11) (4, 46). Tim23 was copurified with tagged Tim21 in the presence and in the absence of preprotein (Fig. 1B, lanes 23 and 24). In the presence of accumulated preprotein, the TOM complex was copurified with tagged Tim21 (Fig. 1B, lane 12), demonstrating that the Tim21-containing complex was competent in carrying the accumulated preprotein and association with the TOM complex. Thus, both Tim23ProtA and Tim21ProtA led to the copurification of the TOM complex in a preprotein-dependent manner but differed strongly in the copurification of motor subunits. These results suggest a model that the following two active preprotein-carrying forms of the TOM-TIM23 supercomplex can be isolated: a TOM-TIM23/Tim21 supercomplex (TOM-TIM23SORT), where the motor subunits are not present in stoichiometric amounts, and a TOM-TIM23-PAM supercomplex that lacks stoichiometric amounts of Tim21 (Fig. 1C).
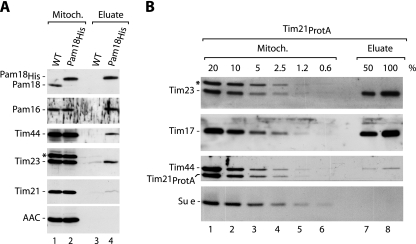
To exclude that the tag at Tim21 interfered with the formation of a TIM23-PAM complex, we performed a reverse experiment by using mitochondria with His-tagged Pam18. Pam16 was copurified with Pam18, as well as Tim44 and Tim23 (Fig. 2A, lane 4). However, only minute amounts of Tim21 were recovered in the eluate (Fig. 2A, lane 4). Considering the proposal of a single-entity presequence translocase containing PAM and Tim21 (27, 33, 41, 42), we performed a Western blot titration of the Tim21ProtA affinity purification to assess the yield of copurification. While Tim23 and Tim17 were efficiently copurified with Tim21ProtA, reflecting the stoichiometric association, only small amounts of Tim44 were recovered with Tim21ProtA (∼2%) (Fig. 2B). Similarly, only ∼1% of Tim21 was copurified with Pam18His. Thus, the large majority of purified presequence translocase complexes contain either Tim21 or PAM. Only a minor fraction of complexes contain both of them. We conclude that a single-entity presequence translocase containing both Tim21 and PAM in stoichiometric amounts was observed only in very small amounts.
FIG. 2.
Copurification of TIM23 and PAM subunits. (A) Purification of TIM23-PAM via Pam18His. Wild-type (WT) and Pam18His mitochondria were lysed with digitonin. Mitochondrial extracts were subjected to Ni-NTA affinity chromatography and analyzed by SDS-PAGE and immunoblotting. Mitoch., 10% mitochondrial extract; eluate, 100%; asterisk, nonspecific band; AAC, ADP/ATP carrier. (B) Tim44 is present in a minor fraction of the TIM23 complex purified via Tim21ProtA. Tim21ProtA mitochondria were solubilized in digitonin buffer, and Tim21-containing TIM23 complexes were purified by IgG affinity chromatography. Bound proteins were eluted by cleavage of Tim21ProtA with tobacco etch virus protease. The samples were analyzed by SDS-PAGE and immunoblotting. Mitoch., 20% to 0.6% of the total mitochondrial extract; Su e, subunit e of F1Fo-ATP synthase.
Accumulation of a matrix-targeted preprotein in TIM23-PAM devoid of Tim21.
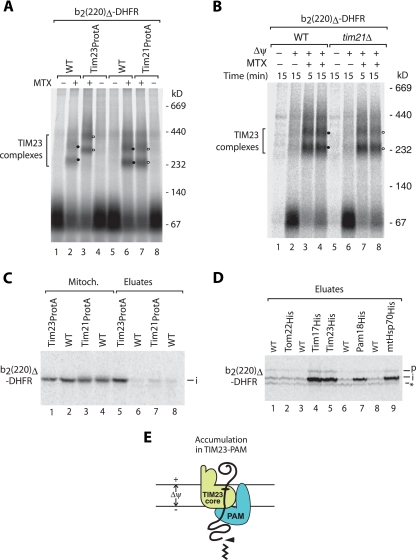
To obtain further evidence that a TIM23-PAM complex lacking Tim21 is engaged in preprotein transport, we accumulated a preprotein directly across the inner membrane. The matrix-targeted preprotein b2(220)Δ-DHFR (4, 50) is efficiently imported into mitoplasts, i.e., mitochondria where the outer membrane was ruptured by hypo-osmotic swelling. We synthesized the preprotein in the presence of [35S]methionine and imported it into mitoplasts. Upon lysis with digitonin, protein complexes were analyzed by blue native electrophoresis. In the presence of methotrexate, b2(220)Δ-DHFR was accumulated in blue native bands (Fig. 3A, lane 2) that were efficiently shifted when mitoplasts carrying a protein A tag at Tim23 were used (Fig. 3A, lane 3). The shift demonstrates that b2(220)Δ-DHFR was accumulated in Tim23-containing complexes. In contrast, no shift was observed with mitoplasts that contained protein A-tagged Tim21 (Fig. 3A, lane 7 versus lane 6), suggesting that Tim21 is not a part of the TIM23 complex engaged with b2(220)Δ-DHFR. To exclude the possibility that a protein A tag at Tim21 interfered with complex formation, we tested mitoplasts lacking Tim21. Also here, the blue native mobility of the TIM23-preprotein complexes was not altered (Fig. 3B). To further substantiate the conclusion that Tim21 is not a stoichiometric component of the TIM23 complex carrying inner membrane-accumulated b2(220)Δ-DHFR, we performed a copurification analysis. b2(220)Δ-DHFR was accumulated in Tim21ProtA and Tim23ProtA mitoplasts in the presence of methotrexate and processed to the intermediate-sized form by the matrix-processing peptidase (4, 50). The mitoplasts were lysed and subjected to affinity purification. In agreement with the blue native analysis, the intermediate form of b2(220)Δ-DHFR was efficiently copurified with Tim23ProtA but not Tim21ProtA (Fig. 3C) (4).
FIG. 3.
Accumulation of matrix-targeted b2(220)Δ-DHFR in TIM23-PAM of mitoplasts. (A) 35S-labeled b2(220)Δ-DHFR was imported into mitoplasts derived from wild-type (WT), Tim23ProtA, and Tim21ProtA mitochondria in the presence or absence of methotrexate (MTX) for 15 min. The mitoplasts were lysed with digitonin and analyzed by blue native electrophoresis and digital autoradiography. Closed and open circles indicate the positions of TIM23-preprotein complexes in wild-type mitochondria and tagged mitochondria, respectively. (B) b2(220)Δ-DHFR was imported into mitoplasts in the presence or absence of Δψ and methotrexate, as indicated. The samples were analyzed by blue native electrophoresis and digital autoradiography. Closed and open circles indicate the positions of TIM23-preprotein complexes in wild-type and tim21Δ mitochondria, respectively. (C) b2(220)Δ-DHFR was imported into mitoplasts derived from Tim23ProtA and Tim21ProtA mitochondria in the presence of methotrexate for 20 min. Mitochondrial extracts were subjected to IgG-Sepharose affinity purification and analyzed by SDS-PAGE and digital autoradiography. Mitoch., 5% mitochondrial extract; eluate, 100%. (D) b2(220)Δ-DHFR was imported into mitoplasts derived from the indicated mitochondria in the presence of methotrexate for 20 min. Mitochondrial extracts were subjected to Ni-NTA affinity purification and analyzed by SDS-PAGE. p, precursor; asterisk, nonspecific band. (E) Schematic representation of the TIM23-PAM complex engaged with a matrix-targeted preprotein. Arrowhead, proteolytic cleavage.
To probe for the association of PAM with the TIM23-preprotein complex, we used yeast strains expressing individual TIM or PAM proteins with a His tag. b2(220)Δ-DHFR was accumulated at the inner membrane of mitoplasts in the presence of methotrexate. Upon lysis with digitonin and affinity chromatography, His-tagged mtHsp70 and His-tagged Pam18 pulled out intermediate-sized b2(220)Δ-DHFR (Fig. 3D, lanes 7 and 9), demonstrating that the motor was associated with the accumulated preprotein. Tim23His and Tim17His efficiently pulled out b2(220)Δ-DHFR (Fig. 3D, lanes 4 and 5), confirming the accumulation in the TIM23 complex. As expected due to the accumulation in the inner membrane, b2(220)Δ-DHFR was not copurified with Tom22His (Fig. 3D, lane 2). Taken together, these results show that b2(220)Δ-DHFR accumulated in mitoplasts in a TIM23-PAM complex that lacked stoichiometric amounts of Tim21 (Fig. 3E).
Sorting signal-dependent shift between different TIM23 forms.
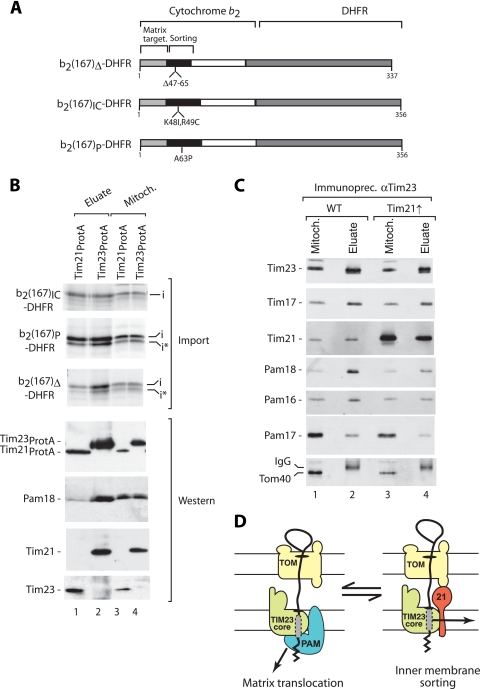
We asked if the inner membrane sorting signal (located behind the matrix-targeting sequence) was important for the accumulation of preproteins in distinct TIM23 complexes (Tim21-containing TIM23 complex versus the Tim21-free form). We used three different forms of b2-DHFR fusion proteins that contained the identical matrix-targeting sequence and C-terminal DHFR but differed only in the sorting signal (Fig. 4A). In b2(167)Δ-DHFR, the sorting signal is completely inactivated, and the preprotein is directed only into the matrix. In two further preproteins, b2(167)IC-DHFR and b2(167)P-DHFR, the sorting signal is partially inactivated only by the replacement of individual amino acid residues (Fig. 4A) (14, 15, 37). These preproteins possess the interesting characteristic that they can use both pathways, inner membrane sorting and matrix translocation: the sorting into the inner membrane is less efficient, and the preproteins are partially redirected to the matrix (14, 15, 37).
FIG. 4.
Point mutations in the sorting signal affect the TIM23-dependent import pathway. (A) Scheme of cytochrome b2-DHFR fusion proteins. Sorting, sorting signal. (B) 35S-labeled b2(167)IC-DHFR, b2(167)P-DHFR, and b2(167)Δ-DHFR were imported into Tim21ProtA and Tim23ProtA mitochondria for 20 min in the presence of methotrexate. Mitochondrial extracts were subjected to IgG-Sepharose chromatography. The samples were analyzed by SDS-PAGE and digital autoradiography (Import) or immunoblotting (Western), as indicated. Mitoch., 10% mitochondrial extract; eluate, 100%. (C) Mitochondria isolated from wild-type (WT) cells or cells overexpressing Tim21 (Tim21↑) were solubilized in digitonin buffer and subjected to immunoprecipitation using anti-Tim23 antibodies. Samples were analyzed by SDS-PAGE and immunoblotting. The yield of copurification from mitochondria of cells overexpressing Tim21 in comparison to the yield from wild-type mitochondria was 46% for Pam18, 62% for Pam16, and 45% for Pam17 (yield of Tim23 purification from mitochondria of cells overexpressing Tim21 set to 100%). (D) Schematic representation of the switch between two forms of TOM-TIM23 supercomplexes with accumulated preprotein. Dashed box, partially inactivated inner membrane sorting signal.
We accumulated the 35S-labeled preproteins in Tim21ProtA mitochondria and Tim23ProtA mitochondria in parallel in the presence of methotrexate. The preproteins were accumulated in both types of mitochondria with a similar efficiency (Fig. 4B, lanes 3 and 4). b2(167)IC-DHFR was copurified with Tim21 with the same yield as with Tim23 (Fig. 4B, lanes 1 and 2). In contrast, b2(167)Δ-DHFR was efficiently isolated via tagged Tim23 but only in part via tagged Tim21 (Fig. 4B, lanes 1 and 2), in agreement with the association of b2(167)Δ-DHFR with both forms of the TIM23 complex during the course of preprotein import (Tim21-containing TIM23SORT and Tim21-free TIM23-PAM) (Fig. 1).
The use of b2(167)P-DHFR allowed an interesting differentiation since this preprotein is processed in two steps by the matrix-processing enzymes (14, 15, 37). The i-form that is generated by the matrix-processing peptidase was copurified with both Tim21ProtA and Tim23ProtA and thus behaved like b2(167)IC-DHFR (Fig. 4B, lanes 1 and 2). A second cleavage product (i*) is formed by the mitochondrial intermediate peptidase after the initial cleavage by the matrix-processing peptidase and thus reflects a further translocation toward the matrix space (14, 15, 37). The i*-form was enriched with tagged Tim23, resembling the behavior of b2(167)Δ-DHFR (Fig. 4B, lanes 1 and 2). Immunoblotting of the eluates demonstrated that the motor subunits like Pam18 were efficiently copurified with tagged Tim23 but present only in tiny amounts in the eluate from Tim21ProtA mitochondria (Fig. 4B, lanes 1 and 2). We conclude that the precursor forms that were further directed onto the matrix-import pathway [b2(167)Δ-DHFR and i*-b2(167)P-DHFR] associated with TIM23-Tim21 with lower efficiency and thus were found mainly in association with TIM23-PAM.
The results obtained so far raised the possibility that the two forms of the presequence translocase are not independent machineries but may be in dynamic exchange with each other. To further analyze this, we used a yeast strain overexpressing Tim21. The resulting mitochondria show a reduced efficiency for import of PAM-dependent proteins (4, 33). We subjected the digitonin-lysed mitochondria to immunoprecipitation with antibodies against Tim23. While Tim17 efficiently interacted with Tim23, the levels of PAM proteins associated with Tim23 (Pam18, Pam16, and Pam17) were decreased by about twofold upon overexpression of Tim21 (Fig. 4C, lane 4 versus lane 2). Popov-Celeketic et al. (33) suggested that Tim21 exerted its effect by selectively removing Pam17 from the translocase, whereas the essential motor proteins would remain permanently associated with TIM23. However, they did not show if the association of other motor subunits with TIM23 was affected by overexpression of Tim21. The results presented here revealed that not only the association of Pam17 with TIM23 but also the association of Pam18 and Pam16 was decreased by overexpression of Tim21. Since the total amounts of Tim23 and Tim17 were not changed by overexpression of Tim21 (Fig. 4C) (4, 33), the ratio between TIM23-PAM and TIM23-Tim21 was shifted toward the latter one. Together with the nearly complete absence of PAM components in Tim21 pulldowns (Fig. 1B and 2B) (4, 46), these results suggest an antagonistic effect of Tim21 on binding of the PAM subunits to the TIM23 complex.
Taken together, we conclude that preproteins can shift between the Tim21-containing TIM23SORT complex and the Tim21-free TIM23-PAM complex (Fig. 4D). At an early stage of import, preproteins that are translocated toward the matrix are associated with TIM23SORT, whereas in a later stage of import, the proteins are found mainly in TIM23-PAM (Fig. 4D).
Dynamic interplay of PAM and Tim21 in membrane sorting.
To further investigate an interplay between the two forms of the TIM23 complex, we used a preprotein that depends on both translocase forms, TIM23SORT and TIM23-PAM. The long preprotein b2(220)-DHFR contains an intact hydrophobic sorting signal and thus is laterally sorted into the inner membrane; in addition, the preprotein contains a folded heme binding domain in the mature b2 part (in front of DHFR) and thus requires the motor activity for import (in order to promote unfolding of the heme binding domain for translocation across the outer membrane) (4, 40, 49). b2(220)-DHFR is processed in the following two steps (Fig. 5A, lane 1): first, the matrix-processing peptidase removes the matrix-targeting sequence to generate the intermediate; second, after lateral sorting of the preprotein into the inner membrane, the hydrophobic sorting signal is removed by the inner membrane protease to generate the mature form that is released into the intermembrane space (4, 16, 31, 49). In the absence of methotrexate, intermediate and mature forms are translocated across the outer membrane and are thus protected against externally added proteinase K (Fig. 5A, lane 3).
FIG. 5.
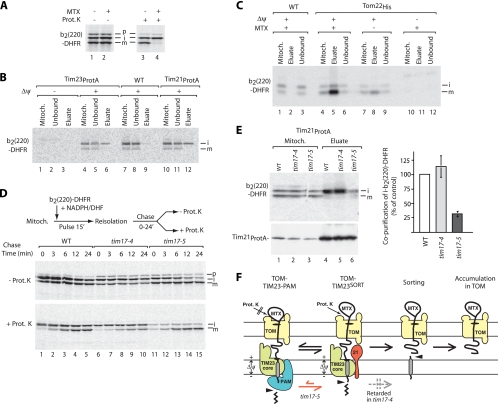
Stage-dependent motor and sorting functions in the import and lateral sorting of a preprotein. (A) 35S-labeled b2(220)-DHFR was imported into isolated wild-type mitochondria in the presence or absence of methotrexate (MTX) and subjected to proteinase K treatment (Prot. K), where indicated. The mitochondria were analyzed by SDS-PAGE and digital autoradiography. p, precursor; m, mature. (B) b2(220)-DHFR was imported into Tim23ProtA and Tim21ProtA mitochondria for 15 min in the presence of methotrexate and in the presence or absence of a Δψ, as indicated. Mitochondrial extracts were subjected to IgG-Sepharose purification and analyzed by SDS-PAGE and digital autoradiography. WT, wild type. (C) b2(220)-DHFR was imported into wild-type or Tom22His mitochondria for 30 min. After solubilization, mitochondria were subjected to Ni-NTA affinity purification and analyzed by SDS-PAGE. Mitoch., 5% mitochondrial extract; eluate, 100%. (D) b2(220)-DHFR was imported in the presence of NADPH and dihydrofolate (DHF) for 15 min into wild-type, tim17-4, and tim17-5 mitochondria. After reisolation, mitochondria were incubated for the indicated time periods (Chase). Samples were treated with proteinase K, where indicated, and analyzed by SDS-PAGE and digital autoradiography. (E) b2(220)-DHFR was imported into wild-type, tim17-4, and tim17-5 mitochondria containing Tim21ProtA for 30 min in the presence of methotrexate. After solubilization, IgG-Sepharose chromatography was performed. The samples were analyzed by SDS-PAGE and digital autoradiography or immunoblotting. The yield of copurification from the mutant mitochondria was compared to that of wild-type mitochondria (control, 100%); standard errors of the means were calculated from three independent experiments. (F) Schematic representation of import and sorting of the b2(220)-DHFR protein. Arrowheads, proteolytic cleavage events on matrix side and intermembrane space side; Prot. K, externally added proteinase K.
In the presence of methotrexate, both processing steps occurred efficiently, yet the intermediate of b2(220)-DHFR/methotrexate was not digested by proteinase K, whereas the mature form was accessible to the protease (Fig. 5A, lanes 2 and 4). We thus imported the methotrexate-bound preprotein into mitochondria containing tagged Tim or Tom proteins in order to determine in which import complexes the preprotein accumulated. When imported into Tim23ProtA and Tim21ProtA mitochondria in the presence of a membrane potential Δψ, the intermediate of b2(220)-DHFR showed the expected association with tagged Tim23 and tagged Tim21 (Fig. 5B, lanes 6 and 12) (4). The mature form of b2(220)-DHFR/methotrexate was not copurified with tagged Tim23 or with tagged Tim21 (Fig. 5B, lanes 6 and 12) but was efficiently copurified with tagged Tom22 (Fig. 5C, lane 5). No copurification with Tom22 was observed when untagged wild-type mitochondria were used (Fig. 5C, lane 2) or when Δψ was dissipated (Fig. 5C, lane 11). We conclude that in the presence of methotrexate, b2(220)-DHFR was inserted via the TOM-TIM23 presequence pathway in a Δψ-dependent manner and underwent both proteolytic processing events to generate the mature protein, which was stably arrested in the TOM complex (Fig. 5F). The different accessibility to externally added proteinase K can be explained by the different accessibility of the intermediate and mature forms of b2(220)-DHFR to PAM. The intermediate spans across outer and inner membranes and is accessible to TIM23-PAM, such that PAM can exert an import-driving activity on the polypeptide chain; it has been shown that the folded DHFR is thus so closely driven toward the outer membrane surface that externally added protease cannot remove it (4, 21, 48, 49). In contrast, the mature form has been released from the inner membrane and is arrested in the TOM complex only. Thus, PAM cannot exert an import-driving activity on the polypeptide chain, and proteinase K can cleave between the folded DHFR and the outer membrane surface.
The transition of b2(220)-DHFR from the intermediate present in the TOM-TIM23 supercomplexes to the mature form engaged only with the TOM complex provided an assay to distinguish between motor and sorting functions with the same preprotein. We combined the following two approaches: (i) reversible accumulation of the intermediate and chase to the mature form (6) and (ii) use of tim17 mutants that differentially affect PAM function and inner membrane sorting. Tim17 is involved in both processes, and the mutants tim17-4 and tim17-5 allow for an experimental separation (4). tim17-4 mutant mitochondria show a defect in inner membrane sorting, whereas tim17-5 mitochondria are impaired in PAM binding to the TIM23 complex, and thus, the import-driving activity is reduced. b2(220)-DHFR was accumulated in isolated mitochondria in the presence of the DHFR substrates dihydrofolate and NADPH (“pulse”); upon reisolation of mitochondria, the substrates were released, and the preprotein was imported to the final destination in the intermembrane space (“chase”) (Fig. 5D) (6). In wild-type mitochondria, the proteinase K-protected mature form was generated in a time-dependent manner in the chase phase (Fig. 5D, lanes 1 to 5). In tim17-4 mitochondria, the generation of the protease-protected mature form was strongly inhibited (Fig. 5D, lanes 6 to 10), consistent with the sorting defect in this mutant. In tim17-5 mitochondria, the protease-protected mature form was generated with a yield close to that of wild-type mitochondria (Fig. 5D, lanes 11 to 15). The analysis of the protease-resistant intermediate form revealed a different pattern. After the pulse phase, wild-type and tim17-4 mitochondria generated protease-resistant intermediate (Fig. 5D, lanes 1 and 6), while in tim17-5 mitochondria, only small amounts of protease-resistant intermediate were observed due to the PAM defect (Fig. 5D, lane 11). As outlined above, the motor activity of PAM exerts an import-driving activity on the polypeptide chain, which spans across TOM and TIM23, such that the folded DHFR (stabilized by substrate) is so closely apposed to the outer membrane that the protease cannot remove it (4, 37, 48). In tim17-4 mitochondria with the sorting defect, PAM interacts with the TIM23 complex (4), and thus, the protease-resistant intermediate was generated in the pulse phase, while in the subsequent chase phase, the formation of the sorted mature form was inhibited (Fig. 5D, lanes 6 to 10), in agreement with the view that PAM acted before inner membrane sorting. In tim17-5 mitochondria, the import-driving activity of PAM was impaired (4), and thus, less intermediate was generated during the pulse phase (Fig. 5D, lane 11 compared to lane 1). The generation of the protease-protected mature form in the chase phase occurred with similar yield in wild-type and tim17-5 mitochondria, despite the significantly smaller amounts of accumulated b2(220)-DHFR at the beginning of the chase phase in tim17-5. This led to the surprising conclusion that tim17-5 mitochondria with defective PAM are more efficient in the conversion of the accumulated preprotein to the mature form.
We thus asked if the motor activity influenced preprotein association with the Tim21-containing TIM23SORT complex. We generated tim17 mutant strains that contained protein A-tagged Tim21. b2(220)-DHFR was accumulated in the presence of methotrexate, and the mitochondria were lysed with digitonin and subjected to affinity purification. In wild-type and tim17-4 mitochondria, the intermediate was copurified with tagged Tim21 (quantification) (Fig. 5E, lanes 4 and 5), indicating an association of the intermediate with TIM23SORT. In tim17-5 mitochondria, however, only small amounts of the intermediate were copurified with Tim21ProtA (quantification) (Fig. 5E, lane 6). At first glance, this result is surprising; however, it fits perfectly well to the high efficiency of conversion of the intermediate to the sorted mature form in tim17-5 mutant mitochondria. Since only the intermediate, but not the mature form, is found in association with TIM23SORT, the rapid formation of mature b2(220)-DHFR leads to smaller amounts of intermediate that is still engaged with the TIM23 translocase (Fig. 5F).
We conclude that the impaired interaction of PAM with the TIM23 complex in the tim17-5 mutant has two opposing effects on the import of b2(220)-DHFR. It reduces the amount of intermediate that is imported by the translocation activity of PAM but accelerates the subsequent conversion of the intermediate to the sorted mature form. Thus, the two forms of the TIM23 translocase do not operate independently but influence the activity of each other (Fig. 5F).
DISCUSSION
Different views have been discussed on the mechanism of protein sorting at the mitochondrial inner membrane, ranging from a single-entity translocase, where TIM23 complex and PAM are permanently associated, to the existence of two TIM23 forms (TIM23-PAM and TIM23-Tim21) that function separately for protein translocation into the matrix rather than protein sorting into the inner membrane. We accumulated b2-DHFR model preproteins at distinct stages of mitochondrial import and monitored the formation of active, preprotein-carrying translocase complexes that were used as snapshots to define the composition of translocases in active mitochondria. Our results indicate that neither a single-entity presequence translocase nor two permanently separated translocase forms are adequate to describe the mechanism of inner membrane protein translocation but suggest that two different modular forms of the presequence translocase are in a dynamic exchange during preprotein translocation and sorting.
The hypothesis of a single-entity presequence translocase predicts that TIM23, Tim21, and PAM are permanently present in one complex and thus would exclude the existence of two forms, TIM23-Tim21 and TIM23-PAM (27, 33, 41, 42). However, several independent findings provide evidence for the existence of two distinct forms of an active TIM23 complex. (i) Using the accumulated b2-DHFR proteins, we demonstrate that mitochondria contain both forms of the TIM23 complex in an active, preprotein-carrying state: TIM23-PAM and TIM23-Tim21. The sorting signal and the import stage of a preprotein are critical for the in organello accumulation in the distinct complexes. (ii) To exclude possible concerns on the functionality of protein A-tagged Tim21 (33), we also used a Pam18His strain. Use of both strains, Tim21ProtA and Pam18His, yielded the identical conclusion that the vast majority of TIM23 complexes did not contain PAM and Tim21 together. Moreover, the TIM23-Tim21 complex purified from a Tim21ProtA strain inserted sorted preproteins into the membrane in a proteoliposome assay in the absence of motor components (46). Mitochondria containing Tim21ProtA were competent in accumulating a matrix-targeted preprotein in the inner membrane, like mitochondria containing Tim23ProtA. (iii) In agreement with these findings, the results of Popov-Celeketic et al. (33) showed that only small, substoichiometric amounts of Tim21 were copurified with Pam16, and Mokranjac et al. (28) reported that they were not able to detect Tim21 together with precipitated Pam16. Similarly, only small, substoichiometric amounts of PAM subunits (Tim44 and Pam18) were copurified with Tim21His and Tim21ProtA (33). (iv) It may be argued that PAM or Tim21 was lost during coprecipitation because the complexes may be detergent labile. The following observations exclude this possibility. PAM and Tim21 were efficiently copurified with TIM23CORE separately, whereas the copurification of both together showed only a very low yield. Since the same detergent (digitonin) was used in the different coprecipitation experiments, the TIM23-PAM and TIM23-Tim21 interactions were stable in the detergent. Moreover, both forms of the TIM23 complex carried the accumulated preprotein and were found in association with the TOM complex under these detergent conditions. We conclude that PAM and Tim21 are not simply lost from TIM23CORE during the purification procedure but are present in two different forms of the translocase. (v) The results obtained with yeast mutant or overexpression strains strongly support the view of a dynamic, competitive situation for association of PAM and Tim21 with TIM23CORE. The partial inactivation of Pam16 in a yeast mutant increased the yield of copurification of Tim21 with TIM23CORE (4), whereas overexpression of Pam17 reduced the amount of Tim21 associated with TIM23CORE (33). We show here that overexpression of Tim21 decreased the copurification of PAM subunits with TIM23CORE. Taken together, we conclude that the components of the mitochondrial presequence translocase and import motor do not function as a permanently associated single entity but consist of several modules that cooperate in a dynamic manner.
We found that both TIM23-PAM and TIM23SORT are involved in the import of matrix-targeted preproteins and inner membrane-sorted preproteins. Tim21 not only is found in association with inner membrane-sorted preproteins but also is a component of the TOM-TIM23 supercomplex that carries a matrix-targeted preprotein yet lacks stoichiometric amounts of PAM. A second form of the TOM-TIM23 supercomplex contains PAM but lacks stoichiometric amounts of Tim21. We observed that preproteins can shift between the TIM23SORT complex and the TIM23-PAM complex. Thus, the two forms of the presequence translocase are not separated throughout the entire import process but are in dynamic exchange with each other. At an early stage of import, matrix-targeted preproteins are preferentially associated with TIM23SORT, whereas in a later stage of import, the proteins are found mainly in TIM23-PAM.
For inner membrane-sorted preproteins with a hydrophobic signal behind the matrix-targeting sequence, the reported studies gave different views. TIM23SORT (containing Tim21) was reported to promote membrane insertion (46), while in organello studies indicated that PAM also was involved when the preprotein contained folded domains (4, 40, 49). Moreover, Popov-Celeketic et al. (33) reported that inner membrane-sorted preproteins were not coisolated with the TIM23 complex. To resolve this puzzling situation, we established an in organello pulse-chase assay to dissect the import of inner membrane-sorted proteins into two stages, accumulation in translocation contact sites and subsequent lateral sorting. The accumulated intermediate form was clearly copurified with TIM23SORT. With tim17 mutants that differentially affected PAM binding and lateral sorting, we show that the PAM activity promotes the accumulation of the preprotein in the import sites but delays the subsequent sorting process. The dynamic switch between PAM form and Tim21 form provides an explanation for a surprising observation. Merlin et al. (25) reported that a b2-DHFR preprotein with a partially inactivated sorting signal was missorted into the matrix; however, upon partial inactivation of PAM, the preprotein was laterally sorted, i.e., the impairment of the sorting signal was relieved by an impairment of the import motor. Thus, both PAM form and Tim21 form of the presequence translocase are involved in the import process of the sorted preprotein and the balance of switching between their activities is important for an efficient sorting process.
Acknowledgments
We thank Agnes Schulze-Specking, Inge Perschil, Birgit Schönfisch, and Klaus Paal for expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Excellence Initiative of the German Federal & State Governments (EXC 294), Landesforschungspreis Baden-Württemberg, Gottfried Wilhelm Leibniz Program, and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Blom, J., M. Kübrich, J. Rassow, W. Voos, P. J. T. Dekker, A. C. Maarse, M. Meijer, and N. Pfanner. 1993. The essential yeast protein MIM44 (encoded by MPI1) is involved in an early step of preprotein translocation across the mitochondrial inner membrane. Mol. Cell. Biol. 13:7364-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bömer, U., M. Meijer, A. C. Maarse, A. Hönlinger, P. J. T. Dekker, N. Pfanner, and J. Rassow. 1997. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 16:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska, A., M. Lind, A. E. Frazier, J. Dudek, C. Meisinger, A. Geissler, A. Sickmann, H. E. Meyer, K. N. Truscott, B. Guiard, N. Pfanner, and P. Rehling. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817-829. [DOI] [PubMed] [Google Scholar]
- 5.Chacinska, A., C. M. Koehler, D. Milenkovic, T. Lithgow, and N. Pfanner. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chacinska, A., P. Rehling, B. Guiard, A. E. Frazier, A. Schulze-Specking, N. Pfanner, W. Voos, and C. Meisinger. 2003. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22:5370-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullin, C., and D. Pompon. 1988. Synthesis of functional mouse cytochromes P-450 P1 and chimeric P-450 P3-1 in the yeast Saccharomyces cerevisiae. Gene 65:203-217. [DOI] [PubMed] [Google Scholar]
- 8.Dekker, P. J. T., F. Martin, A. C. Maarse, U. Bömer, H. Müller, B. Guiard, M. Meijer, J. Rassow, and N. Pfanner. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolezal, P., V. Likic, J. Tachezy, and T. Lithgow. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314-318. [DOI] [PubMed] [Google Scholar]
- 10.D'Silva, P. D., B. Schilke, W. Walter, A. Andrew, and E. A. Craig. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U. S. A. 100:13839-13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Silva, P. R., B. Schilke, W. Walter, and E. A. Craig. 2005. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl. Acad. Sci. U. S. A. 102:12419-12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo, T., H. Yamamoto, and M. Esaki. 2003. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 116:3259-3267. [DOI] [PubMed] [Google Scholar]
- 13.Frazier, A. E., J. Dudek, B. Guiard, W. Voos, Y. Li, M. Lind, C. Meisinger, A. Geissler, A. Sickmann, H. E. Meyer, V. Bilanchone, M. G. Cumsky, K. N. Truscott, N. Pfanner, and P. Rehling. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11:226-233. [DOI] [PubMed] [Google Scholar]
- 14.Geissler, A., A. Chacinska, K. N. Truscott, N. Wiedemann, K. Brander, A. Sickmann, H. E. Meyer, C. Meisinger, N. Pfanner, and P. Rehling. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111:507-518. [DOI] [PubMed] [Google Scholar]
- 15.Geissler, A., T. Krimmer, U. Bömer, B. Guiard, J. Rassow, and N. Pfanner. 2000. Membrane potential-driven protein import into mitochondria: the sorting sequence of cytochrome b2 modulates the Δψ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11:3977-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glick, B. S., A. Brandt, K. Cunningham, S. Müller, R. L. Hallberg, and G. Schatz. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69:809-822. [DOI] [PubMed] [Google Scholar]
- 17.Horst, M., P. Jenö, N. G. Kronidou, L. Bolliger, W. Oppliger, P. Scherer, U. Manning-Krieg, T. Jascur, and G. Schatz. 1993. Protein import into yeast mitochondria: the inner membrane import site protein ISP45 is the MPI1 gene product. EMBO J. 12:3035-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutu, D. P., B. Guiard, A. Chacinska, D. Becker, N. Pfanner, P. Rehling, and M. van der Laan. 2008. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol. Biol. Cell 19:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler, C. M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309-335. [DOI] [PubMed] [Google Scholar]
- 20.Kozany, C., D. Mokranjac, M. Sichting, W. Neupert, and K. Hell. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11:234-241. [DOI] [PubMed] [Google Scholar]
- 21.Krayl, M., J. H. Lim, F. Martin, B. Guiard, and W. Voos. 2007. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol. Cell. Biol. 27:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Q., P. D'Silva, W. Walter, J. Marszalek, and E. A. Craig. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300:139-141. [DOI] [PubMed] [Google Scholar]
- 23.Meinecke, M., R. Wagner, P. Kovermann, B. Guiard, D. U. Mick, D. P. Hutu, W. Voos, K. N. Truscott, A. Chacinska, N. Pfanner, and P. Rehling. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312:1523-1526. [DOI] [PubMed] [Google Scholar]
- 24.Meisinger, C., M. T. Ryan, K. Hill, K. Model, J. H. Lim, A. Sickmann, H. Müller, H. E. Meyer, R. Wagner, and N. Pfanner. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol. Cell. Biol. 21:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlin, A., O. von Ahsen, E. A. Craig, K. Dietmeier, and N. Pfanner. 1997. A mutant form of mitochondrial GrpE suppresses the sorting defect caused by an alteration in the presequence of cytochrome b2. J. Mol. Biol. 273:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Milisav, I., F. Moro, W. Neupert, and M. Brunner. 2001. Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem. 276:25856-25861. [DOI] [PubMed] [Google Scholar]
- 27.Mokranjac, D., and W. Neupert. 2009. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim. Biophys. Acta 1793:33-41. [DOI] [PubMed] [Google Scholar]
- 28.Mokranjac, D., D. Popov-Celeketic, K. Hell, and W. Neupert. 2005. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280:23437-23440. [DOI] [PubMed] [Google Scholar]
- 29.Mokranjac, D., M. Sichting, D. Popov-Celeketic, K. Mapa, L. Gevorkyan-Airapetov, K. Zohary, K. Hell, A. Azem, and W. Neupert. 2009. Role of Tim50 in the transfer of precursor proteins from the outer to inner membrane of mitochondria. Mol. Biol. Cell 20:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neupert, W., and J. M. Herrmann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723-749. [DOI] [PubMed] [Google Scholar]
- 31.Nunnari, J., T. D. Fox, and P. Walter. 1993. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262:1997-2004. [DOI] [PubMed] [Google Scholar]
- 32.Oka, T., and K. Mihara. 2005. A railroad switch in mitochondrial protein import. Mol. Cell 18:145-146. [DOI] [PubMed] [Google Scholar]
- 33.Popov-Celeketic, D., K. Mapa, W. Neupert, and D. Mokranjac. 2008. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, M. T., W. Voos, and N. Pfanner. 2001. Assaying protein import into mitochondria. Methods Cell Biol. 65:189-215. [DOI] [PubMed] [Google Scholar]
- 35.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 36.Schneider, H. C., J. Berthold, M. F. Bauer, K. Dietmeier, B. Guiard, M. Brunner, and W. Neupert. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371:768-774. [DOI] [PubMed] [Google Scholar]
- 37.Schwarz, E., T. Seytter, B. Guiard, and W. Neupert. 1993. Targeting of cytochrome b2 into the mitochondrial intermembrane space: specific recognition of the sorting signal. EMBO J. 12:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stojanovski, D., N. Pfanner, and N. Wiedemann. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80:783-806. [DOI] [PubMed] [Google Scholar]
- 40.Stuart, R. A., A. Gruhler, I. van der Klei, B. Guiard, H. Koll, and W. Neupert. 1994. The requirement of matrix ATP for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur. J. Biochem. 220:9-18. [DOI] [PubMed] [Google Scholar]
- 41.Tamura, Y., Y. Harada, T. Shiota, K. Yamano, K. Watanabe, M. Yokota, H. Yamamoto, H. Sesaki, and T. Endo. 2009. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184:129-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura, Y., Y. Harada, K. Yamano, K. Watanabe, D. Ishikawa, C. Ohshima, S. Nishikawa, H. Yamamoto, and T. Endo. 2006. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truscott, K. N., P. Kovermann, A. Geissler, A. Merlin, M. Meijer, A. J. Driessen, J. Rassow, N. Pfanner, and R. Wagner. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074-1082. [DOI] [PubMed] [Google Scholar]
- 44.Truscott, K. N., W. Voos, A. E. Frazier, M. Lind, Y. Li, A. Geissler, J. Dudek, H. Müller, A. Sickmann, H. E. Meyer, C. Meisinger, B. Guiard, P. Rehling, and N. Pfanner. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Laan, M., A. Chacinska, M. Lind, I. Perschil, A. Sickmann, H. E. Meyer, B. Guiard, C. Meisinger, N. Pfanner, and P. Rehling. 2005. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 25:7449-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Laan, M., M. Meinecke, J. Dudek, D. P. Hutu, M. Lind, I. Perschil, B. Guiard, R. Wagner, N. Pfanner, and P. Rehling. 2007. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9:1152-1159. [DOI] [PubMed] [Google Scholar]
- 47.van der Laan, M., N. Wiedemann, D. U. Mick, B. Guiard, P. Rehling, and N. Pfanner. 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16:2271-2276. [DOI] [PubMed] [Google Scholar]
- 48.Voisine, C., E. A. Craig, N. Zufall, O. von Ahsen, N. Pfanner, and W. Voos. 1999. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97:565-574. [DOI] [PubMed] [Google Scholar]
- 49.Voos, W., B. D. Gambill, B. Guiard, N. Pfanner, and E. A. Craig. 1993. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123:119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voos, W., O. von Ahsen, H. Müller, B. Guiard, J. Rassow, and N. Pfanner. 1996. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 15:2668-2677. [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedemann, N., M. van der Laan, D. P. Hutu, P. Rehling, and N. Pfanner. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]