Abstract
In higher plants, the chloroplast NAD(P)H dehydrogenase (NDH) complex mediates photosystem I (PSI) cyclic and chlororespiratory electron transport. We reported previously that NDH interacts with the PSI complex to form a supercomplex (NDH-PSI). In this study, NDH18 and FKBP16-2 (FK506 Binding Protein 16-2), detected in the NDH-PSI supercomplex by mass spectrometry, were shown to be NDH subunits by the analysis of their knockdown lines. On the basis of extensive mutant characterization, we propose a structural model for chloroplast NDH, whereby NDH is divided into four subcomplexes. The subcomplex A and membrane subcomplex are conserved in cyanobacteria, but the subcomplex B and lumen subcomplex are specific to chloroplasts. Two minor light-harvesting complex I proteins, Lhca5 and Lhca6, were required for the full-size NDH-PSI supercomplex formation. Similar to crr pgr5 double mutants that completely lack cyclic electron flow activity around PSI, the lhca6 pgr5 double mutant exhibited a severe defect in growth. Consistent with the impaired NDH activity, photosynthesis was also severely affected in mature leaves of lhca6 pgr5. We conclude that chloroplast NDH became equipped with the novel subcomplexes and became associated with PSI during the evolution of land plants, and this process may have facilitated the efficient operation of NDH.
INTRODUCTION
In higher plants, photosystem I (PSI) cyclic electron transport consists of both NAD(P)H dehydrogenase (NDH)-dependent and PROTON GRADIENT REGULATION5 (PGR5)-dependent pathways (Joët et al., 2001; Munekage et al., 2004; Shikanai, 2007a). The PGR5-dependent (main) pathway is required for both photosynthesis and photoprotection (Munekage et al., 2002). Additionally, chloroplast NDH helps to prevent the overreduction of stroma, especially under stress conditions (Munekage et al., 2004; Shikanai, 2007a). The existence of NDH in chloroplasts was first suggested by the complete sequencing of the two plastid genomes in tobacco (Nicotiana tabacum) and liverwort (Marchantia polymorpha), in which 11 genes encode the homologs of subunits in the mitochondrial complex I and eubacterial NADH dehydrogenase (Matsubayashi et al., 1987; Shikanai, 2007b). However, the genes encoding three key components, NuoE to G, which function in NADH binding and oxidation in Escherichia coli NDH-1, are missing in the cyanobacterial and higher plant genomes.
Several candidates for the electron donor and electron input module in NDH in chloroplasts and cyanobacteria have been proposed. An NDH subcomplex with a molecular mass of ∼550 kD isolated from pea (Pisum sativum) oxidized NADH (Sazanov et al., 1998). A hydrophilic part of NDH was also shown to contain NADH oxidizing activity (Rumeau et al., 2005). NADPH and ferredoxin (Fd) as well as NADH can be oxidized by the NDH complex in cyanobacteria (Mi et al., 1995). However, NDH purified from cyanobacteria favors NADPH as an electron donor (Matsuo et al., 1998). In contrast with these results, no differences were detected in NADH or NADPH oxidizing activity between the wild type and NDH-less mutants in Arabidopsis thaliana (Sirpiö et al., 2009a). Additionally, Guedeney et al. (1996) proposed that Fd-NADP+ reductase binds to chloroplast NDH. Inconsistently, Fd was required for NDH-dependent plastoquinone (PQ) reduction in our assay using ruptured chloroplasts (Munekage et al., 2004). To resolve this long-debated issue, researchers have tried to identify the missing subunits. So far, four NDH subunits, NdhL to O, have been found in cyanobacteria and chloroplast NDH (Prommeenate et al., 2004; Battchikova et al., 2005; Rumeau et al., 2005; Shimizu et al., 2008). In addition, six subunits specific to higher plants have been identified: PsbP-like protein 2 (PPL2), NDH-DEPENDENT FLOW6 (NDF6), NDF1 (NDH48), NDF2 (NDH45), NDF4, and At CYP20-2 (Ishihara et al., 2007; Ishikawa et al., 2008; Majeran et al., 2008; Sirpiö et al., 2009a, 2009b; Takabayashi et al., 2009). However, these NDH subunits do not contain an NAD(P)H binding motif. Although CRR1 has an NAD(P)H binding domain, it is localized to the stroma and unlikely to be an NDH subunit (Shimizu and Shikanai, 2007). Chloroplast NDH may accept electrons from donors other than NAD(P)H.
It is believed that chloroplast NDH originated from cyanobacterial NDH-1 (Shikanai, 2007b). Proteomics studies revealed that three types of NDH-1 exist in cyanobacteria: NDH-1L, NDH-1M, and NDH-1S, with molecular masses of ∼460, 350, and 200 kD, respectively (Herranen et al., 2004). Mass spectrometry analysis revealed that NDH-1M consists of 13 subunits, including a membrane-embedded arm (NdhA to C, E, G, and L) and a hydrophilic connecting domain (NdhH to K and M to O). NDH-1L includes NdhD1 and NdhF1 in addition to the NDH-1M complex (Prommeenate et al., 2004; Battchikova et al., 2005; Zhang et al., 2005). The NDH-1S complex comprises NdhD3, NdhF3, CupA, and CupS (Ogawa and Mi, 2007) and interacts with NDH-1M to form the functional complex NDH-1MS, which is induced under low CO2 conditions (Zhang et al., 2005). While NDH-1L is involved in respiratory and PSI cyclic electron transport, the NDH-1MS complex is considered to be participated in CO2 uptake in cyanobacteria (Battchikova and Aro, 2007; Ogawa and Mi, 2007). Although several copies of NdhD and NdhF genes were found in cyanobacteral genomes, only NdhD1/D2 and NdhF1 are related to chloroplast ndhD and ndhF genes, respectively. Additionally, the CupA and CupS subunits of the cyanobacteral NDH-1S complex have no counterparts in higher plants. These facts suggest that the structure of chloroplast NDH is similar to the NDH-1L complex in cyanobacteria (Battchikova and Aro, 2007; Ogawa and Mi, 2007; Shikanai, 2007b). However, identification of several novel subunits specific to higher plants and biochemical characterization of chloroplast NDH imply that chloroplast NDH is equipped with additional devices compared with cyanobacterial NDH-1L (Majeran et al., 2008; Peng et al., 2008; Sirpiö et al., 2009a, 2009b; Suorsa et al., 2009; Takabayashi et al., 2009). In particular, a 1000-kD bundle sheath cell-specific NDH complex associated with more than 15 proteins was suggested in maize (Zea mays), and the authors speculate that this novel complex possibly functions in inorganic carbon concentration in addition to PSI cyclic electron transport (Majeran et al., 2008). However, subunits included in the cyanobacterial NDH-1S complex were not discovered in maize.
The x-ray crystal structure of plant PSI reveals that it is composed of a reaction center (RC; PsaA to L and PsaN to P) and light-harvesting complex I (LHCI), which is composed of four different LHC proteins (Lhca1 to 4) (Amunts et al., 2007). The four LHC proteins assemble into two dimers and attach to the PsaF side of the PSI RC (reviewed in Melkozernov et al., 2006; Nelson and Yocum, 2006). Two additional, minor LHCI-like proteins (Lhca5 and Lhca6), with a high degree of similarity to Lhca1 to 4, were identified in the Arabidopsis genome (Jansson, 1999). Recently, Lhca5 was shown to be associated with PSI only in substoichiometric amounts (Ganeteg et al., 2004). Chemical cross-linking studies revealed that Lhca5 interacts with LHCI in the Lhca2/Lhca3 site (Lucinski et al., 2006). The Arabidopsis Lhca6 gene was originally classified as an Lhca2 gene because of their high similarity (Zhang et al., 1994). The scarce information on this protein still cannot resolve the question raised by Jansson (1999): “is the unusual Lhca2 gene (Lhca6) a nonexpressed pseudogene, or does it have a specific function”? The low expression level and different expression pattern under different conditions of Lhca6 compared with Lhca1-4 suggest that Lhca6 would have a distinct function from the major Lhca (Lhca1-4) (Klimmek et al., 2006).
Recently, it was found that thylakoid protein PGR5-Like 1 (PGRL1) was involved in PGR5-dependent PSI cyclic electron transport (DalCorso et al., 2008). PGRL1 and PGR5 interact physically, and this complex further associates with PSI, probably facilitating the operation of PSI cyclic electron transport (DalCorso et al., 2008). In our previous study, we discovered a novel NDH-PSI supercomplex with a molecular mass of >1000 kD in Arabidopsis and its putative subsupercomplex with a slightly lower molecular mass in mutants lacking NdhL or NdhM (Peng et al., 2008). Here, we give evidence that Lhca5 and Lhca6 are required for the formation of this full-size NDH-PSI supercomplex. Furthermore, we found that the interaction of NDH and PSI favors the in vivo function of NDH.
RESULTS
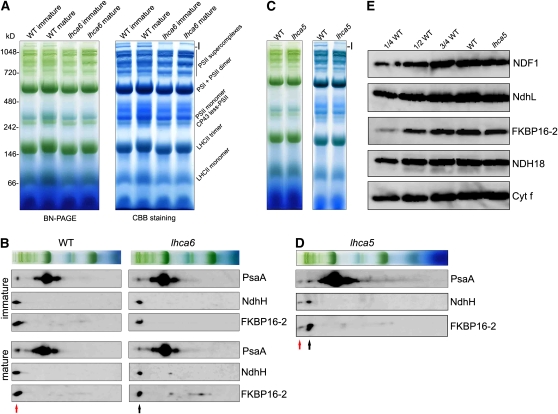
Mass Analysis of the NDH-PSI Supercomplex
The NDH-PSI supercomplex was detected by blue native (BN)-PAGE as a high molecular weight green band, band I, but it was shifted to the smaller molecular weight position of band II in the NdhL-defective ndhl/chlororespiratory reduction 23 (crr23) mutant of Arabidopsis (Peng et al., 2008; Shimizu et al., 2008). To investigate the components of the NDH-PSI supercomplex and its putative subsupercomplex, we excised two bands containing them from BN-PAGE (bands I and II). We digested the proteins in the gel with trypsin and analyzed the extracted peptides by linear ion-trap triple quadrupole (LTQ)-Orbitrap mass analysis, which provides high mass accuracy, high resolution, and high sensitivity. Hundreds of proteins were identified in the NDH-PSI supercomplex corresponding to band I (Table 1; see Supplemental Data Set 1 online). Consistent with our previous conclusion that band I contains the PSI-NDH supercomplex (Peng et al., 2008), the identified proteins were classified mainly into three groups (Table 1). The first group contains almost all of the PSI subunits, including Lhca1-4 and two minor LHCI-like proteins Lhca5 and Lhca6. NDH subunits conserved in the cyanobacterial complex were classified into the second group; these included all the NDH subunits, NdhA-O, except NdhG. In addition, our mass analysis revealed some candidates for novel NDH subunits, including PPL2, NDF1 (NDH48), NDF2 (NDH45), NDF4, and NDF6, which can be classified as genuine NDH subunits (Ishihara et al., 2007; Ishikawa et al., 2008; Sirpiö et al., 2009a; Takabayashi et al., 2009). Several proteins identified in maize and Arabidopsis NDH complexes by proteomics analysis (Majeran et al., 2008; Sirpiö et al., 2009a) were detected and classified into the third group. CYP20-2 (Tlp20), which was recently shown to be an auxiliary subunit of the NDH complex (Sirpiö et al., 2009b), is a peptidyl-prolyl cis/trans isomerase of 20 kD present in the thylakoid lumen (Edvardsson et al., 2003). FKBP16-2, two PsbQ family proteins (PsbQ-F1 and PsbQ-F2), and transmembrane protein NDH18 were also detected (Table 1). FKBP16-2 and NDH18 were shown to be NDH subunits in this study (see later). Besides the PSI and NDH subunits, our mass analysis also found many proteins that were not known to be related to NDH (see Supplemental Data Set 1 online), such as the photosystem II (PSII) and ATPase subunits and other thylakoid membrane proteins, a result that is inconsistent with our previous immunoblot studies (Peng et al., 2008). The samples were inevitably contaminated by other protein complexes owing to the limited resolution of BN-PAGE, and the LTQ-Orbitrap mass analysis was sensitive enough to detect them. It is also possible that some other previously unknown NDH subunits are also included in this group.
Table 1.
Summary of the PSI and NDH Subunits Identified from LTQ-Orbitrap Mass Analysis of the NDH-PSI Supercomplex (Band I) and Subsupercomplex (Band II)
| AGI Code | Protein Name | Morwse Score | Band I Protein Match | Coverage (%) | Morwse Score | Band II Protein Match | Coverage (%) | |
|---|---|---|---|---|---|---|---|---|
| PSI | ATCG00350 | PsaA | 588 | 45 | 26.5 | 474 | 45 | 24.0 |
| Complex | ATCG00340 | PsaB | 1179 | 63 | 34.2 | 975 | 53 | 27.0 |
| ATCG01060 | PsaC | 69 | 6 | 29.6 | 51 | 5 | 29.6 | |
| AT4G02770 | PsaD-1 | 896 | 45 | 73.6 | 769 | 44 | 70.7 | |
| AT1G03130 | PsaD-2 | 848 | 43 | 73.5 | 706 | 44 | 70.6 | |
| AT4G28750 | PsaE-1 | 526 | 26 | 84.6 | 425 | 23 | 69.2 | |
| AT2G20260 | PsaE-2 | 449 | 24 | 84.8 | 407 | 17 | 60.0 | |
| AT1G31330 | PsaF | 558 | 36 | 46.2 | 524 | 36 | 53.4 | |
| AT1G55670 | PsaG | 212 | 9 | 21.9 | 248 | 11 | 21.9 | |
| AT3G16140 | PsaH-1 | 299 | 13 | 60.0 | 193 | 14 | 60.0 | |
| AT1G52230 | PsaH-2 | 378 | 15 | 60.0 | 165 | 14 | 60.0 | |
| AT1G30380 | PsaK | 72 | 10 | 33.8 | 86 | 9 | 19.2 | |
| AT4G12800 | PsaL | 543 | 18 | 27.4 | 278 | 12 | 27.4 | |
| AT1G08380 | PsaO | 131 | 4 | 21.4 | 53 | 3 | 21.4 | |
| AT3G54890 | Lhca1 | 305 | 16 | 24.1 | 336 | 19 | 24.1 | |
| AT3G61470 | Lhca2 | 412 | 18 | 45.9 | 297 | 13 | 36.2 | |
| AT1G61520 | Lhca3 | 736 | 24 | 27.5 | 667 | 25 | 27.5 | |
| AT3G47470 | Lhca4 | 981 | 34 | 68.1 | 666 | 28 | 68.9 | |
| AT1G45474 | Lhca5 | 720 | 32 | 42.6 | 421 | 22 | 36.3 | |
| AT1G19150 | Lhca6 | 499 | 25 | 44.1 | 301 | 17 | 35.9 | |
| NDH | ATCG01100 | NdhA | 716 | 22 | 35.0 | 431 | 14 | 23.1 |
| Complex | ATCG00890 | NdhB | 111 | 8 | 12.1 | 120 | 8 | 9.8 |
| ATCG00440 | NdhC | 93 | 2 | 20.8 | 97 | 3 | 20.8 | |
| ATCG01050 | NdhD | 410 | 14 | 15.4 | 259 | 12 | 14.2 | |
| ATCG01070 | NdhE | 99 | 5 | 8.9 | 71 | 4 | 8.9 | |
| ATCG01010 | NdhF | 1137 | 43 | 32.6 | 966 | 31 | 29.9 | |
| ATCG01110 | NdhH | 2884 | 114 | 81.9 | 0 | 0 | 0.0 | |
| ATCG01090 | NdhI | 897 | 35 | 61.6 | 0 | 0 | 0.0 | |
| ATCG00420 | NdhJ | 603 | 33 | 48.7 | 0 | 0 | 0.0 | |
| ATCG00430 | NdhK | 556 | 30 | 56.4 | 0 | 0 | 0.0 | |
| AT1G70760 | NdhL | 113 | 14 | 33.0 | 0 | 0 | 0.0 | |
| AT4G37925 | NdhM | 780 | 22 | 49.3 | 0 | 0 | 0.0 | |
| AT5G58260 | NdhN | 786 | 29 | 49.3 | 0 | 0 | 0.0 | |
| AT1G74880 | NdhO | 237 | 12 | 43.0 | 0 | 0 | 0.0 | |
| NDH | AT1G15980 | NDF1 (NDH48) | 1293 | 62 | 52.1 | 1228 | 63 | 53.6 |
| Candidates | AT1G64770 | NDF2 (NDH45) | 1374 | 57 | 61.5 | 1116 | 57 | 62.9 |
| AT3G16250 | NDF4 | 34 | 2 | 9.8 | 34 | 3 | 9.8 | |
| AT1G18730 | NDF6 | 360 | 16 | 42.9 | 418 | 18 | 42.9 | |
| AT2G39470 | PPL2 | 1071 | 51 | 60.1 | 838 | 49 | 63.9 | |
| AT1G14150 | PsbQ-F1 | 891 | 34 | 56.8 | 627 | 36 | 56.8 | |
| AT3G01440 | PsbQ-F2 | 813 | 27 | 41.4 | 627 | 28 | 41.4 | |
| AT5G13120 | CYP20-2 | 769 | 33 | 51.0 | 671 | 29 | 51.0 | |
| AT4G39710 | FKBP16-2 | 360 | 15 | 20.7 | 313 | 15 | 20.7 | |
| AT5G43750 |
NDH18 |
252 |
9 |
24.5 |
254 |
9 |
24.5 |
The complete list of proteins identified in bands I and II can be found in Supplemental Data Set 1 online. AGI, Arabidopsis Genome Initiative.
All the PSI subunits detected in band I were identified in the putative subsupercomplex corresponding to band II in ndhl (Table 1), which is consistent with our previous report (Peng et al., 2008). Interestingly, the membrane-embedded NDH subunits NdhA-F were detected in band II, but the hydrophilic subunits, NdhH-O, were absent. NdhG may be also included in band II, since no signals of this subunit were found in either band, possibly for technical reasons (Table 1). These facts imply that NdhL is essential for stabilizing the hydrophilic subunits in chloroplast NDH, and the membrane-embedded NDH subunits stably accumulate even in the absence of the subcomplex consisting of hydrophilic subunits and NdhL in chloroplasts.
NDH Subunits Encoded by At5g43750 and At4g39710
Our proteome analysis of the NDH-PSI supercomplex detected many proteins with unknown function, among which NDH subunits may be included. Recent bioinformatics studies identified several NDH subunits (Ishihara et al., 2007; Ishikawa et al., 2008; Takabayashi et al., 2009) in a strategy based on the phenomenon that the expression of genes encoding NDH subunits is coregulated. To select candidates for novel subunits in our analysis, we used a bioinformatics strategy to search for the genes coexpressed with nucleus-encoded NDH subunit genes in the ATTED-II coexpression database (http://www.atted.bio.titech.ac.jp/). Besides the genes already known to encode NDH subunits, two genes, At4g39710 and At5g43750, were coexpressed with NDH subunit genes with high r values (see Supplemental Table 1 online).
At5g43750 encodes a 212–amino acid protein with a 48–amino acid N-terminal plastid-targeting peptide (predicted by ChloroP; http://www.cbs.dtu.dk/services/ChloroP/) and a transmembrane domain (predicted by TMHMM; http://www.cbs.dtu.dk/services/TMHMM-2.0/) (see Supplemental Figure 1 online). We designated the protein encoded by this gene as NDH18 according to its apparent molecular mass. NDH18 is conserved among higher plants (see Supplemental Figure 1 online), but no homologs were found in cyanobacteria or Chlamydomonas reinhardtii.
At4g39710 was designated as FKBP16-2 and is localized to the lumen side of the thylakoid (He et al., 2004). FKBP16-2 shows significant sequence similarity to FKBP13 (At5g45680), which interacts with the Rieske Fe-S subunit of the cytochrome b6f complex (Gupta et al., 2002). Gm FKBP16-2, Os FKBP16-2, and Zm FKBP16-2 are classified into the same clade with At FKBP16-2 (66 to 73% identity) (Figure 1A), but At FKBP16-2 exhibits only 40 to 50% similarity to FKBP13s, and no proteins closely related to FKBR16-2 were found in Chlamydomonas or cyanobacteria. It is possible that, like NDH18, FKBP16-2 is an NDH subunit specific to chloroplasts.
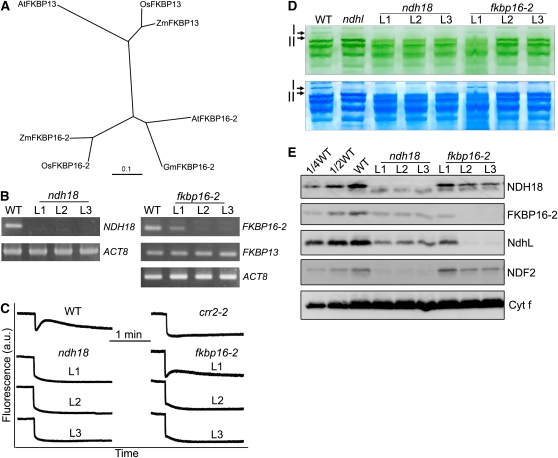
Figure 1.
Characterization of the ndh18 and fkbp16-2 Mutants.
(A) Phylogenetic tree of the FKBP13 and FKBP16-2 proteins. Sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). The sequences are named for each organism. The corresponding amino acid sequences were aligned with the ClustalW program with default settings, and an unrooted tree was constructed using TreeView software.
(B) RT-PCR analysis of NDH18 and FKBP16-2 mRNA. After reverse transcription, the cDNA was analyzed by 30 cycles of amplification with specific primers for NDH18, FKBP16-2, and FKBP13. ACT8 was used as an internal control.
(C) Monitoring of NDH activity by chlorophyll fluorescence. Four-week-old leaves were exposed to AL (50 μmol photons m−2 s−1) for 5 min. After illumination, the subsequent transient increase in chlorophyll fluorescence was monitored as an indicator of NDH activity. a.u.; arbitrary units. The fluorescence levels were standardized by the Fm levels.
(D) Thylakoid protein complexes isolated from the wild type, and RNAi lines (ndh18 and fkbp16-2) were separated by BN-PAGE (top panel) and stained with Coomassie Brilliant Blue (bottom panel). Band I, NDH-PSI supercomplex detected in the wild type; band II, subsupercomplex detected in ndhl. The top part of the gel is compressed.
(E) Immunodetection of NDH subunits in the wild type (including indicated serial dilutions) and ndhl, ndh18, and fkbp16-2 mutants. Immunoblotting was performed with antibodies against NDH18, FKBP16-2, NdhL, and NDF2 proteins. Thylakoid proteins were loaded on an equal chlorophyll basis to SDS-PAGE. Cytf is a loading control. L1, L2, and L3 represent three independent RNAi lines.
To further characterize the function of NDH18 and FKBP16-2, we used RNA interference (RNAi) to decrease mRNA levels of the NDH18 and FKBP16-2 genes. Three independent RNAi lines were characterized in detail for each gene. NDH18 mRNA was undetectable in the three ndh18 lines (L1 to L3) after 30 cycles of RT-PCR (Figure 1B). In two fkbp16-2 RNAi lines (L2 and L3), transcription was also below the detection limit by the same RT-PCR, but the gene expression was mildly suppressed in L1. RT-PCR analysis also showed that the expression of FKBP13 was not affected in fkbp16-2 lines (Figure 1B), indicating that FKBP16-2 was specifically knocked down.
A postillumination rise in chlorophyll fluorescence, which is due to the NDH-dependent reduction of PQ by the stromal electron pool in darkness, is widely used to monitor NDH activity (Burrows et al., 1998; Shikanai et al., 1998). Although this method does not analyze the rate of PSI cyclic electron transport in the light, it reflects NDH activity in vivo and has been used to isolate many mutants specifically defective in NDH activity (Hashimoto et al., 2003). In ndh18 (L1 to L3) and fkbp16-2 (L2 and L3), the transient increase in fluorescence was not detected, indicating that NDH activity was impaired (Figure 1C). Consistent with the result of RT-PCR (Figure 1B), NDH activity was detected in fkbp16-2 L1 but was lower than that in the wild type (Figure 1C). We conclude that NDH18 and FKBP16-2 are essential for NDH activity.
The NDH-PSI supercomplex was analyzed in ndh18 and fkbp16-2 lines by BN-PAGE (Figure 1D). Bands I and II are present at the top of the gel in the wild type and ndhl, respectively. However, both bands were missing in the ndh18 and fkbp16-2 lines, except for the accumulation of band I in fkbp16-2 L1 (Figure 1D). To confirm the absence of the supercomplexes, we excised the region corresponding to bands I and II from the BN gels. The immunoblot results showed that PsaA, NdhH, and NdhL were less than one-eighth of the wild-type levels except for fkbp16-2 L1 (see Supplemental Figure 2 online). Immunoblot analysis using antibodies against FKBP16-2 and NDH18 showed that both proteins were below the detection limit in RNAi lines except for fkbp16-2 L1 (Figure 1E). Consistent with the mRNA level and activity (Figures 1B and 1C) and the supercomplex level (see Supplemental Figure 2 online), fkbp16-2 L1 accumulated one-quarter of the wild-type level of FKBP16-2 (Figure 1E). Furthermore, the accumulation of NdhL and NDF2 was also greatly decreased in the ndh18 and fkbp16-2 lines (Figure 1E). These results demonstrate that NDH18 and FKBP16-2 are novel NDH subunits and are essential for complex stability. Since FKBP16-2 contains the FKBP domain, it may have another role in protein folding or/and in the complex assembly as suggested by Majeran et al. (2008).
Subunit Stability under the Different NDH Mutant Backgrounds
NDH activity was completely lost in ndh18 and fkbp16-2 (Figure 1C, L2 and L3). NdhL accumulation was more severely affected in fkbp16-2 than in ndh18, while that of NDF2 was more severely affected in ndh18 than in fkbp16-2 (Figure 1E). It was also reported that NDF2 accumulates stably in ndhl but unstably in ndhB-defective crr2-2 (Sirpiö et al., 2009a; Takabayashi et al., 2009). This genotype dependency of subunit stability may reflect the different localization of each subunit in the supercomplex. By characterization of subunit stability in different mutant backgrounds, it may be possible to determine the supercomplex structure. For this purpose, we performed a matrix analysis of protein blots using eight genotypes and antibodies against seven NDH subunits (Figure 2A). NdhM, NdhH, NDF1, and NDF2 are hydrophilic proteins (Rumeau et al., 2005; Sirpiö et al., 2009a; Takabayashi et al., 2009), but NdhL and NDH18 are predicted to contain a transmembrane domain (Shimizu et al., 2008; see Supplemental Figure 1 online). In addition, the translation of the membrane subunit NdhD was impaired in crr4-3 (Kotera et al., 2005). PPL2 and FKBP16-2 are localized to the lumen side of the complex (He et al., 2004; Ishihara et al., 2007). Immunoblot analysis showed that the stromal fraction does not contain NDH subunits (NdhL, NDF1, NDF2, NDH18, and FKBP16-2) in the wild type and even in various NDH-defective mutants (see Supplemental Figure 3 online), suggesting that the subunits are stable only when they are associated with thylakoid membranes.
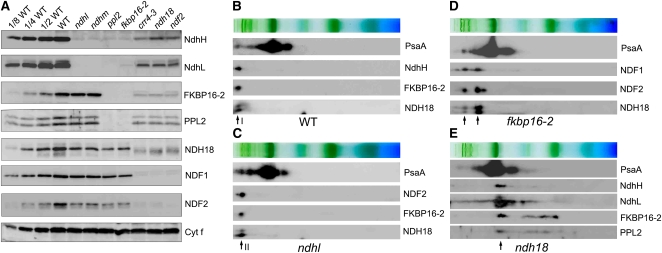
Figure 2.
Stability of Each NDH Subunit in Different NDH Mutant Backgrounds.
(A) Immunoblot of thylakoid membrane proteins, indicated at right, isolated from the wild type (including indicated serial dilutions) and from indicated mutants. Gels were loaded on an equal chlorophyll basis.
(B) to (E) Analysis of thylakoid protein complexes isolated from the wild type (B) and mutants defective in NDH subunits ([C], ndhl; [D], fkbp16-2; [E], ndh18). Complexes were separated by BN-PAGE and further subjected to 2D SDS-PAGE. The proteins were immunodetected with specific antibodies. Positions of band I (B), band II (C), and partially stable subsupercomplexes ([D] and [E]) are indicated by arrows.
Although the accumulation of NdhH and NdhL was almost completely impaired in ndhm and that of NdhH was also in ndhl, the levels of other subunits were more than half of the wild type (Figure 2A). BN-PAGE followed by second-dimensional (2D) SDS-PAGE and immunoblot analysis showed that all the stable subunits were present in band II in ndhl (Figures 2B and 2C), consistent with the mass analysis (Table 1). The results suggest that NdhH and NdhL form a subcomplex whose absence does not greatly affect the stability of other parts of the supercomplex. On the basis of the analogy with bacterial and mitochondrial complex I and cyanobacterial NDH-1 (Shikanai, 2007b) as well as our mass analysis of band II (Table 1), we consider that this putative subcomplex further includes NdhI-K and NdhM-O. The hydrophilic part of this subcomplex is likely to be anchored to the thylakoid membrane via NdhL (subcomplex A in Table 2; see Discussion for more detail), since NdhL has transmembrane domains and is specific to cyanobacteria and chloroplasts (Ogawa, 1992; Shimizu et al., 2008). However, analysis of the NdhL deletion mutant (M9) in Synechocystis sp PCC 6803 revealed that NdhL is not required for the interaction between the hydrophilic and membrane subcomplexes but is essential for NDH activity (Battchikova et al., 2005). These facts imply that the additional transmembrane domain specifically present in chloroplast NdhL may function in stabilizing the subcomplex A on the membrane subcomplex (Shimizu et al., 2008).
Table 2.
Summary of the Chloroplast NDH Subunits
| Subcomplex | Name | Annotation | Molecular Mass (kD) | Reference |
|---|---|---|---|---|
| Membrane | NdhA | ATCG01100 | 40.0 | |
| subcomplex | NdhB | ATCG00890 | 42.0 | |
| NdhC | ATCG00440 | 14.0 | ||
| NdhD | ATCG01050 | 57.0 | ||
| NdhE | ATCG01070 | 11.3 | ||
| NdhF | ATCG01010 | 85.0 | ||
| NdhG | ATCG01080 | 19.0 | ||
| Subcomplex A | NdhH | ATCG01110 | 45.5 | |
| NdhI | ATCG01090 | 20.0 | ||
| NdhJ | ATCG00420 | 20.0 | ||
| NdhK | ATCG00430 | 27.0 | ||
| NdhL | AT1G70760 | 17.0 | Shimizu et al. (2008) | |
| NdhM | AT4G37925 | 22.0 | Rumeau et al. (2005) | |
| NdhN | AT5G58260 | 18.0 | Rumeau et al. (2005) | |
| NdhO | AT1G74880 | 13.0 | Rumeau et al. (2005) | |
| Subcomplex B | NDF1 (NDH48) | AT1G15980 | 48.0 | Takabayashi et al. (2009); Sirpiö et al. (2009a) |
| NDF2 (NDH45) | AT1G64770 | 45.0 | Takabayashi et al. (2009); Sirpiö et al. (2009a) | |
| NDF4 | AT3G16250 | 22.0 | Takabayashi et al. (2009) | |
| NDF6 | AT1G18730 | 18.0 | Ishikawa et al. (2008) | |
| NDH18 | AT5G43750 | 18.0 | This work | |
| Lumen | PPL2 | AT2G39470 | 17.0 | Ishihara et al. (2007) |
| subcomplex | FKBP16-2 | AT4G39710 | 16.0 | This work |
| PsbQ-F1 | AT1G14150 | 17.0 | Majeran et al. (2008) | |
| PsbQ-F2 | AT3G01440 | 17.0 | Majeran et al. (2008) | |
| CYP20-2 |
AT5G13120 |
20.0 |
Majeran et al. (2008); Sirpiö et al. (2009b) |
The chloroplast NDH complex was divided into four subcomplexes. The localization of the subcomplexes is presented in Figure 9.
In ppl2 and fkbp16-2, not only PPL2 and FKBP16-2, but also NdhH and NdhL were absent (Figure 2A), indicating that PPL2 and FKBP16-2 are essential for stabilizing the subcomplex A. Levels of NDH18, NDF1, and NDF2 in ppl2 and fkbp16-2 were one-quarter to one-half of the wild-type levels. In the absence of FKBP16-2, PPL2 is missing and vice versa (Figure 2A), implying that PPL2 and FKBP16-2 form a subcomplex on the lumen side. This lumen subcomplex may include lumen proteins CYP20-2, PsbQ-F1, and PsbQ-F2, which were detected in our mass analysis (Table 1) and also in previous proteomic analyses (Majeran et al., 2008; Sirpiö et al., 2009a). The evidence of CYP20-2 as an auxiliary NDH subunit in Arabidopsis also supports the existence of the lumen subcomplex (Sirpiö et al., 2009b), although there is currently no direct biochemical data to conclude that the lumen subcomplex is present. BN-PAGE analysis detected two kinds of subsupercomplexes in fkbp16-2 (Figure 2D). The larger one includes PSI complex and may be the partially stable subsupercomplex lacking both the subcomplex A and lumen subcomplex. NDH subunits, NDF1, NDF2, and NDH18, were also detected in the smaller subsupercomplex (Figure 2D). The close migration of this subsupercomplex with the main PSI complex makes it difficult to conclude that the partially stable smaller subsupercomplex still includes PSI (Figure 2D). However, we showed that a partially stable subsupercomplex with the similar molecular weight was detected in crr2-2 defective in the accumulation of membrane subunit NdhB, and this subsupercomplex contains Lhca6, which is essential for the formation of the NDH-PSI supercomplex, as well as NDF1 and NDF2 (see below, Figure 6B, and Discussion). These results imply that the smaller partially stable subsupercomplex detected in fkbp16-2 may correspond to the subcomplex B associated with PSI (Table 2; see Discussion for more detail).
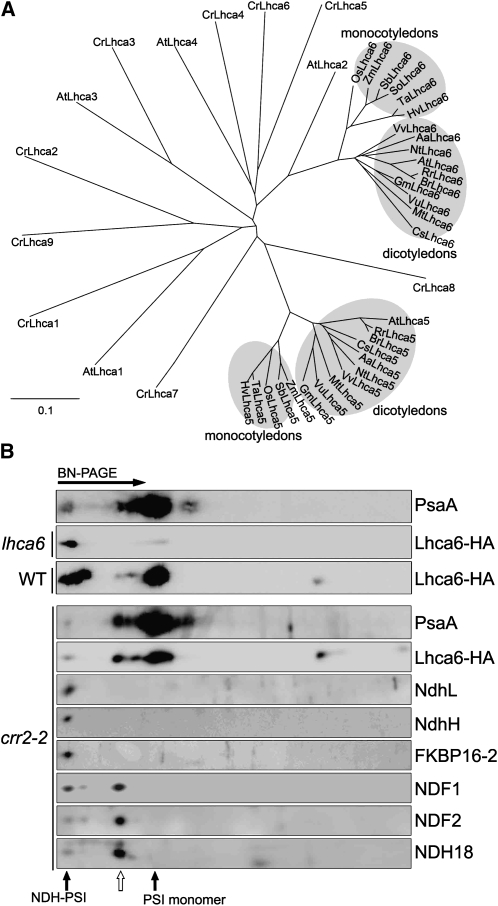
Figure 6.
Characterization of Lhca6.
(A) Phylogenetic tree of Lhca family proteins. Sequences of Lhca5 and Lhca6 were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). The sequences are named for each organism. The sequences of Lhca1-9 of Chlamydomonas and Lhca1-4 of Arabidopsis were also included in the analysis. The corresponding amino acid sequences were aligned with the ClustalW program with default settings, and an unrooted tree was constructed using TreeView software.
(B) Immunodetection of Lhca6 using a monoclonal antibody against the HA tag. The lhca6 plants were transformed with the Os Lhca6 gene fused to the sequence encoding the HA tag. Wild-type and crr2-2 plants were transformed with At Lhca6 cDNA fused to the region encoding the HA tag expressed under the control of the CaMV 35S promoter. Thylakoid complexes isolated from transformants were separated by BN-PAGE and further subjected to 2D SDS-PAGE. The proteins were immunodetected with specific antibodies. The positions of NDH-PSI supercomplex and PSI monomer are indicated by closed arrows. The position of the putative subsupercomplex detected in crr2-2 is indicated by an open arrow.
NDH18, NDF1, and NDF2 were absent in crr4-3, ndh18, and ndf2, yet approximately one-tenth of wild-type levels of NdhH, NdhL, FKBP16-2, and PPL2 accumulated in these mutants (Figure 2A). 2D/SDS-PAGE immunoblot also confirmed the high molecular weight complexes containing PSI corresponding to the supercomplexes detected in wild-type, ndhl, and fkbp16-2 plants were missing in the ndh18 mutant (Figure 2E). The remaining NDH subunits in ndh18 also form a subcomplex with a molecular mass of ∼500 kD, which is smaller than that of the PSI monomer (Figure 2E).
Stoichiometry of the NDH and PSI Subunits in the Supercomplex
To study the structure of the NDH-PSI supercomplex further, we determined the stoichiometry of the NDH and PSI subunits in the supercomplex through the use of antibodies (Figure 3). We purified the His-tagged recombinant PsaA, NDH18, and FKBP16-2 proteins from E. coli and used them as quantitative standards. The NDH-PSI supercomplex was separated from thylakoid membranes by BN-PAGE, denatured in the gel, and then directly used for SDS-PAGE. The levels of PsaA, NDH18, and FKBP16-2 were approximately estimated by the quantitative immunoblotting to be 0.21, 0.20, and 0.19 pmol in the NDH-PSI supercomplex corresponding to thylakoid membranes containing 10 μg chlorophyll, respectively (Figure 3). Since the recovery of proteins from BN gel may depend on the nature of each protein, we cannot conclude the stoichiometry exactly. But the levels of PsaA, NDH18, and FKBP16-2 are estimated to be roughly equimolar (Figure 3), consistent with the idea that NDH18 and FKBP16-2 are subunits of the NDH-PSI supercomplex.
Figure 3.
Stoichiometric Analysis of NDH and PSI in the Supercomplex.
The NDH-PSI supercomplex corresponding to band I was excised from the BN-PAGE and denatured in gel. Dilution series of the purified His-tagged recombinant PsaA (A), NDH18 (B), and FKBP16-2 (C) proteins were used to estimate the amount of NDH subunits and PsaA in the supercomplex. The signal was visualized by an LAS3000 chemiluminescence analyzer (Fuji Film) and analyzed by Imagemaster software (Amersham Pharmacia Biotech). The result is representative of three experiments using independently isolated thylakoid membranes. Control immunoblots confirmed that the antibodies did not recognize the His-tag or Nus-tag. I, NDH-PSI supercomplex isolated from thylakoids containing 10 μg chlorophyll.
Lhca6 and Lhca5 Are Required for the Full-Size NDH-PSI Supercomplex Formation
Our discovery raised a question of how NDH interacts with PSI. Besides NDH18 and FKBP16-2, Lhca6 was also found to be coexpressed with NDH subunit genes (see Supplemental Table 1 online). It is possible that Lhca6 is specifically required for stabilizing the NDH-PSI supercomplex. To study this possibility, we generated Lhca6 RNAi lines of Arabidopsis. For vector construction, we chose the region encoding the 5′-untranslated region and the plastid-targeting signal of Lhca6, which shows low similarity to Lhca2. Several independent lines (lhca6) showed no visibly different phenotype (see Supplemental Figure 4 online). Lhca6 expression was specifically knocked down, but the Lhca2 mRNA level was not affected (Figure 4A).
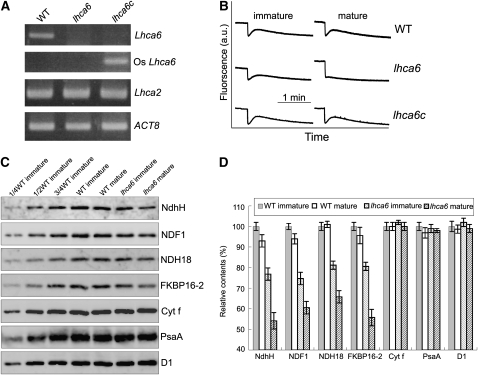
Figure 4.
Characterization of the lhca6 RNAi Lines.
(A) RT-PCR analysis of Lhca6 and Lhca2 mRNA. Total RNA (5 μg) was reverse transcribed, and the resulting cDNA was used in 30 cycles of PCR with specific primers for Lhca6, Os Lhca6, and Lhca2. ACT8 was used as an internal control. “lhca6c” indicates lhca6 RNAi mutant complemented by the introduction of Os Lhca6 gene.
(B) Monitoring of NDH activity by chlorophyll fluorescence as in Figure 1C.
(C) Immunodetection of chloroplast proteins from immature and mature leaves of the wild type and lhca6. The thylakoid membrane proteins were separated by SDS-PAGE and immunodetected with specified antibodies. Thylakoid proteins were loaded on an equal chlorophyll basis. These experiments were repeated three times independently, and similar results were obtained. Results from a representative experiment are shown.
(D) Analysis of thylakoid proteins. Immunoblot results of three independent isolations of thylakoid membranes were analyzed with Imagemaster software (Amersham Pharmacia Biotech). The protein levels in the wild-type mature leaves and lhca6 immature and mature leaves are shown relative to those in the wild-type immature leaves (100%). Means ± sd (n = 3).
To check whether NDH activity was affected in lhca6, we measured the transient increases in chlorophyll fluorescence after actinic light (AL) was turned off. NDH activity was detected in immature leaves of lhca6 (leaf age is shown in Supplemental Figure 4 online), although it seems slightly lower than that in the wild type (Figure 4B). By contrast, mature leaves of lhca6 showed no chlorophyll fluorescence increase (Figure 4B). To confirm that this phenotype is due to the defective expression of Lhca6, we introduced the genomic Lhca6 sequence of rice (Oryza sativa; Os Lhca6) into Arabidopsis lhca6 lines. Owing to its sequence differences in the region encoding the 5′-untranslated region and plastid-targeting signal, the rice gene escaped RNAi and fully restored the transient increase in chlorophyll fluorescence in mature leaves (Figures 4A and 4B, lhca6c).
Immunoblotting analysis provided direct evidence that NDH content was reduced slightly in immature leaves of lhca6 (∼80%) compared with the wild type and more so (∼60%) in mature leaves (Figures 4C and 4D). However, subunits of the other protein complexes, D1, PsaA, and Cytf, were not affected (Figures 4C and 4D), indicating that accumulation of the NDH complex was specifically affected in the absence of Lhca6, especially in mature leaves. Surprisingly, BN-PAGE showed that the NDH-PSI supercomplex corresponding to band I was completely absent in both immature and mature leaves of lhca6 (Figure 5A; see Supplemental Figure 5 online), and NDH subunits were present in a complex of ∼1000 kD (Figure 5B). We conclude that Lhca6 is essential for the full-size NDH-PSI supercomplex formation but not for the activity detected in the chlorophyll fluorescence analysis in immature leaves (Figure 4B).
Figure 5.
Analysis of Thylakoid Protein Complexes from Wild-type, lhca6, and lhca5 Plants.
(A) Thylakoid protein complexes isolated from immature and mature leaves of wild-type and lhca6 plants were separated by BN-PAGE (left) and stained with Coomassie Brilliant Blue (CBB) (right). Band I position is indicated.
(B) Thylakoid membrane complexes separated by BN-PAGE in (A) were further subjected to 12.5% 2D SDS-PAGE, and the proteins were immunodetected with specific antibodies against PsaA, NdhH, and FKBP16-2. Positions of NDH-PSI supercomplex and the smaller NDH-PSI supercomplex are indicated by red and black arrows, respectively.
(C) Thylakoid protein complexes isolated from wild-type and lhca5 plants were separated by BN-PAGE (left) and stained with Coomassie blue (right). Band I position is indicated.
(D) Thylakoid protein complexes isolated from the lhca5 plants were separated by BN-PAGE and further subjected to 2D SDS-PAGE. The proteins were immunodetected with specific protein antibodies against PsaA, NdhH, and FKBP16-2. Positions of the NDH-PSI supercomplex and the smaller NDH-PSI supercomplex are indicated by red and black arrows, respectively.
(E) Immunodetection of chloroplast proteins from the wild type and lhca5. The thylakoid membrane proteins were separated by SDS-PAGE and immunodetected with antibodies against the indicated proteins. Thylakoid proteins were loaded on an equal chlorophyll basis.
Since Lhca5 was also detected in the NDH-PSI supercomplex by mass analysis (Table 1), we analyzed Lhca5 knockout lines of Arabidopsis and found that the NDH-PSI supercomplex corresponding to band I was absent and the levels of NDH subunits were slightly decreased (Figures 5C and 5E). 2D BN/SDS-PAGE and immunoblotting studies showed that the NDH subunits were present mainly in a complex of ∼1000 kD and in trace amounts in the NDH-PSI supercomplex corresponding to band I in lhca5 (Figure 5D). To estimate the levels of residual supercomplex in lhca5 and lhca6 further, we excised the region corresponding to the NDH-PSI supercomplex from BN gels. Immunoblot results showed that signals of PsaA, NdhH, and NdhL were below the detection limit in lhca6 RNAi lines (at least less than one-sixteenth of the wild-type levels), and approximately one-sixteenth of the supercomplex was still present in the lhca5 mutant compared with the wild type (see Supplemental Figure 5 online).
In BN gel, NDH subunits were detected in the 1000-kD complex exclusively in lhca6 and mainly in lhca5 (Figures 5B and 5D). Unexpectedly, this 1000-kD complex still includes PsaA, although the PsaA level is reduced compared with that in the NDH-PSI supercomplex (Figures 5B and 5D). These results can be explained by an idea that the NDH complex interacts with multiple copies of the PSI complex (see Discussion). Further analyses confirmed that this 1000-kD complex also contains Lhca3 and NDH subunits, NDH18 and NDF1 (see Supplemental Figure 6 online), suggesting that the complex observed in lhca5 and lhca6 is a smaller version of the NDH-PSI supercomplex, which contains entire subunits of NDH and at least a single copy of PSI.
Specific Function of Lhca6 in the NDH-PSI Supercomplex
EST databases included 22 and 18 putative orthologs of Arabidopsis Lhca6 and Lhca5, respectively, in flowering plants, but no homologs were found in Chlamydomonas, indicating that Lhca5 and Lhca6 are conserved among flowering plants. Although Lhca6 is homologous to Lhca2 (Jansson, 1999), the Lhca6 orthologs were not clustered with At Lhca2 (Figure 6A). Furthermore, as indicated by the phenotype of lhca6 (Figures 4 and 5), Lhca2 clearly could not complement the function of Lhca6, even though the level of Lhca2 mRNA was much higher than that of Lhca6 (Figure 4A). This fact supports the idea that Lhca6 is not an isoform of Lhca2 and has a different physiological function. Protein alignment revealed that mature Lhca6 has an N-terminal extension (see Supplemental Figure 7 online), implying a specific function of Lhca6 in this region.
Lhca5 is associated with PSI monomer in substoichiometric amounts (Ganeteg et al., 2004). To study whether Lhca6 also binds to PSI monomer, we excised the band corresponding to PSI monomer from the BN gel and used it in LTQ-Orbitrap mass analysis. Although Lhca1-5 were detected in PSI monomer isolated from the wild type, crr2-2, and lhca6, no Lhca6 signal was found in our mass analysis (see Supplemental Data Set 2 online). We also constructed a chimeric gene in which the C-terminal end of Os Lhca6 was fused with HA (influenza hemagglutinin protein epitope) tag (Os Lhca6-HA) under the control of the Os Lhca6 promoter. The construct was then introduced into Arabidopsis lhca6 lines and complemented NDH activity. BN-PAGE and immunoblot studies showed that Lhca6 was associated mainly with the NDH-PSI supercomplex (Figure 6B), indicating that Lhca6 is predominantly present in the NDH-PSI supercomplex. We also transformed wild-type Arabidopsis and crr2-2 with Lhca6-HA tag under the control of the cauliflower mosaic virus (CaMV) 35S promoter. BN-PAGE and immunoblot studies showed that Lhca6 was associated with both the NDH-PSI supercomplex and PSI monomer in the wild type probably due to the result of overaccumulation but exclusively with PSI monomer in crr2-2 (Figure 6B). These results indicate that Lhca6 is stable in the absence of NDH and can interact with PSI monomer in the same way as Lhca5 (Ganeteg et al., 2004). However, PSI monomer is ∼100 times as common as NDH-PSI supercomplex (Peng et al., 2008), indicating that Lhca6 has a much higher affinity for NDH than for PSI monomer. We conclude that Lhca6 is an LHCI specifically required for NDH-PSI supercomplex formation, consistent with the fact that the gene is not conserved in Chlamydomonas, which does not contain chloroplast NDH.
The trace level of Lhca6-HA was detected in the NDH-PSI supercomplex position in crr2-2 probably because of the leaky accumulation of the NDH complex in this mutant (Figure 6B; Hashimoto et al., 2003; Peng et al., 2008). Besides PSI monomer and the NDH-PSI supercomplex, Lhca6 was also associated with a putative subsupercomplex that contains NDF1, NDF2, and NDH18 but does not contain NdhL, NdhH, or FKBP16-2 (Figure 6B). These results suggest that the subcomplex B, including NDF1, NDF2, and NDH18, may be still associated with PSI via Lhca6 even in the absence of the membrane subcomplex including NdhB.
Lhca6 Is Required for Efficient Operation of NDH
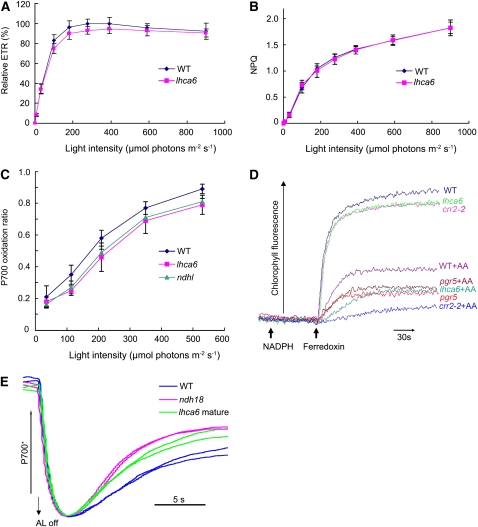
Although lhca6 lines contained 60 to 80% of the wild-type levels of NDH subunits, NDH activity was undetectable in mature leaves (Figure 4B). To study the link between the supercomplex formation and activity, we further analyzed the photosynthetic electron transport in mature leaves of lhca6. We determined two chlorophyll fluorescence parameters, electron transport rate (ETR) and nonphotochemical quenching (NPQ), since they reflect even subtle defects in photosynthetic apparatus (Shikanai et al., 1999). ETR was slightly reduced and NPQ was not affected in mature leaves of lhca6 (Figures 7A and 7B). The P700+ (oxidized RC chlorophyll of PSI) level, which is a sensitive indicator of the capacity for electron acceptance from PSI, in lhca6 mature leaves was similar to that in ndhl, but slightly lower than that in the wild type (Figure 7C). These phenotypes are consistent with those of other Arabidopsis mutants defective in chloroplast NDH (Munekage et al., 2004). We also assayed Fd-dependent PQ reduction activity, which is involved in PSI cyclic electron transport in vivo, using ruptured chloroplasts isolated from mature leaves of lhca6 (Figure 7D). As in crr2-2, PQ reduction activity was slightly lower in lhca6 than in the wild type. Antimycin A inhibits the PGR5-dependent pathway of PQ reduction by Fd (Munekage et al., 2004) and can be used to discriminate NDH-dependent activity from PGR5-dependent activity. Antimycin A decreased the PQ reduction activity slightly less in lhca6 than in crr2-2. The remaining Antimycin A–resistant PQ reduction activity may depend on the smaller version of NDH-PSI supercomplex, which is still present in lhca6 mature leaves (Figures 4 and 5). We also compared the oxidation kinetics of P700 by far-red light (FR) after AL illumination between wild-type, ndh18, and lhca6 mature leaves (Figure 7E). After 2-min illumination of AL (900 μmol photons m−2 s−1) supplemented with FR, AL was turned off and P700+ was transiently reduced by electrons from the PQ pool, and subsequently P700 was reoxidized by background FR. The operation of NDH retards the reoxidation of P700 by transferring electrons from the reduced stromal pool to PQ (Shikanai et al., 1998). The reoxidation of P700 was slower in wild-type leaves than ndh18 and lhca6 mature leaves (Figure 7E), which is consistent with the phenotypes observed in the ΔndhB tobacco mutant (Shikanai et al., 1998). From these results, we conclude that NDH activity was affected in the mature leaves of lhca6 mutant.
Figure 7.
In Vivo and in Vitro Analysis of Electron Transport Activity.
(A) The ETR is depicted relative to the maximum value of ΦPSII × light intensity in the wild type (100%).
(B) Dependence of NPQ of chlorophyll fluorescence on light intensity.
(C) Light intensity dependence of the P700 oxidation ratio (ΔA/ΔAmax) in ndhl, lhca6, and wild-type mature leaves.
(D) Increases in chlorophyll fluorescence by addition of NADPH (0.25 mM) and Fd (5 μM) under weak illumination (1.0 μmol photons m−2 s−1) were monitored in osmotically ruptured chloroplasts (20 μg chlorophyll/mL) of wild-type, lhca6, crr2-2, and pgr5 mature leaves. Ruptured chloroplasts were incubated with 10 μM Antimycin A before measurement. All values are mean ± sd (n = 5) in (A) to (C). This is a representative result of three experiments using thylakoid membranes independently isolated.
(E) Redox kinetics of P700 after termination of AL illumination (900 μmol photons m−2 s−1 for 2 min) under a background of FR. The leaves were illuminated by AL supplemented with FR to store electrons in the stromal pool. After termination of AL illumination, P700+ was transiently reduced by electrons from the PQ pool; thereafter, P700 was reoxidized by background FR. The redox kinetics of P700 was recorded. The P700+ levels were standardized by their maximum levels by exposing FR. The results using two independent plants for each genotype are overlapped.
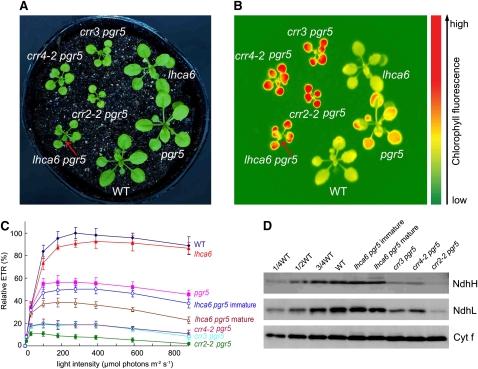
To further investigate the physiological significance of the NDH-PSI supercomplex formation, we constructed the lhca6 pgr5 double mutant. At a light intensity of 50 μmol photons m−2 s−1, lhca6 pgr5 plant growth was impaired, as in crr4-2 pgr5 and crr3 pgr5, but slightly better than in crr2-2 pgr5 (Figure 8A). Among the three crr mutants, the NDH level was most severely affected in crr2-2, resulting in a smaller plant (Figures 8A and 8D; Munekage et al., 2004). Steady state chlorophyll fluorescence captured by a CCD camera was compared among mutants at a light intensity of 100 μmol photons m−2 s−1 (Figure 8B). The chlorophyll fluorescence level was similar between the wild type and lhca6, suggesting that the NDH-PSI supercomplex formation is not essential for photosynthesis (Munekage et al., 2004). However, pgr5 displayed slightly higher chlorophyll fluorescence than the wild type, indicating a mild impairment of photosynthetic electron transport at this light intensity. Although NDH is dispensable for photosynthesis under these conditions, it is essential for efficient photosynthesis in the pgr5 mutant background, thus resulting in the drastic phenotype in the double mutants (Figures 8A and 8B; Munekage et al., 2004). The chlorophyll fluorescence level was high in lhca6 pgr5, suggesting a defect in photosynthesis, as in the other double mutants (Figure 8B).
Figure 8.
Characterization of the lhca6 pgr5 Double Mutant.
(A) Visible phenotype of the double mutants. Seedlings were cultured at 50 μmol photons m−2 s−1 for 3 weeks after germination.
(B) High chlorophyll fluorescence phenotype of lhca6 pgr5. Dark-adapted seedlings of the wild type and knockout mutants were illuminated at 100 μmol photons m−2 s−1 for 1 min and then a chlorophyll fluorescence image was captured by CCD camera. Arrows indicate immature leaves of lhca6 pgr5 emitting less fluorescence than mature leaves.
(C) Light intensity dependence of the relative ETR. ETR is shown relative to the maximum ETR in the wild type (100%); means ± sd (n = 5).
(D) Immunodetection of chloroplast proteins. The thylakoid membrane proteins were separated by SDS-PAGE and immunodetected with antibodies against NdhH, NdhL, and Cytf proteins. Thylakoid proteins were loaded on an equal chlorophyll basis.
To further characterize the photosynthetic activity in lhca6 pgr5, we analyzed the light intensity dependence of ETR (Figure 8C). Although the maximum ETR in pgr5 decreased to ∼60% of the wild-type level, it was reduced to 10 to 20% in the crr4-2 pgr5, crr3 pgr5, and crr2-2 pgr5 double mutants. Although ETR was lower in mature leaves of lhca6 pgr5 than in pgr5, it was higher than in the other double mutants (Figure 8C). ETR was significantly higher in immature leaves than in mature leaves, as reflected in the chlorophyll fluorescence image (Figures 8A and 8B). The NDH subunit levels in lhca6 pgr5 were more than three-quarters of those in the wild type (Figure 8D), and the levels were even higher than those in the lhca6 single mutant (Figure 4D). As expected, the other double mutants accumulated less than half of the wild-type level of NDH subunits (Figure 8D). Although lhca6 pgr5 accumulated >75% of the wild-type level of the NDH complex, plant photoautotrophic growth and photosynthesis were severely impaired in the pgr5 mutant background.
DISCUSSION
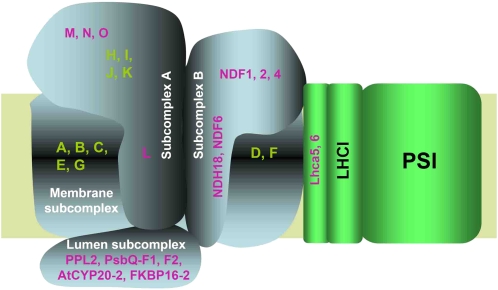
The discovery of several chloroplast-specific NDH subunits (Figure 1; Ishihara et al., 2007; Ishikawa et al., 2008; Majeran et al., 2008; Sirpiö et al., 2009a; Takabayashi et al., 2009) makes it likely that chloroplast NDH has a more complex structure than cyanobacterial NDH. Taking together all the available information, we present a structural model of the NDH-PSI supercomplex (Table 2, Figure 9). On the basis of the analogy with E. coli and cyanobacterial NDH-1 (Herranen et al., 2004; Zhang et al., 2004, 2005; Shikanai 2007b), it is likely that membrane-spanning subunits corresponding to chloroplast NdhA-G form the membrane subcomplex, and hydrophilic subunits corresponding to NdhH-K and NdhM-O form one of the stroma-side subcomplexes (Table 2, Figure 9). Although NdhL contains three transmembrane domains (Shimizu et al., 2008), our results suggest the direct interaction among NdhL and hydrophilic subunits NdhH-K and NdhM-O in chloroplast NDH. We grouped these subunits, including NdhL, into the subcomplex A to distinguish it from subcomplex B, which consists of chloroplast-specific subunits (Table 2, Figure 9).
Figure 9.
A Schematic Model of the NDH-PSI Supercomplex in Chloroplasts.
The chloroplast NDH is divided into four subcomplexes (Table 2). The membrane subcomplex contains of plastid-encoded subunits NdhA-G. The subcomplex A consists of plastid-encoded NdhH-K and nuclear-encoded NdhL-O. The NdhL subunit is a transmembrane protein and is required for stabilizing this subcomplex. The lumen subcomplex may include PPL2, PsbQ-F1, PsbQ-F2, AtCYP20-2, and FKBP16-2 and is essential for stabilizing the subcomplex A, probably via NdhL or other unidentified membrane proteins. The subcomplex B contains NDF1 (NDH48), NDF2 (NDH45), NDF4 (Sirpiö et al., 2009a; Takabayashi et al., 2009), and two transmembrane proteins, NDF6 and NDH18. The subcomplex A partially protects NDF1 from protease attack, suggesting that it also interacts with the subcomplex B (Sirpiö et al., 2009a). The subcomplex B is partially unstable in the absence of the subcomplex A and lumen subcomplexes. Without the membrane subunit NdhB, the subcomplex B still interacts with Lhca6. However, the subcomplex B is totally unstable without the membrane subunit NdhD in the crr4-3 mutant, suggesting that it also interacts with NdhD or/and NdhF. Subunits of the subcomplex B and lumen subcomplex are specific to chloroplast NDH. The two minor LHCI proteins Lhca5 and Lhca6 are required for the full-size NDH-PSI supercomplex formation. The model does not include the information of stoichiometry, which is discussed in the main text in detail.
Our results suggest that lumen proteins PPL2 and FKBP16-2 form a putative subcomplex on the lumen side (Figure 2A). CYP20-2, PsbQ-F1, and PsbQ-F2 may also be included in this lumen subcomplex (Table 2, Figure 9). Subunits of subcomplex A are unstable without subunits of the lumen subcomplex (Figure 2A), suggesting that subcomplex A may interact with the lumen subcomplex. This idea is consistent with the discovery of a 500-kD complex including the subunits of subcomplex A and lumen subcomplex in ndh18 (Figure 2E) and with trace levels of a complex of similar molecular weight in mature leaves of lhca6 (Figure 5B). NdhL and/or other unidentified transmembrane subunits may intermediate in the binding of the two subcomplexes separated by thylakoid membranes. The subunits of the lumen subcomplex do not exist in cyanobacteria, implying that higher plants developed a novel mechanism to stabilize subcomplex A.
NDF1 (NDH48), NDF2 (NDH45), and NDF4 are attached to the membrane subcomplex on the stromal side (Sirpiö et al., 2009a; Takabayashi et al., 2009). NDH18, NDF1, and NDF2 were missing in ndf2 and ndh18 and in NdhD-defective crr4-3 (Figure 2A). These results suggest that the subcomplex including NDF1, NDF2, and NDH18 is associated, directly or indirectly, with the membrane subunit NdhD. This idea is further supported by the similar NDH subunit accumulation profile in crr4-3, ndh18, and ndf2: the complete lack of subcomplex B including NDF1, NDF2, and NDH18, the low-level accumulation of subcomplex A including NdhH and NdhL (<10%), and the milder effect on the lumen subcomplex including PPL2 and FKBP16-2 (<20%) (Figure 2A). We classified NDF1, NDF2, and NDH18 into the subcomplex B, in which all subunits are specific to chloroplasts (Table 2, Figure 9). NDF4 and NDF6 probably belong to this group (Ishikawa et al., 2008; Takabayashi et al., 2009). In the absence of the subcomplex A and lumen subcomplex, the subcomplex B and membrane subcomplex still associate with PSI (larger subsupercomplex in fkbp16-2 mutant), although this subsupercomplex is partially unstable (Figures 2D and 9). The subcomplex B still associates with Lhca6 without the membrane subunit NdhB (Figures 6B) forming a partially stable subsupercomplex whose molecular weight is similar to that of the smaller subsupercomplex detected in fkbp16-2 (Figure 2D). These results imply that the NDH complex interacts with PSI probably via the subcomplex B and also possibly via membrane subunits NdhD and/or NdhF (Figure 9) based on the analogy with cyanobacterial NDH-1L whose membrane complex easily dissociates into the core domain consisting of NdhA-C, E, G, and L and the NdhD/F subcomplex (Battchikova and Aro, 2007).
Immunoblots showed that Lhca6 was mainly detected in the NDH-PSI supercomplex (Figure 6B). In agreement with our results (Table 1; see Supplemental Data Set 2 online), Lhca6 was not detected in the PSI monomer in various plant species by mass spectrometry analysis (Zolla et al., 2007). These facts suggest that Lhca6 is mainly localized to the NDH-PSI supercomplex, although we do not eliminate the possibility that an extremely low level of Lhca6 associates with monomeric PSI since they can interact in the Lhca6 overexpressors (Figure 6B). Furthermore, Lhca6 is specifically detected in species containing chloroplast NDH (Figure 6A), implying that this key gene was acquired during the evolution of land plants, allowing NDH to interact with PSI. By contrast, Lhca5 was found in both the NDH-PSI supercomplex and PSI monomer (Table 1; see Supplemental Data Set 2 online), which is consistent with previous reports (Ganeteg et al., 2004; Storf et al., 2004; Zolla et al., 2007). Lhca6 overaccumulating in thylakoid membranes also can bind to PSI monomer (Figure 6B), suggesting that Lhca6 intermediates directly in the binding of NDH and PSI or indirectly via other proteins, such as Lhca5.
Majeran et al. (2008) reported that the subunits of PSI and NDH complex showed average bundle sheath cell (BSC)/mesophyll cell (MC) accumulation ratios of 1.6 and 3.0, respectively, in Z. mays. Given the apparent role of Lhca5 and Lhca6 in the formation of the NDH-PSI supercomplex, these two minor LHCI proteins should accumulate more in the BSC chloroplasts, which include higher levels of NDH subunits. Consistent with this idea, Lhca6 and Lhca5 showed BSC/MC ratios of 3.5 and 2.5, respectively (Majeran et al., 2008). From these results, we conclude that both Lhca5 and Lhca6 are required for the NDH-PSI supercomplex formation.
Normal state transition occurs in the lhca6 mutant (see Supplemental Figure 8 online; Kouril et al., 2005; Jensen et al., 2007). State transition is also not essential for NDH-PSI supercomplex formation (Peng et al., 2008), excluding the possibility that NDH interacts with PSI via LHCII. Since Lhca5 interacts with PSI on the Lhca2/Lhca3 site (Lucinski et al., 2006), it is likely that NDH associates with PSI via Lhca5/Lhca6 and the LHCI complex.
BN-PAGE showed that NDH subunits accumulate in smaller versions of the NDH-PSI supercomplex (Figure 5) in both lhca5 and lhca6, in which the major photosynthetic protein complexes, including PSI, PSII, and LHCII, were not affected (Figure 5; see Supplemental Figure 5 online). NDH activity was still detected in immature leaves of lhca6 (Figure 4B) in which accumulation of the NDH-PSI supercomplex was completely impaired (Figures 5B and 5D; see Supplemental Figure 5 online), suggesting that the smaller NDH-PSI supercomplex detected in lhca5 and lhca6 contains at least a minimum set of NDH subunits. Immunoblot analysis showed that the smaller NDH-PSI supercomplex contains all the subunits of the PSI complex tested (Figures 5B and 5D; see Supplemental Figure 6 online). The smaller versions of NDH-PSI supercomplex migrated faster in BN gel than the subsupercomplex corresponding to band II detected in ndhl (Figure 5; Peng et al., 2008), suggesting that the size difference between the smaller NDH-PSI supercomplex and the intact NDH-PSI supercomplex is larger than 280 kD (based on our estimation of the molecular mass of the subcomplex A). One possible explanation for the results is that NDH interacts with two copies of the PSI complexes possibly via Lhca5 and Lhca6. Our rough estimation of the NDH/PSI stoichiometry does not exclude this possibility (Figure 3). However, we do not include this information in the model of the supercomplex (Figure 9) since the biochemical information is still lacking especially on the docking sites. The most straightforward approach is coimmunoprecipitation using antibodies against subunits of the supercomplex, but this trial was unsuccessful so far probably because of a low abundance and fragility of the NDH-PSI.
In the absence of Lhca5 and Lhca6, NDH exists in thylakoids as a smaller NDH-PSI supercomplex at a level mildly reduced compared with that in the wild type (Figures 4 and 5). The levels of this smaller NDH-PSI supercomplex depend on the leaf development in lhca6 (Figures 4C and 4D). The pgr5 defect did not decrease the content of NDH subunits (Figure 8D), implying that the smaller NDH-PSI supercomplex may not be sensitive to oxidative stress. The level of protein is determined by the balance of synthesis and degradation, and active synthesis of NDH subunits may overcome its instability more efficiently in immature leaves than in mature leaves.
What is the determinant of the severe phenotype observed in lhca6 pgr5 (Figures 8A to 8C)? At least NDH activity detected in the postillumination rise of chlorophyll fluorescence reflects the level of NDH in immature and mature leaves (Figures 4B to 4D). The in vitro Fd-dependent PQ reduction assay using ruptured chloroplasts also suggests that the smaller NDH-PSI supercomplex still retains some activity (Figure 7D). It is possible that under the certain threshold level the in vivo function of NDH complex is drastically impaired, leading to the phenotype of lhca6 pgr5 (Figure 8). Consistent with this idea, the fluorescence level was lower in immature leaves of lhca6 pgr5 than its mature leaves (Figures 8A and 8B). In this case, Lhca6 is required for the supercomplex formation and consequently for full NDH activity in vivo via its function stabilizing NDH. However, the level of NDH was only mildly affected in lhca6 pgr5, including at least >75% levels of NDH subunits (Figure 8D). It is also probable that the supercomplex formation is required for the efficient operation of NDH activity.
As a conclusion, we cannot clearly explain the discrepancy between the partial impairment of NDH activity in lhca6 (Figures 4B, 7D, and 7E) and the drastic phenotype in lhca6 pgr5 (Figure 8). The problem is related to the difficulty in monitoring NDH-dependent PSI cyclic electron transport in the light, and our methods rely on the measurement in the dark (Figures 4B and 7D) or under low FR light (Figure 7E). Consistent with the mutant phenotype (Munekage et al., 2004), PGR5-dependent PSI cyclic electron transport is predominate in thylakoids in the light and NDH-dependent PSI cyclic electron transport is under the detection limit (Okegawa et al., 2008). Although we cannot evaluate the activity experimentally, the most straightforward discussion for the clear mutant phenotype (Figure 8) is the compensatory contribution of NDH to PSI cyclic electron transport in pgr5. The supercomplex formation via Lhca6 is required for the process, but the exact molecular mechanism remains for future analysis.
We propose that the NDH-PSI supercomplex is a minimal functional unit for the efficient in vivo function of NDH, as evident in the lhca6 pgr5 phenotype. Our findings relate to a long debate on the electron donor to NDH, since it is clear now that the previous biochemical approaches focused on NDH monomer or NDH subcomplex (Guedeney et al., 1996; Sazanov et al., 1998). What was a reason why chloroplast NDH acquired the subcomplex B and lumen subcomplexes and used Lhca5 and Lhca6 to form the supercomplex? This process may have facilitated the novel electron transport in chloroplasts required for stress tolerance. It may be necessary to reevaluate the biochemistry of chloroplast NDH in the supercomplex.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (ecotype Columbia gl1) plants were grown in soil in a growth chamber (50 μmol photons m−2 s−1, 16-h photoperiod, 23°C) for 3 to 4 weeks. The lhca5 mutant was obtained from the RIKEN Bioresource Center (http://www.brc.riken.jp/lab/epd/Eng/catalog/seed.shtml). The ppl2 and ndf2 mutants were kindly provided by Kentaro Ifuku and Tsuyoshi Endo, respectively, of Kyoto University.
Thylakoid Membrane Preparation, BN-PAGE, and Immunoblot Analysis
Chloroplasts and thylakoids were isolated as described (Munekage et al., 2002). BN-PAGE and subsequent 2D/SDS-PAGE immunoblot analysis was performed as described (Peng et al., 2008). For immunoblot analysis, thylakoid proteins were loaded on an equal chlorophyll basis. The signals were detected using an ECL Advance Western Blotting Detection Kit for NdhH (GE Healthcare) or an ECL Plus Western Blotting Detection Kit for the others (GE Healthcare) and visualized by an LAS3000 chemiluminescence analyzer (Fuji Film). Immunoblots were quantified by Imagemaster software (Amersham Pharmacia Biotech) in three independent experiments.
Peptide Preparation for Tandem Mass Spectrometry Analysis
Thylakoid membrane complexes isolated from wild-type and ndhl mutant plants were solubilized and separated by BN-PAGE. Bands I and II (described in Peng et al., 2008) and the PSI monomer (described in Peng et al., 2006) were excised from the gel. Peptide preparation and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses were performed as previously described (Fujiwara et al., 2009). The excised bands were treated twice with 25 mM ammonium bicarbonate in 30% (v/v) acetonitrile for 10 min and 100% (v/v) acetonitrile for 15 min and then dried in a vacuum concentrator. The dried gel pieces were treated with 0.01 mg/mL trypsin (sequence grade; Promega)/50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides in the gel pieces were recovered twice with 20 μL 5% (v/v) formic acid/50% (v/v) acetonitrile. The extracted peptides were combined and then dried in a vacuum concentrator.
MS Analysis and Database Searching
LC-MS/MS analyses were performed on an LTQ-Orbitrap XL-HTC-PAL system. Trypsin-digested peptides were loaded on the column (diameter 75 μm, 15 cm; L-Column, CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics) and were eluted by a gradient of 5 to 45% (v/v) acetonitrile in 0.1% (v/v) formic acid over 70 min. The eluted peptides were introduced directly into the LTQ-Orbitrap XL MS at a flow rate of 300 nL/min and a spray voltage of 2.0 kV. The range of MS scan was m/z 450 to 1500, and the top three peaks were analyzed by MS/MS analysis. MS/MS spectra were compared by the MASCOT server (version 2.2) against TAIR8 (The Arabidopsis Information Resource) with the following search parameters: set-off threshold at 0.05 in the ion score cutoff; peptide tolerance, 10 ppm; MS/MS tolerance, ±0.8 D; peptide charge, 2+ or 3+; trypsin as enzyme allowing up to one missed cleavage; carboxymethylation on cysteines as a fixed modification, and oxidation on Met as a variable modification.
Chlorophyll Fluorescence and P700 analysis
The transient increase in chlorophyll fluorescence after AL had been turned off was monitored as described (Shikanai et al., 1998). An image of chlorophyll fluorescence was captured by a CCD camera after 1 min illumination with AL (100 μmol photons m−2 s−1) as described (Shikanai et al., 1999). Chlorophyll fluorescence was measured with a MINI-PAM portable chlorophyll fluorometer (Walz). ΦPSII was calculated as (Fm′ – Fs)/Fm′, where Fm′ is the maximum fluorescence level in the light, and Fs is the steady state fluorescence level. ETR was calculated as ΦPSII × photon flux density (μmol photons m−2 s−1). NPQ was calculated as (Fm – Fm′)/Fm′. The redox change of P700 was assessed by monitoring absorbance at 830 nm with a PAM101 chlorophyll fluorometer (Walz) equipped with an emitter-detector unit (ED P700DW) as described (Munekage et al., 2004). The redox kinetics of P700 was measured according to previously described methods (Shikanai et al., 1998). Fd-dependent PQ reduction activity was measured in ruptured chloroplasts as described (Endo et al., 1998), with minor modifications (the pH of the assay medium was changed to 8.0). As electron donors, 5 mM maize Fd (Sigma-Aldrich) and 0.25 mM NADPH (Sigma-Aldrich) were used. Antimycin A (Sigma-Aldrich) at 10 mM was added before measurement.
RNAi, Complementation, and Plant Transformation
For RNAi vector construction, short sequences of Arabidopsis Lhca6, NDH18, and FKBP16-2 were cloned into the pHANNIBAL vector (Wesley et al., 2001) between the XbaI-BamHI sites in sense orientation and between the XhoI-KpnI sites in antisense orientation. The primers are listed in Supplemental Table 2 online. The expression cassette was excised with NotI and cloned into the NotI site of the binary vector pART27 (Gleave, 1992). For complementation of the Lhca6 RNAi mutant, 3.7-kb Oryza sativa Lhca6 genomic DNA amplified by primers 5′-CCAAAGCTTTAGGAGTATTCACTGCTCAG-3′ and 5′-AATCTCGAGAATGGGACGTGAATGCCTGC-3′ was cloned into the pGWB-NB1 vector. The genomic sequence of O. sativa Lhca6 used for the complementation was modified to carry the sequence encoding the HA tag (YPYDVPDYAG). The fusion gene was cloned into the pGWB-NB1 vector. For overexperssion of Lhca6-HA in wild-type and crr2-2 plants, the cDNA of Arabidopsis Lhca6 carrying the sequence encoding the HA tag was subcloned into the pBI121 vector under the control of the CaMV 35S promoter. The vectors were transferred into Agrobacterium tumefaciens C58C by electroporation, and the bacteria were used to transform wild-type Arabidopsis or Lhca6 RNAi lines by floral dipping (Clough and Bent, 1998).
Nucleic Acid Preparation and RT-PCR analysis
Total RNAs were isolated from Arabidopsis leaves with an RNeasy Plant Mini Kit (Qiagen). Total RNA (5 μg) was reverse transcribed with a SuperScript III first-strand synthesis system (Invitrogen) in a total volume of 20 μL. The cDNA was used in 30 cycles of PCR. The PCR primers are listed in Supplemental Table 2 online. Each set of primers covered at least one intron sequence to eliminate amplification of the genomic DNA sequence. RT-PCR products were separated in agarose gels and were detected by ethidium bromide staining.
Production of Polyclonal Antisera against NDH18 and FKBP16-2
The nucleotide sequences encoding the soluble parts of NDH18 (amino acids 41 to 131 and 155 to 212) and the mature protein of FKBP16-2 (amino acids 51 to 217) were amplified and cloned into the pET30a vector (Novagen). Expression of the recombinant proteins was induced in Escherichia coli BL21 (DE3) cells by 1 mM isopropylthio-β-galactoside for 2 h, and then the cells were harvested in 300 mM NaCl and 50 mM Tris-HCl, pH 8.0. After incubation for 30 min at 4°C in the presence of 1 mg/mL lysozyme, the inclusion bodies were pelleted from the sonicated cells by centrifugation at 3000g for 30 min. Recombinant proteins were then purified from the inclusion bodies in Ni2+-NTA columns (Qiagen) under denaturing conditions according to the manufacturer's protocol. Polyclonal antisera were raised in a rabbit from purified recombinant protein.
Immunochemical Quantification of NDH Subunits and PSI in the Supercomplex
Antiserum against Arabidopsis PsaA was produced in rabbits using the N-terminal part of recombinant PsaA protein (amino acids 1 to 77) as antigen. The nucleotide sequence encoding this N-terminal part was amplified by PCR using the primers 5′-GGCGAATTCATGATTATTCGTTCGCCGG-3′ and 5′-GATCTCGAGTTGGCCGAAATGGGCAC-3′. The amplified sequence was fused to the Nus-tag in the pET43.1a vector (Novagen). The PsaA-Nus recombinant protein was induced in DE3 cells and purified in a Ni2+-NTA column according to the manufacturer's protocol. The protein contents were determined with a Bio-Rad protein assay kit. The band corresponding to the NDH-PSI supercomplex was excised from BN gel and further denatured in gel as described (Peng et al., 2008) and then directly used for SDS-PAGE together with the recombinant protein. Immunoblot analysis using an ECL Advance Western Blotting Detection Kit (GE Healthcare) was performed according to standard procedures. The signal was visualized by an LAS3000 chemiluminescence analyzer (Fuji Film). Immunoblots from three independent experiments were quantified by Imagemaster software (Amersham Pharmacia Biotech).
Phylogenetic Analysis
Protein sequences of FKBP13 and FKBP16-2 proteins (shown in Supplemental Data Set 3 online) and Lhca family proteins (shown in Supplemental Data Set 4 online) were aligned using the ClustalW program with default settings (http://clustalw.ddbj.nig.ac.jp/top-e.html) and adjusted manually. The phylogenetic tree was constructed using TreeView software (version 1.6.6) (http:/taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At (Arabidopsis thaliana) FKBP13 (AT5G45680), At FKBP16-2 (AT4G39710), Os (Oryza sativa) FKBP13 (OS06G0663800), Os FKBP16-2(OS02G0751600), Zm (Zea mays) FKBP13 (EU958134), Zm FKBP16-2 (EU955427), Gm (Glycine max) FKBP16-2 (DB960344), At NDH18 (AT5G43750), Gm NDH18 (CD394214), Nt (Nicotiana tabacum) NDH18 (EB679832), Os NDH18 (OS01G0929100), Zm NDH18 (DV514173), At Lhca6 (AT1G19150), Rr (Raphanus raphanistrum) Lhca6 (EV549333), Br (Brassica rapa) Lhca6 (EX086756), Vv (Vitis vinifera) Lhca6 (EC925446), Mt (Medicago truncatula) Lhca6 (EV260067), Vu (Vigna unguiculata) Lhca6 (FG880635), Aa (Artemisia annua) Lhca6 (EY082250), Cs (Citrus sinensis) Lhca6 (EY664891), Ta (Triticum aestivum) Lhca6 (CJ883523), Sb (Sorghum bicolor) Lhca6 (CN151166), So (Saccharum officianrum) Lhca6 (CA294625), Hv (Horheum vulgare) Lhca6 (BI953315 and AJ432207), Nt Lhca6 (DW000027), Gm Lhca6 (EH258354), Zm Lhca6 (DV507315), Os Lhca6 (AK067780), Os Lhca2 (AK104651), At Lhca5 (At1g45474), Rr Lhca5 (EV538184), Br Lhca5 (EX087930), Vv Lhca5 (EC934896), Mt Lhca5 (CX519116), Vu Lhca5 (FG880536), Aa Lhca5 (EY103979), Cs Lhca5 (EN184748), Ta Lhca5 (CJ723109), Sb Lhca5 (CN147414), Hv Lhca5 (BE422210 and BI950547), At Lhca4 (AT3G47470), At Lhca3 (AT1G61520), At Lhca2 (AT3G61470), At Lhca1 (AT3G54890), Cr (Chlamydomonas reinhardtii) Lhca1 (AAD03734), Cr Lhca2 (XP_001691031), Cr Lhca3 (XP_001701405), Cr Lhca4 (EDP08012), Cr Lhca5 (XP_001702730), Cr Lhca6 (XP_001698070), Cr Lhca7 (XP_001691959), Cr Lhca8 (EDP08179), and Cr Lhca9 (XP_001692548).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of NDH18.
Supplemental Figure 2. NDH-PSI Supercomplex Content in the ndh18 and fkbp16-2 Lines.
Supplemental Figure 3. Localization Analysis of NDH Subunits in Chloroplasts of the Wild Type, ndhl, ppl2, crr4-3, and ndh18 Mutants.
Supplemental Figure 4. Visible Phenotype of lhca6 Mutant.
Supplemental Figure 5. NDH-PSI Supercomplex Content in lhca5 and lhca6.
Supplemental Figure 6. Analysis of Thylakoid Protein Complex from Wild-type and lhca6 Mature Leaves.
Supplemental Figure 7. Amino Acid Sequence Alignments of Lhca6 and Lhca2.
Supplemental Figure 8. State 1–State 2 Transitions in Wild-Type, stn7, and lhca6 Plants.
Supplemental Table 1. The r Values between NDH Complex-Related Genes with Lhca6, FKBP16-2, and NDH18.
Supplemental Table 2. Primers Used in This work.
Supplemental Data Set 1. The Total Proteins Identified from Bands I and II.
Supplemental Data Set 2. The Total Proteins Identified from PSI Monomer from Wild Type, crr2-2, and lhca6.
Supplemental Data Set 3. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 1A.
Supplemental Data Set 4. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 6A.
Supplementary Material
Acknowledgments
We thank Tsuyoshi Endo (Kyoto University, Kyoto, Japan), Amane Makino (Tohoku University, Sendai, Japan), and Kentaro Ifuku (Kyoto University, Kyoto, Japan) for giving us antibodies. This work was supported by Grant 17GS0316 for Creative Science Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation; GPN0008). This work was also supported by a grant from the Japan Society for the Promotion of Science (JSPS-19-07142) to L.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toshiharu Shikanai (shikanai@pmg.bot.kyoto-u.ac.jp).
Online version contains Web-only data.
References
- Amunts, A., Drory, O., and Nelson, N. (2007). The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447 58–63. [DOI] [PubMed] [Google Scholar]
- Battchikova, N., and Aro, E.-M. (2007). Cyanobacterial NDH-1 complexes: Multiplicity in function and subunit composition. Physiol. Plant. 131 22–32. [DOI] [PubMed] [Google Scholar]
- Battchikova, N., Zhang, P., Rudd, S., Ogawa, T., and Aro, E.-M. (2005). Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 280 2587–2595. [DOI] [PubMed] [Google Scholar]
- Burrows, P.A., Sazanov, L.A., Svab, Z., Maliga, P., and Nixon, P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- DalCorso, G., Pesaresi, P., Masiero, S., Aseeva, E., Schünemann, D., Finazzi, G., Joliot, P., Barbato, R., and Leister, D. (2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132 273–285. [DOI] [PubMed] [Google Scholar]
- Edvardsson, A., Eshaghi, S., Vener, A.S., and Andersson, B. (2003). The major peptidyl-prolyl isomerase activity in thylakoid lumen of plant chloroplasts belongs to a novel cyclophilin TLP20. FEBS Lett. 542 137–141. [DOI] [PubMed] [Google Scholar]
- Endo, T., Shikanai, T., Sato, F., and Asada, K. (1998). NAD(P)H dehydrogenase-dependent, antimycin A-sensitive electron donation to plastoquinone in tobacco chloroplast. Plant Cell Physiol. 39 1226–1231. [Google Scholar]
- Fujiwara, M., Hamada, S., Hiratsuka, M., Fukao, Y., Kawasaki, T., and Shimamoto, K. (2009). Proteome analysis of detergent resistant membranes (DRMs) associated with OsRac1 mediated innate immunity in rice. Plant Cell Physiol. 50 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg, U., Klimmek, F., and Jansson, S. (2004). Lhca5 – An LHC-type protein associated with photosystem I. Plant Mol. Biol. 54 641–651. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Guedeney, G., Corneile, S., Cuine, S., and Peltier, G. (1996). Evidence for an association of ndhB, ndhJ gene products and ferredoxin-NADP reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 378 277–280. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Mould, R.M., He, Z., and Luan, S. (2002). A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. USA 99 15806–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36 541–549. [DOI] [PubMed] [Google Scholar]
- He, Z., Li, L., and Luan, S. (2004). Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134 1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen, M., Battchikova, N., Zhang, P., Graf, A., Sirpiö, S., Paakkarinen, V., and Aro, E.-M. (2004). Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, S., Takabayashi, A., Ido, K., Endo, T., Ifuku, K., and Sato, F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, N., Takabayashi, A., Ishida, S., Hano, Y., Endo, T., and Sato, F. (2008). NDF6: A thylakoid protein specific to terrestrial plants is essential for activity of chloroplastic NAD(P)H dehydrogenase in Arabidopsis. Plant Cell Physiol. 49 1066–1073. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4 236–240. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Bassi, R., Boekema, E.J., Dekker, J.P., Jansson, S., Leister, D., Robinson, C., and Scheller, H.V. (2007). Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 1767 335–352. [DOI] [PubMed] [Google Scholar]
- Joët, T., Cournac, L., Horváth, E.M., Medgyesy, P., and Peltier, G. (2001). Increased sensitivity of photosynthesis to antimycin A induced by inactivation of the chloroplast ndhB gene. Evidence for a participation of the NADH-dehydrogenase complex to cyclic electron flow around photosystem I. Plant Physiol. 125 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimmek, F., Sjödin, A., Noutsos, C., Leister, D., and Jansson, S. (2006). Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330. [DOI] [PubMed] [Google Scholar]
- Kouril, R., Zygadlo, A., Arteni, A.A., de Wit, C.D., Dekker, J.P., Jensen, P.E., Scheller, H.V., and Boekema, E.J. (2005). Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44 10935–10940. [DOI] [PubMed] [Google Scholar]
- Lucinski, R., Schmid, V.H., Jansson, S., and Klimmek, F. (2006). Lhca5 interaction with plant photosystem I. FEBS Lett. 580 6485–6488. [DOI] [PubMed] [Google Scholar]
- Majeran, W., Zybailov, B., Ytterberg, A.J., Dunsmore, J., Sun, Q., and van Wijk, K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7 1609–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, T., Wakasugi, T., Shinozaki, K., Yamaguchi-Shinozaki, K., Zaita, N., Hidaka, T., Meng, B.Y., Ohto, C., Tanaka, M., Kato, A., Maruyama, T., and Sugiura, M. (1987). Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. 210 385–393. [DOI] [PubMed] [Google Scholar]
- Matsuo, M., Endo, T., and Asada, K. (1998). Properties of the respiratory NAD(P)H dehydrogenase isolated from the cyanobacterium Synechocystis PCC6803. Plant Cell Physiol. 39 263–267. [DOI] [PubMed] [Google Scholar]
- Melkozernov, A.N., Barber, J., and Blankenship, R.E. (2006). Light harvesting in photosystem I supercomplexes. Biochemistry 45 331–345. [DOI] [PubMed] [Google Scholar]
- Mi, H., Endo, T., Ogawa, T., and Asada, K. (1995). Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 36 661–668. [Google Scholar]
- Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K., Endo, T., Tasaka, M., and Shikanai, T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429 579–582. [DOI] [PubMed] [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110 361–371. [DOI] [PubMed] [Google Scholar]
- Nelson, N., and Yocum, C.F. (2006). Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57 521–565. [DOI] [PubMed] [Google Scholar]
- Ogawa, T. (1992). Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 99 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, T., and Mi, H. (2007). Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93 69–77. [DOI] [PubMed] [Google Scholar]
- Okegawa, Y., Kagawa, Y., Kobayashi, Y., and Shikanai, T. (2008). Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol. 49 825–834. [DOI] [PubMed] [Google Scholar]
- Peng, L., Ma, J., Chi, W., Guo, J., Zhu, S., Lu, Q., Lu, C., and Zhang, L. (2006). Low PSII accumulation1 is involved in the efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., Shimizu, H., and Shikanai, T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283 34873–34879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Prommeenate, P., Lennon, A.M., Markert, C., Hippler, M., and Nixon, P.J. (2004). Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 279 28165–28173. [DOI] [PubMed] [Google Scholar]
- Rumeau, D., Becuwe-Linka, N., Beyly, A., Louwagie, M., Garin, J., and Peltier, G. (2005). New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov, L.A., Burrows, P.A., and Nixon, P.J. (1998). The plastid ndh genes code for an NADH-specific dehydrogenase: Isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. USA 95 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T. (2007. a). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58 199–217. [DOI] [PubMed] [Google Scholar]
- Shikanai, T. (2007. b). The NAD(P)H dehydrogenase complex in photosynthetic organisms: Subunit composition and physiological function. Funct. Plant Sci. Biotechnol. 1 129–137. [Google Scholar]
- Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K., and Yokota, A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Munekage, Y., Shimizu, K., Endo, T., and Hashimoto, T. (1999). Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol. 40 1134–1142. [DOI] [PubMed] [Google Scholar]
- Shimizu, H., Peng, L., Myouga, F., Motohashi, R., Shinozaki, K., and Shikanai, T. (2008). CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 49 835–842. [DOI] [PubMed] [Google Scholar]
- Shimizu, H., and Shikanai, T. (2007). Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52 539–547. [DOI] [PubMed] [Google Scholar]
- Sirpiö, S., Allahverdiyeva, Y., Holmström, M., Khrouchtchova, A., Haldrup, A., Battchikova, N., and Aro, E.-M. (2009. a). Novel nuclear-encoded subunits of the chloroplast NAD(P)H dehydrogenase complex. J. Biol. Chem. 284 905–912. [DOI] [PubMed] [Google Scholar]
- Sirpiö, S., Holmström, M., Battchikova, N., and Aro, E.-M. (2009. b). AtCYP20-2 is an auxiliary protein of the chloroplast NAD(P)H dehydrogenase complex. FEBS Lett. 583 2355–2358. [DOI] [PubMed] [Google Scholar]
- Storf, S., Stauber, E.J., Hippler, M., and Schmid, V.H.R. (2004). Proteomic analysis of the photosystem I light-harvesting antenna in tomato (Lycopersicon esculentum). Biochemistry 43 9214–9224. [DOI] [PubMed] [Google Scholar]
- Suorsa, M., Sirpiö, S., and Aro, E.-M. (July 21, 2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant http://dx.doi.org/10.1093/mp/ssp052. [DOI] [PubMed]
- Takabayashi, A., Ishikawa, N., Obayashi, T., Ishida, S., Obokata, J., Endo, T., and Sato, F. (2009). Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 57 207–219. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Wang, J., and Goodman, H.M. (1994). Differential expression in Arabidopsis of Lhca2, a PSI cab gene. Plant Mol. Biol. 25 551–557. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Battchikova, N., Jansen, T., Appel, J., Ogawa, T., and Aro, E.-M. (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16 3326–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Battchikova, N., Paakkarinen, V., Katoh, H., Iwai, M., Ikeuchi, M., Pakrasi, H.B., Ogawa, T., and Aro, E.-M. (2005). Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem. J. 390 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla, L., Rinalducci, S., and Timperio, A.M. (2007). Proteomic analysis of photosystem I components from different plant species. Proteomics 7 1866–1876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.