The Arabidopsis thaliana genome encodes >1500 transcription factors, and ∼45% of these belong to families specific to plants (Riechmann et al., 2000). Comparison of the entire complement of transcription factors of Arabidopsis, Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae revealed that families common to all eukaryotes are conserved only in the sequence and structure of the domains defining the respective families and that each eukaryotic lineage has diversified greatly to create novel proteins regulating lineage-specific processes (Riechmann et al., 2000). The zinc finger family of proteins is an example of such diversification and it consists of a large number of proteins that are further classified into distinct subfamilies (see SMART, http://smart.embl-heidelberg.de/smart/show_motifs.pl). The Zn finger domains form multiple finger-like protrusions that can bind metals like zinc. Proteins with Zn finger domains often contain other specialized interaction domains and have been recognized to bind (1) DNA, as in the case of Zn finger transcription factors, such as Dof and WRKY proteins; (2) RNA, as in the case of the microRNA processing factor SERRATE; and (3) proteins, as is thought to be the case for other Zn finger proteins like the B-box proteins (Borden et al., 1995; Rushton et al., 1995; Yanagisawa, 1995; de Pater et al., 1996; Yanagisawa and Schmidt, 1999; Torok and Etkin, 2001; Yang et al., 2006; Fujioka et al., 2007). The B-box family represents a subgroup of Zn finger proteins that contain one or more B-box domains with specialized tertiary structures that are stabilized by binding Zn ions (Klug and Schwabe, 1995). As indicated above, the B-box domain is thought to be involved in protein–protein interactions. It is not yet known how B-box proteins function at the biochemical level, but they may be involved in modulating protein–protein interactions, such as those occurring in transcriptional complexes. Some of the B-box family members contain additional domains (see below), but it remains to be seen if these regions of the protein confer additional discrete functions. In Arabidopsis, the B-box family consists of 32 proteins (Riechmann et al., 2000; Robson et al., 2001; Chang et al., 2008; Kumagai et al., 2008). The purpose of this letter is to establish a uniform terminology for the B-box proteins. There is a growing effort to understand the functions of B-box proteins; hence, it is timely to provide a uniform nomenclature to this family consistent with the format used for other families, like the basic helix-loop-helix and the MYB families (Stracke et al., 2001; Bailey et al., 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003). We provide a uniform designation for proteins containing one or more copies of the B-box domain (e.g., At BBX1/CO).
CONSTANS (CO) was the first B-box protein to be identified in Arabidopsis (Putterill et al., 1995; Robson et al., 2001). In addition to CO, 16 other CO-Like (COL) proteins have since been identified that contain one or two B-box domains at the N terminus and a CCT (CO, COL, TOC1) domain (Strayer et al., 2000) at the C terminus (Ledger et al., 2001; Robson et al., 2001). Other proteins, STH (STH1), STH2, and STO, were identified that had two tandem B-box domains but lacked the C-terminal CCT domain (Holm et al., 2001; Datta et al., 2007). More recent studies have used different names for some of the other members of the B-box family (Chang et al., 2008; Datta et al., 2008; Kumagai et al., 2008). In order to provide a uniform nomenclature for the B-box protein family, we performed a search to identify a complete set of all Arabidopsis genes with B-box motifs. Consistent with the previous reports (Chang et al., 2008; Kumagai et al., 2008), we found a final set of 32 Arabidopsis B-box proteins (Table 1).
Table 1.
Positions of B-Box Domains
| BBX Name | B-Box 1 (B1) Coordinates | B-Box 2 (B2) Coordinates | |
|---|---|---|---|
| AT5G15840 | BBX1 | 20–57 | 63–100 |
| AT5G15850 | BBX2 | 12–49 | 55–92 |
| AT3G02380 | BBX3 | 16–53 | 59–96 |
| AT2G24790 | BBX4 | 8–45 | 51–88 |
| AT5G24930 | BBX5 | 50–87 | 93–130 |
| AT5G57660 | BBX6 | 22–55 | 61–98 |
| AT3G07650 | BBX7 | 5–42 | 48–94 |
| AT5G48250 | BBX8 | 5–42 | 48–87 |
| AT4G15250 | BBX9 | 5–42 | 48–83 |
| AT3G21880 | BBX10 | 5–42 | 48–83 |
| AT2G47890 | BBX11 | 13–50 | 56–91 |
| AT2G33500 | BBX12 | 12–49 | 55–92 |
| AT1G28050 | BBX13 | 9–46 | 52–89 |
| AT1G68520 | BBX14 | 17–54 | |
| AT1G25440 | BBX15 | 17–54 | |
| AT1G73870 | BBX16 | 22–60 | |
| AT1G49130 | BBX17 | 28–65 | |
| AT2G21320 | BBX18 | 5–42 | 56–91 |
| AT4G38960 | BBX19 | 5–42 | 56–91 |
| AT4G39070 | BBX20 | 5–42 | 58–95 |
| AT1G75540 | BBX21 | 5–42 | 60–97 |
| AT1G78600 | BBX22 | 5–42 | 57–94 |
| AT4G10240 | BBX23 | 5–42 | 63–96 |
| AT1G06040 | BBX24 | 5–42 | 57–94 |
| AT2G31380 | BBX25 | 5–42 | 57–94 |
| AT1G60250 | BBX26 | 5–41 | |
| AT1G68190 | BBX27 | 14–51 | |
| AT4G27310 | BBX28 | 5–41 | |
| AT5G54470 | BBX29 | 6–42 | |
| AT4G15248 | BBX30 | 32–68 | |
| AT3G21890 | BBX31 | 31–67 | |
| AT3G21150 |
BBX32 |
5–41 |
B-box coordinates are from start of the first residue of protein.
SPECIFIC B-BOX MOTIFS FOR A GENOME-WIDE SEARCH IN ARABIDOPSIS
A representative B-box from AT1G78600 was used as a query in a PSI-BLAST (Altschul et al., 1997) search against the set of 32 Arabidopsis B-box–containing proteins to identify exact B-box amino acid sequence coordinates within each protein. Of the full set of 32 proteins, 21 had a potential second B-box copy as found by PSI-BLAST analysis. The second B-box motifs are present 5 to 20 residues following the first B-box. In every case, first and second B-box motifs were found in the same orientation. B-box–containing proteins and coordinates of B-box motifs within each protein are shown in Table 1. These results are consistent with previous reports (Putterill et al., 1995; Chang et al., 2008; Kumagai et al., 2008).
Three sets of protein sequence multiple alignments (ClustalW and MUSCLE; Edgar, 2004; Larkin et al., 2007) were constructed with the B-box motif: a set of 32 B-box sequences from the first B-box (N-terminal; called B1 from here on), a set of 21 sequences from the second B-box (termed B2), and a set of 53 sequences containing both B-boxes B1 and B2. A motif was made from each multiple alignment (Figure 1) and used as a position-specific score matrix for PSI-BLAST (Altschul et al., 1997) search against the full set of TAIR 8 (Swarbreck et al., 2008) Arabidopsis proteins. A cutoff E-value of 0.1 for the length of the B-box region was used for prediction of new B-box–containing proteins. No additional proteins beyond the initial 32 were identified with the B1 or B2 queries; the set of 32 B-box genes is therefore considered complete.
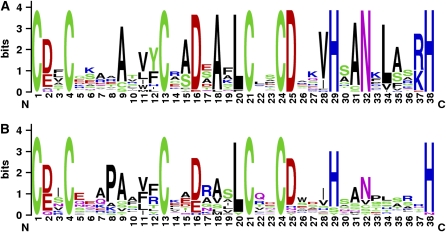
Figure 1.
The B-Box Family of Proteins from Arabidopsis Have Two Distinct B-Box Domains.
Motif logos (Schneider and Stephens, 1990; Crooks et al., 2004) of protein alignments of B-box B1 (A) and B2 (B) show differences. Conservation of residues across all proteins is shown by height of each letter; a position with all like residues has a bit score of 4. The 38 conserved residues of B1 and B2 are shown; see Table 1 for individual coordinates of each B-box motif. B1 is more conserved, containing 12 out of 38 residues that are seen in 95% of the 32 family members. B2 shows seven amino acids with 95% conservation. The conserved residues of B2 are also found conserved in B1.
RENAMING OF B-BOX–CONTAINING PROTEINS BASED ON PHYLOGENETIC ANALYSIS
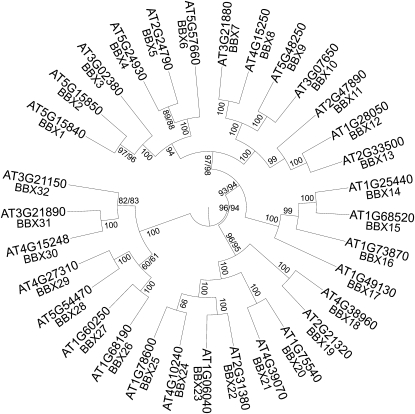
The 32 full-length proteins identified by B-box sequence searches were combined for multiple alignment and phylogeny analyses as a basis for renaming the entire gene family. Proteins were aligned using MUSCLE (Edgar, 2004), alignments were viewed, and minor adjustments were made using SeaView (Galtier et al., 1996). A Bayesian inference phylogenetic tree was built using MrBayes (Huelsenbeck and Ronquist, 2001). Blosum62 (Henikoff and Henikoff, 1992) was used as the amino acid model, and alignment gaps were included in the analysis using the binary (restriction site) model. One million generations were run to create two independent phylogenies using metropolis coupling with four chains (Nchains = 4): one cold and three heated chains (Temp = 0.2) (Figure 2). Clade posterior probabilities differed by at most 0.02 between the two trees (if differences were seen, both values are shown in Figure 2).
Figure 2.
Phylogenetic Analysis of the Arabidopsis B-Box Family.
Full-length proteins of the 32 B-box members were aligned using MUSCLE (maxiters = 16) (Edgar, 2004). Multiple alignments were viewed and realigned using SeaView (Galtier et al., 1996). MrBayes (Huelsenbeck and Ronquist, 2001) was used to construct a Bayesian inference phylogenetic tree with 1,000,000 generations. Bayesian posterior probability values are displayed on the tree as percentages, and if differing values were seen between the two independently run trees, both are displayed. This phylogeny was used to order B-box genes to provide a uniform naming terminology. For full-length protein sequences used and the multiple alignments, see Supplemental Data Set 1 (FASTA format) and Supplemental Figure 1 (alignment) online.
The B-box protein family, now termed BBX, follows a numbering convention starting with the original identified family member CO (Putterill et al., 1995; Robson et al., 2001), continuing by clades in a clockwise rotation on Figure 2 around the phylogenetic tree. CO becomes BBX1, and COL1 to 16 become BBX2 to 17 (Figure 2). Subsequently named proteins, STH, STO, LZF1, and DBB (Holm et al., 2001; Datta et al., 2007, 2008; Chang et al., 2008; Kumagai et al., 2008), are named BBX18 to 32 (Figure 2).
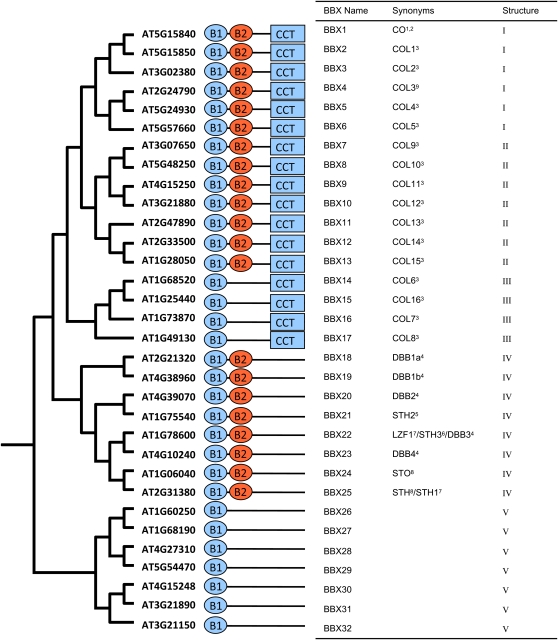
BBX FAMILY STRUCTURE CLASSIFICATION
The Bayesian inference phylogeny from Figure 2 was rooted between the clades of BBX1 to 25 and BBX26 to 32 based on midpoint rooting on the Bayesian majority rule phylogram to give the table arrangement seen in Figure 3. Schematic illustrations for each identified motif present in proteins are shown to the right of the phylogenetic tree (Figure 3). Corresponding to the schematics is the column “Structure Groups” (Figure 3), where like motif configurations found in phylogenetic clades are designated from I to V. Each member of structure group I contains a B-box B1, a B-box B2, and a CCT domain. Structure group II members are similar to group I, both containing B1, B2, and CCT domains; however, differences in their B2 domains were observed (Chang et al., 2008). Structure group III members contain a B1 and CCT. Structure group IV contains B-box B1 and B2, and no CCT domains are found. Finally, structure group V is made up of members with just a single B1 domain.
Figure 3.
Structure Classification of the B-Box Family.
A graphical representation of a phylogeny is shown at left with Arabidopsis Genome Initiative gene names. Schematics of identified protein structures are shown; B-box 1 is labeled as B1, B-box 2 is labeled as B2, and the CCT domain is indicated. Following the structure of each protein is the new BBX naming scheme, synonym/s for proteins, and structure groups. References: 1Koornneef et al. (1991), 2Putterill et al. (1995), 3Robson et al. (2001), 4Kumagai et al. (2008), 5Datta et al. (2007), 6Datta et al. (2008), 7Chang et al. (2008), 8Holm et al. (2001), and 9Datta et al. (2006).
The name CONSTANS is widely used, and we expect that it will continue to be used in the future instead of the BBX1 designation for this locus. Nonetheless, we propose that the scientific community studying the Arabidopsis B-box proteins adopts the designation of the entire B-box family with the suggested uniform nomenclature. Reference to B-box genes (BBX) and proteins (BBX) will follow conventional rules. We suggest that such a designation can be used to describe mutant alleles accordingly, for example, bbx21-1 instead of sth2-1 (Datta et al., 2007) and bbx18-1 instead of dbb1a-1 (Kumagai et al., 2008). Mutants identified first would be designated with one digit (e.g., bbx#-1), subsequent mutants from different labs would be named following a general rule in chronological order, such as bbx#-101 and -102 and bbx#-201 and -202, and so on. Similar approaches have been used previously to unify the nomenclatures for phytochromes (Quail et al., 1994) and phototropins (Briggs et al., 2001) and have been adopted by the scientific community.
B-box proteins from other species would be named following a similar nomenclature scheme. For example, one or more rice proteins clustering with At-BBX1 can be named Os-BBX1a, Os-BBX1b, and so forth. In cases of species-specific B-box proteins, we propose that the naming scheme is continued from BBX33 and onwards (e.g., Os-BBX33 could be a rice-specific B-box protein). Any future orthologs of the hypothetical example Os-BBX33 will follow the rules described above. In cases where two At-BBX proteins can equally be considered as orthologs of one or more proteins from other species, homology-based comparisons can be used to assign the name. For example, two Arabidopsis paralogs may share a single rice ortholog. In such a case, the rice protein will be assigned the name of the Arabidopsis protein with highest homology. This scheme can be expanded for multiple sets of ortholog-paralog combinations. With the continuing availability of new genome sequences, these proposed classification schemes can be followed by the scientific community to provide nomenclature unification while naming proteins of other protein families and other species.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Alignment of 32 B-Box–Containing Full-Length Arabidopsis Proteins.
Supplemental Data Set 1. FASTA Format Multiple Alignment of 32 Arabidopsis B-Box Proteins Aligned with MUSCLE (Edgar, 2004).
Supplementary Material
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, P.C., Martin, C., Toledo-Ortiz, G., Quail, P.H., Huq, E., Heim, M.A., Jakoby, M., Werber, M., and Weisshaar, B. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K.L., Lally, J.M., Martin, S.R., O'Reilly, N.J., Etkin, L.D., and Freemont, P.S. (1995). Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J. 14 5947–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.S., Li, Y.H., Chen, L.-T., Chen, W.-C., Hsieh, W.-P., Shin, J., Jane, W.-N., Chou, S.-J., Choi, G., Hu, J.-M., Somerville, S., and Wu, S.-H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54 205–219. [DOI] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S., Hettiarachchi, C., Johansson, H., and Holm, M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19 3242–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S., Hettiarachchi, G.H.C.M., Deng, X.W., and Holm, M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M.L., Desai, M., Rubio, V., and Holm, M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20 2324–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater, S., Greco, V., Pham, K., Memelink, J., and Kijne, J. (1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, Y., Utsumi, M., Ohba, Y., and Watanabe, Y. (2007). Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 48 1243–1253. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Gouy, M., and Gautier, C. (1996). SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12 543–548. [DOI] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 735–747. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Henikoff, J.G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Hardtke, C.S., Gaudet, R., and Deng, X.W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Klug, A., and Schwabe, J.W.R. (1995). Zinc fingers. FASEB J. 9 597–604. [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Kumagai, T., Ito, S., Nakamichi, N., Niwa, Y., Murakami, M., Yamashino, T., and Mizuno, T. (2008). The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72 1539–1549. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Ledger, S., Strayer, C., Ashton, F., Kay, S.A., and Putterill, J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26 15–22. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Briggs, W.R., Chory, J., Hangarter, R.P., Harberd, N.P., Kendrick, R.E., Koornneef, M., Parks, B., Sharrock, R.A., Schafer, E., Thompson, W.F., and Whitelam, G.C. (1994). Spotlight on phytochrome nomenclature. Plant Cell 6 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. [DOI] [PubMed] [Google Scholar]
- Robson, F., Costa, M.M., Hepworth, S.R., Vizir, I., Pineiro, M., Reeves, P.H., Putterill, J., and Coupland, G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28 619–631. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J., Macdonald, H., Huttly, A.K., Lazarus, C.M., and Hooley, R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 29 691–702. [DOI] [PubMed] [Google Scholar]
- Schneider, T.D., and Stephens, R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Swarbreck, D., al. (2008). The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 36 D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok, M., and Etkin, L.D. (2001). Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 67 63–71. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S. (1995). A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 23 3403–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa, S., and Schmidt, R.J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17 209–214. [DOI] [PubMed] [Google Scholar]
- Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47 841–850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.