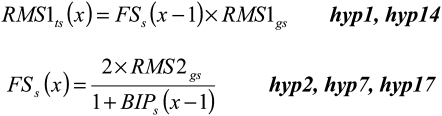Abstract
Bud outgrowth is regulated by the interplay of multiple hormones, including auxin, cytokinin, strigolactones, and an unidentified long-distance feedback signal that moves from shoot to root. The model of bud outgrowth regulation in pea (Pisum sativum) includes these signals and a network of five RAMOSUS (RMS) genes that operate in a shoot-root-shoot loop to regulate the synthesis of, and response to, strigolactones. The number of components in this network renders the integration of new and existing hypotheses both complex and cumbersome. A hypothesis-driven computational model was therefore developed to help understand regulation of shoot branching. The model evolved in parallel with stepwise laboratory research, helping to define and test key hypotheses. The computational model was used to verify new mechanisms involved in the regulation of shoot branching by confirming that the new hypotheses captured all relevant biological data sets. Based on cytokinin and RMS1 expression analyses, this model is extended to include subtle but important differences in the function of RMS3 and RMS4 genes in the shoot and rootstock. Additionally, this research indicates that a branch-derived signal upregulates RMS1 expression independent of the other feedback signal. Furthermore, we propose xylem-sap cytokinin promotes sustained bud outgrowth, rather than acting at the earlier stage of bud release.
INTRODUCTION
Axillary buds are found in the axils of leaves and can either remain in a state of suspended growth or grow out to form branches. The regulation of their outgrowth is a complex process that involves crosstalk among multiple hormones and signals moving within and between the root and shoot (Beveridge, 2000, 2006; McSteen, 2009). It has long been established that the plant hormones auxin and cytokinin are involved in this regulation; strigolactones were recently added to this list (reviewed in Dun et al., 2009; Leyser, 2009; McSteen, 2009).
Genetic studies have identified a number of genes involved in shoot branching in a variety of plant species; many are orthologous. These include several RAMOSUS (RMS) genes in garden pea (Pisum sativum), DECREASED APICAL DOMINANCE (DAD) genes in petunia (Petunia hybrida), MORE AXILLARY GROWTH (MAX) genes in Arabidopsis thaliana, and DWARF (D) and HIGH-TILLERING DWARF (HTD) genes in rice (Oryza sativa) (Beveridge, 2006; Leyser, 2009; McSteen, 2009). Mutants for all of these genes are recessive and exhibit an increased branching phenotype.
Grafting studies demonstrated a function for shoot and rootstock tissue in bud outgrowth and suggested that a subset of genes (RMS1/MAX4/DAD1, RMS5/MAX3, MAX1, and DAD3) were involved in the synthesis of an upward moving signal (Beveridge, 2006) now identified as a strigolactone (Figure 1; Gomez-Roldan et al., 2008; Umehara et al., 2008). Even though it remains possible that the branch-inhibitory hormone is actually a downstream product of strigolactones, here, we will refer to it simply as a strigolactone. RMS5/MAX3/HTD1/D17 and RMS1/MAX4/DAD1/D10 encode CAROTENOID CLEAVAGE DIOXYGENASE7 (CCD7) and CCD8, respectively (Sorefan et al., 2003; Booker et al., 2004; Snowden et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007). This putative carotenoid cleavage function was the key to the discovery that the branch-inhibitory hormone was a strigolactone (Gomez-Roldan et al., 2008; Umehara et al., 2008) because strigolactone biosynthesis was suggested to require CCD enzymes (Matusova et al., 2005). In Arabidopsis, MAX1 encodes a cytochrome P450 family member that is thought to act downstream of CCD7 and CCD8 (Booker et al., 2005) in strigolactone biosynthesis (Gomez-Roldan et al., 2008; Umehara et al., 2008).
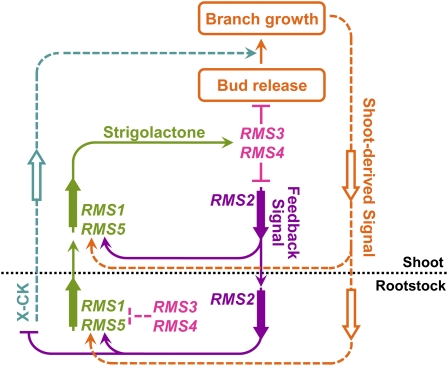
Figure 1.
Model for Bud Outgrowth Regulation in Pea.
RMS1 and RMS5 are involved in the biosynthesis of the upwards-moving strigolactone branching inhibitor produced in the rootstock and shoot. The response to strigolactones requires RMS3 and RMS4 in the shoot to inhibit bud outgrowth and downregulate the feedback signal. The feedback signal, which is partly dependent on RMS2, is made in the shoot and rootstock, moves downwards from shoot to rootstock, enhances expression of RMS1 and RMS5, and reduces X-CK export from the rootstock. The remaining interactions are based on this study. RMS3 and RMS4 in the rootstock, acting independently from long-distance signals, reduce RMS1 and RMS5 expression. When a bud is released to grow, the shoot-derived signal moves downwards and enhances RMS1 and RMS5 expression. X-CK moves upwards from the rootstock and promotes growth of released buds. Flat-ended lines represent repression, and arrowheads represent promotion. Large arrows represent directional flow of signals in the plant. Solid lines represent interactions included in both computational models, based on Beveridge (2006). New interactions based on our findings are represented by dashed lines and are included in the final computational model.
[See online article for color version of this figure.]
In addition to genes involved in strigolactone biosynthesis, grafting experiments indicated other genes are likely to be involved in hormone response (RMS4/MAX2, RMS3, and DAD2; Figure 1 Beveridge et al., 1996; Napoli et al., 1999; Booker et al., 2005; Simons et al., 2007). Consistent with this, the orthologous genes MAX2, RMS4, and D3 were found to encode an F-box protein (Stirnberg et al., 2002; Ishikawa et al., 2005; Johnson et al., 2006), of which some family members function like receptors (e.g., Kepinski and Leyser, 2005). The requirement of RMS4/MAX2/D3 for strigolactone response is also supported by the finding that strigolactone treatments do not reduce branching of the corresponding mutant plants (Gomez-Roldan et al., 2008; Umehara et al., 2008). RMS3 and DAD2 sequences are not yet known. However, the recently identified rice branching gene, D14 (Arite et al., 2009), is a good candidate for RMS3 and/or DAD2 as d14 is insensitive to exogenous strigolactone treatments.
Pea grafting studies led Beveridge (2000) to propose a signal dependent on RMS2 (sequence unknown) that moves in the direction of shoot to root (Figure 1). This was named the feedback signal, and it was later demonstrated to be a positive regulator of RMS1 and RMS5 expression (Foo et al., 2005; Johnson et al., 2006) and a negative regulator for export of xylem-sap cytokinin (X-CK) from roots (Beveridge et al., 1997a; Beveridge, 2000; Morris et al., 2001). While auxin shares similar properties with the feedback signal (Ongaro and Leyser, 2008), it is likely that the feedback signal is distinct from auxin and that both are involved in maintaining homeostasis in the shoot branching regulatory network (discussed in Beveridge, 2000, 2006; Dun et al., 2009; Figure 1). The feedback signal was suggested to be negatively regulated by the perception of strigolactones (Figure 1; Foo et al., 2005) because various mutants unable to produce or to respond to strigolactones have reduced export of X-CK from the roots (Beveridge et al., 1997a; Morris et al., 2001; Foo et al., 2007) and increased expression of strigolactone biosynthesis genes (Foo et al., 2005; Johnson et al., 2006; Arite et al., 2007; Hayward et al., 2009). Such changes in the level of expression of strigolactone biosynthesis genes are likely to affect shoot branching (Johnson et al., 2006; Hayward et al., 2009).
Grafting studies with rms4 and the wild type indicate that X-CK export from roots is essentially shoot regulated (Beveridge et al., 1997b), whereas RMS1 expression is regulated by shoot and root (Foo et al., 2005; Johnson et al., 2006). In general, X-CK and RMS1 expression levels are anticorrelated; however, in double mutants with rms2, such as rms2 rms4, both are increased relative to the wild type (Foo et al., 2005, 2007). Without a method to evaluate different hypotheses, it is not entirely clear how to interpret this result.
In this study, we tested the interactions within the pea strigolactone regulatory network, rather than the molecular mechanism by which strigolactones inhibit bud outgrowth. The recently reported network (e.g., Dun et al., 2009) for the regulation of bud outgrowth in pea is represented in Figure 1. We seek here to evaluate whether this convenient description is in fact adequate to explain essentially all of the data available for this aspect of shoot branching. Computational modeling methods can provide an unbiased assessment of whether a hypothesized network of interactions explains all biological data sets and can help to predict new biological results and generate new hypotheses (discussed in Forger et al., 2005). Since we cannot be certain our line diagram (Figure 1, solid lines) represents a network that captures all experimental data, we decided to create computational models to evaluate and test its accuracy. An additional aim was to use the computational models to help guide our experiments to test key features of the regulatory network. Consequently, it was important to develop a model that explicitly represented the biological hypotheses. This approach allowed us to ensure that our biological experiments were always testing a set of biologically relevant hypotheses. Our continual aim was to evaluate the models against all available and relevant biological data sets. In other words, once our hypotheses computationally conformed to all past results, we then tested them with new biological experimentation. In this process, experiments were sometimes needed to help us articulate completely new hypotheses that might enable the model to capture the data.
In this article, we present an improved model for the regulation of axillary bud outgrowth in pea. The model accounts for all new and published data sets, which includes over 100 data points for traits affected by long-distance signaling. These include branching phenotype, RMS1 gene expression, and levels of X-CK exported from the root of plants of different graft combinations. Importantly, the modeling method supported an iterative process of hypothesis development, evaluation, and experimentation that led to a new understanding of the network regulating bud outgrowth in pea.
RESULTS
Initial Computational Models
To ascertain whether the published network (Figure 1, solid lines) accounted for the branching phenotype, RMS1 expression, and X-CK export from the rootstock in different grafting situations, we created a computational model and evaluated it against all published experimental data sets. First, detailed hypotheses were determined based on the literature (Table 1; hypotheses reviewed in Beveridge, 2006 and Dun et al., 2009). This study was focused on shoot branching and long-distance signaling in grafted wild-type and rms single and double mutant plants with intact shoot tips. The function for apical auxin in the current bud outgrowth regulatory network is not different between rms mutant and wild-type plants (Beveridge, 2006) and is only apparent in auxin-depleted (e.g., decapitated) plants. Due to our focus on branching in intact plants, we have not specifically incorporated a role for auxin in this study. We also have not included detail on the molecular mechanism of strigolactone function, which might involve local or short-distance auxin transport and interactions with shoot-derived cytokinin (Leyser, 2009; Shimizu-Sato et al., 2009).
Table 1.
Model Hypotheses
| Hyp. No. | Model |
|||
|---|---|---|---|---|
| First | Final | Hypothesis | Reference | |
| 1 | X | X | RMS1 acts in the rootstock and shoot | ab |
| 2 | X | X | RMS2 acts in the rootstock and shoot | a |
| 3A | X | RMS3 acts in the shoot only | bc | |
| 3B | X | RMS3 acts in the rootstock and shoot | Figure 2, Supplemental Figures 2 and 4 | |
| 4A | X | RMS4 acts in the shoot only | abc | |
| 4B | X | RMS4 acts in the rootstock and shoot | de | |
| 5 | X | X | RMS5 acts in the rootstock and shoot | b |
| 6 | X | X | RMS1 regulates, or is required for, a signal that moves (a strigolactone or product) | abf |
| 7 | X | X | RMS2 controls or is required for the transport or synthesis of a signal that moves, named the feedback signal | adeg |
| 8 | X | X | RMS3 is involved in signal perception and/or transduction and does not move | bcgh |
| 9 | X | X | RMS4 is involved in signal perception and/or transduction and does not move | bcgh |
| 10 | X | X | RMS5 regulates, or is required for, a signal that moves (a strigolactone or product) | bf |
| 11 | X | X | RMS1 and RMS5 together are required for strigolactones to be produced | bf |
| 12 | X | X | Strigolactones move upward only | fi |
| 13 | X | X | The feedback signal moves downward only | gijk |
| 14 | X | X | The amount of feedback signal enhances RMS1 and RMS5 expression in the rootstock and shoot | de |
| 15 | X | X | RMS3 and RMS4 are required together for perception and/or transduction of strigolactones | bcfgh, Figure 2, Supplemental Figures 2 and 4 |
| 16 | X | X | The amount of strigolactone signal transduction determines amount of inhibition of bud release | def |
| 17 | X | X | The feedback signal in the shoot and rootstock is produced at a constant rate that is reduced only in the shoot by the amount of RMS3 and RMS4 perception and/or transduction of strigolactones | dk |
| 18 | X | X | When two shoots are present, each receives the same concentration of strigolactones as transported from the rootstock | This study |
| 19 | X | X | When two rootstocks are present, each receives the same concentration of the feedback signal as transported from the shoot | This study |
| 20 | X | X | For signals that move down, the signal in the rootstock is equal to the source plus the contribution from each shoot | This study |
| 21 | X | X | For signals that move up, the signal in the shoot is equal to the source plus the contribution from each rootstock | This study |
| 22 | X | X | When two shoots are present, the concentration of the down signal is important, so the concentration making it to the rootstock is the average of the concentration from each shoot | This study |
| 23 | X | X | When two rootstocks are present, the concentration of the up signal is important, so the concentration making it to the shoot is the average of the concentration from each rootstock | This study |
| 24 | X | X | Signals moving between rootstock(s) and shoot(s) take time to move and hence experience a delay from the time they are produced in their source to when they arrive in their destination | This study |
| 25 | X | X | Shoot- and rootstock-derived feedback signal reduces xylem sap cytokinin synthesis or export from the rootstock | bgjk |
| 26 | X | X | The likely pathway for signals moving upwards (strigolactones) and downwards (feedback signal) is the xylem and phloem, respectively, so signals moving downwards would experience a greater proportion of loss or metabolism than those moving upwards | This study |
| 27 | X | rms2 is a leaky mutation; when mutated, some functional gene product is produced | d | |
| 28 | X | RMS3 and RMS4 act constitutively in the rootstock to reduce RMS1 and RMS5 expression in the rootstock only | Supplemental Figures 2 and 4 | |
| 29 | X | Growing branches (those that are not inhibited) produce a signal, named the branch-derived feedback signal | Figures 3 to 5 | |
| 30 | X | The branch-derived feedback signal moves downwards only | Figures 3 to 5 | |
| 31 | X | The branch-derived feedback signal enhances RMS1 and RMS5 expression in tissues to which it is transported | Figures 3 to 5 | |
| 32 |
X |
The amount of X-CK, relative to that in the wild type, determines the length of the branch; higher levels cause longer branches; lower levels cause shorter branches |
Figures 6 to 8 |
|
Hyp. No. refers to the hypothesis number as referred to in the text. References:
In order to have a direct and obvious relationship between the proposed network (Figure 1) and the computational model, a minimum set of hypotheses was determined from the network (Table 1) and represented as algebraic equations (Figure 2; see Methods and Supplemental Methods online). For example, provided the RMS1 genotype is wild-type, the amount of RMS1 transcript is dependent on the amount of the feedback signal (Table 1, hyp14); accordingly, the two model components, RMS1 genotype and the amount of feedback signal, were multiplied to give the amount of RMS1 transcript (Figure 2). The values of 0 or 1 were given for the genotype (mutant or wild type), with the outcome being that no product is produced in mutant plants. In a more complex example, we hypothesized that the level of the feedback signal increases where the amount of strigolactone decreases (Table 1, hyp17). As above, here and throughout the model we have used an algebraic equation to reflect the expected relationship and trends (Figure 2). In this way, the computational model showed trends and relationships but was not intended to predict actual values. Using such a modeling approach, we avoided the use of a multitude of parameters. This is because the relationships among the model outputs were compared against the relationships within the biological data, rather than comparing exact values (see Methods and Supplemental Figures 1 and 2 online). The final model has only three parameters described as hypotheses 26 and 27 (Table 1). These parameters can cover a very wide range of values without affecting the modeling outcome (see Supplemental Table 1 online). Inputs by the user are the genotypes and the graft combination, which changes the number of shoot and/or root compartments (see Supplemental Methods online).
Figure 2.
Calculation of RMS1 Transcript Level in the Shoot and the Level of Feedback Signal in Shoot as Example Equations.
The equations were developed using the principles outlined in the Methods and Supplemental Methods online and are based on the hypotheses listed in Table 1. RMS1gs and RMS2gs are the shoot RMS1 and RMS2 genotype, respectively; BIPs is the branching inhibitor (strigolactone) as perceived in the shoot by RMS3 and RMS4; RMS1ts is the level of RMS1 transcript in the shoot; FSs is the level of feedback signal in the shoot; x is the iteration number.
The output from the model was the branching phenotype (measured as level of perceived branching inhibition signal), RMS1 expression, and rootstock X-CK for intact plants and for each of the various possible graft combinations among the genotypes, including I-grafts (one shoot and one rootstock) and two-shoot and two-rootstock grafts. This output was compared against available experimental data. The data from grafting studies included 73 branching phenotypes, 20 rootstock X-CK values, and RMS1 gene expression in rootstock and shoot for 14 graft combinations (see Supplemental Tables 2 to 4 and Supplemental Figures 1 and 2 online).
Our initial computational model (based on the published network; solid lines in Figure 1; Table 1, hyp1-26) captured all previously reported branching phenotypes (listed in Supplemental Tables 2 to 4 online) and most trends in rootstock X-CK content (see Supplemental Figure 1 online). However, as the model did not include a separate function for RMS4 in the rootstock (Table 1, hyp4A), the changes in RMS1 expression in the rootstock of grafts between rms4 and the wild type (Foo et al., 2005) were not captured by the model (see Supplemental Figure 2F online). We therefore implemented a new hypothesis in the model: that RMS4 in the rootstock regulates RMS1 expression independent of the RMS2-mediated feedback signal (Table 1, hyp28), thereby repressing RMS1 expression in the rootstock without enhancing rootstock X-CK export. This also required the modification of hypotheses 3 and 4 (Table 1, hyp3B and 4B) to incorporate RMS3 and RMS4 action in rootstocks. However, because the model implemented the hypothesis that RMS2 is required for the feedback signal (Table 1, hyp7), the normal levels of X-CK export from wild-type rootstocks grafted to rms2 rms4 double mutant scions (Foo et al., 2007) and the higher than wild-type RMS1 expression in rms2 rms4 and rms2 rms5 plants (Foo et al., 2005) were not captured by this model (see Supplemental Figures 1 and 2A online). As such, the hypothesis that rms2 mutant plants produce some functional product (Table 1, hyp27; Foo et al., 2005) was also implemented. This enhanced intermediate model (incorporating hyp1-28 [with hyp3B and 4B] in Table 1) captured the published branching and X-CK export data as well as maintaining the shoot phenotype relationships that were represented in the original model (model not shown). However, in this intermediate model, the higher than wild-type RMS1 expression in rms2 rms5 double mutant plants (Foo et al., 2005) was still not captured (data not shown). We then set out to further evaluate and test the model hypotheses (Table 1), particularly those pertaining to RMS2 and the feedback signal, using a range of experimental approaches commencing with strigolactone response experiments.
Bud Outgrowth Response of rms Mutants to Strigolactone Treatment Fits Model
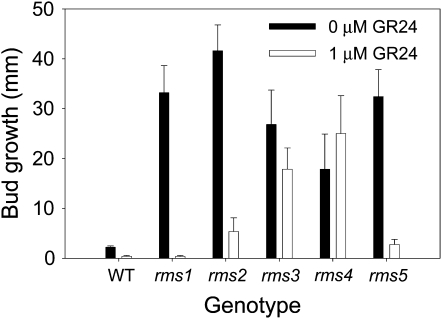
To check that the model placed RMS2 and RMS3 correctly on the strigolactone network (Figure 1), GR24 (synthetic strigolactone; Akiyama et al., 2005) was applied directly to buds and their growth measured. As found previously, GR24 inhibited the growth of rms1 and rms5 buds and had no effect on the growth of rms4 buds (Figure 3; Gomez-Roldan et al., 2008; Brewer et al., 2009). As expected from the model (Figure 1), GR24 inhibited the growth of rms2 buds (P < 0.001) but did not restore outgrowth inhibition to rms3 buds (Figure 3). The growth of wild-type buds was also reduced significantly (P < 0.001). These results indicate that the network correctly places RMS3 as being likely required for strigolactone response (Table 1, hyp8 and 15) and RMS2 as being involved in regulating strigolactone biosynthesis (Table 1, hyp7 and 14).
Figure 3.
Strigolactone Application Inhibits Growth of Torsdag Wild-Type, rms1, rms2, and rms5 Buds but Has No Effect on the Growth of rms3 or rms4 Buds.
Buds at the uppermost expanded node (12-15) of 33-d-old plants were treated with 0 or 1 μM GR24 and their growth measured after 7 d. Data are means ± se (n = 11 to 15).
Feedback: New Biological Data Supports Early Models
To understand feedback regulation further, we tested whether strigolactone levels, rather than simply the ability to respond to strigolactones (rms4/RMS4 genotype), may be important as predicted by the model implementation (Table 1, hyp17). For example, the model predicted that wild-type shoots should have enhanced strigolactone biosynthesis gene expression when grafted to strigolactone deficient rootstocks (see Supplemental Figure 2C online). RMS1 expression was monitored via real-time PCR in the scion and rootstock epicotyls of grafts between rms5 or rms4 and the wild type. An increase in RMS1 expression was observed in wild-type scions when grafted to rms5 rootstocks relative to wild-type self-grafted plants, even though both graft combinations do not branch (see Supplemental Figure 3 online). This is consistent with feedback upregulation of RMS1 expression in the shoot due to a lack of strigolactone supply from the rootstock. Conversely, relative to wild-type self-grafted plants, a decrease in RMS1 expression was observed in a wild-type scion grafted to an rms4 rootstock (see Supplemental Figure 2E online). This is consistent with functional feedback downregulation in the shoot and is possibly due to an increased strigolactone supply from the rms4 roots (Johnson et al., 2006). These experiments support the model hypothesis that the feedback signal is downregulated by the amount of RMS4-mediated strigolactone response in the shoot (Figure 1, Table 1, hyp17).
Our intermediate model (model not shown; Table 1, hyp1-28) predicted that rms3 and rms4 rootstocks should inhibit branching in wild-type scions more effectively than wild-type rootstocks, even though intact and self-grafted rms3 and rms4 plants are highly branched. Using plants grown in short days to enhance branching in wild-type shoots, this was confirmed for both rms3 and rms4 rootstocks and could be attributed to high RMS1 expression in mutant rootstocks (Johnson et al., 2006; see Supplemental Figures 2E, 2F, and 4 online). These data support the hypotheses in the model that RMS3 and RMS4 both act in strigolactone response and feedback regulation (Table 1, hyp3B, 4B, 8, 9, 15, 17, and 28).
Developmental Regulation of RMS1 and RMS5 Expression: Unpredicted Data
A major part of the model is that rms2 mutants branch due to perturbed regulation of strigolactone biosynthesis genes (Table 1, hyp7, 14, 11, and 16). This is supported by gene expression studies (mostly in young tissues; Foo et al., 2005; Johnson et al., 2006) and the response of rms2 plants to exogenously supplied strigolactone (Figure 3). We therefore sampled a broader range
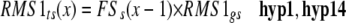 |
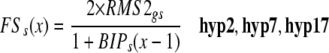 |
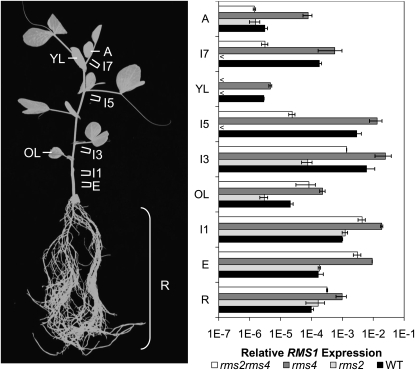
of tissues to determine whether rms2 mutation always results in low RMS1 expression (Table 1, hyp7 and 14). As strigolactones likely move acropetally in shoots (Table 1, hyp12; Foo et al., 2001; Gomez-Roldan et al., 2008), low RMS1 expression would be expected in rms2 mutant plants throughout the stem axis and root. Real-time PCR was performed on cDNA derived from 17-d-old (slightly older than in previous experiments) wild-type, rms2, rms4, and rms2 rms4 plants with six to seven expanded leaves. RMS1 expression differences between wild-type and rms2 mutants were not maintained in all tissues (Figure 4). In these relatively tall plants, we observed relatively low RMS1 expression in upper internodes of the rms2 mutant compared with the wild type, as reported previously (Foo et al., 2005). This difference became less apparent at lower positions with, unexpectedly, no difference observed between rms2 and the wild type at internode 1 or below (Figure 4). By contrast, as expected, RMS1 expression in the rms4 mutant was consistently higher in all tissues examined relative to the wild type (Figure 4).
Figure 4.
RMS1 Expression Relative to the Internal Reference Gene 18S in Different Tissues of 17-d-Old Wild-Type, rms2, rms4, and rms2 rms4 Plants with Six to Seven Leaves Expanded.
Tissues harvested are as shown (left) and are whole roots (R); the uppermost 12 mm of epicotyl (E), internode 1 (I1), internode 3 (I3), internode 5 (I5), and internode 6 or 7 (I7); oldest true leaf (at node 3; OL); youngest fully expanded leaf (at node 6 or 7; YL); and the apex (inside the unexpanded leaves at node 9 or 10; A). Data are averages ± se of two biological replicates of 12 plants, except for leaves (eight plants) and roots (four plants). The < indicates data points where the transcript abundance was below the limit of detection.
Another effect of tissue type on genotypic differences was observed for comparisons between rms2 rms4 double mutant plants and the wild type. RMS1 expression in rms2 rms4 double mutant plants was increased compared with the wild type in lower tissues, as in previous studies (Foo et al., 2005), but this difference was reversed in the young aerial tissues of these 17-d-old plants (Figure 4).
These results were not captured in the models developed to this point (see Supplemental Figures 2A and 2B online; intermediate model not shown) and were not easily explained based on these data. In particular, the near-wild-type RMS1 expression in lower internodes and roots of rms2 plants is not consistent with previous observations that RMS1 expression is low in rms2 plants and our suggestion that strigolactone deficiency is the cause of shoot branching in these plants. To evaluate this potential evidence against low RMS1 expression being the cause of branching in rms2 plants, another experiment was conducted to examine the regulation of RMS1 expression in rms2 mutants throughout development.
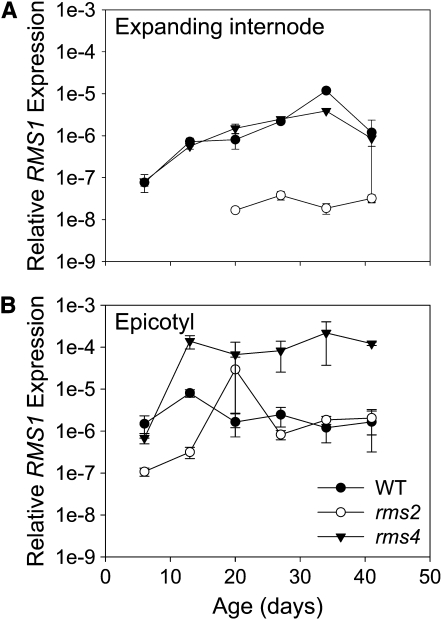
RMS1 expression was monitored in the uppermost expanding internode and epicotyl of wild-type, rms2, and rms4 plants of different ages (Figure 5). RMS1 expression in the uppermost expanding internode was always lower in rms2 plants compared with the wild type (Figure 5A) regardless of plant age. By contrast, RMS1 expression in epicotyls differs between rms2 and the wild type only at early time points (Figure 5B). The time from which RMS1 expression was not reduced in rms2 epicotyls corresponds to the time when a significant branch (10.58 ± 1.02 cm at day 19) was growing at node 2 (above the epicotyl). Obviously, these high RMS1 expression results were not captured by the intermediate computational model (data not shown; Table 1, hyp1-28).
Figure 5.
RMS1 Expression Relative to 18S in the Expanding Internode and the Epicotyl of Wild-Type, rms2, and rms4 Plants of Different Ages Harvested on the Same Day.
RMS1 transcript abundance was below the level of detection in the expanding internode (A) of rms2-1 plants for 6-d and 13-d data points. Data for the epicotyl are shown in (B). Data are averages ± se of two biological replicates of five to seven plants.
The occurrence of a branch coinciding with increased RMS1 expression in rms2 plants led us to hypothesize that growing branches produce a signal that moves only downwards into the main stem and enhances RMS1 expression (Table 1, hyp29-31). In support of these hypotheses and a developmentally dependent strigolactone deficiency in rms2 plants, rms2 buds can respond to exogenous strigolactone (Figure 3). The reduction in RMS1 expression early in development, consistent with reduced strigolactone levels at that stage, may explain why intact rms2 plants mainly branch at the basal nodes that are established early in development (Beveridge et al., 1994; Ferguson and Beveridge, 2009). As the plant ages and begins to branch, RMS1 expression in the epicotyl of rms2 mutants increases to wild-type levels (Figure 5B). This presumably enhances strigolactone biosynthesis and transport up the shoot (Table 1, hyp11 and 12), thereby suppressing further aerial branching. Indeed, rms2 plants do not exhibit much aerial branching compared with rms3 or rms4 plants unless the basal branches are removed (Beveridge et al., 1996; Ferguson and Beveridge, 2009).
Hypotheses related to branches causing upregulation of RMS1 expression at positions below the branch (Figure 1, dashed lines; Table 1, hyp29-31) were implemented in a new model. This new model fully reflected the new and old data (see Supplemental Figures 1, 2, and 5 online). We were therefore in a position to test these hypotheses experimentally. In particular, the model now predicted that RMS1 would be expressed at wild-type levels in rms2 shoot tissue below the site of branches and would have low expression levels in rms2 shoots where branching was inhibited by grafting to wild-type rootstocks (see Supplemental Figures 5D and 5E online).
Verification of New Long-Distance Regulation Hypotheses (hyp29-31)
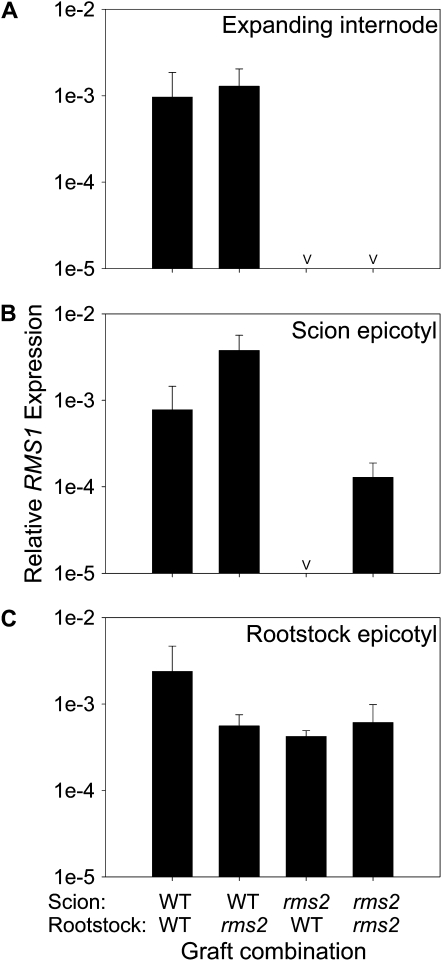
We conducted an independent experiment to evaluate and test the new model incorporating the hypotheses on branches regulating RMS1 expression (Table 1, hyp29-31). We used rms2 and wild-type grafting, which causes branching inhibition in rms2 shoots (Beveridge et al., 1994), and quantified RMS1 expression in scion epicotyls to observe whether these nonbranched rms2 shoots would now have low RMS1 expression, like nonbranched young rms2 seedlings. RMS1 expression was severely depleted in the young expanding internodes of rms2 scions whether they were grafted to the wild type (nonbranched) or not (branched) (Figure 6A). As predicted, RMS1 expression was also severely depleted in epicotyls of rms2 scions grafted to the wild type and was closer to wild-type levels in rms2 self-grafts (Figure 6B). These results support the new hypothesis that branches in an rms2 scion cause an upregulation of RMS1 expression in the stem (Table 1, hyp29-31).
Figure 6.
RMS1 Expression Relative to 18S in the Expanding Internode, Scion Epicotyl, and the Rootstock Epicotyl of Wild-Type and rms2 Grafts 17 d after Grafting.
Data are means ± se of three biological replicates of 8 to 11 plants. The “v” indicates data points where the transcript abundance was below the limit of detection. As found previously (Beveridge et al., 1994), rms2 self-grafts were the only graft combination that branched; all other combinations had near-wild-type branching phenotypes. Expanding internode (A), scion epicotyl (B), and the rootstock epicotyl (C).
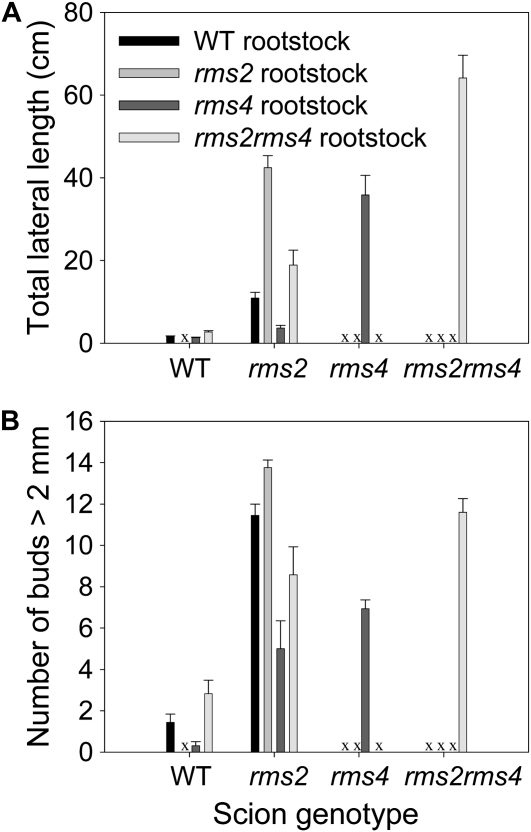
Next, we sought to test if bud outgrowth could be inhibited by elevating RMS1 expression in basal tissues of rms2 plants. Double mutant rms2 rms4 epicotyls have high RMS1 expression (Foo et al., 2005; Figure 4), and the model predicted that this would be maintained when grafted to rms2 scions. As predicted, rms2 rms4 rootstocks substantially reduced total lateral bud outgrowth (reported as total lateral length, which is the sum of the lengths of all lateral buds and branches on the main stem; P < 0.001; Figure 7A) and the number of buds >2 mm in length (P < 0.001; Figure 7B) in rms2 scions compared with rms2 scions grafted to rms2 rootstocks. As expected, rms2 rms4 double mutant self-grafts branched more than rms2 self-grafts (P < 0.001), and rms4 rootstocks were more effective inhibitors of branching in rms2 scions than wild-type rootstocks (Figure 7; Beveridge et al., 1996). These results indicate that branching can be inhibited in an rms2 plant by increasing RMS1 expression in the rootstock; hence, branches may inhibit other branches through this mechanism (Table 1, hyp29-31). This further supports the hypothesis that the reduced RMS1 expression in young tissues of rms2 plants contributes to their increased basal branching phenotype.
Figure 7.
Branching Phenotype of Grafts among rms2, rms4, rms2 rms4, and Wild-Type Seedlings Measured 34 d after Grafting.
Data are means ± se (n = 11 to 17). The “x” indicates grafts not done. Total lateral length (A); number of buds or branches >2 mm in length (B).
High X-CK Might Promote Sustained Bud Growth
The differences observed between the ability of wild-type, rms4, and rms2 rms4 rootstocks to inhibit bud release and subsequent branch growth (Figure 7) revealed an additional intriguing finding. Mutant rms2 rms4 rootstocks displayed an increased ability to inhibit the initial release of buds in rms2 scions compared with wild-type rootstocks (Figure 7B), whereas wild-type rootstocks were better than rms2 rms4 rootstocks at reducing their subsequent growth (Figure 7A). These results suggest that X-CK content, which is relatively high in rms2 rms4 rootstocks compared with the wild type (Foo et al., 2007), might promote sustained branch growth (Table 1, hyp32). By contrast, strigolactones may predominantly inhibit the initial release of a bud (Table 1, hyp16). Consistent with this, despite strigolactone insensitivity (Figure 3; Gomez-Roldan et al., 2008), not all rms4 buds grow out into branches, possibly due to the low X-CK (Beveridge et al., 1997b) being limiting. However, strigolactone-insensitive rms2 rms4 double mutant plants (see text below) form more and longer branches (Figure 7; Murfet and Symons, 2000; Foo et al., 2007), possibly due to the higher X-CK content (Foo et al., 2007) sustaining their outgrowth. Mutant rms2 rms4 plants do not respond to strigolactone treatment; after 7 d, rms2 rms4 buds treated directly with 1 μM GR24 grew 11.19 ± 3.71 mm, while comparable buds treated with 0 μM GR24 grew 9.17 ± 1.70 mm. This function for X-CK in promoting sustained branch growth, but not bud release, and strigolactone in inhibiting bud release is consistent with the bud outgrowth phenotypes of all other graft combinations (e.g., with rms2 rootstocks); such graft combinations increase X-CK supply but do not increase bud release in wild-type shoots (Beveridge et al., 1996; Beveridge, 2000).
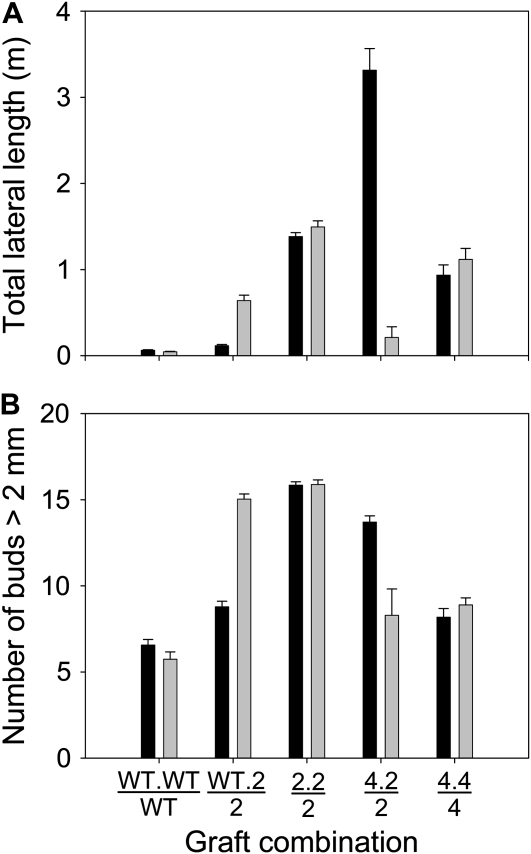
A two-shoot grafting experiment was used to investigate the hypothesis that increased X-CK export from roots promotes sustained branch growth after initial release (Table 1, hyp32). For this, rms2 scions and rootstocks were used due to their enhanced X-CK content (Foo et al., 2007). rms4 strigolactone-signaling mutant scions, which have released buds, were used to measure the response to increased X-CK. The computational model incorporating the effect of X-CK on bud growth (Table 1, hyp1-32) predicted that the branches on these rms4 shoots (grafted to rms2 rootstock and shoot) should be longer than in controls grafted to rms4. To reduce factors such as competition between shoots of unequal origin (Bangerth, 1989), we developed a two-scion grafting technique in which two equal scions were wedge-grafted to one rootstock. Total lateral outgrowth and the number of branches in the rms4 scion grafted with an rms2 scion and rms2 rootstock were indeed increased in comparison to rms4 self-grafted plants (P < 0.001; Figures 8A and 8B). This result supports the hypothesis that high X-CK promotes sustained branch growth after buds have already been released to grow (Table 1, hyp32), such as in a strigolactone-response mutant plant. When this hypothesis was implemented, the computational model (Table 1, hyp1-32) captured the branch length data for the grafts presented here.
Figure 8.
Branching Phenotype of Two-Scion Grafts among rms2, rms4, and Wild-Type Seedlings Measured 57 d after Grafting.
Data are means ± se (n = 17 to 33). Notation is scion.scion/rootstock. Total lateral length of scions (A); number of buds or branches >2 mm in length (B). 2, rms2; 4, rms4.
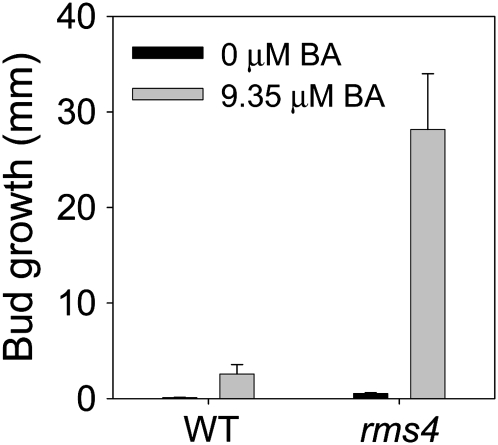
In a more direct test of the hypothesis that X-CK promotes sustained bud growth (not initial bud release; Table 1, hyp32), we supplied 0 or 9.35 μM 6-benzylaminopurine (a cytokinin) to the vascular stream of wild-type and rms4 strigolactone response mutant plants. In the wild type, this X-CK treatment caused little outgrowth (Figure 9). By contrast, it caused the released buds to grow considerably longer in rms4 plants (P < 0.001; Figure 9). Again, this supports the hypothesis that X-CK affects the growth rather than initial release of buds (Table 1, hyp32).
Figure 9.
rms4 Buds Are More Responsive to Cytokinin Than Are Wild-Type Buds.
Bud growth at node 3 was measured 8 d after 0 or 9.4 μM 6-benzylaminopurine (BA) was supplied to the vascular stream below node 3 when the plants had three to four leaves expanded. Growing buds at other nodes were not removed. Data are means ± se (n = 10).
The two-shoot plants also enabled us to evaluate predictions of the model (Table 1, hyp1-32) on signaling between different shoots. The model predicted that feedback signaling induced in one scion can upregulate strigolactone production in the rootstock to reduce branching in the other scion (Table 1, hyp12, 13, 14, 16, 18, and 21). Indeed, when grafted with rms2 rootstocks and rms2 scions, rms4 and wild-type scions both reduced branching in the rms2 scions (Figure 8A). The rms4 scion displayed a greater ability to inhibit bud release than did the wild-type scion (Figure 8B). According to our hypotheses (Table 1, hyp14, 16, 17, 25, and 32), and consistent with the implemented model output, the reduction of branching in the rms2 scion was due to an increase in strigolactone supply and a decrease in X-CK (both of which are more greatly affected in these grafts with rms4 rootstocks than with wild-type rootstocks due to feedback regulation). Conversely, we observed that bud outgrowth was enhanced in a wild-type scion by grafting it with an rms2 scion and an rms2 rootstock (Figure 8B). The computational model predicts that strigolactone supply to the wild-type scion is reduced in this graft combination accounting for the bud release observed.
Implications for Our Understanding of Bud Outgrowth
Collectively, our data support three new concepts of bud outgrowth regulation: (1) RMS4 in the rootstock negatively regulates RMS1 expression independent of the feedback signal in the rootstock (Table 1, hyp28). (2) Growing branches upregulate RMS1 expression in tissues below the site of branch growth, regardless of the RMS genotype (Table 1, hyp29-31). (3) X-CK exported from the rootstock to the shoot promotes branch length in buds that have already been released to grow (Table 1, hyp32).
By incorporating these new hypotheses based on new results presented here together with results from previous studies, we have developed a model of the genetics and physiology of long-distance branch regulation in pea. The model accurately predicts branching phenotypes, trends in rootstock X-CK content, and trends in RMS1 expression in the shoot and rootstock (above and below branches) in the wild type and each of the rms branching mutant lines whether they are intact or grafted.
DISCUSSION
Previous studies describing branching in pea have failed to account for all of the phenotypes and available data. The network proposed here (Figure 1), which is supported by a computational model, conserves key features of the previously published networks (Beveridge, 2000) and also explains all relevant new and published data. The model development involved an iterative process of experimental analysis and model improvement. Through this process, we made three main findings. First, RMS3 and RMS4 function in the rootstock and constitutively reduce RMS1 expression. Second, growing branches upregulate RMS1 expression in tissues below the site of branch growth via an RMS2-independent long-distance signal and thus can inhibit the outgrowth of axillary buds above via strigolactones. This mechanism contributes substantially to our understanding of the inhibition of axillary buds by other larger branches. Third, X-CK exported from the rootstock to the shoot increases the growth (but not release) of uninhibited buds (Figure 1, dashed lines). This also supports the notion of stages of bud outgrowth (reviewed in Dun et al., 2006). The computational model was essential in guiding our experiments and facilitated us reaching this point of increased understanding of shoot branching.
RMS1 Expression Is Feedback Regulated by Two Independent Signals
Computational modeling of the feedback signaling in pea plants demonstrated the requirement for two feedback signals. In addition to the previously described RMS2-dependent feedback signal, an RMS2-independent (branch-derived) feedback signal is required. Both of these feedback signals were required for the model to capture the RMS1 expression trends in grafts between rms2 and the wild type (see Supplemental Figures 5D and 5E online) and in the double mutants rms2 rms4 and rms2 rms5 (see Supplemental Figure 5B online). Without the inclusion of both signals, the expression of RMS1 in rms2 plants could not be explained.
The regulation of RMS1 expression by both of the feedback signals appears to play an important role in bud outgrowth. This is clearly demonstrated by the analysis of RMS1 expression in the rms2 mutant (Figures 4 to 6). rms2 plants branch mainly at basal nodes, with little aerial branching, albeit usually more than the wild type (Beveridge et al., 1994; Ferguson and Beveridge, 2009). This is in contrast with the rms4 strigolactone-response mutant that branches strongly at both basal and aerial nodes (Ferguson and Beveridge, 2009). The rms2 branching phenotype can be explained by the lack of the RMS2-dependent feedback signal, which results in low RMS1 expression, and presumably low strigolactone levels, in young seedlings early in development (Figures 4 and 5). The growing branches cause increased RMS1 expression in older rms2 plants (Figures 4 to 6), which presumably results in increased strigolactones moving up the stem to suppress aerial branching.
The importance of the branch-derived feedback signal becomes apparent when the growing basal branches are removed from rms2 plants. If the growing basal branches are removed from rms2 shoots, buds at proximal higher nodes grow out into branches (Beveridge et al., 1996; Ferguson and Beveridge, 2009). This is presumably due to removal of the branch-derived signal and, hence, depleted RMS1 expression and strigolactone levels. By contrast, while a response to basal branch removal is also observed in rms4 shoots (Ferguson and Beveridge, 2009), branch length, rather than bud release, is increased. As discussed later, this could be due to limited X-CK.
The long-distance feedback signaling is intriguing and prompts the search for the mobile compound(s) responsible. There are a number of similarities between auxin function and feedback signaling in that both promote strigolactone biosynthesis gene expression (Sorefan et al., 2003; Foo et al., 2005) and negatively regulate X-CK export from roots. However, at least in pea, auxin and the RMS2-dependent feedback signal differ in their magnitude of effects (discussed in Dun et al., 2009). By contrast, it is likely that auxin is a more dominant regulator of strigolactone biosynthesis gene expression in Arabidopsis than the feedback signal is (Hayward et al., 2009). By showing that two long-distance signals are required for long-distance feedback regulation in pea, we can postulate that one signal is auxin and the other, which we term the feedback signal (Figure 1), is currently unknown (Dun et al., 2009; Hayward et al., 2009).
A likely candidate for the branch-derived signal is auxin, as growing branches are known to export auxin into the main stem (Morris, 1977; Li and Bangerth, 1999; Ongaro and Leyser, 2008). By contrast, it should be noted that the signal we refer to as the feedback signal (Figure 1) is produced regardless of bud development and is therefore less likely to be correlated with stem auxin levels (Foo et al., 2007). If the branch-derived signal is auxin, it implies that rms2 mutant plants can respond to the relevant auxin concentration; hence, rms2 is unlikely to be an auxin response mutant.
Interestingly, the levels of indole-3-acetic acid (an auxin) are up to 4.4-fold higher in young apical rms2 mutant shoots compared with wild-type shoots, even when branching is inhibited by grafting to a wild-type rootstock (Beveridge et al., 1994). Thus, in these young rms2 tissues, indole-3-acetic acid levels are inversely proportional to RMS1 expression (Figures 4 to 6). The relationship between feedback, auxin content in whole internodes of rms2 plants, auxin supply to the main stem from branches, and RMS1 expression therefore remains a little unclear. The role of auxin transport needs to be investigated in this context. A computational model was recently published that proposed that strigolactones inhibit shoot branching by affecting auxin transport properties and, hence, competition between auxin sources (shoot tip and axillary bud; Prusinkiewicz et al., 2009).
Previously, it was demonstrated that the long-distance feedback signal is upregulated in situations where strigolactone response is reduced, such as by rms4 mutation (Foo et al., 2005; Johnson et al., 2006). Here, we confirm that this is also the case when strigolactone levels (and, hence, response) are altered (see Supplemental Figures 2E and 3 online). Consistent with this, treatment with the strigolactone analog GR24 causes downregulation of D10 (Os RMS1) gene expression in wild-type or strigolactone-deficient rice plants, but not in strigolactone-response mutant plants (Umehara et al., 2008). Future studies should investigate whether this strigolactone-induced feedback downregulation can occur over a long distance and whether it can also act more directly to regulate RMS1 expression (Hayward et al., 2009).
The modeling process also supported the notion of local RMS2-independent feedback regulation of RMS1 expression, particularly in the rootstock. Future studies of strigolactone response should test whether local feedback regulation of RMS1 expression is mediated directly via local strigolactone signaling. For strigolactone-responsive shoots, the model also supports a correlation between levels of RMS1 expression and branching phenotypes and suggests that RMS2 is not an absolute requirement for strigolactone biosynthesis (Figures 4 to 7; see Supplemental Figure 4 online).
X-CK Promotes Sustained Branch Growth
It is likely that local cytokinin biosynthesis may be an important regulator of bud outgrowth (Böhner and Gatz, 2001; Tanaka et al., 2006; Ferguson and Beveridge, 2009; Shimizu-Sato et al., 2009). However, a function of root-derived X-CK in mediating bud outgrowth has previously been considered unlikely as cytokinin overproducing rootstocks do not appear to promote branching in wild-type nonbranching shoots (Faiss et al., 1997). Here, we provide several lines of evidence that suggest that the supply of X-CK to axillary buds is important for their growth, once released.
The computational model incorporating all hypotheses discussed herein, including a role for X-CK in promoting sustained growth of released buds (Table 1, hyp1-32), predicted the longer branch lengths of particular plants. The increased branching phenotype of rms2 rms4 double mutants compared with the single mutant plants correlates well with the elevated X-CK caused by the rms2 mutation and the defective strigolactone response caused by the rms4 mutation (Figure 7; Foo et al., 2007). Importantly, we found that increasing X-CK supply to the shoot, either by grafting or a X-CK treatment method, increased bud growth in rms4 strigolactone response mutant plants, but had little effect on wild-type plants (Figures 8 and 9). Previous grafting studies have shown that increased X-CK has no effect on bud release in wild-type shoots (Beveridge et al., 1996; Beveridge, 2000), presumably because wild-type buds are not yet responsive to X-CK as they are not yet released. By comparing the number of buds released to grow into branches and the length of branches (Figures 7 and 8), it appears that strigolactones are involved in bud release, and X-CK is involved in promoting sustained branch growth (Figure 1). Limited X-CK levels might explain why all rms4 buds do not grow into long branches (Beveridge et al., 1996) and why branches at aerial nodes are able to grow longer in response to the removal of basal branches (Ferguson and Beveridge, 2009). This study provides a clear hypothesis that needs evaluation via localized hormone measurements and a thorough investigation of different modes of cytokinin delivery. It is very important to note that despite alterations in X-CK export from roots, overall shoot cytokinin content is unchanged in pea rms2 and strigolactone biosynthesis and response mutant plants (Foo et al., 2007). However, cellular differences in cytokinin levels may be revealed in future studies.
The function of X-CK in branch growth, but not in the initial release of buds to grow, supports the conceptualization of bud outgrowth as a number of distinct developmental stages (Dun et al., 2006). The developmental identity of individual axillary buds will be an important feature of future models aimed at representing the response to auxin perturbing treatments. In these instances, buds at older, basal nodes may have a different response to these treatments than the buds at younger nodes (Ferguson and Beveridge, 2009).
Modeling
Many modeling approaches rely on the availability of highly detailed quantitative knowledge about the system (Bolouri and Davidson, 2002; de Jong, 2002). Unfortunately, this renders some approaches difficult to apply to some biological systems due to a shortage of biological information. Some methods deal with the lack of detailed information by searching for parameters that give the model the best fit to biological data (Locke et al., 2005). This allows the network structure (e.g., interactions between signals) to remain the focus. However, the development of suitable equations still requires knowledge of the nature of interactions. This information is not yet available for shoot branching. More abstract modeling approaches can require less detailed information and have been successfully used to model regulatory networks (Espinosa-Soto et al., 2004; Wenden et al., 2009). Here, we developed a modeling method that is well suited to the genetic and physiological data available for branching in pea. Moreover, the approach we have used is not dependent on modifying several parameters to reach the desired output. Rather, the output is compared directly with the experimental data in much the same way as one would compare experiments that use different conditions or genetic backgrounds, but for which the central points of comparison remain the same. In the model, there are only three variable parameters; they are explicitly based on hypotheses (Table 1, hyp26, 27) and can take on a wide range of values without affecting the model outcome (see Supplemental Table 1 online).
Through modeling we have successfully demonstrated that the hypotheses implemented (Table 1, hyp1-32) as a set are indeed valid explanations of the broad data sets against which the model was evaluated. This is now a solid framework through which we may continue to evaluate new hypotheses or modify existing ones. This hypothesis-driven modeling approach has proved invaluable in deriving new hypotheses and insightful biological experiments to test them. The beauty of this approach is that each and every aspect of the model is intrinsically linked to biological hypotheses. In contrast with empirical models that can capture only correlations, this model captures the mechanisms explicitly.
This hypothesis-driven modeling approach does not seek to find all possible networks that may explain the data sets or system. Having created a single model that adequately captures all existing biological data sets, we have mathematically and computationally articulated one potential solution for the network. As mentioned above, the most useful feature of this model is its explicit link to relevant biological hypotheses. New hypotheses or new relationships can be easily evaluated in the context of this existing model framework.
Many features of the pea branching network appear to be conserved in other species (Beveridge, 2006; Foo et al., 2007; Simons et al., 2007; Hayward et al., 2009), meaning this computational model may have broad relevance. We can also consider modifying the model to test whether changing the relative importance of interactions in the system will allow us to essentially convert the pea network to one for Arabidopsis or rice, for example. In this case, particular hypotheses may be removed, such the long-distance feedback signal (Table 1, hyp2, 7, 13, 14, 17, 25, and 27), to determine if this is a better explanation of the biological data generated using Arabidopsis. A future application of this modeling approach will include incorporation of molecular data on auxin signaling and response, in addition to stages of bud outgrowth. In the context of auxin and long-distance signaling, this will add considerable further complexity to the model. Our correlative studies on stem auxin depletion after decapitation demonstrate that buds at basal nodes can respond to decapitation well before the corresponding depletion in apical auxin supply could have reached the node (Morris et al., 2005). Considerable biological experimentation is clearly also required.
METHODS
Plant Material and Growth Conditions
For all experiments, unless otherwise specified, pea (Pisum sativum) plants were grown as described by Ferguson and Beveridge (2009). The wild-type cultivar was Torsdag (L107), and the mutant lines introduced into Torsdag were rms1-2 (rms1-2T), rms2-1 (K524), rms3-1 (K487), rms4-1 (K164; Beveridge et al., 1996), rms5-3 (BL298; Morris et al., 2001), and rms2-1 rms4-1 (BL24; Murfet and Symons, 2000).
Nodes were numbered acropetally from the first scale leaf, and lengths of lateral branches and buds at each node were recorded to the nearest 0.1 mm using digital calipers and summed to form the total lateral length for the scion (scions in the case of two-scion grafts).
A Student's t test was used to test for statistical significance in experiments.
Grafting
Grafts were performed using the epicotyl-epicotyl wedge graft technique on 7-d-old seedlings as described by Beveridge et al. (1994). The two-scion grafting technique was based on the epicotyl-epicotyl wedge graft technique, but with two scions wedge grafted to the one rootstock, grown at one per 2-liter pot.
Hormone Treatments
Ten microliters of solution containing 0 or 1 μM GR24 (synthetic strigolactone) with 2% polyethylene glycol 1450, 50% ethanol, and 0.2% acetone was applied directly to test axillary buds. All other growing axillary buds at nodes below were removed to encourage the growth of the test bud.
Solutions (1.5 mL) containing 0 or 9.35 μM 6-benzylaminopurine and 5.2% ethanol were supplied to the vascular stream of the main stem as described by Gomez-Roldan et al. (2008).
Real-Time PCR Gene Expression Analyses
Total RNA was isolated and quantified as described by Ferguson and Beveridge (2009). cDNA was synthesized in a 20-μL reaction volume with 0.9 to 2.4 μg total RNA (depending which experiment), 250 ng random hexamers (Invitrogen), 0.5 mM deoxynucleotide triphosphate (Invitrogen), 2.5 mM oligo(dT) (Promega), 5 mM DTT (Invitrogen), 1× first-strand buffer, and 100 units Superscript III reverse transcriptase (Invitrogen) incubated at 52°C for 60 min and 70°C for 15 min. cDNA was checked via PCR and gene expression analyses performed as described by Ferguson and Beveridge (2009).
For the expression profile (Figure 4), each reaction contained 3 μL SYBR Green 2× Master Mix (Applied Biosystems), 10 ng of cDNA, and 200 nM of each gene-specific primer pair, with a final volume of 6 μL. For the age profile (Figure 5) and rms2 grafting (Figure 6) experiments, each reaction contained 5 μL SYBR Green 2× Master Mix (Applied Biosystems), 16 ng of cDNA, and 400 nM of each gene-specific primer pair, with a final volume of 10 μL. These reagents were aliquoted to each well of the 384-well plate by an Eppendorf epMotion 5075 liquid handler.
Primer sequences used were as follows: RMS1 forward (5′-AAGGAGCTGTGCCCTCAGAA-3′) and RMS1 reverse (5′-ATTATGGAGATCACCACACCATCA-3′) (Foo et al., 2005); 18S forward (5′-ACGTCCCTGCCCTTTGTACA-3′) and 18S reverse (5′-CACTTCACCGGACCATTCAAT-3′) (Ozga et al., 2003). Error bars represent biological standard error.
Modeling
The modeling method followed the general approach of hypothesis and model formulation, verification, and calibration (Haefner, 2005) and is similar to that reported by Wenden et al. (2009). Hypotheses were formulated and translated to mathematical terms in the program L-Studio version 4.18 (University of Calgary, Canada). Although not required for this study, L-Studio is based on the principals of the rule-based formalism L-systems (Lindenmayer systems), which lend themselves to visualization of plant structure (Prusinkiewicz, 2004). Algebraic equations were used to represent network interactions affecting each model component using a limited number of parameters (see Supplemental Table 1 online). As an input to the model, for each graft combination, genotypes of each gene in the shoot(s) and rootstock(s) were assigned a value of 1 for the wild-type state or 0 (or slightly greater than 0) for the null (or leaky) mutant state. All model components were initially assigned a randomly selected value between 0 and 1. The algebraic equations were then iterated until a steady state was reached for the branching inhibition output (example in Supplemental Figure 6 online). In order to compare the model output to the biological data sets, all data sets were divided into classes. For example, graft combinations were divided into three classes for their branching phenotype: a wild-type branching phenotype, rms branching phenotype, and an intermediate branching phenotype. Model output was compared with the biological data by checking that the groupings were correct for branching inhibition, RMS1 expression, and X-CK levels.
See Supplemental Methods online for further details on the formulation of hypotheses and algebraic equations and the comparison of the model output to biological data.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: AY557341 for RMS1 (Térèse haplotype), AY557342 for RMS1 (Raman haplotype), and U43011 for 18S.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Model and Biological Classes of Rootstock X-CK.
Supplemental Figure 2. RMS1 Expression Model Output and Biological Data Prior to Branching.
Supplemental Figure 3. RMS1 Expression Relative to 18S in Grafts between the Wild Type and rms5.
Supplemental Figure 4. Branching Phenotypes of Grafts between the Wild Type and rms3.
Supplemental Figure 5. RMS1 Expression Model Output and Biological Data after Branching.
Supplemental Figure 6. Output of Branching Inhibition over 100 Iterations for a WT/WT I-Grafted Plant from Final Computational Model.
Supplemental Table 1. Model Parameters.
Supplemental Table 2. Branching Phenotypic Classes of Epicotyl-Epicotyl I-Grafted Plants.
Supplemental Table 3. Branching Phenotypic Classes of Two-Shoot Grafted Plants.
Supplemental Table 4. Branching Phenotypic Classes of Two-Rootstock Grafted Plants.
Supplemental Methods.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Catherine Rameau for kindly supplying GR24, Kerry Condon, Skye Thomas-Hall, Tanya Brcich, Fiona Filardo, Alice Hayward, Benedicte Wenden, and Bob Simpson for assistance with harvesting and/or real-time PCR, Michael Renton for initial work on modeling, and Philip Brewer, Geoffrey Dun, and particularly Michael Mason for excellent comments on the manuscript. This work was supported by the Australian Research Council Centre of Excellence for Integrative Legume Research and by Australian Postgraduate Awards (scholarship to E.A.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christine A. Beveridge (c.beveridge@uq.edu.au).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827. [DOI] [PubMed] [Google Scholar]
- Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., Sakakibara, H., and Kyozuka, J. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., and Kyozuka, J. (2009). d14, a strigolactone insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50 1416–1424. [DOI] [PubMed] [Google Scholar]
- Bangerth, F. (1989). Dominance among fruits/sinks and the search for a correlative signal. Physiol. Plant. 76 608–614. [Google Scholar]
- Beveridge, C.A. (2000). Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 32 193–203. [Google Scholar]
- Beveridge, C.A. (2006). Axillary bud outgrowth: sending a message. Curr. Opin. Plant Biol. 9 35–40. [DOI] [PubMed] [Google Scholar]
- Beveridge, C.A., Murfet, I.C., Kerhoas, L., Sotta, B., Miginiac, E., and Rameau, C. (1997. b). The shoot controls zeatin riboside export from pea roots. Evidence from the branching mutant rms4. Plant J. 11 339–345. [Google Scholar]
- Beveridge, C.A., Ross, J.J., and Murfet, I.C. (1994). Branching mutant rms-2 in Pisum sativum. Grafting studies and endogenous indole-3-acetic acid levels. Plant Physiol. 104 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge, C.A., Ross, J.J., and Murfet, I.C. (1996). Branching in pea. Action of genes Rms3 and Rms4. Plant Physiol. 110 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge, C.A., Symons, G.M., Murfet, I.C., Ross, J.J., and Rameau, C. (1997. a). The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol. 115 1251–1258. [Google Scholar]
- Böhner, S., and Gatz, C. (2001). Characterisation of novel target promoters for the dexamethasone-inducible/tetracycline-repressible regulator of TGV using luciferase and isopentyl transferase as sensitive reporter genes. Mol. Gen. Genet. 264 860–870. [DOI] [PubMed] [Google Scholar]
- Bolouri, H., and Davidson, E.H. (2002). Modelling transcriptional regulatory networks. Bioessays 24 1118–1129. [DOI] [PubMed] [Google Scholar]
- Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H., and Leyser, O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signalling molecule. Curr. Biol. 14 1232–1238. [DOI] [PubMed] [Google Scholar]
- Booker, J., Sieberer, T., Wright, W., Williamson, L., Willett, B., Stirnberg, P., Turnbull, C., Srinivasan, M., Goddard, P., and Leyser, O. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8 443–449. [DOI] [PubMed] [Google Scholar]
- Brewer, P.B., Dun, E.A., Ferguson, B.J., Rameau, C., and Beveridge, C.A. (2009). Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, H. (2002). Modelling and simulation of genetic regulatory systems: A literature review. J. Comput. Biol. 9 67–103. [DOI] [PubMed] [Google Scholar]
- Dun, E.A., Brewer, P.B., and Beveridge, C.A. (2009). Strigolactones: Discovery of the elusive shoot branching hormone. Trends Plant Sci. 14 364–372. [DOI] [PubMed] [Google Scholar]
- Dun, E.A., Ferguson, B.J., and Beveridge, C.A. (2006). Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 142 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Soto, C., Padilla-Longoria, P., and Alvarez-Buylla, E.R. (2004). A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16 2923–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiss, M., Zalubìlová, J., Strnad, M., and Schmülling, T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12 401–415. [DOI] [PubMed] [Google Scholar]
- Ferguson, B.J., and Beveridge, C.A. (2009). Roles for auxin, cytokinin and strigolactone in regulating shoot branching. Plant Physiol. 149 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, E., Bullier, E., Goussot, M., Foucher, F., Rameau, C., and Beveridge, C.A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, E., Morris, S.E., Parmenter, K., Young, N., Wang, H., Jones, A., Rameau, C., Turnbull, C.G.N., and Beveridge, C.A. (2007). Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiol. 143 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, E., Turnbull, C.G.N., and Beveridge, C.A. (2001). Long-distance signalling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger, D., Drapeau, M., Collins, B., and Blau, J. (2005). A new model for circadian clock research? Mol. Syst. Biol. 1 2005.0014. [DOI] [PMC free article] [PubMed]
- Gomez-Roldan, V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 189–194. [DOI] [PubMed] [Google Scholar]
- Haefner, J.W. (2005). Modelling biological systems: Principles and Applications, 2nd ed. (New York: Springer Science and Business Media), pp. 18–23.
- Hayward, A., Stirnberg, P., Beveridge, C., and Leyser, O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 151 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I., and Kyozuka, J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46 79–86. [DOI] [PubMed] [Google Scholar]
- Johnson, X., Brcich, T., Dun, E.A., Goussot, M., Haurogné, K., Beveridge, C.A., and Rameau, C. (2006). Branching genes are conserved across species: Genes controlling a novel signal in pea are co-regulated by other long-distance signals. Plant Physiol. 142 1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2009). The control of shoot branching: an example of plant information processing. Plant Cell Environ. 32 694–703. [DOI] [PubMed] [Google Scholar]
- Li, C.J., and Bangerth, F. (1999). Autoinhibition of indoleacetic acid transport in the shoots of two-branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol. Plant. 106 415–420. [Google Scholar]
- Locke, J.C.W., Southern, M.M., Kozma-Bognár, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1 2005.0013. [DOI] [PMC free article] [PubMed]
- Matusova, R., Rani, K., Verstappen, F.W.A., Franssen, M.C.R., Beale, M.H., and Bouwmeester, H.J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen, P. (2009). Hormonal regulation of branching in grasses. Plant Physiol. 149 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, D.A. (1977). Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 136 91–96. [DOI] [PubMed] [Google Scholar]
- Morris, S.E., Cox, M.C., Ross, J.J., Krisantini, S., and Beveridge, C.A. (2005). Auxin dynamics are not correlated with the initial growth of axillary buds. Plant Physiol. 138 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S.E., Turnbull, C.G.N., Murfet, I.C., and Beveridge, C.A. (2001). Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfet, I.C., and Symons, G.M. (2000). The pea rms2-1 rms4-1 double-mutant phenotype is transgressive. Pisum Genet. 32 59–60. [Google Scholar]
- Napoli, C.A., Beveridge, C.A., and Snowden, K.C. (1999). Reevaulating concepts of apical dominance and the control of axillary bud outgrowth. Curr. Top. Dev. Biol. 44 127–169. [DOI] [PubMed] [Google Scholar]
- Ongaro, V., and Leyser, O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59 67–74. [DOI] [PubMed] [Google Scholar]
- Ozga, J.A., Yu, J., and Reinecke, D.M. (2003). Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol. 131 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz, P. (2004). Modeling plant growth and development. Curr. Opin. Plant Biol. 7 79–83. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz, P., Crawford, S., Smith, R.S., Ljung, K., Bennett, T., Ongaro, V., and Leyser, O. (2009). Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 106 17431–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato, S., Tanaka, M., and Mori, H. (2009). Auxin-cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69 429–435. [DOI] [PubMed] [Google Scholar]
- Simons, J.L., Napoli, C.A., Janssen, B.J., Plummer, K.M., and Snowden, K.C. (2007). Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 143 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden, K.C., Simkin, A.J., Janssen, B.J., Templeton, K.R., Loucas, H.M., Simons, J.L., Karunairetnam, S., Gleave, A.P., Clark, D.G., and Klee, H.J. (2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan, K., Booker, J., Haurogné, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., and Leyser, O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., van de Sande, K., and Leyser, H.M.O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Takei, K., Kojima, M., Sakakibara, H., and Mori, H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45 1028–1036. [DOI] [PubMed] [Google Scholar]
- Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., Magome, H., Kamiya, Y., Shirasu, K., Yoneyama, K., Kyozuka, J., and Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200. [DOI] [PubMed] [Google Scholar]
- Wenden, B., Dun, E.A., Hanan, J., Andrieu, B., Weller, J.L., Beveridge, C.A., and Rameau, C. (2009). Computational analysis of flowering in pea (Pisum sativum). New Phytol. 184 153–167. [DOI] [PubMed] [Google Scholar]
- Zou, J., Zhang, S., Zhang, W., Li, G., Chen, Z., Zhai, W., Zhao, X., Pan, X., Xie, Q., and Zhu, L. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48 687–696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.