Abstract
All-trans retinoic acid (RA) induces transforming growth factor beta (TGF-β)-dependent autocrine growth of mouse embryonic fibroblasts (MEFs). We have used chromatin immunoprecipitation to map 354 RA receptor (RAR) binding loci in MEFs, most of which were similarly occupied by the RARα and RARγ receptors. Only a subset of the genes associated with these loci are regulated by RA, among which are several critical components of the TGF-β pathway. We also show RAR binding to a novel series of target genes involved in cell cycle regulation, transformation, and metastasis, suggesting new pathways by which RA may regulate proliferation and cancer. Few of the RAR binding loci contained consensus direct-repeat (DR)-type elements. The majority comprised either degenerate DRs or no identifiable DRs but anomalously spaced half sites. Furthermore, we identify 462 RAR target loci in embryonic stem (ES) cells and show that their occupancy is cell type specific. Our results also show that differences in the chromatin landscape regulate the accessibility of a subset of more than 700 identified loci to RARs, thus modulating the repertoire of target genes that can be regulated and the biological effects of RA.
Retinoic acid (RA), the naturally active vitamin A metabolite, exerts a wide range of effects on vertebrate development and adult tissue homeostasis by regulating cell proliferation, differentiation, and apoptosis (8, 34, 42). RA activates three members of the nuclear receptor (NR) superfamily, RARα, RARβ, and RARγ, that function as ligand-dependent transcriptional regulators by binding, usually as heterodimers with rexinoid receptors (RXRs α, β, and γ) to RA response elements (RAREs) located in target genes (16, 17). Many RAREs are formed by a direct repeat (DR) of the consensus sequence 5′-RGKTCA-3′ separated by 1, 2, or 5 nucleotides (for a review, see references 3, 4, and 6), and a large number of DR2-type elements are present within Alu repeats (27, 58).
RARs and RXRs exhibit the conserved structure of NRs, with N-terminal activation function 1 (AF-1), a central DNA binding domain, and a C-terminal ligand binding domain (LBD) that harbors ligand-dependent AF-2 (38, 40; for a review, see references 2, 41, and 51). The ability of RARs to modulate the expression of target genes results from a complex and dynamic interaction with coactivator/corepressor complexes (54). A general model proposes that unliganded RARs occupy regulatory elements at their target genes and repress their expression. Ligand binding leads to a conformational change in LBD structure, releasing the corepressor complexes and allowing recruitment of coactivator complexes with histone acetyl- and methyltransferase activities and activation of target genes. A third scenario is ligand-dependent repression involving the recruitment of proteins such as NRIP1 (RIP140), PRAME, or TRIM24 (TIF1a) that interact with the liganded receptors to repress the transcription of target genes (11, 14, 30). An additional level of control contributing to cell specificity could occur at the DNA binding step, as RAR/RXR heterodimers may bind to distinct sets of regulatory elements in different cell types. This may be regulated by cell or tissue type differences in the epigenetic organization of the chromatin in which the regulatory elements are present (22).
We have previously shown that RA activates the transforming growth factor beta (TGF-β) signaling pathway to induce morphological changes and serum-independent autocrine growth of Taf4lox/− mouse embryonic fibroblasts (MEFs) (15). Transcriptome analysis showed that around 1,000 genes are activated or repressed by RA in these cells. Among the activated genes are TGF-β ligands and connective tissue growth factor, which is rapidly induced by RA to induce autocrine growth (15, 28). While this study identified the genes and pathways that are activated by RA, we could not discriminate between the direct and indirect targets of the RARs.
To identify RA-regulated genes directly, we used chromatin immunoprecipitation (ChIP) coupled with array hybridization (ChIP-chip) to identify 354 target loci bound by the RARs in MEFs, only a subset of which are induced or repressed by RA. Furthermore, we also mapped 462 RAR binding loci in undifferentiated embryonic stem (ES) cells and show that RAR occupancy of target loci is cell type specific, as only a minority of the MEF loci were occupied in ES cells. We propose that cell-specific occupancy of target promoters by RARs is an important mechanism contributing to the distinct effects of RA seen in different cell types.
MATERIALS AND METHODS
Generation and culture of tagged RAR-expressing cells.
Taf4lox/− MEFs, as previously described (35), were infected with pBABE retroviruses encoding RARα or RARγ with a hemagglutinin (HA)-3×Flag tag at the C terminus, and populations of cells stably expressing the tagged RARs were selected with puromycin. Cells were grown at 5% CO2 in Dulbecco's minimal essential medium supplemented with GlutaMAX and 10% fetal calf serum. For the generation of ES cells expressing tagged RAR, recombination targeting vectors containing HA-3×Flag or SBP (streptavidin binding peptide)-3×Flag (for RARα and RARγ, respectively), followed by a neomycin resistance cassette flanked by FRT sites, were constructed. Four-kilobase homology arms corresponding to the regions immediately upstream and downstream of the stop codon were cloned on either side of these elements. The construct was electroporated into SV/129 ES cells, and the neomycin-resistant clones were screened for homologous recombination by PCR and for expression of the recombinant proteins by Western blotting using the previously described anti-RARα and -RARγ antibodies (5). All cells were treated with 1 μM RA for the indicated times.
ChIP, ChIP-chip, and ChIP-seq.
ChIP experiments were performed according to standard protocols (data not shown). All ChIP was performed in triplicate and analyzed by triplicate quantitative PCR (qPCR). For ChIP-chip, the total input chromatin and ChIPed material were hybridized to the extended promoter array from Agilent covering the kb −5 to +2 regions of around 17,000 cellular promoters. Data were analyzed with ChIP Analytics from Agilent (see the supplemental material). ChIP-seq was performed using an Illumina GAII sequencer, and the raw data were analyzed by the Illumina Eland pipeline. Peak detection was performed using the MACS software (63), and the peaks were annotated using GPAT (26; data not shown). The antibodies used were against HA (12CA5), pan-RAR (Sc-773), pan-RXR(Sc-774), H3K4me3 (04-745; Upstate), H3K9me3 (07-442; Upstate), H3K27me3 (07-449; Upstate), H3K9Ac (07-352; Upstate), and RNA polymerase II (Pol II; Sc-9001). Flag ChIP was performed with anti-Flag M2 affinity gel (A2220; Sigma). Real-time PCRs were performed on a Roche LightCycler using Roche SYBR green mix. The sequences of the primers used are available on request.
Electrophoretic mobility shift assays (EMSAs).
EMSAs were performed essentially as previously described (15). After electrophoresis, the gels were dried and exposed to autoradiographic film or a PhosphorImager plate.
RESULTS
Identification of RAR target genes in MEFs.
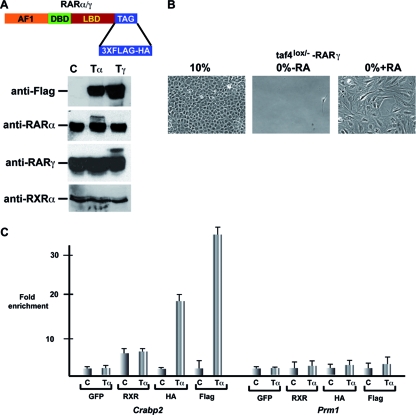
To identify RAR target genes in Taf4lox/− MEFs, we used retroviral vectors to generate cell lines that stably express RARα and RARγ carrying a 3×Flag-HA tandem affinity purification (TAP) tag at the C terminus. The presence of the TAP tag does not affect RAR function in transient expression assays or activation of RAR target genes in the MEFs (data not shown). Western blotting on extracts of the infected cells showed that the exogenous TAP-tagged RARs made up only around 10 to 20% of the total RARα or RARγ (Fig. 1A). Cells expressing exogenous RARs also displayed RA-induced morphological changes and serum-independent growth (Fig. 1B), consistent with the idea that their presence does not affect the proper RA response of the cells. ChIP assay using either Flag or HA antibodies showed that tagged RARs and endogenous RXR were efficiently recruited to the DR2-type RARE in the cellular RA binding protein 2 gene (Crabp2) promoter, while no enrichment was seen at the control protamine 1 gene (Prm1) promoter (Fig. 1C and data not shown).
FIG. 1.
Expression of tagged RARs in MEFs and ES cells. (A) Schematic representation of RAR with a C-terminal 3×Flag-HA tag and Western blot assay showing expression of tagged RARs in total extracts from cells expressing tagged RARα or RARγ or control cells (Tα, Tγ, and C, respectively). In the upper panel, tagged RARs were detected with anti-Flag antibody, and in the lower panels, endogenous and tagged RARs were detected with isotype-specific antibodies and endogenous RXR was detected with a pan-RXR antibody. (B) Phase-contrast microscopy after 5 days of culture in 10% or 0% serum showing that RA treatment of Taf4lox/− MEFs expressing tagged RARγ leads to serum-independent growth. (C) ChIP-qPCR on the Crabp2 RARE and the Prm1 promoter in cells expressing tagged RARα and control cells treated for 2 h with RA. The antibodies used are shown below the graph. The value obtained with each antibody and the control cells was assigned a value of 1, and fold enrichment relative to this value is indicated. GFP, green fluorescent protein.
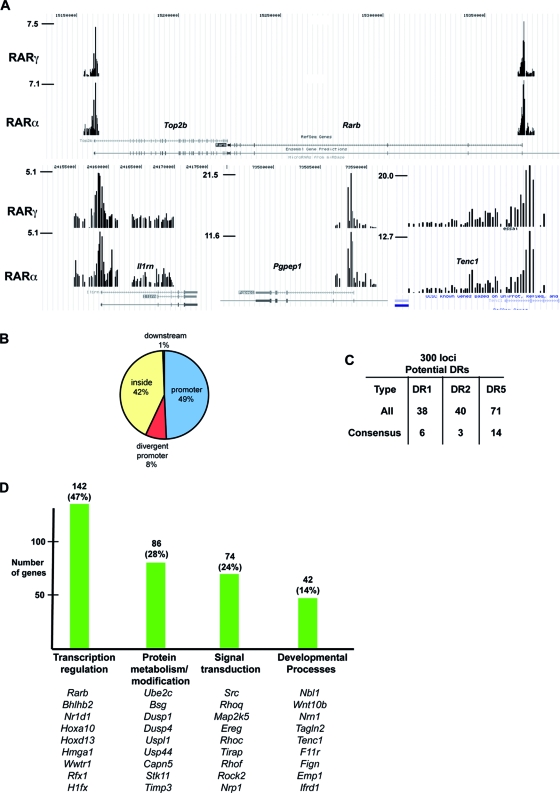
We used these cells to perform ChIP-chip experiments with the Agilent extended promoter array comprising the kb −5 to +2 regions of around 17,000 cellular promoters. Duplicate tandem Flag ChIP-chip and HA ChIP-chip assays were performed on cells expressing RARγ that had been treated with RA for 2 h and compared with a tandem Flag ChIP-chip on untagged cells as a control. These experiments identified 354 loci bound by RARγ after Flag and HA ChIP in tagged cells but not in control cells (see Table S1 and Fig. S1A in the supplemental material). For example, occupancy of the characterized DR5 element in the Rarb promoter was observed along with occupancy of several other known targets such as Crabp2, Bhlhb2 (Stra13 [56]), and the ubiquitin-conjugating enzyme E2C gene (Ube2c) (64) (Fig. 2A; data not shown). Among the identified loci are previously characterized target genes known to be RA regulated, but not as direct targets, such as the ceruloplasmin (Cp), matrix metallopeptidase 2 (Mmp), cathepsin D (Ctsd), or type 1 alpha collagen (Col1a1) gene (for a review, see reference 4). Most of the occupied regions are, however, in novel target genes such as the topoisomerase II beta gene (Top2b), which is transcribed convergently to Rarb, or the interleukin-1 receptor antagonist (Il1rn), pyroglutamyl-peptidase I (Pgpep1), and tensin like C1 domain-containing phosphatase (Tenc1) gene promoters (Fig. 2A). Analysis of 200 of the most sharply defined RAR binding sites showed that 49% were located upstream of the transcription start site (TSS), while 42% were located downstream, principally in the first intron (Fig. 2B).
FIG. 2.
Identification of RAR binding sites in MEFs. (A) Graphic representation of tandem Flag ChIP-chip results on cells expressing tagged RARα and RARγ in the UCSC web browser (http://genome.ucsc.edu/) at the indicated loci. The values on the y axis show the normalized immunoprecipitate/input ratio. (B) Pie chart representation of the locations of RAR binding sites relative to the TSS. (C) Summary of the presence of classical DR-type RAR binding motifs at ChIPed loci. The total number of potential sites with one mismatch is shown along with the number of consensus sites. (D) Summary of gene ontology analysis of target genes. The number of genes in each category is shown along with representative examples.
Gene ontology analysis (http://david.abcc.ncifcrf.gov/) showed that the largest class of target genes are involved in transcription regulation, for example, transcription factors such as Hoxa10 and Hoxd13 and the RAR coactivators Hmga1 (39), Trim16 (12), and Top2b (23). One of the main functions of RARs is therefore to regulate other transcriptional networks (Fig. 2D). RARs also bind a significant number of genes involved in signal transduction, protein modification, and metabolism, as well as developmental regulatory genes.
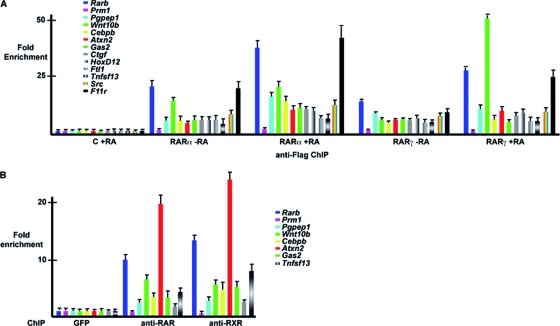
Very similar results were observed in ChIP-chip experiments with cells expressing tagged RARα (Fig. 2A), as the vast majority of sites were similarly bound by RARα and RARγ (Fig. 1A; see Table S1 and Fig. S1A in the supplemental material). ChIP-qPCR confirmed specific binding of RARα and RARγ to promoters in the absence of RA (Fig. 3A). RA substantially increased RARα binding on the Rarb, Pgpep1, Cebpb, and F11 receptor (F11r) promoters, while the strongest increase in RARγ binding was seen at the Wnt10b locus, but little or no effect was observed on the other sites. RAR and RXR occupancy was also verified using pan-RAR and pan-RXR antibodies against the endogenous proteins in MEFs that do not express the tagged RARs (Fig. 3B; see also below). Together, the above results show that most, if not all, of the sites were occupied in the absence of RA by both RARα and RARγ but that the influence of RA is promoter specific.
FIG. 3.
Binding of RARs to target loci. (A) qPCR on the indicated loci after tandem Flag ChIP on cells expressing tagged RARα or RARγ in the absence of RA or after 2 h of RA treatment. The value obtained for each amplicon in the control cells was set to 1, and fold enrichment relative to this value is indicated. Oligonucleotide primers are designed at the center of the ChIP peak at each locus. (B) ChIP-qPCR at the indicated loci using pan-RAR and pan-RXR antibodies. The value obtained for each amplicon after control ChIP with anti-GFP antibody was set to 1, and fold enrichment relative to this value is indicated.
Characterization of RAR binding motifs.
To identify the RAR binding motifs, the 1,200 nucleotides flanking the highest-scoring oligonucleotide in each peak were analyzed for the presence of DR1, -2, and -5 elements that had either a perfect match to the known consensus [5′-RGKTCA-(n)1,2,5-RGKTCA-3′] or a single mismatch (see Table S2 in the supplemental material). While 149 potential DRs were identified, only 23 were fully consensus sites (Fig. 2C). This was confirmed by a more restricted analysis of the 300 nucleotides around the top 100 most sharply defined peaks that identified only 7 consensus elements (see Table S2 in the supplemental material). This analysis also revealed that 71 of these sites contained at least one consensus half site with a majority containing several anomalously spaced half sites but no obvious DR1, -2, or -5.
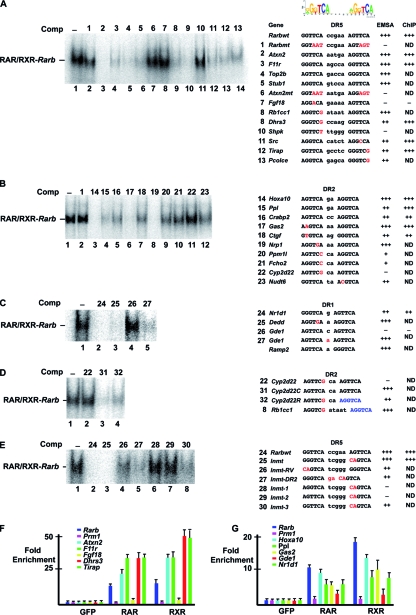
We verified RAR binding to several of the novel DRs by competition EMSAs in vitro or ChIP-qPCRs using primers centered on the DR element. In the EMSAs, the retarded RAR/RXR complex formed using an oligonucleotide comprising Rarb DR5 was efficiently competed by consensus DR5 elements from the Atxn2, F11r, Top2b, and STIP1 homology and U-box-containing protein 1 (Stub1) genes but not by mutated Rarb or Atxn2 oligonucleotides (Fig. 4A, lanes 1 to 7). Efficient competition was also seen with DR5 elements with a single mismatch in the 5′ or 3′ half sites from the dehydrogenase/reductase (SDR family) member 3 (Dhrs3), Toll-interleukin-1 receptor domain-containing adaptor protein (Tirap), Rous sarcoma oncogene (Src), RB1-inducible coiled-coil 1 (Rb1cc1), and procollagen C-endopeptidase enhancer protein (Pcolce) genes (Fig. 4A, lanes 9, 10, and 12 to 14). In contrast, little or no competition was observed with the potential DR5 element from the fibroblast growth factor 18 (Fgf18) or sedoheptulokinase (Shpk) gene (lanes 8 and 11), both of which contain a single mismatch with respect to the consensus.
FIG. 4.
Characterization of DR-type RAR binding sites. (A to E) Competition EMSAs. EMSAs were performed using Rarbwt DR5 as a radioactively labeled probe and a 100-fold excess of the unlabeled competitor (Comp) oligonucleotides shown above each lane. The sequences of the DRs within the oligonucleotides are indicated at the right of each panel. Mismatches with the consensus sequence are shown in red. For the chimeric DR, a color code for each half site is used. The results of competition EMSAs and ChIP assays are summarized on the right. (F and G) ChIP-qPCR on the indicated loci using amplicons centered on the DR element.
These results were confirmed by ChIP-qPCR using RAR and RXR antibodies, where occupancy of the Rarb, Atxn2, Dhrs3, Tirap, and F11r loci was observed (Fig. 4F), suggesting that these elements bind the RAR/RXR both in vitro and in MEFs. In contrast, although the Fgf18 and Shpk loci are clearly occupied in MEFs (see Fig. S1A in the supplemental material), no occupancy was observed using the amplicon centered on the potential DRs (Fig. 4F). The potential DRs that were identified do not coincide well with the peak detected by ChIP-chip, indicating that the RAR binds not to these DRs but rather another sequence at these loci.
Consensus or single-mismatch DR2 elements from the homeobox a10 (Hoxa10), periplakin (Ppl), Crabp2, growth arrest-specific 2 (Gas2), Ctgf, neuropilin 1 (Nrp1), and nudix (nucleoside diphosphate-linked moiety X)-type motif 6 (Nudt6) genes efficiently competed the formation of the retarded complex in vitro (see appropriate lanes in Fig. 4B) and were occupied in MEFs (Fig. 4G and not shown). In contrast, competition with the DR2 elements from the protein phosphatase 1-like (Ppm1l) and FCH domain only 2 (Fcho2) genes were much less efficient and no competition was seen with the cytochrome P450, family 2, subfamily d, polypeptide 22 (Cyp2d22) element.
For DR1 elements, competition was observed using those of the chromatin-modifying protein 1A (Chmp1a), calcitonin receptor activity-modifying protein 2 (Ramp2), NR subfamily 1, group D, member 1 (Nr1d1), and death effector domain-containing (Dedd) genes (Fig. 4C and data not shown). Surprisingly, however, no significant competition was observed with the consensus DR1 motif from the glycerophosphodiester phosphodiesterase 1 (Gde1) element. We noted that the spacer nucleotide in this element was C, while it was A in the Ramp2 and Dedd elements that compete. Exchange of the C in Gde1 for an A resulted in efficient competition (lane 5). Thus, half-site sequences and spacer nucleotides can play important roles in the formation of a complex with RAR/RXR. As observed in vitro, the Nr1d1 locus, but not Gde1, was occupied in cells (Fig. 4G). Similar to Fgf18, this indicates that binding to the Gde1 locus must be mediated by another element.
The above results indicate that many single mismatches do not abrogate RAR/RXR binding even when located at highly conserved positions such as the G in position 2 or 5′-TCA-3′. In contrast, in other elements, the equivalent mutations lead to a loss of binding. For example, in Cyp2d22 and Rb1cc1, 5′-TCA-3′ is mutated to 5′-TCG-3′ but only Rb1cc1 binds RAR/RXR. In Cyp2d22, replacing the G with an A to create a consensus leads to a strong increase in competition (Cyp2d22C, Fig. 4D, lanes 2 and 3). We exchanged the 3′ half site of Cyp2d22 with that of Rb1cc1 (i.e., changing T to G at the third position). The resulting Cyp2d22R was a significantly better competitor than native Cyp2d22 although less efficient than the full consensus (lanes 2 to 4). Hence, the exact sequence of one half site plays a critical role in compensating for mismatches in the other half site.
A more striking example of this is found upon analysis of the indolethylamine N-methyltransferase (Inmt) gene binding site. At this locus, we did not find any consensus or single-mismatch DR element. However, visual inspection identified a DR5 with two mismatches that efficiently competes in an EMSA (Fig. 4E). Inverting the 5′ and 3′ half sites (Inmt-RV) led to a decrease in competition, showing that the mismatches are less well tolerated in the 5′ position, where RXR normally binds (lane 4). Similarly, transforming the DR5 to a DR2 also decreases competition (lane 5). Strikingly, however, replacing 5′-GGG-3′ in the 5′ half site with 5′-AGG-3′ or 5′-AGT-3′ leads to a strong reduction in competition (lanes 6 to 7), while 5′-GGT-3′ competes, but less well than native 5′-GGG-3′ (lanes 3 and 8). These results show that in nonconsensus elements, mutations in one half site can be compensated for by the exact sequence of the other half site. In the case of Inmt, a G in the first position seems critical to compensate for mutations in the second half site.
Association of RAR binding sites with other transcription factors.
Many RAR-bound loci did not contain identifiable DR1, -2, and -5 elements but contained consensus half sites. Multiple alignments of the regions adjacent to these half sites did not reveal the presence of an additional consensus sequence for another transcription factor that would bind together with RAR/RXR (see Fig. S2A in the supplemental material). Analysis of a larger 300 nucleotides around the peaks identified numerous consensus sites for SP1 and AP2. However, they were also commonly found in control promoter sets and were not enriched in the RAR-bound set.
It has recently been shown in MCF7 cells that almost 60% of the RAR-bound loci were also bound by estrogen receptor (ER) (21). We therefore examined the 354 RAR-bound loci for ER binding sites by using the consensus 5′-RGGTCWnnnWGACMY-3′ (61) but identified only 3 estrogen response elements (EREs) in the MEF data set. Similarly, Hua et al. reported a significant overlap between RAR and FOXA1 binding sites. Using the consensus 5′-TRTTTRYWYW-3′, we observed 46 potential FOXA1 sites. Thus, assuming that they are all bona fide binding sites, only 12% of the RAR-bound loci show an associated FOXA1 site. Similar results were obtained upon the analysis of 211 of the ES RAR-bound loci (see below), where we found 44 potential FOXA1 sites (20%) but no EREs.
RA regulation of target gene expression.
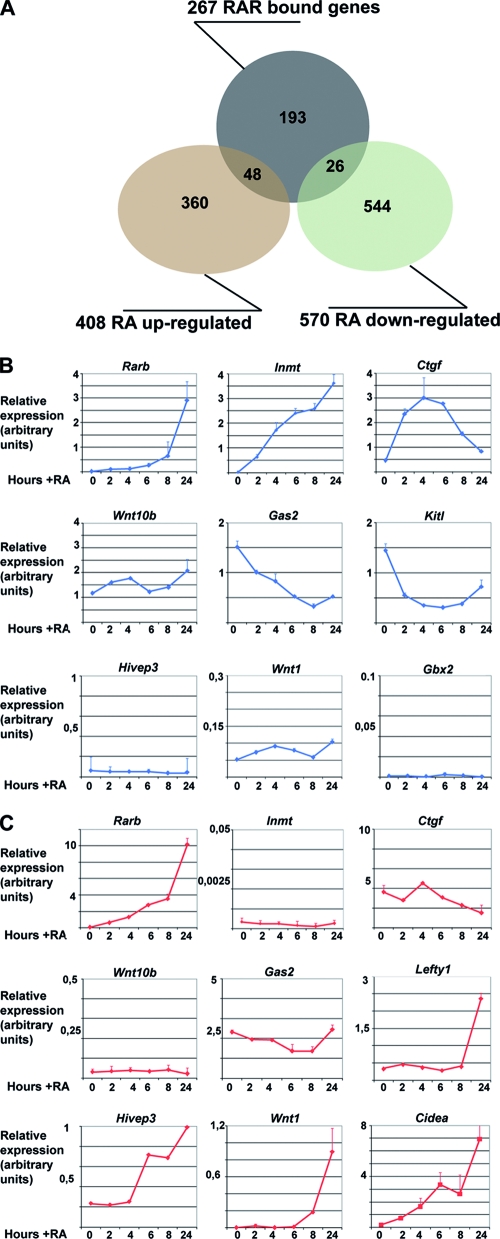
We have identified 354 RAR-bound loci, but taking into account the divergent promoters and genes with close neighbors, there are 383 potential RA-regulated genes. Comparison with our previous transcriptome data shows that 267 of the 383 genes were present on the Affymetrix array, but only 48 genes were upregulated and 26 were repressed by RA (Fig. 5A; see Table S1 in the supplemental material). As the transcriptome analysis was performed after 24 h of RA treatment, we verified gene expression to determine whether we had missed many transiently regulated genes. Reverse transcription coupled to qPCR identified several categories of genes. Rarb and Inmt are not expressed in the absence of RA and are strongly induced over a 24-h period (Fig. 5B). Others, such as Ctgf, show significant expression in the absence of ligand but are transiently stimulated by RA. In contrast, genes such as Gas2 or Kit ligand (Kitl) are repressed by RA. However, the expression of wingless-related MMTV integration site 10b (Wnt10b), like that of many other genes (data not shown), is not affected by RA. These observations indicate that only around 27% of the RAR-bound genes are regulated by RA in these cells under the experimental conditions used.
FIG. 5.
Expression of RAR target genes. (A) Venn diagram of RAR-bound versus RAR-regulated genes. The numbers refer only to the ChIPed genes that are represented on the Affymetrix array as described in the text. (B) Expression of the indicated target genes in MEFs in the absence or presence of RA over a 24-h period as measured by qPCR and normalized to the ribosomal Rplp0 transcript. (C) Normalized expression in ES cells in the absence or presence of RA for the indicated times as measured by qPCR. In panels B and C, the relative expression at each time point is shown in arbitrary units.
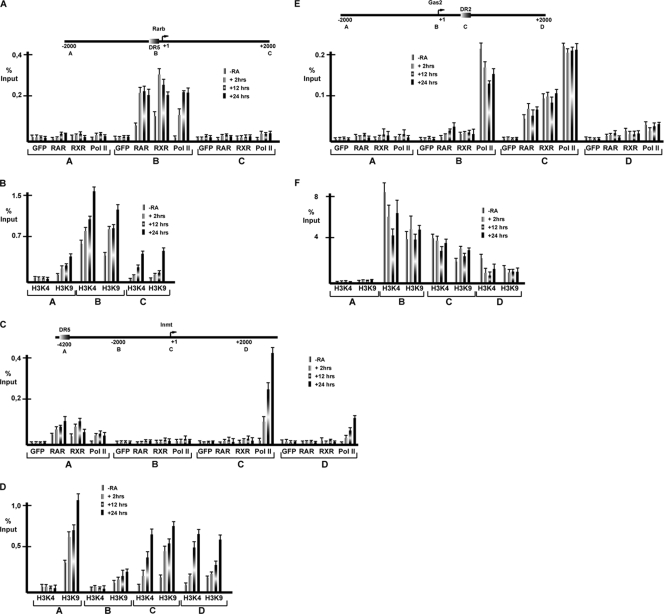
We investigated several promoters more closely by using primer pairs designed to amplify the TSS and regions located at −2 kb and +2 kb relative to the TSS. RAR/RXR occupancy of Rarb DR5 was observed in the absence of RA and was strongly increased after 2 to 24 h with RA (Fig. 6A). In contrast, Pol II was observed only in the presence of ligand. This result differs from that observed previously in P19 cells, where some Pol II was present at Rarb in the absence of RA (31, 46). Interestingly, significant H3K4me3 and H3K9ac covalent histone modifications tightly associated with transcriptional activity (57) were observed in the absence of RA and strongly increased after RA treatment (Fig. 6B). Lower levels of both of these markers were also observed downstream of the TSS.
FIG. 6.
ChIP at RA-regulated genes. (A and B) Schematic representation of the Rarb promoter with DR5 located close to the TSS. The localization of each amplicon is shown. ChIP was performed with the antibodies indicated below the graph, GFP, RAR, RXR, Pol II, H3K4me3, and H3K9ac. Results are expressed as percentages of the input. (C and D) Schematic representation of the Inmt promoter. The nonconsensus DR5 RAR binding site is located 4.2 kb upstream of the TSS. ChIP was performed as described above. (E and F) Schematic representation of the Gas2 promoter with the DR2 located 300 bp downstream of the TSS.
At the Inmt promoter, RAR/RXR binding to the nonconsensus DR5 located 4.2 kb upstream of the TSS was observed in the absence of RA and increased by RA (Fig. 6C). No Pol II was observed at the TSS in the absence of RA, but it was strongly recruited in its presence. Interestingly, a minor but significant RA-dependent recruitment of Pol II at the RAR binding site was also observed. H3K4me3 was seen only in the presence of RA at the TSS and in the downstream region (Fig. 6D). Thus, unlike Rarb, where H3K4me3 was seen in the absence of RA, this marker is fully RA dependent at Inmt. H3K9ac was observed at the TSS, in the downstream region, and also at the RAR binding site and was stimulated in each region by RA.
We were unable to detect such striking changes at RA-repressed promoters. At the Gas2 promoter, RAR and RXR occupancy of the downstream DR2 element could be observed in the absence of RA and was not altered in its presence (Fig. 6E). Pol II occupancy of this promoter was observed in the absence of ligand, consistent with its constitutive expression; however, no significant changes in Pol II promoter occupancy were observed in the presence of RA. Similarly, high levels of the H3K4me3 and H3K9ac markers were observed in the absence of RA (Fig. 6F), but a transient reduction was seen in the presence of RA concomitant with the transient repression (Fig. 5B). Thus, while the strong de novo activation is reflected by Pol II recruitment and the appearance and/or increase of histone modifications associated with transcription, at repressed genes, only minor and transient changes were seen.
Cell-specific binding of RAR to its cognate elements.
We have previously noted that the repertoire of RA-regulated genes in MEFs is significantly different from that observed in other cell types such as F9 or ES cells (15). To examine the basis for this specificity, we generated ES cells in which the genes encoding RARα and RARγ were modified by homologous recombination to introduce a TAP tag at the C terminus (Fig. 7A). Western blot analysis of extracts from the tagged ES cells showed somewhat lower expression of tagged than untagged RAR, probably due to the presence of the neomycin resistance cassette in the 3′ untranslated region.
FIG. 7.
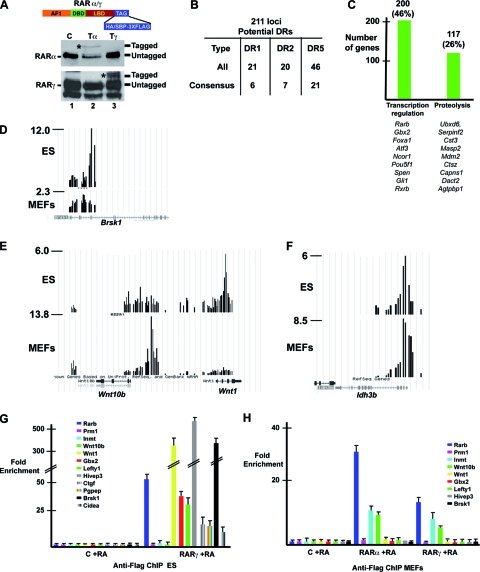
Identification of RAR binding sites in ES cells. (A) Schematic representation of RARs with a C-terminal 3×Flag-SBP or 3×Flag-HA tag and Western blot assay showing the expression of tagged and endogenous RARs in extracts of recombined or control ES cells. (B) Summary of classical DR-type RAR binding motifs at ChIPed ES cell loci as described in the legend to Fig. 1C. (C) Summary of gene ontology analysis of target genes. (D to F) Graphic representation of tandem Flag ChIP-chip results on ES cells and MEFs expressing tagged RARγ in the UCSC web browser at the indicated loci. The y axis shows the normalized immunoprecipitate/input ratio. (G and H) Flag ChIP-qPCR at the indicated loci in tagged and control MEFs and ES cells.
We compared duplicate tandem Flag ChIP-chip on ES cells expressing tagged RARγ to that on untagged cells to identify 462 bound loci. Among these are many previously identified genes, such as the Rarb, caudal-type homeobox 1 (Cdx1), left-right determination factor 1 (Lefty1), and pluripotency factor Pou5f1 (Oct4) genes, as well as a series of novel genes (see Table S3 and Fig. S1B in the supplemental material). Analysis of the 211 genes with the sharpest peaks indicated that, as observed in MEFs, the majority of the binding sites contained not consensus DR elements but either degenerate DRs or no identifiable DRs but anomalously spaced consensus half sites (Fig. 7B; see Table S3 in the supplemental material). Gene ontology analysis indicated that the largest functional classes of target genes were related to transcription regulation and proteolysis (Fig. 7C).
Comparison with MEFs showed that only 58 loci were bound in both cell types (see Table S3 in the supplemental material). For example, the brain serine/threonine kinase 1 (Brsk1) locus is occupied in ES cells but not in MEFs (Fig. 7D). At the Wnt10b-Wnt1 locus, RAR binds to a site upstream of the Wnt10b gene in MEFs not occupied in ES cells, whereas in ES cells, RAR occupies a site within the Wnt1 gene (Fig. 7E). Similarly, the HoxA and HoxB loci are differentially occupied in MEFs and ES cells (see Fig. S1C in the supplemental material). In contrast, the retinitis pigmentosa isocitrate dehydrogenase 3 (NAD+) beta (Idh3b) locus (18) was comparably occupied in both cell types (Fig. 7F).
These results were verified by ChIP-qPCR. In ES cells, RAR occupancy of the Lefty1, human immunodeficiency virus type I enhancer binding protein 3 (Hivep3; also know as Schnurri-3 [Shn3]), gastrulation brain homeobox 2 (Gbx2), Brsk1, and cell death-inducing DNA fragmentation factor alpha subunit-like effector A (Cidea) loci, but not of the Wnt10b and Inmt loci, was observed (7G). In contrast, in MEFs, occupancy of the Inmt and Wnt10b loci was observed, but not of the Lefty1, Gbx2, Hivep3, and Brsk1 loci, while the Rarb and Ctgf loci are occupied in both cell types (Fig. 7G and H and 3A [discussed above]). These results show clearly that binding of RAR to many of its cognate elements is cell type specific.
We assessed the expression and RA regulation of several genes in the different cell types. In ES cells, Rarb is not expressed in the absence of RA and is progressively and strongly induced by RA (Fig. 5B and C). In contrast, Inmt is neither expressed nor induced by RA in ES cells. Wnt10b and Gas2 were expressed at low levels in ES cells and were not regulated by RA. Hivep3 and Wnt1 are strongly upregulated by RA in ES cells, while they are neither expressed nor RA regulated in MEFs. Gbx2 is also not RA regulated in MEFs.
Chromatin modifications at target loci.
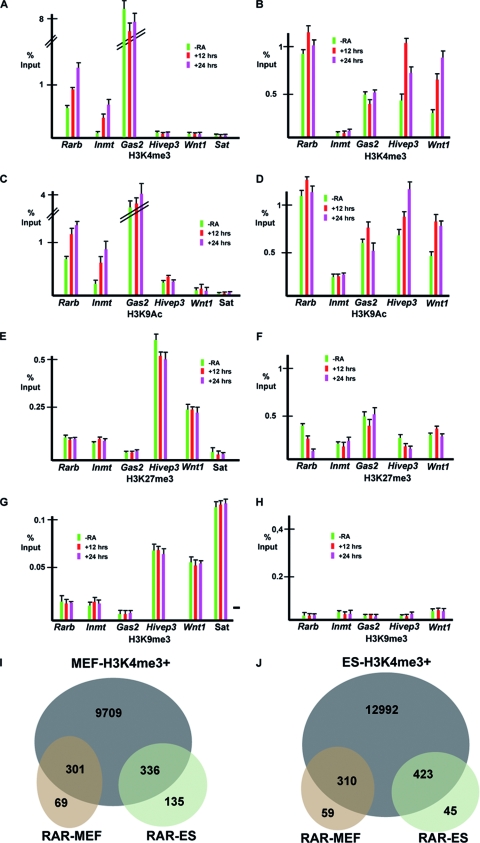
We compared the chromatin states of these loci in the two cell types. In MEFs, the highest level of H3K4me3 is observed at the constitutively active Gas2 promoter, while it is strongly increased by RA at Rarb and Inmt (Fig. 8A). In contrast, no significant H3K4me3 was observed at Hivep3 and Wnt1 in the presence or absence of RA. In ES cells, a high level of H3K4me3 was seen at Rarb that was only mildly increased by RA (Fig. 8B). H3K4me3 was also observed at Hivep3 and Wnt1 and significantly increased by RA. In contrast, there was no H3K4me3 at Inmt in the presence or absence of RA, while at Gas2, levels were much lower in ES cells than in MEFs. Similar results were seen for H3K9ac (Fig. 8C and D), although it is worth noting that low but significant levels (compared to major satellite as a negative control) were seen at Hivep3 in MEFs and at Inmt in ES cells, where the corresponding genes were not expressed.
FIG. 8.
Chromatin at RAR-bound loci. (A to H) ChIP-qPCRs using the indicated antibodies at loci in MEFs (A, C, E, and G) and ES cells (B, D, F, and H). Amplicons are centered on the TSS. (I and J) Venn diagrams comparing RAR-bound loci and their H34Kme3 status in MEFs (I) and ES cells (J). The total number of loci with H3K4me3 in MEFs and H3K4me3 in addition to the bivalent loci is shown as an intersection with the RAR loci bound in MEFs or ES cells.
In MEFs, a significantly higher level of the repressive H3K27me3 marker was observed at Hivep3 and Wnt1 than at the others (Fig. 8E). In ES cells, H3K27me3 at Rarb and Hivep3 was decreased by RA, while it was unaffected at the other loci (Fig. 8E). Unlike MEFs, however, the low or inactive Gas2 and Inmt promoters did not show H3K27me3 levels significantly higher than those of the expressed genes. In MEFs, the Hivep3 and Wnt1 promoters also showed higher levels of H3K9me3 than did the others, while in ES cells, none of the promoters showed a high level of this marker (Fig. 8G and H).
Together, these results show that the Hivep3 and Wnt1 promoters are in a repressed state in MEFs, suggesting that this chromatin conformation may restrict RAR/RXR binding to these loci. In ES cells, the Inmt locus shows no significant H3K4me3, in agreement with the lack of its expression, and neither this promoter nor the Gas2 promoter showed higher levels of the repressive H3K9 and H3K27me3 markers. Thus, these promoters are not in a repressed heterochromatin state that would account for the lack of RAR binding in ES cells.
These results are similar, but not identical, to those obtained in the global mapping of H3K4me3 in ES cells and MEFs in the absence of RA (36). In this study, differential H3K4me3 at Rarb and Inmt was observed in MEFs, but Hivep3 and Wnt1 had H3K4me3, while we clearly do not detect this marker at these loci. This perhaps reflects differences between individual MEF lines. In contrast, in ES cells, our results are in agreement with those of Mikkelsen et al.
These observations prompted us to make a global comparison of RAR binding and H3K4me3 in MEFs and ES cells. We performed H3K4me3 ChIP-seq on our MEFs and used these data and the Mikkelsen ES cell data for comparison (see Table S4 in the supplemental material). This analysis shows that the vast majority of genes bound by RAR in MEFs show significant H3K4me3 in the absence of RA, only a minority are in a repressed state (Fig. 8I; see Fig. S1D in the supplemental material). Similarly, although the Wnt1 and Hivep3 loci are in a repressed state in MEFs, the majority of the genes bound selectively in ES cells by RAR are in an active state in MEFs. A similar situation exists in ES cells, where most RAR-bound genes, whether in MEFs or in ES cells, show H3K4me3 and are in an active state or are bivalent promoters marked by both K4 and K27 trimethylation (Fig. 8J). Together, these results indicate that while the heterochromatinization in MEFs of some genes active in ES cells may explain the loss of RAR binding, this is not a general mechanism to account for the cell-specific association of RAR with its binding loci.
DISCUSSION
RAR binding and gene expression.
We describe the identification of 354 RAR-bound loci in the regions flanking 383 potential target genes in MEFs. These sites obviously do not correspond to the full repertoire, as the arrays cover an extended region around the TSS and many NR binding sites are known to be located far from their regulated genes (9, 43). For example, we did not find sites around the Enpp1-Enpp3 locus, which is rapidly induced by RA in these cells (15). The RAR binding sites regulating their expression may therefore be located further upstream or downstream of the TSSs.
We initiated this study to better understand the previously described RA-TGF-β cross talk (15). Our data confirm that RAR binds directly to a DR2 element in the Ctgf promoter, mediating RA induction of its expression and the resulting autocrine growth. Furthermore, we observe RAR binding to the gene encoding the TGF-β3 ligand, which is also strongly induced upon RA treatment. The direct activation of these two genes by RA is therefore central to the reported cross talk. Aside from these two genes, the other RAR-bound components of the TGF-β pathway, with the exception of Skil, are either weakly or not RA regulated in MEFs.
In addition to the TGF-β pathway, we observed RAR bound to several genes involved in other pathways of cell cycle regulation and transformation, such as tumor necrosis factor (ligand) superfamily member 13 (Tnfsf13; also called APRIL, a proliferation-inducing ligand [47]) cyclin D1 (Ccnd1), metastasis marker nuclear protein 1 (Nupr1), growth factor Fgf18, and the tyrosine kinase Src. Furthermore, RARs bind to the gene encoding Src-related Shc1(A) and cortactin (Cttn), an SRC substrate involved in metastasis (60). These results suggest novel pathways by which RA may regulate cell proliferation and cancer. It is also striking that RARs can potentially regulate genes involved in degenerative pathologies such as Atxn2 and dystrophia myotonica protein kinase (Dmpk) or Paget's disease through sequestosome 1 (Sqstm1) and tumor necrosis factor receptor superfamily member 11b (Tnfrsf11b, osteoprotegerin) (48). On the other hand, RARs also occupy the regulatory regions of WNK lysine-deficient protein kinase 4 (Wnk4) and odd-skipped-related 1 (Osr1), which act together to regulate salt transport and blood pressure (52). We also note that RARs bind to the Rho-GTPase-encoding genes Rhoc, Rhod, Rhof, Rhoq, Rhobtb2, Rhobtb3, and Rnd3 (Rhoe), as well as the downstream effector kinase gene Rock2 and the substrate Cofilin2 (Cfln2) gene. These genes act in the pathways that regulate actin dynamics and vesicle trafficking. RARs thus have the potential to control multiple cellular functions, and it is therefore important to determine if, when, and where RA may regulate these genes.
In ES cells, RAR is also found to occupy genes with a wide variety of functions. In addition to Pou5F1, RAR may also regulate other genes involved in pluripotency, such as Lin28 (45) or Utf1 (44), left-right determination through Lefty1 and Lefty2, and also Ier2 (20). RAR occupies the regulatory regions of genes encoding proteins involved in several other signaling pathways such as Gdf3 (32) and Nodal, Notch4 and its mediator Zfp64 (55), and the Ptch1, Gli1, Zic2, and Tectonic (Tect1) components of the sonic hedgehog pathway (50), as well as Fgf1, Fgf4, and Fgf8. RAR regulation of one or another of these genes and their associated pathways contributes to the differentiation of ES cells in vitro and developmental processes in vivo (7, 13, 33, 42).
Although our results show RAR occupancy of genes with diverse functions, the majority of MEF RAR target genes are not RA regulated under the conditions used. The lack of RA response may, in some cases, be due to the fact that the genes are already expressed and cannot be further stimulated, but in many others, low or no basal expression is observed and yet there is no stimulation by RA. However, some of the genes not regulated in MEFs are RA regulated in other cell types or tissues. For example, Gde1 is RA regulated in Wilms’ tumor cells (64), while Myc is regulated in various cell types but not in MEFs (4). It remains to be determined whether all RAR-bound genes can be RA regulated in the appropriate cell type and under the appropriate conditions or whether there are instances in which RAR is always a silent partner. The observation that only a subset of RAR-bound genes are RA regulated is in keeping with what has been observed for other transcription factors. For example, only a small subset of CREB-bound genes are regulated by cyclic AMP (62) and only a subset of ER-bound genes by are regulated estrogen (61).
In contrast to what may have been expected from a model in which unliganded RAR recruits repressor complexes to silence target genes, a large majority of genes bound by unliganded RAR show significant H3K4me3 and expression. Although this may be explained by heterogeneity in the cell population where RAR is not bound in cells where the genes are active, our results rather suggest a model where unliganded RARs actively silence only a small subset of their target genes.
Sequence diversity of RAR binding sites.
Our results show that the majority of the RAR-bound loci do not contain consensus DR1, -2, or -5 elements. At the bound MEF loci, we identified 149 potential DRs. EMSA analysis showed, however, that the only mismatch consistently found not to affect interaction was 5′-TCA-3′ to 5′-TCG-3′. Taking this criterion into account, only 27 bone fide DR elements in MEFs and 42 in ES cells can be identified with confidence. Although elements with other mismatches can bind RAR/RXR in vitro, we cannot readily predict the binding properties of DRs.
A closer investigation of the MEF and ES cell bound loci revealed that many did not contain readily identifiable DR1, -2, or -5 elements, even considering multiple mismatches, but rather comprised one or several anomalously spaced consensus half sites. There is no correlation between the presence and absence of recognizable DR1, -2, or -5 elements and gene regulation. Several genes with consensus DR elements are not RA regulated in MEFs, while genes with no identified DR are RA regulated. Furthermore, functional anomalously spaced half sites have previously been documented (24, 49). Multiple alignments of these half sites did not reveal the systematic presence of cryptic half sites or the presence of a consensus sequence for another transcription factor that would bind together with RAR/RXR. Our results therefore show that the majority of RAR binding sites comprise either degenerate DRs with multiple mismatches; no DR1, -2, or -5; or anomalously spaced consensus half sites. A similar observation was made for PML-RARα binding sites, the majority of which lack consensus DRs (19). This was ascribed to a relaxed specificity of the PML-RAR chimera but clearly also seems to be the case with the native RAR. Alternatively, the paucity of DR elements at the identified loci could be explained if they were not, in fact, the primary RAR binding sites but were detected as a consequence of DNA loop formation due to interactions between RARs bound at more distal sites and factors bound at the proximal promoter. Formaldehyde cross linking could capture such interactions, allowing the detection of RAR at the promoter even although the authentic binding sites are in distal enhancer regions. Such interactions have been observed with other NRs (9, 59).
While this work was in progress, Hua et al. reported a significant overlap between RAR and ER binding in MCF7 breast cancer cells. In contrast, we did not find significant numbers of palindromic EREs in the RAR-bound loci in MEFs and no EREs in the 211 analyzed ES cell loci. Similarly, we found potential FOXA1 sites in only 12 to 20% of the RAR-bound loci, much less than the 50% seen in MCF7 cells. Our observations are similar to those recently reported for ER binding sites in MCF7 cells (61), where, in contrast to a previous report (10), only 6 to 10% of the ER binding loci contain FOXA1 binding sites.
Chromatin topology contributes to cell-specific RAR occupancy.
Comparison of RAR-bound loci in MEFs and ES cells indicates that the majority are occupied in a cell-specific manner. We identified a subset of loci that are bound in ES cells but are in an inactive state in MEFs that is characterized by repressive chromatin markers and/or increased DNA methylation (our unpublished data). The compaction into a repressed heterochromatin state may account for the absence of RAR binding to these loci in MEFs. However, most ES cell-specific loci are associated with genes that are expressed and marked with H3K4me3 in MEFs. Similarly, most genes that are selectively bound in MEFs are marked by H34Kme3 and/or H3K27me3 in ES cells and are thus expressed or in a bivalent state. Thus, while chromatin condensation may explain the lack of RAR binding to a subset of sites in MEFs, there must be other mechanisms that regulate the accessibility of the majority of the loci in both MEFs and ES cells to RAR.
RA exerts distinct effects in different cell types. In ES, F9, or HL60 cells, RA induces cell cycle arrest and differentiation (1, 37, 53). In contrast, in MEFs or hepatocytes, RA promotes cell proliferation (15, 25, 29). It has been proposed that differential interaction of RAR with distinct sets of coregulatory complexes may explain the observed cell-specific effects of RA (54). Our results indicate that cell-specific binding of RAR to its genomic sites is a major mechanism regulating the repertoire of target genes that can be regulated and hence the biological effects of RA.
Supplementary Material
Acknowledgments
We thank, C. Rochette-Egly for the anti-RAR antibodies and expression vectors, J. Greenblatt and M Rudnicki for the backbone of the targeting vector, Y. Anno for help with bioinformatics, and C. Bole-Feysot for ChIP-chip labeling and hybridization.
This work was supported by grants from the CNRS, the INSERM, the Association pour la Recherche contre le Cancer, the Ligue Nationale et Départementale Région Alsace contre le Cancer, the INCA, and the ANR Regulome project grant. I.D. is an équipe labéllisée of the Ligue Nationale contre le Cancer. L.D. and G.A. were supported by the EuTRACC program of the European Union and the French ANR.
Footnotes
Published ahead of print on 2 November 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altucci, L., M. D. Leibowitz, K. M. Ogilvie, A. R. de Lera, and H. Gronemeyer. 2007. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 6:793-810. [DOI] [PubMed] [Google Scholar]
- 2.Bain, D. L., A. F. Heneghan, K. D. Connaghan-Jones, and M. T. Miura. 2007. Nuclear receptor structure: implications for function. Annu. Rev. Physiol. 69:201-220. [DOI] [PubMed] [Google Scholar]
- 3.Balmer, J. E., and R. Blomhoff. 2005. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J. Steroid Biochem. Mol. Biol. 96:347-354. [DOI] [PubMed] [Google Scholar]
- 4.Balmer, J. E., and R. Blomhoff. 2002. Gene expression regulation by retinoic acid. J. Lipid Res. 43:1773-1808. [DOI] [PubMed] [Google Scholar]
- 5.Bastien, J., S. Adam-Stitah, T. Riedl, J. M. Egly, P. Chambon, and C. Rochette-Egly. 2000. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J. Biol. Chem. 275:21896-21904. [DOI] [PubMed] [Google Scholar]
- 6.Bastien, J., and C. Rochette-Egly. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1-16. [DOI] [PubMed] [Google Scholar]
- 7.Bibel, M., J. Richter, K. Schrenk, K. L. Tucker, V. Staiger, M. Korte, M. Goetz, and Y. A. Barde. 2004. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 7:1003-1009. [DOI] [PubMed] [Google Scholar]
- 8.Blomhoff, R., and H. K. Blomhoff. 2006. Overview of retinoid metabolism and function. J. Neurobiol. 66:606-630. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 11.Cavaillès, V., S. Dauvois, F. L'Horset, G. Lopez, S. Hoare, P. J. Kushner, and M. G. Parker. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, B. B., J. Bell, A. Raif, A. Bohlken, J. Yan, B. Roediger, A. Poljak, S. Smith, M. Lee, W. D. Thomas, M. Kavallaris, M. Norris, M. Haber, H. L. Liu, D. Zajchowski, and G. M. Marshall. 2006. The estrogen-responsive B box protein is a novel regulator of the retinoid signal. J. Biol. Chem. 281:18246-18256. [DOI] [PubMed] [Google Scholar]
- 13.Dequéant, M. L., and O. Pourquie. 2008. Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9:370-382. [DOI] [PubMed] [Google Scholar]
- 14.Epping, M. T., L. Wang, M. J. Edel, L. Carlee, M. Hernandez, and R. Bernards. 2005. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122:835-847. [DOI] [PubMed] [Google Scholar]
- 15.Fadloun, A., D. Kobi, L. Delacroix, D. Dembele, I. Michel, A. Lardenois, J. Tisserand, R. Losson, G. Mengus, and I. Davidson. 2008. Retinoic acid induces TGFbeta-dependent autocrine fibroblast growth. Oncogene 27:477-489. [DOI] [PubMed] [Google Scholar]
- 16.Germain, P., P. Chambon, G. Eichele, R. M. Evans, M. A. Lazar, M. Leid, A. R. De Lera, R. Lotan, D. J. Mangelsdorf, and H. Gronemeyer. 2006. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol. Rev. 58:712-725. [DOI] [PubMed] [Google Scholar]
- 17.Germain, P., P. Chambon, G. Eichele, R. M. Evans, M. A. Lazar, M. Leid, A. R. De Lera, R. Lotan, D. J. Mangelsdorf, and H. Gronemeyer. 2006. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 58:760-772. [DOI] [PubMed] [Google Scholar]
- 18.Hartong, D. T., M. Dange, T. L. McGee, E. L. Berson, T. P. Dryja, and R. F. Colman. 2008. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat. Genet. 40:1230-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoemme, C., A. Peerzada, G. Behre, Y. Wang, M. McClelland, K. Nieselt, M. Zschunke, C. Disselhoff, S. Agrawal, F. Isken, N. Tidow, W. E. Berdel, H. Serve, and C. Muller-Tidow. 2008. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood 111:2887-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, S. K., and I. B. Dawid. 2009. FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc. Natl. Acad. Sci. U. S. A. 106:2230-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua, S., R. Kittler, and K. P. White. 2009. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137:1259-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John, S., P. J. Sabo, T. A. Johnson, M. H. Sung, S. C. Biddie, S. L. Lightman, T. C. Voss, S. R. Davis, P. S. Meltzer, J. A. Stamatoyannopoulos, and G. L. Hager. 2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29:611-624. [DOI] [PubMed] [Google Scholar]
- 23.Ju, B. G., V. V. Lunyak, V. Perissi, I. Garcia-Bassets, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 2006. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312:1798-1802. [DOI] [PubMed] [Google Scholar]
- 24.Kato, S., H. Sasaki, M. Suzawa, S. Masushige, L. Tora, P. Chambon, and H. Gronemeyer. 1995. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 15:5858-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khetchoumian, K., M. Teletin, J. Tisserand, M. Mark, B. Herquel, M. Ignat, J. Zucman-Rossi, F. Cammas, T. Lerouge, C. Thibault, D. Metzger, P. Chambon, and R. Losson. 2007. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat. Genet. 39:1500-1506. [DOI] [PubMed] [Google Scholar]
- 26.Krebs, A., M. Frontini, and L. Tora. 2008. GPAT: retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics 9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laperriere, D., T. T. Wang, J. H. White, and S. Mader. 2007. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leask, A., and D. J. Abraham. 2004. TGF-beta signaling and the fibrotic response. FASEB J. 18:816-827. [DOI] [PubMed] [Google Scholar]
- 29.Ledda-Columbano, G. M., M. Pibiri, F. Molotzu, C. Cossu, L. Sanna, G. Simbula, A. Perra, and A. Columbano. 2004. Induction of hepatocyte proliferation by retinoic acid. Carcinogenesis 25:2061-2066. [DOI] [PubMed] [Google Scholar]
- 30.Le Douarin, B., A. L. Nielsen, J. M. Garnier, H. Ichinose, F. Jeanmougin, R. Losson, and P. Chambon. 1996. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701-6715. [PMC free article] [PubMed] [Google Scholar]
- 31.Lefebvre, B., C. Brand, P. Lefebvre, and K. Ozato. 2002. Chromosomal integration of retinoic acid response elements prevents cooperative transcriptional activation by retinoic acid receptor and retinoid X receptor. Mol. Cell. Biol. 22:1446-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, A. J., and A. H. Brivanlou. 2006. GDF3 at the crossroads of TGF-beta signaling. Cell Cycle 5:1069-1073. [DOI] [PubMed] [Google Scholar]
- 33.Maden, M. 2007. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 8:755-765. [DOI] [PubMed] [Google Scholar]
- 34.Mark, M., N. B. Ghyselinck, and P. Chambon. 2006. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 46:451-480. [DOI] [PubMed] [Google Scholar]
- 35.Mengus, G., A. Fadloun, D. Kobi, C. Thibault, L. Perletti, I. Michel, and I. Davidson. 2005. TAF4 inactivation in embryonic fibroblasts activates TGFbeta signalling and autocrine growth. EMBO J. 24:2753-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongan, N. P., and L. J. Gudas. 2007. Diverse actions of retinoid receptors in cancer prevention and treatment. Differentiation 75:853-870. [DOI] [PubMed] [Google Scholar]
- 38.Nagpal, S., S. Friant, H. Nakshatri, and P. Chambon. 1993. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 12:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagpal, S., C. Ghosn, D. DiSepio, Y. Molina, M. Sutter, E. S. Klein, and R. A. Chandraratna. 1999. Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J. Biol. Chem. 274:22563-22568. [DOI] [PubMed] [Google Scholar]
- 40.Nagpal, S., M. Saunders, P. Kastner, B. Durand, H. Nakshatri, and P. Chambon. 1992. Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell 70:1007-1100. [DOI] [PubMed] [Google Scholar]
- 41.Nagy, L., and J. W. Schwabe. 2004. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 29:317-324. [DOI] [PubMed] [Google Scholar]
- 42.Niederreither, K., and P. Dolle. 2008. Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 9:541-553. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen, R., T. A. Pedersen, D. Hagenbeek, P. Moulos, R. Siersbaek, E. Megens, S. Denissov, M. Borgesen, K. J. Francoijs, S. Mandrup, and H. G. Stunnenberg. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22:2953-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimoto, M., S. Miyagi, T. Yamagishi, T. Sakaguchi, H. Niwa, M. Muramatsu, and A. Okuda. 2005. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 25:5084-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okita, K., M. Nakagawa, H. Hyenjong, T. Ichisaka, and S. Yamanaka. 2008. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322:949-953. [DOI] [PubMed] [Google Scholar]
- 46.Pavri, R., B. Lewis, T. K. Kim, F. J. Dilworth, H. Erdjument-Bromage, P. Tempst, G. de Murcia, R. Evans, P. Chambon, and D. Reinberg. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18:83-96. [DOI] [PubMed] [Google Scholar]
- 47.Planelles, L., J. P. Medema, M. Hahne, and G. Hardenberg. 2008. The expanding role of APRIL in cancer and immunity. Curr. Mol. Med. 8:829-844. [DOI] [PubMed] [Google Scholar]
- 48.Ralston, S. H., A. L. Langston, and I. R. Reid. 2008. Pathogenesis and management of Paget's disease of bone. Lancet 372:155-163. [DOI] [PubMed] [Google Scholar]
- 49.Raouf, A., V. Li, I. Kola, D. K. Watson, and A. Seth. 2000. The Ets1 proto-oncogene is upregulated by retinoic acid: characterization of a functional retinoic acid response element in the Ets1 promoter. Oncogene 19:1969-1974. [DOI] [PubMed] [Google Scholar]
- 50.Reiter, J. F., and W. C. Skarnes. 2006. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 20:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renaud, J. P., and D. Moras. 2000. Structural studies on nuclear receptors. Cell. Mol. Life Sci. 57:1748-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson, C., and D. R. Alessi. 2008. The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J. Cell Sci. 121:3293-3304. [DOI] [PubMed] [Google Scholar]
- 53.Rochette-Egly, C., and P. Chambon. 2001. F9 embryocarcinoma cells: a cell autonomous model to study the functional selectivity of RARs and RXRs in retinoid signaling. Histol. Histopathol. 16:909-922. [DOI] [PubMed] [Google Scholar]
- 54.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 55.Sakamoto, K., Y. Tamamura, K. Katsube, and A. Yamaguchi. 2008. Zfp64 participates in Notch signaling and regulates differentiation in mesenchymal cells. J. Cell Sci. 121:1613-1623. [DOI] [PubMed] [Google Scholar]
- 56.Sun, H., and R. Taneja. 2000. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 97:4058-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taverna, S. D., H. Li, A. J. Ruthenburg, C. D. Allis, and D. J. Patel. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vansant, G., and W. F. Reynolds. 1995. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc. Natl. Acad. Sci. U. S. A. 92:8229-8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19:631-642. [DOI] [PubMed] [Google Scholar]
- 60.Weaver, A. M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welboren, W. J., M. A. van Driel, E. M. Janssen-Megens, S. J. van Heeringen, F. C. Sweep, P. N. Span, and H. G. Stunnenberg. 2009. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28:1418-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, X., D. T. Odom, S. H. Koo, M. D. Conkright, G. Canettieri, J. Best, H. Chen, R. Jenner, E. Herbolsheimer, E. Jacobsen, S. Kadam, J. R. Ecker, B. Emerson, J. B. Hogenesch, T. Unterman, R. A. Young, and M. Montminy. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U. S. A. 102:4459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nussbaum, R. M. Myers, M. Brown, W. Li, and X. S. Liu. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zirn, B., B. Samans, C. Spangenberg, N. Graf, M. Eilers, and M. Gessler. 2005. All-trans retinoic acid treatment of Wilms tumor cells reverses expression of genes associated with high risk and relapse in vivo. Oncogene 24:5246-5251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.