Abstract
The acquisition of nutrients requires tight regulation to ensure optimal supply while preventing accumulation to toxic levels. Ammonium transporter/methylamine permease/rhesus (AMT/Mep/Rh) transporters are responsible for ammonium acquisition in bacteria, fungi, and plants. The ammonium transporter AMT1;1 from Arabidopsis thaliana uses a novel regulatory mechanism requiring the productive interaction between a trimer of subunits for function. Allosteric regulation is mediated by a cytosolic C-terminal trans-activation domain, which carries a conserved Thr (T460) in a critical position in the hinge region of the C terminus. When expressed in yeast, mutation of T460 leads to inactivation of the trimeric complex. This study shows that phosphorylation of T460 is triggered by ammonium in a time- and concentration-dependent manner. Neither Gln nor l-methionine sulfoximine–induced ammonium accumulation were effective in inducing phosphorylation, suggesting that roots use either the ammonium transporter itself or another extracellular sensor to measure ammonium concentrations in the rhizosphere. Phosphorylation of T460 in response to an increase in external ammonium correlates with inhibition of ammonium uptake into Arabidopsis roots. Thus, phosphorylation appears to function in a feedback loop restricting ammonium uptake. This novel autoregulatory mechanism is capable of tuning uptake capacity over a wide range of supply levels using an extracellular sensory system, potentially mediated by a transceptor (i.e., transporter and receptor).
INTRODUCTION
Plants as primary biomass producers need to acquire a wide spectrum of inorganic nutrients from the soil. Due to the changes in availability, the uptake for each of the nutrients needs to be fine-tuned in order to allow optimal growth and to prevent accumulation to toxic levels.
As the fourth most abundant element in living organisms, nitrogen is essential for plant growth and development. Only certain prokaryotic organisms are able to fix N2; thus, plants that are not associated with N2-fixing symbiotic microorganisms depend directly on their ability to absorb nitrogen as NO3−, NH4+, or urea from the soil. In the cell, NH4+ derives either from NO3− reduction, is directly taken up from the soil, or is produced during photorespiration or amino acid catabolism. NH4+ is then assimilated to produce the N-transport amino acids (i.e., Glu, Gln, Asp, and Asn) and indirectly all other N-containing molecules.
In bacteria, fungi, and plants, high-affinity uptake of ammonium is mediated by transporters belonging to the ammonium transporter/methylamine permease/rhesus (AMT/MEP/Rh) superfamily (Loqué and von Wirén, 2004; von Wirén and Merrick, 2004; Ludewig et al., 2007). The AMT/MEP ammonium transporter genes were identified simultaneously in yeast and plants by screening expression cDNA libraries in a yeast mutant deficient in ammonium uptake (Marini et al., 1994; Ninnemann et al., 1994). The Arabidopsis thaliana genome encodes six members of the family, which fall into two clades, AMT1 and AMT2. In addition to their function in ammonium uptake from soil, AMTs are most likely involved in ammonium transport through the plant, ammonia retrieval in roots and leaves, and in the supply of nitrogen to pollen (Sohlenkamp et al., 2002; Yuan et al., 2007b, 2009).
The analysis of crystal structures showed that bacterial AMTs form a trimeric complex, with each monomer being composed of 11 transmembrane helices (TMH) that form a noncontinuous channel through which the substrate can pass (Khademi et al., 2004; Andrade et al., 2005; Lupo et al., 2007; Javelle et al., 2008). Expression of plant AMT1 in Xenopus laevis oocytes demonstrated transport of charged NH4+ or cotransport of NH3 with a proton (Ludewig et al., 2002, 2003; Mayer et al., 2006). AMT1 homologs from Arabidopsis and tomato (Solanum lycopersicum) are highly selective for ammonium over potassium and can transport the methylated form methylammonium (MeA) (Ludewig et al., 2002, 2003).
AMT/MEP ammonium transporters require a productive interaction between subunits of the trimer to function. Allosteric regulation is mediated by a cytosolic C-terminal domain (Marini et al., 2000; Loqué et al., 2007; Neuhäuser et al., 2007; Severi et al., 2007). The AMT/MEP C-terminal domain is highly conserved in >700 AMT homologs from cyanobacteria to higher plants with no obvious occurrence of cases lacking this domain (Loqué et al., 2007). Previous results indicated that AMT1;1 exists in active and inactive states, probably regulated by phosphorylation of residues in the C terminus (Loqué et al., 2007). Indeed, phosphoproteomic studies identified phosphorylated residues in the C terminus of AMT1;1 (Nühse et al., 2004; Benschop et al., 2007; Hem et al., 2007). More specifically, a highly conserved Thr residue in the soluble C terminus of the Arabidopsis ammonium transporter AMT1;1 was found to be phosphorylated (MAGMDMpTRHGGFA) (Nühse et al., 2004).
Mutation of Thr-460 led to a nonfunctional and trans-inactivated trimeric complex (Loqué et al., 2007). However, the signals that induce phosphorylation of T460 in AMT1;1 have not yet been identified.
Here, we show that phosphorylation of the critical Thr residue T460 in the trans-activation domain of the C terminus of AMT1;1 is triggered specifically by ammonium, providing a link between ammonium levels in the environment and modification of the conformation of AMT transporters. Moreover, resupply of ammonium and, thus, phosphorylation correlates with a decrease in ammonium uptake. The results suggest that the allosteric regulation mechanism in AMTs is part of a feedback mechanism to modulate the uptake of ammonium into the root. The conservation of allosteric regulation by the C terminus in archaebacteria implies that the basic mechanism of this regulatory system was developed early during evolution (Loqué et al., 2009).
RESULTS
Ammonium Induces Phosphorylation of T460 in Arabidopsis AMT1;1
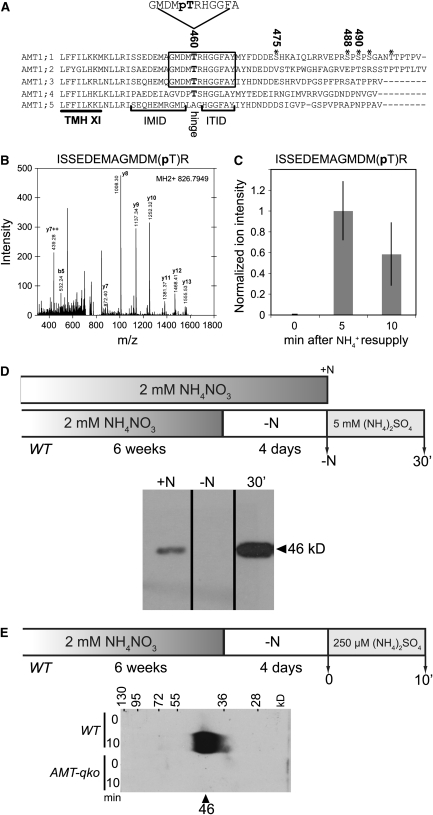
T460 in the cytosolic C terminus of the Arabidopsis AMT1;1 is important for the allosteric regulation of transport activity of the trimeric AMT1;1 complex when expressed in yeast (Loqué et al., 2007). T460 had been found to be phosphorylated in Arabidopsis cell cultures grown on media containing high levels of ammonium; we thus hypothesized that ammonium may trigger phosphorylation of T460 (Nühse et al., 2004). The location of T460 in the hinge between the intramolecular interaction domain (IMID) and the inter-trans-interaction domain (ITID) places it in a crucial position for regulating the activity of the AMT1;1 transporter complex (Figure 1A) (Loqué et al., 2007). Since AMT1;1 expression is induced by nitrogen starvation, we tested AMT phosphorylation in seedlings after resupply of ammonium to nitrogen-starved roots. Using phosphoproteomics, a peptide containing T460 in the C terminus was found to be phosphorylated in plasma membrane fractions of Arabidopsis seedlings (Figure 1B). Five minutes after NH4+ resupply (10 mM NH4Cl), the normalized ion intensity of peptide ISSEDEMAGMDM(pT)R increased, indicating elevated abundance of phosphorylation at T460 (Figure 1C). To characterize the signals that trigger phosphorylation of T460 in planta in more detail, a phospho-specific antiserum (AMT1-P) was raised against a peptide covering T460. Dot blots showed that the serum is specific for the phosphorylated peptide (see Supplemental Figure 1 online). Due to the conservation of the region around T460 with other Arabidopsis AMTs, the antiserum is expected to detect phosphorylation of T460, T472, and T464 in the three paralogs AMT1;1, AMT1;2, and AMT1;3, respectively; but not of AMT1;4 or AMT1;5, (Figure 1A; Loqué et al., 2007). The antiserum detected a polypeptide with the expected mass of 46 kD in microsomal fractions of wild-type plants grown in the presence of 2 mM NH4NO3 (Figure 1D; see Supplemental Figure 2A online). The apparent molecular mass of the phosphorylated polypeptide is similar to the one detected with an antiserum (‘AMT1;1 loop 2/3′) raised against a nonphosphorylated peptide located in the loop between TMH2 and TMH3 of AMT1;1 (Figure 3A; Loqué et al., 2006). By contrast, phosphorylation remained undetectable in plants starved for nitrogen for 4 d. The absence of the signal cannot be caused by the absence of AMT1 because nitrogen starvation increases transcript and protein levels of AMT1;1, AMT1;2, and AMT1;3 (Figure 3A; Gazzarrini et al., 1999; Loqué et al., 2006; Yuan et al., 2007a, 2007b). The simplest hypothesis is that ammonium triggers phosphorylation leading to inactivation of AMT1. To directly test for phosphorylation induced by ammonium, 10 mM NH4+ was supplied to nitrogen-starved roots in the form of (NH4)2SO4. NH4+ led to phosphorylation of AMT1 in roots within <30 min (Figure 1D; see Supplemental Figure 2A online). High levels of phosphorylated AMT1 were detected also after treatment with 250 μM (NH4)2SO4 for only 10 min (Figure 1E; see Supplemental Figure 2B online). No response was found in the Arabidopsis quadruple mutant (AMT-qko) carrying T-DNA insertions in AMT1;1, AMT1;2, and AMT1;3 and AMT2;1 (Figure 1E; see Supplemental Figure 2B online), demonstrating the specificity of the antiserum (Yuan et al., 2007b). To further confirm the specificity of the antiserum for the phosphorylated peptide in planta, root microsomal fractions were treated with alkaline phosphatase (CIP; see Supplemental Figure 3 online). CIP treatment led to a loss of a reaction with the antiserum, confirming that the antiserum detects the phosphorylated form of T460 in vivo.
Figure 1.
Induction of Phosphorylation by Ammonium.
(A) Alignment of the C terminus of five members of the AMT1 family. Displayed is the C-terminal part of TMH 11 and the cytosolic C terminus with IMID and ITID domains. The peptide sequence against which the AMT1;1-P phospho-antiserum was raised is indicated above the alignment. Asterisks indicate residues found to be phosphorylated in phosphoproteomic studies.
(B) Tandem mass spectrometry (MS/MS) fragmentation spectra for the peptide ISSEDEMAGMDM(pT)R as detected by CID fragmentation in an LTQ mass spectrometer.
(C) Normalized ion intensity of ISSEDEMAGMDM(pT)R peptide. Phosphorylation levels were measured by the ion intensities of phosphopeptides normalized to the time course of ion intensities of nonphosphopeptides of the same protein. Normalized ion intensities of all time points for each peptide were then divided by the maximum value across all time points to standardize the values to a scale of between 0 and 1. Mean value ± sd (n = 3).
(D) and (E) Phosphorylation of Thr redidues T460/T472/T464 in AMT1;1, AMT1;2, and AMT1;3, respectively.
(D) Wild-type plants were grown hydroponically on 2 mM NH4NO3 for 6 weeks and transferred either to 2 mM NH4NO3 medium (+N) or to nitrogen-depleted medium (−N) for 4 d. 5 mM (NH4)2SO4 was applied for 30 min (30′) to –N plants.
(E) Wild-type and AMT-qko plants were grown for 6 weeks in the presence of 2 mM NH4NO3 and transferred for 4 d to –N media. Roots were collected before (0) and 10 min (10′) after resupply of 250 μM (NH4)2SO4. Root microsomal fractions were prepared and protein gel blots were performed using the AMT1-P antiserum.
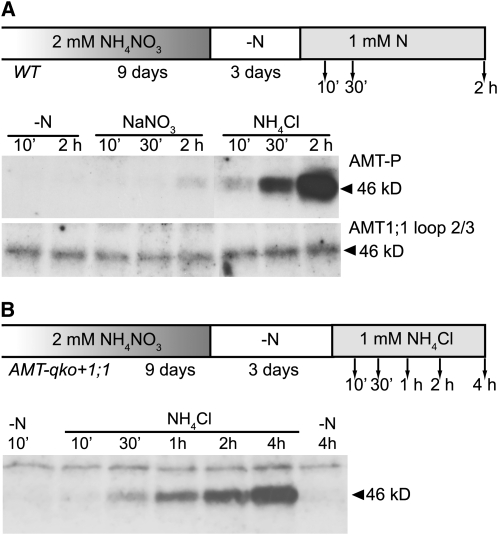
Figure 3.
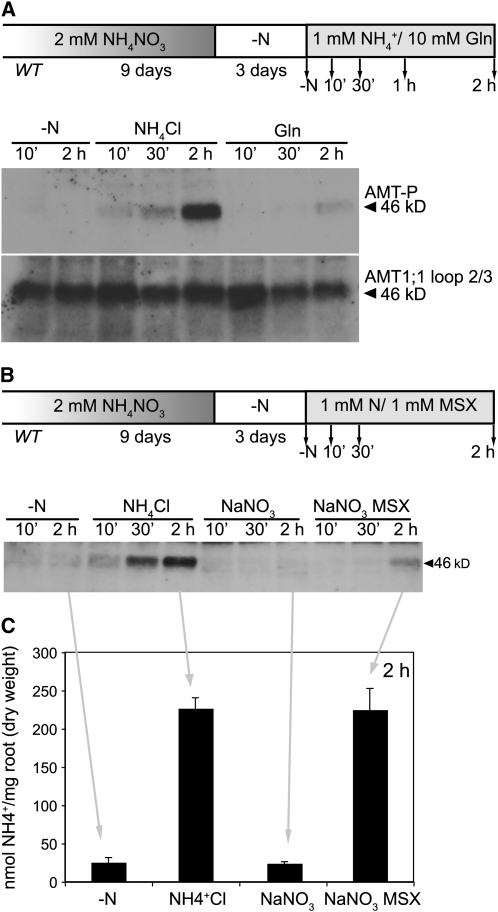
AMT1;1 Is Phosphorylated in Response to NH4+.
(A) Protein gel blot analysis of microsomal fractions of plants grown for 9 d in the presence of 2 mM NH4NO3 and transferred for 3 d to nitrogen-free medium. Nitrogen-free medium (−N), 1 mM of NaNO3, or 1 mM of NH4Cl were supplied, and roots were collected after 10 min, 30 min, and 2 h. Protein gel blots were performed using the AMT1-P or the ‘AMT1;1 loop 2/3′ antiserums (n = 3).
(B) Protein gel blot analysis of total protein extract of AMT-qko+1;1 plants grown for 9 d in the presence of 2 mM NH4NO3 and transferred for 3 d to nitrogen-free medium. Nitrogen-free medium (−N) or 1 mM NH4Cl were supplied, and roots were collected after 10 min, 30 min, 1 h, 2 h, and 4 h. Protein gel blots were performed using the AMT1-P antiserum (n = 1).
Together, the loss of phosphorylation during starvation and the induction during ammonium supply suggest that NH4+ triggers phosphorylation and that the phosphorylation status is sustained during continuous ammonium supply.
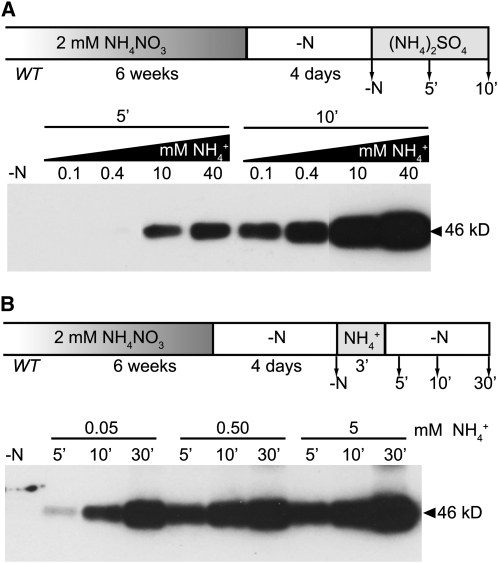
T460 Phosphorylation Is Concentration and Time Dependent
To characterize time and concentration dependence of NH4+-triggered phosphorylation, nitrogen-deprived plants were resupplied with 0.1 to 40 mM ammonium and roots were harvested 5 and 10 min after resupply. Phosphorylation levels were analyzed by protein gel blotting (Figure 2A; see Supplemental Figure 4A online). After 5 min of ammonium supply, phosphorylation was readily detectable. Interestingly, even low NH4+ levels (50 μM) were sufficient to trigger phosphorylation (Figure 2B; see Supplemental Figure 4B online). Phosphorylation levels increased steadily between 0.1 and 40 mM NH4+, demonstrating that the ammonium-induced phosphorylation is time and concentration dependent (Figure 2A). When monitored over an extended time period, phosphorylation increased linearly and saturated after 60 min (see Supplemental Figure 5 online).
Figure 2.
Phosphorylation Is Time and Concentration Dependent.
Time course of AMT1 phosphorylation of wild-type plants grown for 6 weeks in 2 mM NH4NO3 and nitrogen-starved (−N) for 4 d prior to ammonium addition.
(A) Plants were exposed to NH4+ [as (NH4)2SO4] and roots were collected after 5 and 10 min.
(B) Plants were exposed to NH4+ [as (NH4)2SO4] for 3 min and transferred back to nitrogen-free medium (−N). Samples were collected 5, 10, and 30 min after transfer. Protein gel blots were performed using the AMT1-P antiserum.
To test whether phosphorylation of T460 can be induced by a transient pulse, roots of nitrogen-starved plants were subjected to a pulse of 50, 500, or 5000 μM NH4+ for 3 min, washed, and transferred back to nitrogen-free medium (−N; Figure 2B; see Supplemental Figure 4B online). Roots were collected 5, 10, or 30 min after the initiation of the pulse (2, 7, and 27 min after retransfer to –N). Transient exposure to 50 μM NH4+ for 3 min was sufficient to induce phosphorylation. Phosphorylation increased over the tested period of 30 min (see Supplemental Figure 6 online). After 30 min, phosphorylation of AMT1 proteins induced by the ammonium pulse reached levels slightly lower than those found for continuous exposure to 50 μM NH4+ (see Supplemental Figure 7 online). AMT1;1 protein levels remained constant during the treatment. Thus, ammonium signaling occurs within a scale of minutes and transient exposure to ammonium is sufficient to trigger a robust and sustained response; higher concentrations led to a larger fraction of AMT1 being phosphorylated, and phosphorylation activity continued to increase even after removal of the signal.
NH4+ supply leads to a decrease of AMT1;1 mRNA levels and inhibition of NH4+ influx (Rawat et al., 1999). To control for AMT1;1 protein levels during resupply of NH4+, wild-type plants were grown in axenic culture for 9 d in the presence of 2 mM NH4NO3 before transfer to nitrogen deficiency for 3 d. Subsequently, nitrogen was resupplied in the form of 1 mM NH4Cl or 1 mM NaNO3. Microsomal fractions of roots were collected after 10, 30, or 120 min and analyzed by protein gel blots with antisera against the AMT1;1 cytosolic loop or its T460 phosphorylated form (Figure 3A; see Supplemental Figure 8A online). AMT1;1 protein levels remained constant during the 2 h of treatment, whereas phosphorylation levels increased only during NH4+ application.
The antiserum is expected to react with AMT1;1, 1;2, and 1;3. To specifically examine if AMT1;1 is phosphorylated in response to exposure to NH4+, plants expressing only AMT1;1 (AMT-qko+1;1; generated from the AMT-qko quadruple AMT knockout line by backcrossing with the wild type) were analyzed (Yuan et al., 2007b). AMT-qko+1;1 plants were grown in axenic culture, treated as described above for the wild type, and roots were collected after 10, 30, 60, 120, or 240 min of exposure to ammonium (Figure 3B; see Supplemental Figure 8B online). The steady increase of phosphorylated peptide over time confirmed that AMT1;1 was phosphorylated in response to ammonium resupply.
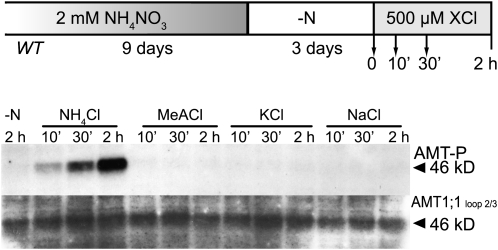
Specificity for Ammonium as the Signal
Four lines of evidence suggest that ammonium is the signal triggering phosphorylation of T460. Phosphorylation was (1) present in plants or cell cultures cultivated in media containing high levels of NH4+, (2) lost during nitrogen depletion, (3) induced by exposure to a pulse of 50 μM ammonium, and (4) increased in a concentration-dependent manner. However, other potential signals, such as general cation responses, acidification of the cytosol, or plasma membrane depolarization, could act as signals. Since other ions, such as NO3−, also lead to a depolarization of the plasma membrane but not to phosphorylation (Figure 3A; Wang et al., 1994; Miller et al., 2001), and since membrane depolarization by 50 μM ammonium is comparatively low (Ayling, 1993), we conclude that plasma membrane depolarization by ammonium is probably insufficient for triggering AMT1-phosphorylation at T460. To analyze the specificity of the response to ammonium, various monovalent cations were tested, including the AMT1 substrate analog methylammonium. Nitrogen-starved plants were exposed to 500 μM NH4Cl, methylammonium chloride (MeACl), potassium chloride, or sodium chloride (Figure 4; see Supplemental Figure 8C online). Phosphorylation was detected in microsomal fractions of plants within 10 min of exposure to NH4Cl, which strongly increased within 2 h, while Na+, MeA+, or K+ did not induce detectable phosphorylation (n = 2 independent experiments; a very weak response was observed in a third experiment after 30 min). These data strongly support the hypothesis that ammonium is the primary factor triggering T460 phosphorylation.
Figure 4.
Phosphorylation Is Specific for NH4+.
Wild-type plants were grown for 9 d in 2 mM NH4NO3 and were nitrogen-starved for 3 d (−N) preceding supply with 500 μM NH4Cl, MeACl, KCl, NaCl, or control media (−N). Microsomal fractions from roots were prepared. Protein gel blots were performed using the AMT1-P and the ‘AMT1;1 loop 2/3′ antisera (n = 2).
Effect of Gln and l-Methionine Sulfoximide on AMT1 Phosphorylation
To investigate whether phosphorylation can be triggered by the ammonium assimilation product Gln, nitrogen-deficient wild-type seedlings were exposed to 1 mM NH4Cl or 10 mM Gln (Figure 5A; see Supplemental Figure 8D online). In contrast with NH4+, which showed phosphorylation already after 10 min of exposure and then increased further over 1 h, Gln did not lead to a significant accumulation of the phosphorylated AMT1s within the first 30 min. However, weak phosphorylation was detected after 2 h. Possibly, the low level of Gln-induced phosphorylation is the consequence of contamination by ammonium or partial deamination of Gln. AMT1;1 protein levels remained stable during the 2-h treatment (Figure 5A). Glu did not initiate any detectable phosphorylation after 2 h. To corroborate the finding that newly synthesized organic nitrogen compounds derived from NH4+ assimilation do not serve as signal, NH4+ and l-methionine sulfoximide (MSX) were added simultaneously. MSX, a Gln synthetase inhibitor, impairs the production of intracellular Gln (Rhodes et al., 1986; Jackson et al., 1993). If Gln or downstream products were the signal triggering phosphorylation, addition of MSX should result in a reduction of the AMT1 phosphorylation. Interestingly, AMT1 phosphorylation was unaffected by the addition of MSX (see Supplemental Figure 9 online), and MSX alone did not trigger any phosphorylation even after 4 h of application (see Supplemental Figure 10 online). Thus, phosphorylation appears to be induced by ammonium itself rather than its assimilation products.
Figure 5.
Effect of Intracellularly Generated Ammonium.
AMT1 phosphorylation of seedlings treated with Gln, NaNO3, or NaNO3 + MSX. Plants were grown for 9 d in presence of 2 mM NH4NO3, transferred for 3 d to nitrogen-free medium (−N), and collected at different time points. Total proteins were prepared from roots.
(A) Roots were exposed to 1 mM NH4Cl or 10 mM Gln. Microsomal proteins were prepared from roots. Protein gel blots were performed using the AMT1-P and the ‘AMT1;1 loop 2/3′ antisera (n = 4).
(B) Roots were exposed to 1 mM NaNO3 ± MSX, 1 mM NH4Cl, or nitrogen-free medium (−N). Total protein extracts were prepared from roots. Protein gel blots were performed using the AMT1-P antiserum (n = 3).
(C) Ammonium concentration in roots after 2-h treatment with nitrogen-free medium (−N), 1 mM NaNO3 ± MSX, or 1 mM NH4Cl. Data shown are mean ± se. Each measurement was performed in triplicate (n = 50 to 60 seedlings/measurement). Results are shown for one representative experiment (n = 2).
Apoplasmic NH4+ Serves as a Phosphorylation Signal
To determine whether phosphorylation is triggered by endogenous NH4+, intracellular NH4+ was generated by adding MSX to the roots in the presence of 1 mM NaNO3. NO3− is first reduced to NO2− and then to NH4+. In the presence of MSX, NH4+ cannot be assimilated and intracellular NH4+ increases (Rhodes et al., 1986). Wild-type seedlings were grown for 9 d in the presence of 2 mM NH4NO3, transferred for 3 d to nitrogen-free medium, and then exposed to 1 mM NH4Cl, 1 mM NaNO3, or 1 mM NaNO3 + 1 mM MSX for 10 min, 30 min, or 2 h. Roots were harvested to monitor AMT1 phosphorylation, and root NH4+ concentrations were measured for the 2-h time point (Figures 5B and 5C; see Supplemental Figure 8E online). No significant phosphorylation was detected in roots exposed to nitrogen-free medium, NaNO3, or NaNO3 + MSX within the first 30 min (Figure 5B). Low levels of phosphorylation were detected 2 h after exposure to NaNO3 + MSX. This weak phosphorylation is probably caused by cytosolic NH4+ leaking from the cell due to high cytoplasmic NH4+ accumulation (Jackson et al., 1993). Indeed, after 2 h of exposure, roots of seedlings treated with NH4Cl or NaNO3 + MSX contained similar amounts of NH4+ (∼225 nmol/mg) and nine times more NH4+ compared with roots exposed to nitrogen-free media or to NaNO3 (Figure 5C). Despite similar intracellular NH4+ levels, the phosphorylation level of AMT1 was ∼5 times higher when cells were exposed to extracellular NH4+ than when they were exposed to intracellular NH4+ derived from nitrate assimilation. Thus, most probably, an extracellular NH4+ sensing mechanism is responsible for triggering phosphorylation of T460.
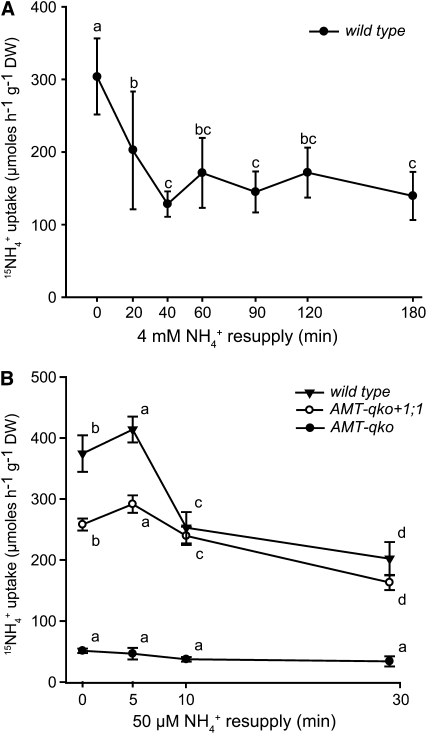
Feedback Inhibition of Ammonium Uptake by Ammonium
The activity of AMT1;1, when expressed in yeast, depended on the communication between subunits of the trimer through allosteric activation by the C terminus (Loqué et al., 2007). Since ammonium induces phosphorylation of the critical residue T460 in the C terminus, we hypothesized that ammonium-triggered phosphorylation causes feedback inhibition of ammonium uptake through AMTs. To test this hypothesis, Arabidopsis plants were grown hydroponically under the same conditions as described above. After ammonium deprivation, roots were exposed to 4 mM NH4+ and short-term uptake of 15N-ammonium was measured. Within 40 min of exposure to 4 mM NH4+,15NH4+ uptake decreased by ∼60% in wild-type plants (Figure 6A). Since phosphorylation was induced even by exposure to low levels of ammonium, 15N-ammonium uptake was determined after resupply of 50 μM NH4+ to wild-type or AMT-qko+1;1 plants (Figure 6B). After a slight initial increase of uptake rates within the first 5 min, the uptake rate decreased by ∼40% in the wild type and by ∼30% in AMT-qko+1;1 plants. A similar decrease (∼40%) was observed for plants supplied with 300 μM ammonium (see Supplemental Figure 11 online). 15NH4+ uptake rates in AMT-qko were ∼7 times lower and remained stable over the time of the experiment (Figure 6B).
Figure 6.
Ammonium Uptake Is Repressed by Ammonium Resupply.
(A) Uptake of 15N-labeled ammonium into roots of wild-type plants. Six-week-old plants were precultured hydroponically under continuous supply of 2 mM ammonium nitrate (+N) and then deprived of nitrogen for 4 d (−N) prior to resupply of 4 mM NH4+ [2 mM (NH4)2SO4] for the indicated times. Bars indicate means ± sd (n = 6 to 8 plants). Significant differences are indicated by different letters (one-way analysis of variance, Fisher's LSD, P < 0.05).
(B) Uptake of 15N-labeled ammonium into roots of wild-type, AMT-qko+1;1, and AMT-qko plants. Six-week-old plants were precultured hydroponically under continuous supply of 2 mM ammonium nitrate (+N) and then deprived of nitrogen for 4 d (−N) prior to resupply of 50 μM NH4+ [25 μM (NH4)2SO4] for indicated times. Bars indicate means ± sd (n = 10 plants). Significant differences are indicated by different letters for each genotype (two-way analysis of variance, Fisher's LSD, P < 0.05).
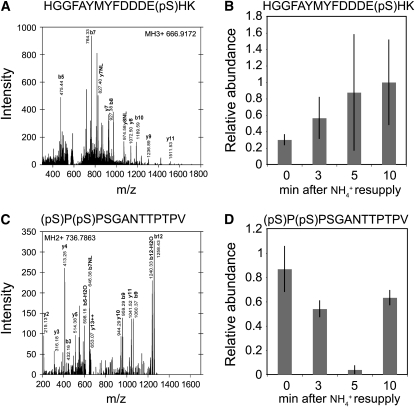
NH4+ Triggers Phosphorylation of Other Sites in the C Terminus
Besides phosphorylation of T460, the mass spectrometric analyses detected two other AMT1;1 phosphopeptides in the C terminus in planta (Figures 7A and 7C). One of the peptides [HGGFAYMYFDDDE(pS)HK] was phosphorylated at position S475; the phosphorylation level of this peptide showed a trend to increase upon ammonium supply; however, the trend was not statistically significant (Figure 7B). The other peptide [(pS)P(pS)PSGANTTPTPV] was phosphorylated at the positions S488 and S490. Interestingly, phosphorylation levels for these two Ser residues correlated inversely with exposure to ammonium (3 mM) (Figure 7D). In a comparative 15N-labeling experiment study, dephosphorylation induced by the presence of NH4+ was confirmed for S488 and S490 (see Supplemental Figure 12 online). Although these domains of the C terminus were not essential for AMT1;1 function when expressed in yeast (Loqué et al., 2007), phosphorylation may play a different role in regulating AMT activity in planta, for example, by integrating other signals. Analysis of deletion mutants in planta will be required to analyze the role and interplay of the different phosphorylation sites.
Figure 7.
Relative Abundance of C-Terminal Phosphopeptides during Ammonium Resupply after Nitrogen Starvation.
(A) Fragmentation spectra of HGGFAYMYFDDDE(pS)HK as detected by CID fragmentation in an LTQ mass spectrometer.
(B) Normalized ion intensity of HGGFAYMYFDDDE(pS)HK peptide. Phosphorylation levels were measured by the ion intensities of phosphopeptides normalized to the time course of ion intensities of nonphosphopeptides of the same protein. Normalized ion intensities of all time points for each peptide were then divided by the maximum value across all time points to standardize the values to a scale of between 0 and 1. Mean value ± sd (n = 5).
(C) Fragmentation spectra of (pS)P(pS)PSGANTTPTPV as detected by CID fragmentation in an LTQ mass spectrometer.
(D) Normalized ion intensity of (pS)P(pS)PSGANTTPTPV peptide. Phosphorylation levels were measured as described in Figure 7B. Mean value ± sd (n = 3).
DISCUSSION
Due to changing nutrient availabilities, plants have to be able to acclimate the properties of nutrient uptake systems to a wide range of nutrient concentrations (Glass, 2002). Moreover, plants have to coordinate uptake and metabolic conversion at varying nutrient supplies and have to promote rapid shutdown of the transporters to prevent accumulation of toxic levels of the nutrient or its products.
Ammonium is one of the two major inorganic forms of nitrogen used by plants and is taken up preferentially over nitrate (Gazzarrini et al., 1999). However, when supplied as the sole nitrogen source over periods of days, ammonium causes toxicity in a wide range of organisms, including Arabidopsis (see Supplemental Figure 13 online; Cooper and Plum, 1987; Britto and Kronzucker, 2002; Hess et al., 2006). It is thus conceivable that ammonium itself inhibits uptake at least during short-term exposure, possibly to prevent ammonium toxicity. The finding that mutants such as T460D as well as most other modifications of the conserved domain of the C terminus of AMT1 inactivate the transporter suggests that phosphorylation of T460 in AMT1;1 may lead to inhibition of uptake (Loqué et al., 2007; Neuhäuser et al., 2007). This hypothesis is consistent with the observed downregulation of the transport by ammonium itself (Figure 6; Rawat et al., 1999).
Previous work had shown that at least under certain conditions (i.e., in Arabidopsis cell cultures grown on high concentrations of ammonium), AMT1;1 is phosphorylated at position T460 (Nühse et al., 2004). T460 is located in a critical position in a hinge region of the trans-activating C terminus, more specifically between a domain that interacts with the loops of its own subunit (IMID) and a domain that couples to the neighboring subunit (ITID; Loqué et al., 2007, 2009). Our data show that phosphorylation of T460 was undetectable when the nutrient solution was depleted for ammonium but T460 became phosphorylated in response to ammonium resupply (Figures 1 to 4). Phosphorylation increased both with time and ammonium concentrations. Interestingly, phosphorylation occurred already at external ammonium levels as low as 50 μM (Figure 2B) (i.e., under conditions that do not necessarily induce ammonium toxicity). The response to comparatively low ammonium levels may suggest the involvement of a yet unknown high-affinity ammonium receptor.
Phosphorylation was detected in <5 min; however, during this period, a small transient increase in the uptake was observed (Figures 6; see Supplemental Figure 11 online). This transient peak was relatively small and might be caused by (1) transient overcompensation for membrane potential in response to an initial depolarization after addition of ammonium, (2) a rapid change in the phosphorylation status of the additional phosphorylation sites identified by phosphoproteomics that could transiently enhance AMT activity, or (3) rapid induction of cytosolic Gln synthetase activity by ammonium supply, which thus creates a larger sink for ammonium compared with uninduced plants. A tight coupling of ammonium transport and GS activity metabolism has been demonstrated in Escherichia coli (Javelle et al., 2005).
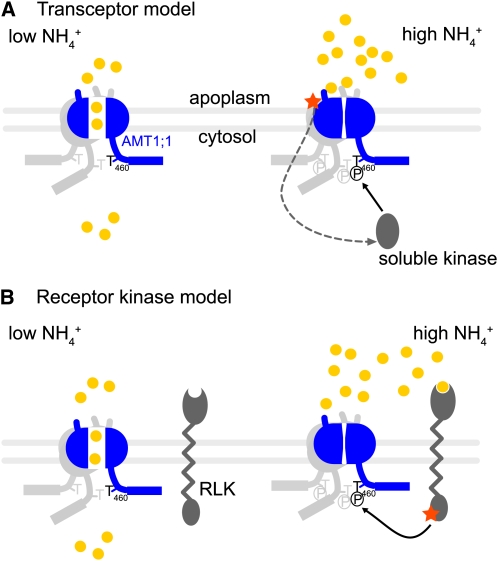
Ammonium itself appears to function as the primary signal, since other monovalent cations or the assimilation products Gln and Glu were not effective in inducing phosphorylation. The inhibitor of ammonium assimilation MSX, which induced intracellular accumulation of NH4+ in roots, also did not lead to AMT1 phosphorylation (Figure 5B; see Supplemental Figure 9 online). This observation suggested that apoplasmic rather than intracellular ammonium acts as the signal. Extracellular signaling requires the activity of a cell-surface receptor (e.g., a receptor kinase). Alternatively, AMT1 could serve as a transceptor (transporter and receptor; Holsbeeks et al., 2004) with a dual function in transport and signaling by recruiting a protein kinase upon binding or translocating ammonium (Figure 8). The transceptor hypothesis is supported by the finding that the yeast AMT homolog Mep2 probably functions as both sensor and transporter. Indeed, the Δmep2 mutant displays defects in pseudohyphal differentiation observed only in the presence of NH4+, a phenotype not observed in cells defective in MEP1 or MEP3 (Lorenz and Heitman, 1998). Moreover, in a recent study, the substitution of two conserved His residues located in the pore of Mep2 led to uncoupling of transport and sensing functions (Boeckstaens et al., 2008). Similarly, the bacterial AmtB homologs have been shown to function in both transport and signaling (Tremblay and Hallenbeck, 2009). When E. coli is exposed to ammonium, GlnK is recruited to AmtB, leading to shutdown of the transporter and affecting the PII signaling cascade, which regulates gene expression (Coutts et al., 2002; Tremblay and Hallenbeck, 2009). Furthermore, GlnK binds α-ketoglutarate and ATP/ADP, integrating both carbon availability and energy status to regulate ammonium uptake (Forchhammer, 2008; Berg et al., 2009). Interestingly, the only plant PII/GlnK homolog identified so far localizes to chloroplasts and thus does not appear to be available to interact with plasma membrane AMTs (Hsieh et al., 1998; Mizuno et al., 2007).
Figure 8.
Hypotheses for T460 Phosphorylation by NH4+ in Plants.
(A) Transceptor model: At low ammonium concentration, the transporter senses the extracellular NH4+ status and T460 is not phosphorylated. NH4+ can enter the cell through the transporter. When ammonium levels increase, the transporter senses the high NH4+ external status. A kinase is recruited and phosphorylation shuts down NH4+ import via the allosteric trans-regulatory mechanism.
(B) Receptor kinase model: At low ammonium concentration, a receptor kinase senses the extracellular NH4+ status and T460 is not phosphorylated. When ammonium increases, the receptor kinase senses the high NH4+ external status and triggers phosphorylation of T460. The transporter is converted to the shut conformation.
The identification of proteins that interact with the C terminus AMT1 may provide a means to identify protein kinases or other proteins involved in ammonium signaling. The amino acids surrounding T460 do not constitute a motif identified so far as a preferred sequence for a certain class of kinases nor is it conserved in other proteins known to be phosphorylated (Nühse et al., 2004; Vlad et al., 2008).
Phosphoproteomics identified two Ser residues, S488 and S490, that were phosphorylated in the absence of ammonium and showed a decrease in phosphorylation levels after ammonium resupply. S488 and S490 are located in the domain of the cytosolic C terminus of the plant AMTs, which is not conserved in the bacterial homologs. Expression of C-terminal mutated variants in yeast showed that the domains downstream of Tyr-469 (Y469) are dispensable for AMT1;1 function (Loqué et al., 2007). Combinations of phosphorylation sites in the C terminus of the plasma membrane H+/ATPase appear to regulate ATPase activity in an additive manner (Fuglsang et al., 2007). The use of multiple phosphorylation sites may constitute a mechanism for integration of multiple signaling pathways. It is thus conceivable that the additional phosphorylation sites found in the C terminus of AMT1;1 also contribute to the regulation of the protein activity in planta. Further phosphorylation sites were detected (S475 in this study and S492 and T496 in Nühse et al., 2004; Benschop et al., 2007; Hem et al., 2007), adding more complexity to the model (Figures 1A and 7). Future work, specifically protein interaction and coexpression studies, will be used to address the question of whether receptor kinases or soluble factors are involved in the phosphorylation of residues in the C terminus of AMT1 (Figure 8).
Our study identified a feedback loop regulating ammonium transport capacity, which operates via an allosteric regulatory mechanism on AMT1 trimers and leads to a decrease in ammonium uptake when extracellular ammonium levels increase. In general, single negative feedback loops serve specific signaling functions and thus create distinct responses: they can generate a basal homeostat, limit the output of a signal, serve in acclimation, or generate transients (Brandman and Meyer, 2008). The type of response depends on the initial conditions and the properties of the feedback loop. More complex responses are typically created in signaling networks by interlacing multiple feedback and feed-forward loops. The high dynamic range of the ammonium response may suggest that the ammonium feedback loop functions primarily in homeostasis and acclimation; however, more detailed information on the overall network structure will be required to define its role. Such homeostat/acclimation mechanisms are probably important in soils, where ammonium levels can change either due to spatial variation, sudden exposure to animal excretion-derived ammonium, or ammonium derived from organic matter decomposition. This mechanism may reflect an adaptation of plants to a wider range of nutrient levels. The extraordinary conservation of the C terminus and the allosteric regulation even in archaebacteria suggests that the principal regulatory machinery developed early during evolution and has been retained in organisms from different kingdoms (Loqué et al., 2009). There is no evidence that bacteria or yeast also use phosphorylation-dependent regulation; thus, alternative regulatory input mechanisms must have been set in place, such as those identified in E. coli (Coutts et al., 2002).
Feedback loops operating via allosteric trans-regulation and involving phosphorylation may not be restricted to the AMT family. A similar allosteric regulation has recently been suggested for osmotic regulation of compatible solute transporters in Corynebacterium (Krämer and Ziegler, 2009). It will be interesting to explore whether similar phospho-dependent allosteric regulation within oligomers exists in the case of other nutrient acquisition systems. Phosphorylation has been reported to regulate the activity of a variety of transporters, channels, and pumps (Fuglsang et al., 2003; Lee et al., 2007). The C-terminal domain of PIP2 aquaporins interacts with adjacent subunits in a tetramer, and phosphorylation of the C-terminal S274 enhances transport activity (Törnroth-Horsefield et al., 2006). Interestingly, CHL1 (also named NRT1;1) simultaneously functions as a dual affinity nitrate uptake transporter (Liu and Tsay, 2003; Ho et al., 2009). The affinity of CHL1 is regulated by phosphorylation. When Thr-101 is unphosphorylated, CHL1 works as a low affinity transporter and modulates the expression of genes regulated by high levels of nitrate. When phosphorylated, CHL1 is converted into a high-affinity transporter and regulates the expression of genes affected by low levels of nitrate. Thus, similar as for nitrate uptake, AMT transporter activity is regulated by ammonium. It is conceivable that plant AMTs also function as receptors, as has been shown in the case of AmtB in E. coli (Coutts et al., 2002; Tremblay and Hallenbeck, 2009).
METHODS
Plant Materials
The generation of the AMT-qko quadruple AMT knockout line and of the AMT-qko+1;1 has been described by Yuan et al. (2007b). Wild-type plants used here were Arabidopsis thaliana ecotype Columbia-0.
Plant Growth Conditions
For hydroponic growth experiments, seeds were germinated and precultured for 1 week on a glass wool/plastic fiber mix moistened with tap water under a plastic cover. After 1 week, tap water was substituted with a full hydroponic growth medium, which was renewed weekly for 6 weeks, containing 2 mM NH4NO3, 1 mM KH2PO4, 1 mM MgSO4, 250 μM K2SO4, 250 μM CaCl2, 100 μM Na-Fe-EDTA, 50 μM KCl, 50 μM H3BO3, 5 μM MnSO4, 1 μM ZnSO4, 1 μM CuSO4, and 1 μM NaMoO4 (pH adjusted to 6.0 with KOH). Plants were then transferred to hydroponic growth medium without nitrogen for 4 d; the medium was changed every 2 d. All ammonium and salt treatments were conducted at mid-day with freshly prepared nitrogen-free medium preincubated at 22°C. (NH4)2SO4, NH4Cl, MeACl, KCl, or NaCl were added and mixed to the incubation solution prior to the treatment. Ammonium pulse treatments were conducted by transferring 4-d-old nitrogen-starved plants to fresh nutrient solution containing 50, 500, or 5000 μM NH4+ for 3 min, followed by a 2-min wash in nitrogen-free hydroponic medium; then plants were either harvested or transferred to new nitrogen-depleted hydroponic medium until sampling. Roots were immediately harvested in liquid nitrogen. Plants were grown under nonsterile conditions in a growth cabinet under the following conditions: 10/14-h light/dark at 22°C and 50% relative humidity.
For axenic culture experiments, Arabidopsis seeds were surface sterilized and germinated on full nutrient (FN) medium (2 mM NH4NO3, 1 mM KH2PO4, 1 mM MgSO4, 250 μM K2SO4, 250 μM CaCl2, 100 μM Na-Fe-EDTA, 50 μM KCl, 50 μM H3BO3, 5 μM MnSO4, 1 μM ZnSO4, 1 μM CuSO4, 1 μM NaMoO4, 1 mM MES, 1% [w/v] sucrose, and 1.2% [w/v] agar, pH 5.8 [KOH]) on plates kept vertically (Chaudhuri et al., 2008). After 9 d, seedlings were transferred to the medium lacking nitrogen. Three days after nitrogen starvation, seedlings were transferred to fresh medium supplemented with 1 mM NH4Cl, 10 mM Gln, 10 mM Glu, or 1 mM MSX. Alternatively, roots were treated with 1 mM NaNO3 in the presence or absence of MSX. Roots were collected at different time points and frozen in liquid nitrogen. Plants were grown under sterile conditions in a growth cabinet under the following conditions: 16/8-h light/dark at 22°C.
For plasma membrane preparation, Arabidopsis seedlings were grown in axenic liquid culture for 9 d in medium supplemented with 3 mM NH4NO3 before transfer to nitrogen-deficient conditions for 2 d. Subsequently, nitrogen was resupplied in the form of 10 mM NH4Cl. Seedlings were harvested, and plasma membrane fractions as well as cytosolic protein fractions were prepared as described below.
For 15N-labeling, Arabidopsis seedlings were grown in FN liquid medium supplemented with either 2 mM 15NH415NO3 or 14NH414NO3 as the sole nitrogen source. After 7 d, seedlings were transferred to FN medium containing either 400 μM of K15NO3 or K14NO3 as nitrogen source and were grown for three more days. 15N-labeled seedlings were exposed to 2 mM NH4Cl in FN medium for 10 min, seedlings were combined (equal mass) with untreated 14N-grown seedlings, and plasma membranes were prepared. In a reciprocal experiment, 14N-grown seedlings were exposed to 2 mM NH4Cl in FN medium for 10 min, seedlings were combined (equal mass) with untreated 15N-grown seedlings, and plasma membranes were prepared (for concept of reciprocal experiments, see Lanquar et al., 2007; Kierszniowska et al., 2009).
Protein Isolation
Total proteins from roots were prepared by homogenization in 50 mM HEPES adjusted to pH 7.2 with NaOH, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 10% (w/v) glycerol, 1% (v/v) Triton X-100, 150 mM NaCl, 5 mM DTT, 1× Complete Protease Inhibitor Cocktail, and 0.5× PhosStop phosphatases inhibitor cocktail (Roche Applied Science). Samples were then centrifuged for 10 min, 1000g at 4°C, and supernatants were recovered. For preparation of microsomes, roots were ground in liquid nitrogen and resuspended in buffer containing 250 mM Tris-Cl, pH 8.5, 290 mM sucrose, 25 mM EDTA, 5 mM β-mercaptoethanol, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.5× Complete Protease Inhibitor Cocktail, and 0.5× PhosStop Phosphatase Inhibitor Cocktail (Roche Applied Science). After centrifugation at 10,000g for 15 min, supernatants were filtered through Miracloth (Calbiochem) and recentrifuged at 100,000g for 45 min. The sediment containing the microsomes was resuspended in storage buffer (400 mM mannitol, 10% glycerol, 6 mM MES/Tris, pH 8, 4 mM DTT, 2 mM phenylmethylsulfonyl fluoride, and 1× phosphatase inhibitor cocktail 1 and 2 [Sigma-Aldrich P2850 and P5726]). Protein concentration was estimated by a modified Bradford method (Stoscheck, 1990). For the phosphatase treatment, 10 μg of microsomal membrane proteins were incubated for 45 min at 37°C with either 5 units of calf intestinal phosphatase alkaline (CIP; New England Biolabs) or with 1× Phosphatase Inhibitor Cocktail 1 and 2 (Sigma-Aldrich P2850 and P5726).
Plasma Membrane Preparation and Phosphopeptide Enrichment
Plasma membranes of seedlings were isolated using a two-phase partitioning system (Niittyla et al., 2007). Plasma membrane vesicles were inverted to inside-out using Brij58, and proteins were digested in solution using trypsin. Phosphopeptides were enriched over titanium dioxide (Titansphere; 5 μm) (Olsen et al., 2006).
Protein Gel Blot Analysis
Microsomal membrane proteins or total proteins (12 to 15 μg per lane) were denatured in loading buffer (62.5 mM Tris-HCl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 2.5% [v/v] β-mercaptoethanol, and 0.01% [w/v] bromophenol blue) at 40°C for 30 min, separated on 4 to 20% gradient SDS-PAGE gels (precast gels; Bio-Rad), and transferred to polyvinylidene fluoride membranes. An affinity-purified polyclonal antiserum (AMT1-P) raised against a peptide corresponding to AMT1;1 with phosphorylated T460 (n-GMDNorleucinpTRHGGFA-c; BioGenes) was used at 1/2000 dilution; ‘AMT1;1 loop 2/3′ antiserum was used as described (Loqué et al., 2006); the secondary antibody peroxidase-conjugated anti-rabbit IgG (Pierce Biotechnology) was used at 1/10,000 to 1/20,000 dilutions. Blots were developed using SuperSignal West Pico or Femto Chemiluminescent Substrate (Pierce Biotechnology). Phosphorylation was quantified using ImageJ (http://rsbweb.nih.gov/ij/) using plot lane (Belin et al., 2006).
Ammonium Content
After harvesting, roots were washed for 1 min in 1 mM CaSO4 and immediately frozen. Samples were freeze-dried and ground, and 50 to 100 mg were homogenized in 1 mL of cold 10 mM formic acid. After centrifugation at 13,000g, the supernatant was frozen overnight at −20°C. Thirty microliters of supernatant was mixed with cold 3 mM o-phthalaldehyde, 10 mM β-mercaptoethanol, and 10 mM potassium phosphate buffer, pH 6.8, incubated at 80°C for 15 min, and immediately cooled on ice. Fluorescence was measured at 470 nm after excitation at 410 nm using a fluorescence spectrophotometer (Infinite; Tecan) (Loqué et al., 2005).
15N-Ammonium Uptake
Plants were cultured and nitrogen-starved for 4 d as described above. Two sets of experiments were performed: ammonium was resupplied by transferring plants to fresh nutrient solution containing 4 mM NH4+ [2 mM (NH4)2SO4] or 50 μM NH4+ [25 μM (NH4)2SO4] for different periods of time. For the 4 mM NH4+ supply experiment, ammonium uptake into Arabidopsis roots was conducted after rinsing roots in 1 mM CaSO4 for 1 min, followed by incubation for 6 min in nutrient solution containing 200 μM ammonium [100 μM (15NH4)2SO4, 95 atom% 15N] as the sole nitrogen source, followed by a wash in 1 mM CaSO4. In the second set of experiments (50 μM NH4+ supply), in order to reduce the effect of phosphorylation, the uptake period was reduced to 2 min in nutrient solution containing 200 μM ammonium [75 μM (15NH4)2SO4 (99 atom% 15N) and 25μM (NH4)2SO4], and the first 1-min wash in 1 mM CaSO4 was omitted. In both experiments, after harvest, roots were stored at −70°C before freeze-drying. Samples were ground, and ∼1.6 mg powder was used for 15N determination by isotope mass spectrometry (Thermo-Finnigan).
Liquid Chromatography–MS/MS and Data Analysis
Tryptic peptide mixtures were analyzed by liquid chromatography–MS/MS using nanoflow HPLC (Proxeon Biosystems) and a linear ion trap instrument (LTQ-Orbitrap; Thermo Scientific) as mass analyzer. Peptides were eluted from a 75-μm analytical column (Reprosil C18; Dr. Maisch GmbH) on a linear gradient running from 10 to 30% acetonitrile in 50 min and sprayed directly into the LTQ mass spectrometer. Proteins were identified by MS/MS by information-dependent acquisition of fragmentation spectra of multiple-charged peptides. Additional fragmentation using the Pseudo MSn method (Schroeder et al., 2004) was used if peptides displayed a loss of phosphoric acid (neutral loss, 98 D) upon MS/MS fragmentation. Fragment MS/MS spectra from raw files were extracted as DTA files and then merged to peak lists using default settings of DTASuperCharge version 1.19 (msquant.sourcforge.net) with a tolerance for precursor ion detection of 50 ppm.
Fragmentation spectra were searched against a nonredundant Arabidopsis protein database (TAIR8, version 2008-04; 31,921 entries; www.Arabidopsis.org) using the Mascot algorithm (version 2.2.0; Matrix Science). The database contained the full Arabidopsis proteome and commonly observed contaminants (human keratin, trypsin, and lysyl endopeptidase); thus, no taxonomic restrictions were used during the automated database search. The following search parameters were applied: trypsin as cleaving enzyme, peptide mass tolerance 10 ppm, MS/MS tolerance 0.8 D, and one missed cleavage allowed. Carbamidomethylation of Cys was set as a fixed modification, and Met oxidation and phosphorylation of Ser, Thr, and Tyr were chosen as variable modifications. Only peptides with a length of more than five amino acids were considered.
In general, peptides were accepted without manual interpretation if they displayed a Mascot score of >32 (as defined by Mascot P < 0.01 significance threshold), peptides with a score of >24 were manually inspected, requiring a series of three consecutive y or b ions to be accepted; in addition, mass accuracy and delta scores were taken into account when single peptides were accepted.
Phosphorylation sites were assigned using MSQuant version 1.4.1 as described (Olsen et al., 2006). Briefly, for each peptide, different combinations of phosphorylation sites were scored (PTM score), and the highest scoring match was accepted if the sum of mascot score and PTM score was higher than 34. If no distinction could be made on the basis of the score difference between different site assignments, the phosphorylation site was marked as ambiguous. Currently, phosphorylation sites with P values within three orders of magnitude would be considered equally likely.
Quantitative analysis of changes in phosphorylation status was performed based on normalized ion intensities as described (Niittyla et al., 2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AMT1;1 (At4g13510), AMT1;2 (At1g64780), AMT1;3 (At3g24300), AMT1;4 (At4g28700), AMT1;5 (At3g24290), and AMT2;1 (At2g38290).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Antiserum AMT-P Specifically Recognizes the Phosphorylated (T460) Peptide GMDMTRHGGFAY.
Supplemental Figure 2. Ponceau S Staining as Loading Control for Protein Gel Blots.
Supplemental Figure 3. CIP Treatment Leads to a Loss of a Reaction with the Antiserum.
Supplemental Figure 4. Ponceau S Staining as Loading Control for Protein Gel Blots.
Supplemental Figure 5. Kinetics of AMT1 Phosphorylation after NH4+ Supply.
Supplemental Figure 6. Concentration Dependence of AMT1 Phosphorylation.
Supplemental Figure 7. AMT1 Phosphorylation Level Resulting from Constant Exposure to NH4+ versus Transient Exposure to NH4+.
Supplemental Figure 8. Control for Protein Loading.
Supplemental Figure 9. Effect of NH4+ + MSX on AMT1 Phosphorylation.
Supplemental Figure 10. Effect of MSX on AMT1 Phosphorylation.
Supplemental Figure 11. Ammonium Uptake Is Repressed by Ammonium Resupply.
Supplemental Figure 12. NH4+ Induces the Decrease of Phosphorylation of S488 and S490.
Supplemental Figure 13. Arabidopsis Is Sensitive to High NH4Cl Concentrations.
Supplementary Material
Acknowledgments
This work was made possible by grants from the National Science Foundation Arabidopsis 2010 program (MCB-0618402) and the Department of Energy (DE-FG02-04ER15542) to W.B.F. and by the Deutsche Forschungsgemeinschaft, Bonn to N.v.W. We thank Elke Dachtler for excellent technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wolf B. Frommer (wfrommer@stanford.edu).
Online version contains Web-only data.
References
- Andrade, S.L., Dickmanns, A., Ficner, R., and Einsle, O. (2005). Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 102 14994–14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayling, S.M. (1993). The effect of ammonium ions on membrane potential and anion flux in roots of barley and tomato. Plant Cell Environ. 16 297–303. [Google Scholar]
- Belin, C., de Franco, P.O., Bourbousse, C., Chaignepain, S., Schmitter, J.M., Vavasseur, A., Giraudat, J., Barbier-Brygoo, H., and Thomine, S. (2006). Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 141 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop, J.J., Mohammed, S., O'Flaherty, M., Heck, A.J., Slijper, M., and Menke, F.L. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6 1198–1214. [DOI] [PubMed] [Google Scholar]
- Berg, J., Hung, Y.P., and Yellen, G. (2009). A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods 6 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckstaens, M., André, B., and Marini, A.M. (2008). Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J. Biol. Chem. 283 21362–21370. [DOI] [PubMed] [Google Scholar]
- Brandman, O., and Meyer, T. (2008). Feedback loops shape cellular signals in space and time. Science 322 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto, D.T., and Kronzucker, H.J. (2002). NH4+ toxicity in higher plants: a critical review. J. Plant Physiol. 159 567–584. [Google Scholar]
- Chaudhuri, B., Hormann, F., Lalonde, S., Brady, S.M., Orlando, D.A., Benfey, P., and Frommer, W.B. (2008). Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 56 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A.J., and Plum, F. (1987). Biochemistry and physiology of brain ammonia. Physiol. Rev. 67 440–519. [DOI] [PubMed] [Google Scholar]
- Coutts, G., Thomas, G., Blakey, D., and Merrick, M. (2002). Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer, K. (2008). P(II) signal transducers: novel functional and structural insights. Trends Microbiol. 16 65–72. [DOI] [PubMed] [Google Scholar]
- Fuglsang, A.T., Borch, J., Bych, K., Jahn, T.P., Roepstorff, P., and Palmgren, M.G. (2003). The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: Involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J. Biol. Chem. 278 42266–42272. [DOI] [PubMed] [Google Scholar]
- Fuglsang, A.T., Guo, Y., Cuin, T.A., Qiu, Q., Song, C., Kristiansen, K.A., Bych, K., Schulz, A., Shabala, S., Schumaker, K.S., Palmgren, M.G., and Zhu, J.K. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W.B., and von Wirén, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, A. (2002). Nutrient Absorption by Plant Roots: Regulation of Uptake to Match Plant Demand. (New York: Marcel Dekker).
- Hem, S., Rofidal, V., Sommerer, N., and Rossignol, M. (2007). Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem. Biophys. Res. Commun. 363 375–380. [DOI] [PubMed] [Google Scholar]
- Hess, D.C., Lu, W., Rabinowitz, J.D., and Botstein, D. (2006). Ammonium toxicity and potassium limitation in yeast. PLoS Biol. 4 e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.H., Lin, S.H., Hu, H.C., and Tsay, Y.F. (2009). CHL1 function as a nitrate sensor in plants. Cell 138 1184–1194. [DOI] [PubMed] [Google Scholar]
- Holsbeeks, I., Lagatie, O., Van Nuland, A., Van de Velde, S., and Thevelein, J.M. (2004). The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29 556–564. [DOI] [PubMed] [Google Scholar]
- Hsieh, M.H., Lam, H.M., van de Loo, F.J., and Coruzzi, G. (1998). A PII-like protein in Arabidopsis: Putative role in nitrogen sensing. Proc. Natl. Acad. Sci. USA 95 13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, W.A., Chaillou, S., Morot-Gaudry, J.-F., and Volk, R.J. (1993). Endogenous ammonium generation in maize roots and its relationship to other ammonium fluxes. J. Exp. Bot. 44 731–739. [Google Scholar]
- Javelle, A., Lupo, D., Ripoche, P., Fulford, T., Merrick, M., and Winkler, F.K. (2008). Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB. Proc. Natl. Acad. Sci. USA 105 5040–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle, A., Thomas, G., Marini, A.M., Kramer, R., and Merrick, M. (2005). In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem. J. 390 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi, S., O'Connell III, J., Remis, J., Robles-Colmenares, Y., Miercke, L.J., and Stroud, R.M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305 1587–1594. [DOI] [PubMed] [Google Scholar]
- Kierszniowska, S., Walther, D., and Schulze, W.X. (2009). Ratio-dependent significance thresholds in reciprocal 15N-labeling experiments as a robust tool in detection of candidate proteins responding to biological treatment. Proteomics 9 1916–1924. [DOI] [PubMed] [Google Scholar]
- Krämer, R., and Ziegler, C. (2009). Regulative interactions of the osmosensing C-terminal domain in the trimeric glycine betaine transporter BetP from Corynebacterium glutamicum. Biol. Chem. 390 685–691. [DOI] [PubMed] [Google Scholar]
- Lanquar, V., Kuhn, L., Lelievre, F., Khafif, M., Espagne, C., Bruley, C., Barbier-Brygoo, H., Garin, J., and Thomine, S. (2007). 15N-metabolic labeling for comparative plasma membrane proteomics in Arabidopsis cells. Proteomics 7 750–754. [DOI] [PubMed] [Google Scholar]
- Lee, S.C., Lan, W.Z., Kim, B.G., Li, L., Cheong, Y.H., Pandey, G.K., Lu, G., Buchanan, B.B., and Luan, S. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 104 15959–15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.H., and Tsay, Y.F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué, D., Ludewig, U., Yuan, L., and von Wirén, N. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué, D., Lalonde, S., Looger, L.L., von Wirén, N., and Frommer, W.B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446 195–198. [DOI] [PubMed] [Google Scholar]
- Loqué, D., Mora, S.I., Andrade, S.L., Pantoja, O., and Frommer, W.B. (2009). Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. J. Biol. Chem. 284 24988–24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55 1293–1305. [DOI] [PubMed] [Google Scholar]
- Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., Ishiyama, K., Takahashi, H., and von Wirén, N. (2006). Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48 522–534. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C., and Heitman, J. (1998). The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., Neuhauser, B., and Dynowski, M. (2007). Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 581 2301–2308. [DOI] [PubMed] [Google Scholar]
- Ludewig, U., von Wirén, N., and Frommer, W.B. (2002). Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277 13548–13555. [DOI] [PubMed] [Google Scholar]
- Ludewig, U., Wilken, S., Wu, B., Jost, W., Obrdlik, P., El Bakkoury, M., Marini, A.M., André, B., Hamacher, T., Boles, E., von Wirén, N., and Frommer, W.B. (2003). Homo- and hetero-oligomerization of ammonium transporter-1 NH4 uniporters. J. Biol. Chem. 278 45603–45610. [DOI] [PubMed] [Google Scholar]
- Lupo, D., Li, X.D., Durand, A., Tomizaki, T., Cherif-Zahar, B., Matassi, G., Merrick, M., and Winkler, F.K. (2007). The 1.3-Å resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc. Natl. Acad. Sci. USA 104 19303–19308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, A.M., Springael, J.Y., Frommer, W.B., and André, B. (2000). Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 35 378–385. [DOI] [PubMed] [Google Scholar]
- Marini, A.M., Vissers, S., Urrestarazu, A., and André, B. (1994). Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13 3456–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M., Dynowski, M., and Ludewig, U. (2006). Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem. J. 396 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A.J., Cookson, S.J., Smith, S.J., and Wells, D.M. (2001). The use of microelectrodes to investigate compartmentation and the transport of metabolized inorganic ions in plants. J. Exp. Bot. 52 541–549. [PubMed] [Google Scholar]
- Mizuno, Y., Berenger, B., Moorhead, G.B., and Ng, K.K. (2007). Crystal structure of Arabidopsis PII reveals novel structural elements unique to plants. Biochemistry 46 1477–1483. [DOI] [PubMed] [Google Scholar]
- Neuhäuser, B., Dynowski, M., Mayer, M., and Ludewig, U. (2007). Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 143 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittyla, T., Fuglsang, A.T., Palmgren, M.G., Frommer, W.B., and Schulze, W.X. (2007). Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6 1711–1726. [DOI] [PubMed] [Google Scholar]
- Ninnemann, O., Jauniaux, J.C., and Frommer, W.B. (1994). Identification of a high affinity NH4+ transporter from plants. EMBO J. 13 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse, T.S., Stensballe, A., Jensen, O.N., and Peck, S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, J.V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127 635–648. [DOI] [PubMed] [Google Scholar]
- Rawat, S.R., Silim, S.N., Kronzucker, H.J., Siddiqi, M.Y., and Glass, A.D. (1999). AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: Evidence for regulation by root glutamine levels. Plant J. 19 143–152. [DOI] [PubMed] [Google Scholar]
- Rhodes, D., Deal, L., Haworth, P., Jamieson, G.C., Reuter, C.C., and Ericson, M.C. (1986). Amino acid metabolism of Lemna minor L.: I. Responses to methionine sulfoximine. Plant Physiol. 82 1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, M.J., Shabanowitz, J., Schwartz, J.C., Hunt, D.F., and Coon, J.J. (2004). A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76 3590–3598. [DOI] [PubMed] [Google Scholar]
- Severi, E., Javelle, A., and Merrick, M. (2007). The conserved carboxy-terminal region of the ammonia channel AmtB plays a critical role in channel function. Mol. Membr. Biol. 24 161–171. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp, C., Wood, C.C., Roeb, G.W., and Udvardi, M.K. (2002). Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130 1788–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck, C.M. (1990). Quantitation of protein. Methods Enzymol. 182 50–68. [DOI] [PubMed] [Google Scholar]
- Törnroth-Horsefield, S., Wang, Y., Hedfalk, K., Johanson, U., Karlsson, M., Tajkhorshid, E., Neutze, R., and Kjellbom, P. (2006). Structural mechanism of plant aquaporin gating. Nature 439 688–694. [DOI] [PubMed] [Google Scholar]
- Tremblay, P.L., and Hallenbeck, P.C. (2009). Of blood, brains and bacteria, the Amt/Rh transporter family: Emerging role of Amt as a unique microbial sensor. Mol. Microbiol. 71 12–22. [DOI] [PubMed] [Google Scholar]
- Vlad, F., Turk, B.E., Peynot, P., Leung, J., and Merlot, S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55 104–117. [DOI] [PubMed] [Google Scholar]
- von Wirén, N., and Merrick, M. (2004). Regulation and function of ammonium carriers in bacteria, fungi, and plants. In Topics in Current Genetics: Molecular Mechanisms Controlling Transmembrane Transport, E. Boles and R. Krämer, eds (Berlin: Springer), pp. 95–120.
- Wang, M.Y., Glass, A., Shaff, J.E., and Kochian, L.V. (1994). Ammonium uptake by rice roots (III. Electrophysiology). Plant Physiol. 104 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., Graff, L., Loqué, D., Kojima, S., Tsuchiya, Y.N., Takahashi, H., and von Wirén, N. (2009). AtAMT1;4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 50 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., Takahashi, H., and von Wirén, N. (2007. b). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19 2636–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., Loqué, D., Ye, F., Frommer, W.B., and von Wirén, N. (2007. a). Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol. 143 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.