Abstract
The intricate biogenesis of multimeric organellar enzymes of dual genetic origin entails several levels of regulation. In Saccharomyces cerevisiae, mitochondrial cytochrome c oxidase (COX) assembly is regulated translationally. Synthesis of subunit 1 (Cox1) is contingent on the availability of its assembly partners, thereby acting as a negative feedback loop that coordinates COX1 mRNA translation with Cox1 utilization during COX assembly. The COX1 mRNA-specific translational activator Mss51 plays a fundamental role in this process. Here, we report that Mss51 successively interacts with the COX1 mRNA translational apparatus, newly synthesized Cox1, and other COX assembly factors during Cox1 maturation/assembly. Notably, the mitochondrial Hsp70 chaperone Ssc1 is shown to be an Mss51 partner throughout its metabolic cycle. We conclude that Ssc1, by interacting with Mss51 and Mss51-containing complexes, plays a critical role in Cox1 biogenesis, COX assembly, and the translational regulation of these processes.
Translational regulation is a fundamental mechanism used to control the accumulation of key proteins in a large variety of biogenetic and physiological processes in both prokaryotic and eukaryotic cells (20, 23). Translational autoregulation is a particular form of regulation exerted by the protein being translated. It is a well-established control mechanism for bacteriophage and prokaryotic systems (15), and it has also been reported in eukaryotes (4). Usually, the newly synthesized protein binds to its own mRNA to repress translation (20). However, repression can also be exerted by nascent chains interacting with the ribosome (49).
Translational autoregulation also occurs in semiautonomous eukaryotic organelles of ancestral bacterial origin, namely, mitochondria and chloroplasts. During evolution, these organelles have retained a few genes in their own genomes, which are transcribed within the organelle, and the mRNAs are translated on organellar ribosomes. Most proteins synthesized within the organelles are part of large multimeric enzyme complexes devoted to energy production. These complexes are formed by subunits of dual genetic origin, nuclear and organellar, and assemble in the organellar membranes. Interestingly, intraorganellar translation of certain subunits has been proposed to be regulated by the availability of their assembly partners (1, 39, 54, 55). A distinctive characteristic of these systems is the involvement of ternary factors, mRNA-specific translational activators whose availability would be regulated by the specific gene products. The players and mechanisms involved remain largely unknown.
We have focused on the characterization, in the yeast Saccharomyces cerevisiae, of an assembly-controlled translational regulatory system that operates during the biogenesis of cytochrome c oxidase (COX), the terminal enzyme of the mitochondrial respiratory chain. The three subunits forming the COX catalytic core (1, 2, and 3) are encoded in the mitochondrial DNA (mtDNA), and the remaining eight subunits are encoded in the nuclear DNA. Subunits 1 and 2 coordinate the heme A and copper prosthetic groups of the enzyme. COX biogenesis requires the assistance of a large number of ancillary factors acting at all the levels of the process (11). COX assembly is thought to be linear, consisting of the sequential addition of subunits to an initial seed formed by the mtDNA-encoded subunit 1 (Cox1) in both mammalian and yeast cells (11).
The concerted accumulation of COX subunits is regulated by posttranslational degradation of most unassembled Cox1 and the other highly hydrophobic core subunits (27). Recently, we along with others have proposed an additional level of regulation, namely, an assembly-controlled synthesis of Cox1 (1, 2, 39, 56). In S. cerevisiae, COX1 mRNA translation is under the control of Mss51 and Pet309 (8, 30). Mss51 is a key element of the regulatory system. Mss51 acts on the 5′ untranslated region (UTR) of COX1 mRNA to promote translation initiation (39, 56) and additionally acts on a target in the protein coding sequence of COX1 mRNA, perhaps to promote elongation (39). Mss51 and newly synthesized Cox1 form a transient complex (2, 39) that is stabilized by Cox14 (2). We have postulated that these interactions downregulate Cox1 synthesis when COX assembly is impaired by trapping Mss51 and limiting its availability for COX1 mRNA translation (2). According to this model, the release of Mss51 from the ternary complex and its availability for Cox1 synthesis probably occur when Cox1 acquires its prosthetic groups or interacts with other COX subunits, a step possibly catalyzed by Shy1, a protein involved in maturation and/or assembly of Cox1 (2, 10, 34). Coa1 could also participate in Cox1 maturation and stabilize the ternary Cox1/Mss51/Cox14 complex until it interacts with Shy1 (34, 40). Further studies are required to understand how Mss51 is recycled from its posttranslational function to become available for COX1 mRNA translation and to fully clarify how this regulatory mechanism operates.
In this study, we have analyzed protein-interacting partners of Mss51 in the wild type and a collection of COX assembly mutants. We found that the native molecular weight (MW) of Mss51 is dependent on both the status of COX assembly and the synthesis of Cox1. The mitochondrial Hsp70 (mtHsp70) chaperone Ssc1 interacts with Mss51 and with several high-molecular weight Mss51-containing complexes involving the COX1 mRNA translational apparatus, Cox1, and several Cox1 assembly factors. Mutants defective in Cox1 maturation or in other aspects of COX biogenesis accumulate distinct ratios of these complexes. In this way, Cox1 regulates its own translation through the action of Mss51 and Ssc1.
MATERIALS AND METHODS
Yeast strains and media.
All S. cerevisiae strains used are listed in Table 1. Wild-type cells and some COX mutants were transformed with integrative plasmids containing MSS51 (pSG91/ST9) or the mss51 suppressor, mss51 with the mutation T167R (mss51T167R; pSG91/ST6) in the integrative plasmid YIp351 as reported (2). The mss51, mss51 cox15, and mss51 cox14 null mutants were transformed with the integrative plasmid pG96/ST13 (2) containing a fusion gene expressing Mss51-glutathione S transferase (GST). The compositions of the growth media have been described elsewhere (36). The following media were used routinely to grow yeast: YPD (2% glucose, 1% yeast extract, 2% peptone), YP-Gal (2% galactose, 1% yeast extract, 2% peptone), YPEG (2% ethanol, 3% glycerol, 1% yeast extract, 2% peptone), WO-EG (2% ethanol, 3% glycerol, 0.67% yeast nitrogen base), and WO-Gal (2% galactose, 0.67% yeast nitrogen base).
TABLE 1.
Genotypes and sources of yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| W303-1A | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303Δmss51 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmss51::HIS3 | 1 |
| W303Δpet309 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δpet309::HIS3 | 14 |
| W303Δcox10 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox10::HIS3 | 37 |
| W303Δcox11 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox11::HIS3 | 51 |
| W303Δshy1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 | 1 |
| W303Δshy1 rev | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3 mss51F199I | 1 |
| W303Δcox15 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox15::HIS3 | 13 |
| W303Δcox1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 cox1 | 56 |
| W303Δcox5a | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox5a::HIS3 | 14 |
| W303Δoxa1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δoxa1::HIS3 | 16 |
| W303Δcox14 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox14::TRP1 | 2 |
| W303Δcox14 Δmss51 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox14::TRP1 Δmss51::HIS3 | 2 |
| W303Δcox11 Δmss51 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox11::KanMX Δmss51::HIS3 | This work |
| W303Δcoa1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcoa1::KanMX4 | 40 |
| ssc1-2 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 can1-100 GAL2+met2-Δ1 lys2-Δ2 ssc1-2::LEU2 | 21 |
| mdj1-5 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δmdj1::HIS3 mdj1-5-URA3 | 53 |
| W303Δshy1 Δmdj1 | MATaade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δshy1::URA3, Δmdj1::KanMX | This work |
Department of Human Genetics, Columbia University, New York, NY.
Sucrose gradients.
The sedimentation properties of Mss51 in sucrose gradients were analyzed essentially as described previously (2). Mitochondria prepared by the method of Herrmann et al. (19) (4 mg of protein) or spheroplast extracts (equivalent of six times the optical density at 600 nm) from the different strains were solubilized in 400 μl of extraction buffer (20 mM HEPES, pH 7.4, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1% digitonin, 1.2 mM MgCl2, and 150 mM KCl) on ice for 30 min. Deviations from these extraction conditions are explained in the text. The clarified extract obtained by centrifugation at 200,000 × average g force (gav) for 15 min was mixed with hemoglobin (Hb) and lactate dehydrogenase (LDH) and applied to 5 ml of linear sucrose gradient (either 7 to 20% or 20 to 40%) containing 20 mM HEPES, 0.5 mM PMSF, 0.1% digitonin, 1.2 mM MgCl2, and 150 mM KCl. Following centrifugation in a Beckman 55Ti rotor, the gradients were collected in 14 equal fractions. For most 7 to 20% gradients, the centrifugation time was 12 h at 28,000 rpm. When indicated, we prepared “expanded” gradients by centrifuging for 15 h at 45,500 rpm. All 20 to 40% sucrose gradients were centrifuged for 12 h at 28,000 rpm. Each fraction was subsequently assayed for Hb by absorption at 409 nm and for LDH activity by measuring NADH-dependent conversion of pyruvate to lactate. The distribution of Mss51 and other proteins was assayed by Western blot analysis. The mass of Mss51 was determined from the positions of the respective peaks relative to those of the markers (31). All the gradients were performed at least in triplicate using independent mitochondrial preparations. The gradients reported are representative of each strain because the patterns observed were reproducible.
GST pulldown, protein purification, and identification by MS.
Mss51 fused with the 26-kDa GST with an intercalated thrombin site was expressed from an integrative plasmid (pG96/ST13) in a strain carrying a null mutant allele of mss51 as previously reported (aW303Δmss51/ST13) (2). This strain was respiratory competent and grew on nonfermentable carbon sources with a doubling time similar to that of the parental wild-type strain (2). Mitochondria were prepared from the aW303Δmss51/ST13 strain by the method of Herrmann et al. (19). Mitochondrial proteins (24 mg) were solubilized in 20 mM HEPES, pH 7.4, 0.5 mM PMSF, 1% digitonin, 1.2 mM MgCl2, and 150 mM KCl, and the extracts were loaded in six sucrose gradients. After centrifugation, 14 equal fractions were collected from each gradient; the equivalent fractions were pooled and tested for Mss51 distribution by Western blot analyses. Three fractions around each peak were pooled and used for GST pulldown experiments. Each set of pooled fractions was incubated in a rotator with glutathione-Sepharose beads for 4 h at 4°C. After centrifugation at 1,500 rpm for 5 min, the beads were washed three times with cold phosphate-buffered saline (PBS). The Mss51-GST fusion protein was eluted with 10 mM reduced glutathione-50 mM Tris-base (pH 8.0) and concentrated using Vivaspin 500 columns. An aliquot of the concentrate was separated by blue native polyacrylamide gel electrophoresis (PAGE) as described previously (45) and stained with Coomassie blue to confirm the native size and integrity of the purified Mss51-containing complexes. Analytical 12% sodium dodecyl sulfate (SDS)-PAGE followed by Coomassie blue staining was used to visualize the individual protein components of the Mss51-containing complexes. Replicas of both kinds of gels were used for protein transfer to a polyvinylidene difluoride membrane and immunodetection with anti-GST (Santa Cruz Biotechnology, CA) and anti-Mss51 (2) antibodies and with antibodies specific to Cox1 and Cox2 (Molecular Probes, OR), Cox14 (2), ShyI (32), Mdj1 (25), and Ssc1 (7). To obtain material for protein identification by mass spectrometry (MS), the concentrated GST pulldowns were separated on preparative native and denaturing PAGE systems and stained with Coomassie blue. The native complexes and the denatured protein components were excised from the gel, dried, and sent for protein identification by MS to Midwest Bio Services, LLC (Overland Park, KS), who used a DECA XP Plus ion trap mass spectrometer (ThermoFinnigan). Full MS and tandem MS (MS/MS) spectra were recorded, and Sequest software was used to match MS/MS spectra to the S. cerevisiae protein database.
In vivo and in organello mitochondrial protein synthesis.
Mitochondrial gene products were labeled with [35S]methionine (7 mCi/mmol; Amersham, Piscataway, NJ) in whole cells at 30°C in the presence of cycloheximide (1). For in organello translation, mitochondria were prepared by the method of Herrmann et al. (19) and labeled with [35S]methionine as described previously (18). Equivalent amounts of total cellular or mitochondrial proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to Kodak X-OMAT X-ray film.
Sedimentation properties of newly synthesized mitochondrial products.
In vivo mitochondrial protein synthesis of wild-type W303-1A cells or cells carrying a null allele of cox14 were labeled with [35S]methionine at 30°C for 15 min in the presence of cycloheximide as described previously (1, 2). After synthesis, the cells were collected by centrifugation and submitted to a rapid cell wall digestion using 10 mg/ml zymolyase and subsequent lysis in the presence of 20 mM HEPES, pH 7.4, 0.5 mM PMSF, 1% digitonin, 1.2 mM MgCl2, and 150 mM KCl. The insoluble material was pelleted by centrifugation at 200,000 × g for 15 min, and the extract was loaded into a 7 to 20% sucrose gradient calibrated with Hb and LDH. After centrifugation at 28,000 rpm for 12 h in a Beckman T55i rotor, the gradient was collected in 14 fractions. Equivalent amounts of each fraction were separated by PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to an X-ray film.
Pulldown of newly synthesized mitochondrial products with Mss51-GST.
Mitochondria were prepared from an mss51 null mutant with a chromosomally integrated plasmid expressing the Mss51-GST fusion protein (2) and the same strain carrying an additional cox14 null allele. Mitochondria were labeled with [35S]methionine for 30 min, as described previously (2), and extracted with 1% digitonin, 20 mM HEPES, pH 7.4, 1.2 mM MgCl2, 150 mM KCl, and 0.5 mM PMSF. The extract was clarified by centrifugation at 50,000 × gav for 30 min and incubated with glutathione-Sepharose beads for 4 h at 4°C. After centrifugation at 1,500 rpm for 5 min, the supernatant was collected, and the beads were washed three times with PBS. Mitochondria corresponding to 20 μg of protein, equivalent volumes of the membrane pellet after digitonin extraction, and the supernatant from the glutathione-Sepharose beads were separated on a 17.5% polyacrylamide gel by SDS-PAGE. The amount of washed beads, however, corresponded to ∼500 μg of the starting mitochondria.
Mss51 solubility in isolated mitochondria.
A sample of mitochondria at 4 mg/ml was sonically irradiated and centrifuged at 50,000 × g for 30 min to separate the soluble and membrane proteins. The membrane pellet was suspended in buffer at a protein concentration of 1 mg/ml and added to a final concentration of 100 mM Na2CO3 (pH 11.3) and 50 mM EDTA. After 30 min on ice, the sample was centrifuged at 100,000 × g for 15 min at 4°C to separate the soluble membrane-extrinsic proteins from the insoluble membrane-intrinsic proteins. Equivalent volumes of each fraction were analyzed by Western blotting using antiserum against Mss51. Detection of ShyI and cytochrome b2 (Cyt b2) in the different fractions served to control the experiment.
Mss51 purification.
Mss51 fused to a thrombin-cleavable trigger factor tag (TF-Mss51) was purified by the GeneScript Corporation (Piscataway, NJ). Approximately 5 mg of purified tagged protein per liter of bacterial culture was obtained. The trigger factor (bacterial chaperone) tag was included to facilitate correct protein folding, thus enabling efficient soluble protein production of the Mss51 membrane protein.
Estimation of mitochondrial concentration of Mss51 and Ssc1.
To estimate the mitochondrial concentration of Mss51, a standard curve was obtained relating known amounts of the purified protein Mss51 fused to a trigger factor tag (TF-Mss51) to the signal detected by Western blot analysis. The concentration of Mss51p in wild-type mitochondria was calculated upon correction for the additional molecular weight of the trigger factor and expressed in pmol of Mss51 per milligram of mitochondrial protein. Similarly, the mitochondrial concentration of Ssc1 was estimated by using purified Ssc1 provided by A. Azem (George Wise Faculty of Sciences, Department of Biochemistry, Tel Aviv University, Ramat Aviv, Israel).
Mss51-Ssc1 binding assays.
Equimolar amounts of purified TF-Mss51 and purified Ssc1 were mixed in the presence or absence of 1 mM ATP-Mg or ADP-Mg in a buffer containing 20 mM HEPES, pH 7.4, 150 mM KCl, and 0.5 mM PMSF. Following 30 min of incubation at ambient room temperature, the mix was supplemented with prewashed Talon Cobalt-Sepharose beads (Clontech) and incubated in a rotator for 1 h at 4°C. TF-Mss51 was bound to the beads by virtue of a His tag present in the trigger factor N terminus. After binding, the Sepharose resin was washed twice with cold PBS. The bound material was subsequently eluted from the beads by thrombin digestion at a site located between the TF tag and Mss51. The eluted material was analyzed by Western blotting. A sample containing Ssc1 but no TF-Mss51 was used as a control and eluted by addition of Laemmli sample buffer. To analyze the effect of an Ssc1 substrate peptide on the Ssc1-Mss51 interaction in vitro, we used a peptide consisting of a portion of the matrix-targeting sequence of chicken aspartate aminotransferase (CALLLSAPRR) as described previously (28).
Mitochondrial respiratory chain enzyme spectrophotometric measurements.
Mitochondria were prepared from strains grown in medium containing 2% galactose, according to the method of Faye et al. (9) except that zymolyase 20T (ICN Biochemicals Inc., Aurora, OH) instead of glusulase was used for the conversion of cells to spheroplasts. Mitochondria prepared from the different strains were used for spectrophotometric assays carried out at 24°C. KCN-sensitive COX activity was assayed with 50 μg of mitochondria which were permeabilized with potassium deoxycholate, as described previously (1), by following the oxidation of 50 μM reduced cytochrome c at 550 nm in a medium containing 20 mM KH2PO4 (pH 7.4). The addition of 0.3 mM KCN inhibited the reaction. Antimycin A-sensitive NADH cytochrome c reductase activity was assayed in 25 μg of mitochondria permeabilized with potassium, deoxycholate as described previously (1), by measuring at 550 nm the reduction of oxidized 50 μM cytochrome c using 0.4 mM NADH as the electron donor in a medium containing 20 mM KH2PO4 (pH 7.4) and 2 mM EDTA. The addition of 0.4 μM antimycin A inhibited the reaction.
Mitochondrial cytochrome spectra.
Mitochondria were prepared from the wild-type strain W303-1A and the Δmss51 and the ssc1.2 mutant strains growing in the presence of galactose at the permissive temperature of 23°C. Isolated mitochondria were extracted at a protein concentration of 5 mg/ml in 20 mM Tris-HCl, pH 7.5, 1 M KCl, 1% potassium deoxycholate, conditions that quantitatively solubilize all mitochondrial cytochromes (50). Samples of the extract were either oxidized with ferricyanide or reduced with sodium dithionite, and the difference spectra were measured at room temperature using a UV-2401PC Shimadzu spectrophotometer.
Miscellaneous procedures.
Standard procedures were used for the preparation and ligation of DNA fragments and for transformation and recovery of plasmid DNA from Escherichia coli (44). Yeast cells were transformed by the method of Schiestl and Gietz (46). The one-step gene insertion method (42) was used to integrate linear plasmids at the URA3 or LEU2 locus of yeast nuclear DNA. Protein concentration was measured with Folin's phenol reagent (29). Proteins were separated by SDS-PAGE in the buffer system of Laemmli (26), and Western blots were treated with antibodies against the appropriate proteins, followed by a second reaction with anti-mouse or anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Sigma, St. Louis, MO). A SuperSignal chemiluminescent substrate kit (Pierce, Rockford, IL) was used for the final detection.
Statistical analysis.
All experiments (sucrose gradients, enzymatic assays, and Western blot quantifications) were done at least in triplicate using independent mitochondrial preparations. For the enzymatic assays, data are presented as means ± standard deviations (SD) of absolute values. The values obtained for wild-type and mutant strains were compared by a Student t test, and a P value of <0.05 was considered significant. For quantification of Western blot signals, the images were digitalized, and densitometry was performed using the histogram function of the Adobe Photoshop program. The values measured in three independent assays did not differ by more than 5%.
RESULTS
Extraction of high-molecular-weight complexes containing Mss51.
To better understand the functions of Mss51, we have investigated its native MW and identified its interacting partners in wild-type cells and a collection of strains carrying mutations in COX assembly genes.
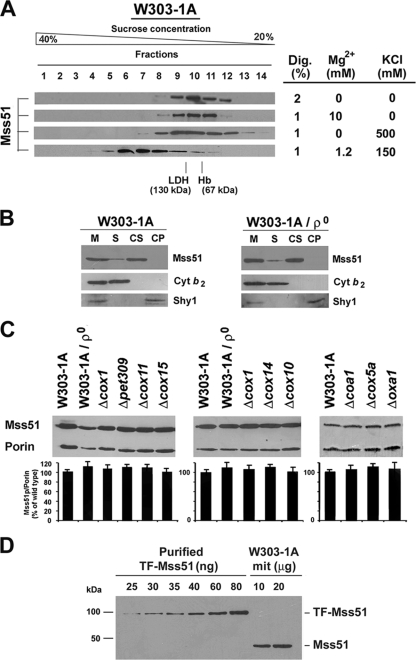
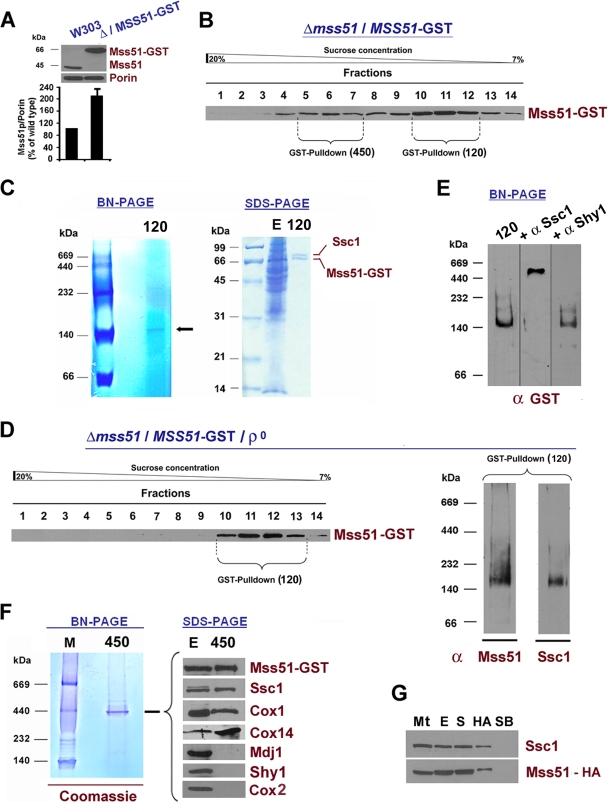
The reported native MW of Mss51 in wild-type mitochondria varies depending on the extraction conditions (2, 40). To standardize these conditions, we tested several KCl, MgCl2, and digitonin concentrations. When the mitochondrial extract was prepared with a digitonin concentration of up to 1% in the presence of 150 mM KCl and 1.2 mM MgCl2 and loaded in a linear 20 to 40% sucrose gradient, most Mss51 was detected in a peak of approximately 450 kDa, which was slightly asymmetrical, suggesting the existence of higher-MW material (Fig. 1A). Under these conditions, a very small portion (less than 5%) of total Mss51 was extracted as a complex of approximately 120 kDa (Fig. 1A). Increasing the concentration of KCl to 0.5 M, MgCl2 to 10 mM, or detergent to 2% resulted in disruption of the 450-kDa complex, and most Mss51 was found in the 120-kDa complex (Fig. 1A). In light of these results, we decided to perform all our experiments, except as otherwise indicated, using 1% digitonin mitochondrial extracts prepared in the presence of potassium (150 mM KCl) and magnesium (1.2 mM MgCl2) close to physiological concentrations.
FIG. 1.
Effect of Cox1 synthesis, maturation, and stability on Mss51 steady-state levels and native MW. (A) Sedimentation properties of Mss51 extracted from wild-type (W303-1A) mitochondria in a linear 20 to 40% sucrose gradient. Several extraction conditions (table at right) were tested. (B) Mss51 solubility in isolated W303-1A and [rho0] mitochondria. A sample of mitochondria (M) was sonicated and centrifuged to separate the soluble (S) and membrane proteins. The membrane pellet was extracted with Na2CO3 (pH 11.3) and centrifuged to separate the soluble membrane-extrinsic (CS) from the insoluble membrane-intrinsic proteins (CP). Equivalent volumes of each fraction were analyzed by Western blotting using antiserum against Mss51, Shy1, and Cyt b2. (C) Western blot analyses of Mss51 steady-state concentrations in wild-type cells, cells devoid of mtDNA [rho0], or cells carrying null alleles of the indicated COX assembly genes (Table 1). An antibody against porin was used to normalize the signals for protein loading. Quantification of the signals is shown in the lower panel. The bars indicate the mean ± SD from at least three independent sets of measurements. (D) Concentration of Mss51p in wild-type mitochondria. Western blots of the indicated amounts of W303-1A mitochondria (mit) and purified TF-Mss51p. The density of the signals was used to estimate the concentration of Mss51p in the mitochondrial samples.
The mitochondrial membrane association and steady-state levels of Mss51 are independent of both Cox1 synthesis and the status of COX assembly.
Mss51 physically interacts with Cox1 and several COX assembly factors. We have tested whether Mss51 loses its association with the mitochondrial inner membrane and becomes less stable in the absence of its interacting partners. In wild-type mitochondria, Mss51 is loosely associated with the matrix side of the inner membrane and remains associated with the membrane after brief sonication but is solubilized with alkaline carbonate (2) (Fig. 1B). Mss51 was extracted from [rho0] (Fig. 1B), cox15, and cox14 mitochondria (data not shown) under similar conditions, suggesting that in all cases it is a peripheral inner membrane protein. The soluble Cyt b2 and the membrane-intrinsic protein Shy1 were monitored as controls (Fig. 1B).
The steady-state level of Mss51 in cells devoid of mitochondrial DNA ([rho0] cells) and hence of COX1 mRNA and Cox1 protein was equivalent to that of wild-type cells (Fig. 1C). Similarly, cells with null mutations in cox1, pet309, and cox14 (described above); cox10 and cox15 (involved in heme A biosynthesis); cox11 (involved in copper delivery to Cox1); oxa1 (necessary for Cox1 membrane insertion); and cox5a (a COX subunit that interacts with Cox1 early in the assembly process) accumulate wild-type levels of Mss51 (Fig. 1C). To estimate the mitochondrial concentration of Mss51, a standard curve was obtained relating known amounts of TF-Mss51 (purified Mss51 fused to a trigger factor tag) to the signal detected by Western blot analysis (Fig. 1D). Upon correction for the additional trigger factor molecular weight, the concentration of Mss51 in wild-type mitochondria was calculated to be approximately 2.5 μg (55.5 pmol) of Mss51 per mg of mitochondrial protein. This value is in the range of other COX assembly factors, such as Sco1 (27 pmol/mg protein) (3), and significantly lower than the ∼275 pmol/mg protein that we estimated for the mtHsp70 chaperone Ssc1 (data not shown).
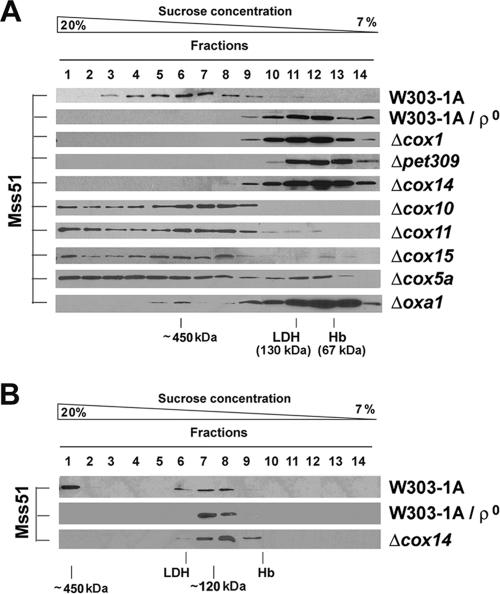
The native sizes of Mss51 depend on both COX1 mRNA translation and the status of COX assembly.
The native MW of Mss51 in wild-type and mutant cells was estimated by sedimentation of mitochondrial extracts in a 7 to 20% sucrose gradient to better appreciate differences in the 120- to 450-kDa area. In wild-type cells, the pattern of Mss51 distribution was as mentioned above (Fig. 2A). In the absence of COX1 mRNA and/or Cox1 ([rho0], Δpet309, and Δcox1 cells), all Mss51 was detected in the 120-kDa complex (Fig. 2A). Similarly, in Δcox14 cells in which Cox1 is normally synthesized but is unstable, Mss51 is detected almost exclusively in the 120-kDa complex as reported (40). The same result was obtained for Δoxa1 cells (Fig. 2A) in which newly synthesized Cox1 is mostly degraded upon failing to be inserted into the membrane (2). The small proportion of 450-kDa complex formed in the absence of Oxa1 is taken to represent the small portion of newly synthesized Cox1 that is inserted into the membrane under these conditions. The pattern of similar extracts in an extended 7 to 20% sucrose gradient showed that in these strains no Mss51 is detected as a 45-kDa monomer but is always complexed with other protein(s) in a 120-kDa complex (Fig. 2B).
FIG. 2.
Effect of Cox1 synthesis, maturation, and stability on Mss51 native MW. (A) Sedimentation properties of Mss51 extracted from mitochondria isolated from the indicated strains in a linear 7 to 20% sucrose gradient (A) or an extended linear 7 to 20% sucrose gradient (B) analyzed as explained under Materials and Methods.
In Δcox10, Δcox15, and Δcox11 cells in which Cox1 cannot be matured by the addition of either heme A or copper prosthetic groups, Cox1 synthesis is downregulated in the absence of COX assembly (2), and Cox1 steady-state levels are 10 to 15% of the wild type (2). In these mutants, Mss51 was detected in the 450-kDa peak, which was consistently found to be highly asymmetrical (Fig. 2A), suggesting the existence of abundant higher-MW complexes. In Δcox5a cells, Cox1 synthesis is also downregulated (2) although 15% of COX activity is retained because the enzyme contains the isoform Cox5b. Mss51 extracted from this strain sedimented similarly to Mss51 extracted from mutants defective in Cox1 maturation although more material was detected in the 120-kDa peak (Fig. 2A).
We conclude that Mss51 is a highly stable protein, but its native size depends on translation and stability of Cox1 as well as on the status of COX assembly.
The 120-kDa Mss51-containing complex is involved in COX1 mRNA translation.
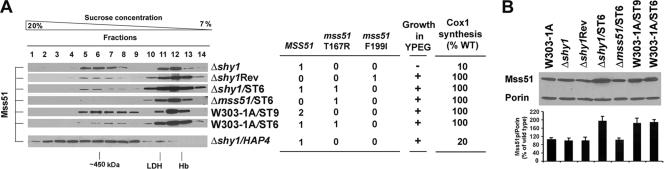
We previously reported that a single copy of the mss51F199I and mss51T167R alleles or an additional copy of wild-type MSS51 partially suppresses the respiratory defect of shy1 mutants by increasing Cox1 translation (1). In most COX-deficient strains, mutant or additional copies of MSS51 do not suppress the COX assembly defect but significantly increase Cox1 synthesis (2). These data suggested that defective COX assembly may inactivate or reduce the effective concentration of Mss51 as a COX1 mRNA translational activator, thus downregulating Cox1 synthesis. The distribution of Mss51 in sucrose gradients loaded with Δshy1 mitochondrial extracts was different from all COX mutants reported in Fig. 2A, with equal amounts in the 450- and the 120-kDa complexes and no high-MW tail (Fig. 3A). Newly synthesized Cox1 could be particularly susceptible to proteolytic degradation in the absence of Shy1. Alternatively, the higher-MW complexes could be more unstable in this strain, thus inducing an accumulation of Mss51 in the 120-kDa complex. In contrast, in the shy1 revertant strain carrying one copy of mss51F199I, in an mss51 null mutant carrying one copy of mss51T167R, and in a shy1 mutant carrying an extra mss51T167R allele, most Mss51 was detected in the 120-kDa complex (Fig. 3A). The steady-state level of Mss51 in shy1 mutants and revertants was similar to that of wild-type cells while it was doubled in shy1 mutant and wild-type cells carrying a second MSS51 allele integrated into their genomes (Fig. 3B). Similar to shy1 revertants, most Mss51 accumulates in the 120-kDa complex in a wild-type strain expressing a second copy of MSS51, particularly when the second copy is the mss51T167R allele (Fig. 3A). Because these strains synthesize Cox1 efficiently and have a respiratory ability indistinguishable from wild-type cells carrying a single copy of MSS51, our results support the possibility that the 120-kDa Mss51 complex is either competent to support COX1 mRNA translation or is the source of Mss51 required for such a function. Our data also provide information about the suppression mechanism of shy1 mutants by MSS51. The suppressor mss51F199I and mss51T167R alleles code for proteins that fully support translation but probably have a lowered affinity for Cox1 or other components of the 450-kDa complex and thus accumulate in the 120-kDa complex. Similarly, an excess of Mss51 in a wild-type strain results in accumulation of the 120-kDa complex, suggesting that the amount of Mss51 in the 450-kDa complex is tightly titrated by other components of the complex, probably by Cox1 levels. In summary, our results strongly suggest that shy1 suppression is due to the increased ability of Mss51 mutant proteins to be released from the 450-kDa complex. They presumably become more readily available for COX1 mRNA translation, an effect that is similarly produced by increasing the concentration of Mss51. Overexpression of HAP4, the catalytic subunit of the nuclear transcriptional activator HAP complex, also suppresses shy1 mutations (10). The suppression mechanism involved an increase in the amount of Cox1-interacting nuclear-encoded COX subunits and was additive to the MSS51 effect (10). In the overexpressor strain, Mss51 accumulates in the 450-kDa complex and extensively in the heavier fractions (Fig. 3A), further supporting a mechanism of suppression different from that of MSS51.
FIG. 3.
Quantitative and qualitative changes in Mss51 affect its native MW. (A) Sedimentation properties of Mss51 extracted from mitochondria isolated from the indicated strains in a linear 7 to 20% sucrose gradient. ST9 and ST6 are integrative plasmids expressing MSS51 and mss51T167R, respectively. (B) Steady-state levels of Mss51 in the strains used in panel A estimated by Western blot analysis as described in the legend of Fig. 1A. The bars indicate the mean ± SD from at least three independent sets of measurements. WT, wild type.
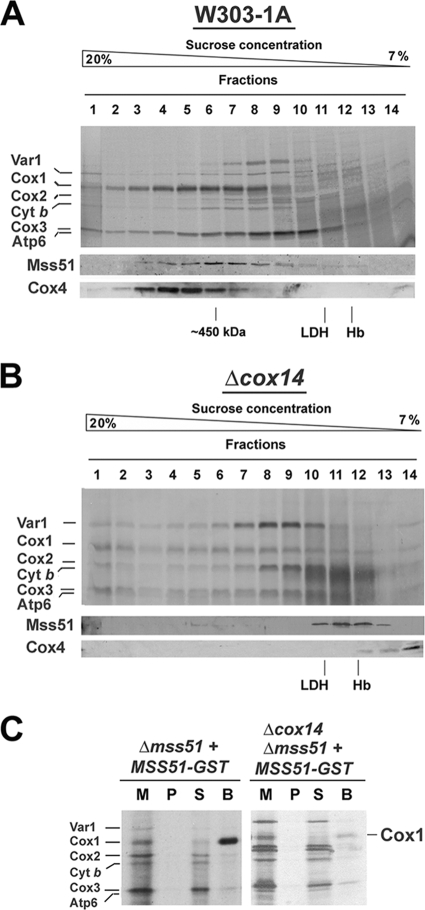
Mss51 interacts with newly synthesized Cox1 in wild-type cells and in COX assembly mutants.
Mss51 interacts with newly synthesized Cox1. We asked whether this interaction occurs within the 450-kDa complex and how it is affected in the absence of Cox1 stability in a cox14 null mutant. In vivo 35S labeling of mitochondrial translation products followed by their sedimentation in a sucrose gradient revealed that in the two strains, all mitochondrial products appear in high-MW complexes (Fig. 4A and B). Specifically, Cox1 is distributed in wild-type cells around a 450-kDa peak, cosedimenting with Mss51 (Fig. 4A). However, the Cox1 distribution is highly asymmetric, with a significant amount of material in the heavy fractions forming an extensive tail (Fig. 4A). In the absence of Cox14, Cox1 is normally synthesized and distributed although only traces of Mss51 are detected in the heavy fractions, and it mostly accumulates in the 120-kDa peak (Fig. 4B). This suggests that the Mss51-Cox1 interaction occurs but is highly unstable in the absence of Cox14.
FIG. 4.
Mss51 interacts with newly synthesized Cox1 prior to its maturation and stabilization in the membrane. Sedimentation properties in a 7 to 20% sucrose gradient of 35S-labeled newly synthesized mitochondrial proteins from wild-type W303-1A (WT) (A) and Δcox14 cells (B). The mitochondrial translation products are identified on the left. In the lower panel, the same samples were used for Western blot analyses using antibodies against Mss51 and Cox4. (C) Pulldown of 35S-labeled newly synthesized Cox1 using extracts from Δmss51 and Δmss51 Δcox14 strains expressing Mss51-GST. Mitochondria (M) (20 μg) and equivalent volumes of the membrane pellet after digitonin extraction (P) and of the bead-unbound material (S) were separated by SDS-PAGE and processed as described for panel A. The amount of washed beads (B) corresponded to ∼500 μg of the starting mitochondria.
The interaction of Mss51 with newly synthesized Cox1 in the absence of Cox14 was also tested in Δmss51 and Δcox14 Δmss51 strains expressing fully functional Mss51 fused to GST (2). Mitochondria were labeled in organello with [35S]methionine, extracted with digitonin, and adsorbed onto glutathione-Sepharose beads. The proteins that were recovered from the beads indicated a selective enrichment of labeled Cox1 in the wild-type strain (Fig. 4C) as reported previously (2). A significant but poorer enrichment of Cox1 was detected in the absence of Cox14 (Fig. 4C), probably due to instability of the Mss51-Cox1 interaction.
The mitochondrial Hsp70 chaperone Ssc1 is present in both the 450-kDa and 120-kDa Mss51-containing complexes.
To identify the functional partners of Mss51 and to gain insights into how it transitions through the several functions in which it is involved, we purified both the 450-kDa and the 120-kDa complexes. For this purpose, we used mitochondria isolated from a Δmss51 strain expressing Mss51-GST (2). In this strain, the steady-state level of Mss51 expressed from an integrated plasmid is double that in a wild-type strain (Fig. 5A). As explained above, the excess of Mss51 favors the accumulation of the protein in the 120-kDa complex. For this reason, Mss51-GST sediments in a sucrose gradient in the two major peaks of 450 and 120 kDa (Fig. 5B). The sizes of these complexes are slightly larger than in a wild-type strain, as expected for the inclusion of the large GST tag on Mss51.
FIG. 5.
The mtHsp70 chaperone Ssc1 is present with Mss51 in both a 450-kDa and a 120-kDa complex. (A) Quantification of Mss51 in a Δmss51 strain expressing Mss51-GST from an integrative plasmid, performed as explained in the legend of Fig. 1C. (B) Sedimentation properties of Mss51-GST extracted from Δmss51/MSS51-GST mitochondria in a linear 7 to 20% sucrose gradient. The indicated fractions were pooled and submitted to GST pulldown. (C) GST pulldown analysis of pooled fractions 10 to 12, containing a 120-kDa complex. The pulled-down material was separated by blue native (BN) or SDS denaturing PAGE and Coomassie blue stained. Gel sections containing the bands detected in both types of gels were excised and submitted to MS analysis. The identified proteins are labeled in the margins. E corresponds to total extract. (D) Sedimentation properties of Mss51-GST extracted under native conditions from mitochondria isolated from the same strain as in panel B but devoid of mitochondrial DNA. The indicated fractions (10 to 13) were pooled and submitted to GST pulldown. In the right-hand panel, the pulled-down material was separated by BN-PAGE, processed for Western blot analyses, and probed with antibodies against Mss51 and Ssc1. (E) The 120-kDa complex in panel B was incubated with or without an excess of a rabbit polyclonal anti-Ssc1 antibody for 2 h. An antibody against Shy1 was used as a negative control. The samples were separated by BN-PAGE, and Western blots were probed with a mouse monoclonal anti-GST antibody. (F) GST pulldown of pooled fractions 5 to 7, containing a 450-kDa complex were separated by BN-PAGE and Coomassie blue stained (left). The 450-kDa complex was also separated by SDS-PAGE and processed for Western blot analyses using antibodies against the indicated proteins. E corresponds to total extract. (G) Mitochondria (Mt) isolated from a Δmss51 strain expressing MSS51-HA were extracted, and the extract (E) was used for immunoprecipitation assays using anti HA-Sepharose-4B. The unbound material in the supernatant (S) and the material bound to the beads (HA) were separated by centrifugation and analyzed by Western blotting using antibodies against Ssc1 and Mss51. Plain beads (SB) were used to control for unspecific binding.
The three main fractions that were part of the 120-kDa peak (fractions 10 to 12) were pooled and used for a large-scale GST pulldown assay. The material adsorbed to the GST beads was run in a blue native gel, and an approximately 120-kDa complex was detected (Fig. 5C, left). The material was also separated in an SDS gel and stained with Coomassie blue (Fig. 5C, right). Two bands were detected, cut, and submitted to MS analyses by Midwest BioServices (Overland Park, KS). The bands were identified as Mss51 (with GST) and Ssc1 (Fig. 5C). The same results were obtained by analyzing the Mss51-containing 120-kDa complex in a strain devoid of mitochondrial DNA (Δmss51/MSS51-GST-[rho0]) (Fig. 5D). Western blot analyses failed to detect other proteins in this complex including Cox1 and Cox14 (data not shown). The Mss51-Ssc1 interaction was confirmed by a gel retardation assay after incubation of mitochondrial extracts with an excess of anti-Ssc1 antibody (Fig. 5E), in which an anti-Shy1 antibody was used as a negative control (Fig. 5E). Ssc1 is the more abundant mtHsp70 chaperone. The existence of this complex suggests cooperation between its two components to support Cox1 biogenesis.
Subsequently, the 450-kDa complex was also pulled down from fractions 5 to 7 of the gradient shown in Fig. 5B, separated in a blue native gel, and stained with Coomassie blue (Fig. 5F, left). The Coomassie-stained 450-kDa band was excised and analyzed by MS, which showed the consistent presence in three independent attempts of Mss51, Cox1, Cox14, and Ssc1 (data not shown). Shy1 and Coa1 were not detected in these complexes. The MS results also provided some hits for other proteins including ATPase subunits α and β, the hsp60 chaperone, and Cox2, although not consistently. To confirm the MS results, the pulled down 450-kDa complex was separated by SDS-PAGE and used for Western blot analyses. Mss51, Ssc1, Cox1, and Cox14 were detected while we failed to detect Cox2, Shy1 and Mdj1 (Fig. 5F, right). Additionally, although the pulldown efficiency was low, the interaction of Mss51 with Ssc1 was also detected in a Δmss51 strain carrying a chromosomally integrated MSS51-hemagglutinin (HA) gene (Fig. 5G) in which the Mss51 sedimentation pattern is similar to that of a wild-type strain (data not shown).
Mutations in Ssc1 affect Mss51 native MW and significantly reduce the efficiency of COX biogenesis.
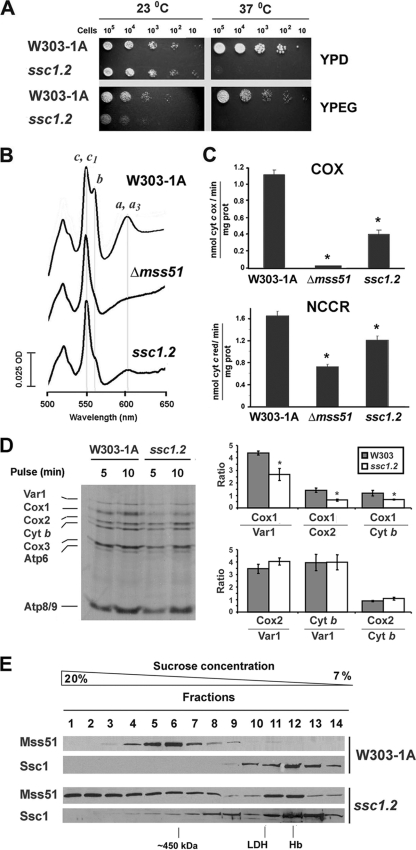
SSC1 is a gene essential for vegetative growth (6). To further explore the role of Ssc1 in Mss51 metabolism, we have analyzed Cox1 synthesis and Mss51 native MW in the ssc1.2 mutant strain (21) which carries a temperature-sensitive allele of SSC1. At 37°C ssc1.2 cells are defective in the import and folding of precursor forms of mitochondrial proteins affecting cell growth (21) (Fig. 6A). We have now observed that although at the permissive temperature (23°C) ssc1.2 cells grow in fermentable carbon sources at a wild-type rate, as reported previously (21), they grow poorly in respiratory medium (YPEG) (Fig. 6A). The respiratory defect at 23°C could not be accounted for by a loss of mtDNA because we used freshly purified cultures virtually free of [rho0] cells. Analyses of total mitochondrial cytochromes in this strain showed a mild decrease in cytochrome b, part of the bc1 complex, and more marked reduction of heme a+a3, the prosthetic groups of COX (Fig. 6B). Consistent with these observations, NADH cytochrome c reductase activity, which measures the activity of the first portion of the mitochondrial respiratory chain, was mildly reduced to ∼80% while COX activity was lowered to ∼35% of wild-type values (Fig. 6C).
FIG. 6.
Mutations in SSC1 alter COX biogenesis and Mss51 sedimentation properties. (A) Wild-type strain W303-1A and a strain carrying the temperature sensitive mutant allele ssc1.2 were grown overnight in liquid YPD medium at the permissive temperature of 23°C. Tenfold serial dilutions of the two strains were plated on solid YPD or YPEG medium and incubated at both 23°C and the nonpermissive temperature of 37°C. Pictures were taken after 3 days of incubation. (B) Total mitochondrial cytochrome spectra from mitochondria isolated from the wild-type strain, a null mutant Δmss51, and the ssc1.2 mutant cells grown at 23°C. (C) COX and NADH cytochrome c reductase (NCCR) were measured spectrophotometrically in mitochondria isolated from cells grown at 23°C. (D) In vivo mitochondrial protein synthesis in W303-1A and ssc1.2 cells performed at 23°C. Newly synthesized mitochondrial proteins are identified on the margin. In the right panel, the signals were quantified by densitometry and plotted at the indicated ratios. The bars indicate the mean ± SD from three independent sets of measurements. (E) Sedimentation properties of Mss51 and Ssc1 from wild-type and ssc1.2 mitochondria isolated from cells grown at 23°C in a linear 7 to 20% sucrose gradient. The samples were processed as described in the legend of Fig. 1A.
We investigated whether mitochondrial protein synthesis could be affected in the ssc1.2 mutant grown at 23°C. Consistently, ssc1.2 cells were able to perform mitochondrial protein synthesis normally albeit at a slightly lower rate than wild-type cells (Fig. 6D). The synthesis alteration particularly affected Cox1 (Fig. 6D). This could be a direct effect or, as in bona fide COX mutants, a reflection of Cox1 translational downregulation secondary to the COX defect observed in ssc1.2 cells.
To explore the possibility that the presence of Ssc1 in Mss51-containing complexes could be disrupted in the ssc1.2 mutant, we analyzed the sedimentation properties of both proteins in wild-type and mutant mitochondria extracted from cells grown at 23°C. In wild-type cells, most Ssc1 is part of a 100-kDa complex (Fig. 6E), and only traces were detected in higher-MW complexes when the film was overexposed (data not shown), while most Mss51 was detected in the 450-kDa complex (Fig. 6E). Contrary to wild-type cells, ssc1.2 cells showed an Mss51 sedimentation pattern similar to COX assembly mutants defective in Cox1 maturation or assembly. Mss51 distributed around the 450-kDa peak and around the 120-kDa peak and was mainly detected in the heavy-MW fractions as a large tail of the 450-kDa complex (Fig. 6E). The distribution pattern of Ssc1 in the ssc1.2 mutant was also modified from wild-type cells. At the permissive temperature, a considerable portion of Ssc1 was detected in the 100-kDa complex, but a significant amount was detected in heavier-MW fractions (Fig. 6E). We conclude that the ssc1-2 mutation affects Mss51 native MW, thus supporting the requirement of the mtHsp70 chaperone for proper Mss51 function and normal efficiency of COX biogenesis.
Mss51 and Ssc1 interact with the mitochondrial translational apparatus together with Cox14 and the cochaperone Mdj1.
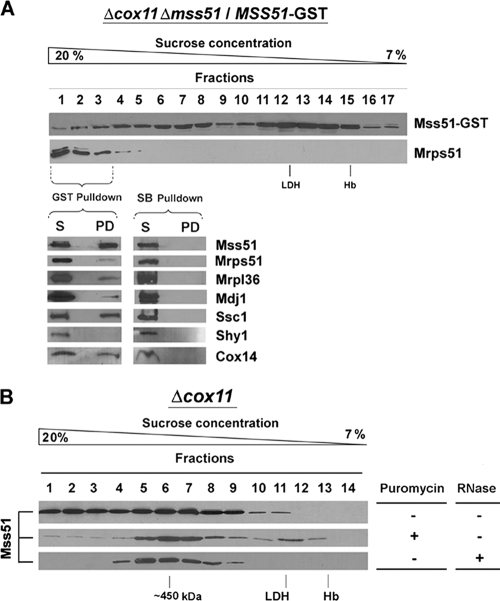
Mss51 interacts with both the COX1 mRNA and with the newly synthesized Cox1 (2, 39). We hypothesized that it could perform its functions by transiently interacting with the COXI mRNA translational machinery. The Mss51 material in the high-MW tail detected in sucrose gradients is particularly prominent in strains deficient in Cox1 maturation and/or assembly, such as Δcox11 (Fig. 2A). In these strains, Mss51 could be trapped with the COX1 mRNA translational machinery with other Cox1 biogenetic factors, including Ssc1. To test these possibilities, we searched for Mss51-interacting partners in complex(es) heavier than 450 kDa using the double Δcox11 Δmss51 mutant expressing GST-tagged Mss51. In this strain, the steady-state level of Mss51 is twice that of a Δcox11 strain (data not shown), and an excess of the protein accumulated in the 120-kDa complex (Fig. 7A), as mentioned earlier. Otherwise, Mss51-GST distributed on a 7 to 20% gradient with a pattern relatively similar to that of Mss51 extracted from Δcox11 cells (Fig. 2A and 7A), with an extensive amount of material in the heaviest fractions. Mrps51, a component of the small ribosomal subunit, cosedimented in these fractions (Fig. 7A). The three heaviest fractions were pooled and used in a GST pulldown assay. The efficiency of the assay was approximately 60% because ∼40% of Mss51-GST was detected in the supernatant corresponding to the unbound material (Fig. 7A, lower panel). Proteins from the small (Mrps51) and large (Mrpl36) ribosomal subunits were detected in the pulldown material (Fig. 7A, lower panel). A significant amount of Cox14 and Ssc1 was also pulled down at a proportion close to that of Mss51 (Fig. 7A). A small portion of Mdj1 was also consistently pulled down. Mdj1 is a mitochondrial DnaJ type chaperone that cooperates with Ssc1 in its folding functions (53). Consistent with our results, Mdj1 and Ssc1 were previously reported to form a complex with nascent mitochondrial polypeptide chains, probably to exert a chaperone function during ongoing translation and thus control the productive folding of the newly synthesized mitochondrial proteins (53). Shy1, which is part of large complexes and interacts with Cox1-containing subassemblies downstream from the roles of Mss51 in COX biogenesis (2, 34), was detected exclusively in the supernatant but not in the pulldown material (Fig. 7A). None of the analyzed proteins, including Mss51, were detected bound to non-GST-conjugated Sepharose beads (Fig. 7A).
FIG. 7.
Mss51 interacts with the mitochondrial translational apparatus. (A) Sedimentation properties in a linear 7 to 20% sucrose gradient of Mss51-GST and a protein from the small ribosomal subunit (Mrps51) extracted from mitochondria isolated from the Δcox11 Δmss51 strain expressing Mss51-GST. The first three fractions of the gradient were pooled and used for GST pulldown. Plain Sepharose beads (SB) were used as a control. The lower panel shows a Western blot analysis of the pulldown material (PD) and the unbound material in the supernatant (S). The blots were probed with antibodies against the indicated proteins. (B) Sedimentation properties of Mss51 extracted after a 5-min treatment with 50 μg/ml puromycin or in the presence of 600 U/ml RNase A from Δcox11 mitochondria.
The nature of the interaction of Mss51 and the other partner proteins with the mitochondrial ribosomes was further characterized in a Δcox11 strain expressing nontagged Mss51. The accumulation of Mss51 in the heavier fractions was sensitive to both 50 μg/ml puromycin, which induces the release of unfinished polypeptide chains, and to high RNase concentrations (600 U/ml), which fully disrupt the integrity of the ribosomes (38). Under these conditions most Mss51 from Δcox11 mitochondria accumulated in the 450-kDa complex (Fig. 7B), suggesting that the interaction of Mss51-Ssc1-Cox14 with the translational apparatus occurs before newly synthesized Cox1 is released from the ribosome and that such an interaction possibly occurs through the nascent polypeptide.
The 120-kDa and 450-kDa complexes remain stable in the presence of ATP.
The various functions performed by Ssc1, as in all other chaperones of the Hsp70 family, rely on their ability to bind to unfolded segments of proteins in an ATP-dependent, reversible manner.
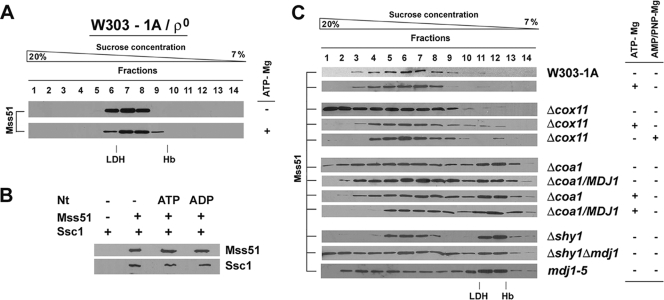
In order to study whether the interaction of Mss51 with Ssc1 in the 120-kDa complex was dependent on ATP binding and/or hydrolysis, we explored the effect of ATP addition and depletion on the Mss51 native MW in a [rho0] strain. To this end, we prepared both mitochondrial extracts in buffer containing 1 mM to 5 mM ATP and equimolar Mg2+ concentrations and extracts from mitochondria treated with 10 U/ml apyrase to deplete ATP in the matrix (Fig. 8A). In all cases, the sedimentation patterns of Mss51 were similar (Fig. 8A).
FIG. 8.
Nature of binding of Ssc1 to its partners in the Mss51-containing complexes. (A) Sedimentation properties of Mss51 extracted in the presence or absence of 1.2 mM MgCl2 plus 1 mM ATP (ATP-Mg) from mitochondria isolated from a [rho0]strain. (B) In vitro binding of recombinant Ssc1 and Mss51. Ssc1 and His-TF-Mss51 were mixed and incubated in the presence or absence of the indicated nucleotides (Nt). The complex was purified with cobalt-Sepharose beads, followed by thrombin digestion to elute Mss51 and interacting Ssc1, as described in Materials and Methods. The proteins were detected by Western blotting using antibodies against Mss51 and Ssc1, respectively. (C) Sedimentation properties of Mss51 extracted in the presence or absence of 1 mM ATP-Mg or AMP-PNP-Mg from mitochondria isolated from wild-type, Δcox11, Δcoa1, Δshy1, Δshy1 Δmdj1, and mdj1-5 strains, overexpressing MDJ1 when indicated.
We further examined the nature of the direct binding between Mss51 and Ssc1 in vitro. Equimolar amounts of purified TF-Mss51 and Ssc1 proteins were mixed in the absence or presence of 1 mM Mg-ADP or Mg-ATP. Following purification of the Mss51-Ssc1 complex (see Materials and Methods), the interacting proteins were detected by Western blotting. Mss51 and Ssc1 were found to interact similarly under all the conditions tested (Fig. 8B). Moreover, the in vitro Mss51-Ssc1 interaction was not affected by the preincubation of Ssc1 with a peptide substrate (see Materials and Methods) in the presence of either ADP or ATP (data not shown). Our results suggest that Mss51 does not bind to the substrate binding cleft of Ssc1 and thus is not an Ssc1 client protein.
The nature of the direct or indirect interaction of Mss51 with Ssc1 in the 450-kDa complex was probed in wild-type mitochondrial extracts prepared in buffer containing 1 to 5 mM ATP and equimolar Mg concentrations. The presence of Mg-ATP did not change the Mss51 sedimentation pattern in sucrose gradients around the 450-kDa peak (Fig. 8C). These results suggest that under our experimental conditions, Ssc1 could bind to Mss51 or other components of the 450-kDa complex with high affinity, thus resulting in rapid rebinding even in the presence of ATP in the extract. Alternatively, the Ssc1 binding could involve a direct interaction with Mss51 as in the 120-kDa complex, through an Ssc1 domain other than the substrate binding domain.
The association of Mss51 with the translational apparatus is regulated by ATP binding and the action of Mdj1.
The presence of Mdj1 in the high-MW complexes containing Ssc1, Mss51, Cox14, and ribosomal subunits suggested that the interaction of these proteins with the COX1 mRNA translational apparatus could be modulated by ATP binding to Ssc1 and subsequent hydrolysis. Mdj1 is known to bind to the ATP-bound conformation of Ssc1 to activate its ATPase activity (53).
To test the effect of ATP on the association of Mss51 with the translational apparatus, we explored the sedimentation properties of Mss51 in Δcox11 mitochondrial extracts prepared in the presence or absence of 1 mM Mg-ATP. The presence of Mg-ATP resulted in a dramatic reduction of the higher-MW material (Fig. 8C). Similar results were obtained when ATP was replaced by the nonhydrolyzable ATP analog AMP-PNP (5′-adenylyl-beta, gamma-imidodiphosphate). Our results suggest that the presence of ATP and not its hydrolysis is sufficient to induce the release of the Ssc1/Mss51/Cox1/Cox14 complex from its interaction with the translational apparatus.
Recent results have independently linked Mdj1 to Cox1 biogenesis. MDJ1 was identified as a high-copy-number suppressor of Δcoa1 (40). We confirmed this result and additionally showed that MDJ1 overexpression failed to suppress the respiratory deficiency of shy1 and cox14 cells (data not shown). To gain an understanding of the mechanism of Δcoa1 suppression by MDJ1 overexpression, we explored the native MW of Mss51 in Δcoa1 cells and Δcoa1 cells overexpressing MDJ1. The sedimentation pattern in sucrose gradients of Mss51 extracted from Δcoa1 mitochondria did resemble the pattern of Δcox5a mutants (Fig. 8C). The Ssc1/Mss51/Cox1/Cox14 450-kDa complex is formed in the absence of Coa1, further supporting the absence of Coa1 in this complex. These results are apparently at variance with the previously reported interaction of Mss51/Coa1/Cox14 (34, 40). However, we cannot eliminate the possibility that transient interactions involving Coa1 could occur, for example, to facilitate some step of Cox1 maturation co- or posttranslationally.
Overexpression of MDJ1 in the Δcoa1 strain slightly changed the pattern of Mss51 distribution, reducing the amount accumulated in the heavier fractions (Fig. 8C), probably enough to provide Cox1 for downstream COX assembly steps and bypass, albeit poorly, the absence of Coa1. A similar result was observed when the Δcoa1 mitochondrial extracts were prepared in the presence of 1 mM Mg-ATP. This effect acted synergistically with MDJ1 overexpression (Fig. 8C).
To further confirm the involvement of Mdj1 in this process, we introduced a null mdj1 mutation in Δshy1 cells. The Mss51 sedimentation pattern in Δshy1 cells displays an approximately equivalent distribution between the 120- and 450-kDa complexes, with no detectable material accumulated in the heavier fractions (Fig. 3A and 8C). Deletion of mdj1 in this strain resulted in a significant accumulation of Mss51 in the heavy fractions. Similar results were obtained using the mdj1-5 mutant strain that carries a temperature-sensitive allele of mdj1 and grows poorly in respiratory medium even at the permissive temperature (53).
Taken together, these results suggest that the association of Mss51-Ssc1-Cox1-Cox14 with the translational apparatus is regulated by the presence of ATP and the chaperoning action of Mdj1.
DISCUSSION
Here, we have further characterized the players and mechanism underlying a translational autoregulatory system operating during the biogenesis of mitochondrial COX in the yeast S. cerevisiae. By virtue of this system, translation of COX1 mRNA on mitochondrial ribosomes is autoregulated by newly synthesized, unassembled Cox1 by trapping the specific COX1 mRNA translation factor Mss51 in high-MW complexes, thus restricting its availability for further translation. This system would allow coordination of Cox1 synthesis with its utilization during assembly. Here, the matrix mtHsp70 chaperone Ssc1 is shown to interact with several high-MW Mss51-containing complexes, which are proposed to serve in coordinating the translational regulation of COX biogenesis.
Ssc1 is required for mitochondrial protein import and folding and plays significant roles in both protein quality control and the prevention of heat-induced protein aggregation (52). Ssc1 also functions in the biosynthesis of mitochondrially synthesized proteins and the assembly of at least the ribosomal protein Var1 and ATPase subunits (19). Specific partner proteins or cochaperones regulate Ssc1 activity (52). Mge1 catalyzes nucleotide exchange, while Mdj1 increases the low intrinsic ATPase activity of Ssc1. Although only Mge1 assists Ssc1 in protein import, Mge1 and Mdj1 are required for efficient folding chaperone function (52). Additionally, Ssc1 and Mdj1 form a complex with nascent polypeptide chains on mitochondrial ribosomes, probably to facilitate their proper folding during translation (53). Our pulldown experiments of heavy-MW complexes containing the Cox1-specific chaperones Mss51 and Cox14, ribosomal subunits, and the two chaperones Ssc1 and Mdj1 are in agreement with these results and point toward cooperation of general and specific chaperones in the folding and stabilization of newly synthesized Cox1.
Beyond its interaction with nascent Cox1 polypeptides in the translational apparatus, Ssc1 remains bound to Mss51/Cox1/Cox14 in a 450-kDa complex in which Mdj1 was not detected. The 450-kDa complex, abundant in wild-type cells, could represent a Cox1-containing complex serving as a reservoir of stable Cox1 ready to be matured and/or to progress in the COX assembly process when required. At this stage, Cox1 synthesis is completed, and the protein is presumably inserted into the mitochondrial membranes with the assistance of the Oxa1 machinery (17). The accumulation of Mss51 in the 120-kDa complex in an oxa1 null mutant suggests that Cox1 insertion into the mitochondrial inner membrane is necessary for the proper formation of the 450-kDa complex. In this complex, Ssc1 could remain bound to Cox1 to maintain its proper folding during insertion or to present it to matrix-localized proteolytic systems in the case of unproductive folding that would prevent membrane insertion. Consistently, cells lacking the Mba1 ribosomal receptor or harboring a mutant Oxa1 variant, in which Cox1 is more exposed to the hydrophilic matrix milieu than wild-type cells, display a more prominent association of Cox1 and Ssc1 (38). The release of Ssc1/Mss51/Cox1/Cox14 from its complex with the translational machinery is promoted by the presence of ATP and does not seem to necessarily require ATP hydrolysis: it was equally promoted by ATP and AMP-PNP in Δcox11 mitochondria although it was significantly enhanced by Mdj1 overexpression in Δcoa1 cells. These results suggest that Mdj1 could act in this scenario as a chaperone rather than an enhancer of the Ssc1 chaperoning activity.
The role of Ssc1 in COX1 mRNA translational regulation that we are describing here constitutes a new Ssc1 task in mitochondrial biogenesis. The requirement of Ssc1 for proper Mss51 function is supported by the observation that mutations in Ssc1 affect Mss51 native MW and significantly reduce the efficiency of COX biogenesis. On the other hand, Ssc1 and Mss51 can also form a very stable binary complex. This complex accumulates in wild-type cells expressing either two copies of the wild-type MSS51 gene or fully functional mutant forms of MSS51 originally identified as shy1 suppressors (1, 2). In the absence of COX1 mRNA and/or Cox1, all Mss51 is bound to Ssc1 in the 120-kDa binary complex, which seems to be the reservoir of Mss51 when not engaged in its functions in Cox1 biogenesis. It may be the source of Mss51 that is competent for translation. Notably, a physical interaction of Mss51-Ssc1 had been reported in three affinity purification/MS high-throughput studies (5, 12, 24).
The study of the interactions involving Ssc1, Mss51, and other Cox1 biogenetic factors in cells carrying different lesions in the COX assembly process has provided an understanding of the functional significance of these interactions. The results obtained helped refine our model of Cox1 translational autoregulation coupled to the availability of its assembly partners during COX biogenesis. We envision an Mss51/Ssc1 cycle ruling the regulatory system. When assembly is limited by lack of Cox1 maturation or absence of assembly partners, Mss51 is sequestered not only in the 450-kDa complex (Fig. 9) but also in a significant amount in the high-MW translational complex (Fig. 9) The amount of Mss51 available for activation of translation by interacting with the COX1 5′ UTR would then be limited, as previously proposed (2, 39, 56). Our results do not eliminate the possibility that the interaction of Mss51 with Cox1 itself could be necessary for elongation of the nascent polypeptide and that in addition it could regulate Cox1 synthesis (39).
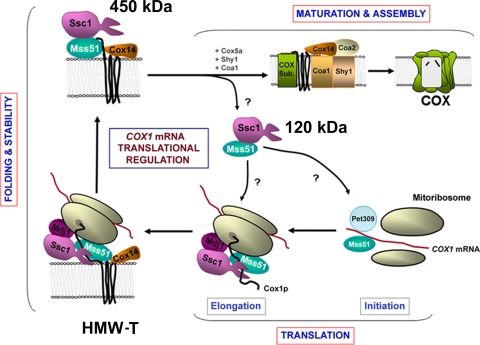
FIG. 9.
Model depicting the role of the mtHsp70 chaperone Ssc1 on translational regulation of COX biogenesis by interacting with Mss51 and Mss51-containing complexes. See explanation in the text. HMW-T, high-MW translational complex.
In our model depicted in Fig. 9, a small portion of the total amount of Mss51 is present in the 120-kDa binary complex with Ssc1 in wild-type cells. When required for COX assembly, Mss51 molecules contained in this pool would interact with the 5′ UTR of the COX1 mRNA to promote its translation (39, 56). During Cox1 synthesis, Mss51 would interact with the nascent polypeptide, probably promoting its elongation (39) together with the chaperones Ssc1 and Mdj1, which would ensure the proper folding of the nascent polypeptide. Cox14 is envisioned to be added to the high-MW translational complex at a later step because Cox1 is synthesized at wild-type rates in the Δcox14 mutant (2). Its incorporation to the complex would serve to stabilize the Ssc1/Mss51/Cox1/Cox14 complex. The release of Mss51 from this complex is possibly catalyzed by the incorporation of the COX assembly factors Shy1 and Coa1, thus allowing Cox1 to proceed to downstream events in the COX assembly process (1, 2, 34, 39, 40). In this way, Mss51 becomes available for new rounds of translation. It remains open whether Mss51 is released from the 450-kDa complex bound to Ssc1 or whether a new Ssc1 molecule binds Mss51 following its release.
At present, we do not know whether Mss51 plays a role in COX1 mRNA translational activation complexed to Ssc1. Using a yeast three-hybrid assay, we had shown that a portion of Mss51 corresponding to the 140 N-terminal amino acids of the mitochondrially imported protein had the ability to interact with the 5′ UTR of COX1 mRNA (56). This portion of Mss51 consists of a hydrophilic domain, which probably protrudes into the mitochondrial matrix where the interaction occurs in vivo. Ssc1 could interact with this portion of the protein to maintain it in a state competent for RNA binding. Similarly, we do not currently know whether the Mss51/Ssc1 complex is eventually disrupted and both proteins reunite with the translational apparatus to support Cox1 elongation and folding or if they remain complexed through the process. Further investigation, currently being implemented in the lab, is required to answer these questions.
Our current results, however, suggest that Mss51 does not interact with Ssc1 as one of its classical client proteins. Both extracted 120-kDa and 450-kDa complexes are stable in the presence of ATP. In vitro binding assays using recombinant proteins also demonstrated a direct Mss51-Ssc1 interaction whose efficiency was not significantly affected by the inclusion of ATP or ADP into the assay or by preincubation of Ssc1 with a substrate peptide. Because classical Ssc1-substrate complexes are unstable in the presence of ATP and because their formation is prevented by preincubation of Ssc1 with a substrate peptide, our results suggest that Mss51 does not interact with the Ssc1 substrate binding domain. Several mitochondrial proteins, including Mge1 (33) and Mdj1 (43), have been reported to interact with the ATPase domain of Ssc1 to act as cochaperones by modulating Ssc1 activity. The mitochondrial inner membrane protein Tim44, a component of the mitochondrial import machinery, also interacts with the ATP binding domain of Ssc1 in an ATP-regulated manner (41, 47) and together with Mge1 forms the protein translocation motor (48). The yeast mitochondrial endonuclease Endo.Sce1 also binds to the ATP binding domain of Ssc1, preferentially to the ADP-bound conformation, forming a heterodimer (35). Within this complex, Ssc1 was shown to be involved in modulating the enzymatic activity of the endonuclease (22, 35). The Ssc1-Mss51 interaction in the 120-kDa complex resembles the Endo.Sce1-Ssc1 binding in that neither case seems to involve the Ssc1 substrate binding domain. The precise role of Ssc1 in the Mss51 120-kDa complex remains to be characterized although our results suggest that it could be required to maintain Mss51 competent for translation, as discussed earlier.
In conclusion, the S. cerevisiae mitochondrial Hsp70 chaperone Ssc1 is involved in the COX assembly-controlled translational autoregulation of Cox1 by interacting with the COX1 mRNA translational factor Mss51 and high-MW complexes containing Mss51 and Cox1. The partnership of Ssc1 with Mss51 could mediate the coordination of the translational and posttranslational Mss51 functions. To date, chaperones of the Hsp70 family have not been involved in organellar translational autoregulatory processes. The key involvement of Ssc1 in the process described here represents a new level of sophistication among the organellar strategies for coordinating the synthesis of structural proteins with their utilization for assembly during the biogenesis of multimeric complexes.
Acknowledgments
We are in debt to M. Azem, E. Craig, T. Endo, T. D. Fox, H. Herrmann, J. Marszalek, C. Suzuki, A. Tzagoloff, and D. Winge for providing reagents.
This research was supported by NIH-RO1 grant GM071775A (to A.B.) and MDA Research Grant (to A.B.). I.S. is supported by NIH NRSA fellowship 1F31-GM081975, and D.H. is supported by AHA fellowship 0815083E.
Footnotes
Published ahead of print on 26 October 2009.
REFERENCES
- 1.Barrientos, A., D. Korr, and A. Tzagoloff. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 21:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beers, J., D. M. Glerum, and A. Tzagoloff. 2002. Purification and characterization of yeast Sco1p, a mitochondrial copper protein. J. Biol. Chem. 277:22185-22190. [DOI] [PubMed] [Google Scholar]
- 4.Chu, E., D. M. Koeller, J. L. Casey, J. C. Drake, B. A. Chabner, P. C. Elwood, S. Zinn, and C. J. Allegra. 1991. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U. S. A. 88:8977-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, S. R., P. Kemmeren, X. C. Zhao, J. F. Greenblatt, F. Spencer, F. C. Holstege, J. S. Weissman, and N. J. Krogan. 2007. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics 6:439-450. [DOI] [PubMed] [Google Scholar]
- 6.Craig, E. A., J. Kramer, and J. Kosic-Smithers. 1987. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc. Natl. Acad. Sci. U. S. A. 84:4156-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, E. A., J. Kramer, J. Shilling, M. Werner-Washburne, S. Holmes, J. Kosic-Smithers, and C. M. Nicolet. 1989. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol. Cell. Biol. 9:3000-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decoster, E., M. Simon, D. Hatat, and G. Faye. 1990. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 224:111-118. [DOI] [PubMed] [Google Scholar]
- 9.Faye, G., C. Kujawa, and H. Fukuhara. 1974. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J. Mol. Biol. 88:185-203. [DOI] [PubMed] [Google Scholar]
- 10.Fontanesi, F., C. Jin, A. Tzagoloff, and A. Barrientos. 2008. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 17:775-788. [DOI] [PubMed] [Google Scholar]
- 11.Fontanesi, F., I. C. Soto, D. Horn, and A. Barrientos. 2006. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291:C1129-C1147. [DOI] [PubMed] [Google Scholar]
- 12.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 13.Glerum, D. M., I. Muroff, C. Jin, and A. Tzagoloff. 1997. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 272:19088-19094. [DOI] [PubMed] [Google Scholar]
- 14.Glerum, D. M., and A. Tzagoloff. 1997. Submitochondrial distributions and stabilities of subunits 4, 5, and 6 of yeast cytochrome oxidase in assembly defective mutants. FEBS Lett. 412:410-414. [DOI] [PubMed] [Google Scholar]
- 15.Gold, L. 1988. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 57:199-233. [DOI] [PubMed] [Google Scholar]
- 16.Hell, K., J. Herrmann, E. Pratje, W. Neupert, and R. A. Stuart. 1997. Oxa1p mediates the export of the N and C termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 418:367-370. [DOI] [PubMed] [Google Scholar]
- 17.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell, K., A. Tzagoloff, W. Neupert, and R. A. Stuart. 2000. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275:4571-4578. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, J. M., R. A. Stuart, E. A. Craig, and W. Neupert. 1994. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 127:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershey, J. W. B., M. B. Mathews, and N. Sonenberg. 1996. Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Kang, P. J., J. Ostermann, J. Shilling, W. Neupert, E. A. Craig, and N. Pfanner. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348:137-143. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, K., T. Shibata, and F. Ito. 2004. Roles of the HSP70-subunit in a eukaryotic multi-site-specific endonuclease, Endo.SceI: autophosphorylation and heat stability. Biosci. Biotechnol. Biochem. 68:2557-2564. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 24.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 25.Kubo, Y., T. Tsunehiro, S. Nishikawa, M. Nakai, E. Ikeda, A. Toh-e, N. Morishima, T. Shibata, and T. Endo. 1999. Two distinct mechanisms operate in the reactivation of heat-denatured proteins by the mitochondrial Hsp70/Mdj1p/Yge1p chaperone system. J. Mol. Biol. 286:447-464. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Langer, T., M. Kaser, C. Klanner, and K. Leonhard. 2001. AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem. Soc. Trans. 29:431-436. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Q., P. D'Silva, W. Walter, J. Marszalek, and E. A. Craig. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300:139-141. [DOI] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14:4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, R. G., and B. N. Ames. 1961. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J. Biol. Chem. 236:1372-1379. [PubMed] [Google Scholar]
- 32.Mashkevich, G., B. Repetto, D. M. Glerum, C. Jin, and A. Tzagoloff. 1997. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem. 272:14356-14364. [DOI] [PubMed] [Google Scholar]
- 33.Miao, B., J. E. Davis, and E. A. Craig. 1997. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J. Mol. Biol. 265:541-552. [DOI] [PubMed] [Google Scholar]
- 34.Mick, D. U., K. Wagner, M. van der Laan, A. E. Frazier, I. Perschil, M. Pawlas, H. E. Meyer, B. Warscheid, and P. Rehling. 2007. ShyI couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizumura, H., T. Shibata, and N. Morishima. 1999. Stable association of 70-kDa heat shock protein induces latent multisite specificity of a unisite-specific endonuclease in yeast mitochondria. J. Biol. Chem. 274:25682-25690. [DOI] [PubMed] [Google Scholar]
- 36.Myers, A. M., L. K. Pape, and A. Tzagoloff. 1985. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobrega, M. P., F. G. Nobrega, and A. Tzagoloff. 1990. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 265:14220-14226. [PubMed] [Google Scholar]
- 38.Ott, M., M. Prestele, H. Bauerschmitt, S. Funes, N. Bonnefoy, and J. M. Herrmann. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25:1603-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierrel, F., M. L. Bestwick, P. A. Cobine, O. Khalimonchuk, J. A. Cricco, and D. R. Winge. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rassow, J., A. C. Maarse, E. Krainer, M. Kubrich, H. Muller, M. Meijer, E. A. Craig, and N. Pfanner. 1994. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127:1547-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 43.Rowley, N., C. Prip-Buus, B. Westermann, C. Brown, E. Schwarz, B. Barrell, and W. Neupert. 1994. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77:249-259. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Schagger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, H. C., J. Berthold, M. F. Bauer, K. Dietmeier, B. Guiard, M. Brunner, and W. Neupert. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371:768-774. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, H. C., B. Westermann, W. Neupert, and M. Brunner. 1996. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 15:5796-5803. [PMC free article] [PubMed] [Google Scholar]
- 49.Tenson, T., and M. Ehrenberg. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108:591-594. [DOI] [PubMed] [Google Scholar]
- 50.Tzagoloff, A., A. Akai, R. B. Needleman, and G. Zulch. 1975. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J. Biol. Chem. 250:8236-8242. [PubMed] [Google Scholar]
- 51.Tzagoloff, A., N. Capitanio, M. P. Nobrega, and D. Gatti. 1990. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 9:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voos, W., and K. Rottgers. 2002. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta 1592:51-62. [DOI] [PubMed] [Google Scholar]
- 53.Westermann, B., B. Gaume, J. M. Herrmann, W. Neupert, and E. Schwarz. 1996. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell. Biol. 16:7063-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wollman, F. A., L. Minai, and R. Nechushtai. 1999. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411:21-85. [DOI] [PubMed] [Google Scholar]
- 55.Wostrikoff, K., and D. Stern. 2007. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. U. S. A. 104:6466-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zambrano, A., F. Fontanesi, A. Solans, R. L. de Oliveira, T. D. Fox, A. Tzagoloff, and A. Barrientos. 2007. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 18:523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]