Abstract
TGA2 and NONEXPRESSER OF PR GENES1 (NPR1) are activators of systemic acquired resistance (SAR) and of the SAR marker gene pathogenesis-related-1 (PR-1) in Arabidopsis thaliana. TGA2 is a transcriptional repressor required for basal repression of PR-1, but during SAR, TGA2 recruits NPR1 as part of an enhanceosome. Transactivation by the enhanceosome requires the NPR1 BTB/POZ domain. However, the NPR1 BTB/POZ domain does not contain an autonomous transactivation domain; thus, its molecular role within the enhanceosome remains elusive. We now show by gel filtration analyses that TGA2 binds DNA as a dimer, tetramer, or oligomer. Using in vivo plant transcription assays, we localize the repression domain of TGA2 to the N terminus and demonstrate that this domain is responsible for modulating the DNA binding activity of the oligomer both in vitro and in vivo. We confirm that the NPR1 BTB/POZ domain interacts with and negates the molecular function of the TGA2 repression domain by excluding TGA2 oligomers from cognate DNA. These data distinguish the NPR1 BTB/POZ domain from other known BTB/POZ domains and establish its molecular role in the context of the Arabidopsis PR-1 gene enhanceosome.
INTRODUCTION
Plant defense against pathogen attack involves global transcriptional reprogramming (Dangl and Jones, 2001; Durrant and Dong, 2004). Among the induced genes are the pathogenesis-related (PR) genes, which are activated both at the site of infection and in uninfected parts of the plant in response to the pathogen-induced accumulation of salicylic acid (SA) (Ryals et al., 1996). Local and distal SA accumulations are mandatory for the deployment of a systemic long-lasting and broad-spectrum plant disease resistance response called systemic acquired resistance (SAR) (Durrant and Dong, 2004; Pieterse and Van Loon, 2004; Ryals et al., 1996). Exogenous application of SA, termed chemical SAR, triggers PR gene induction and SAR deployment (Ward et al., 1991).
In Arabidopsis thaliana, genetic, biochemical, and molecular approaches have established that SA-dependent activation of the SAR marker gene PR-1 is mediated by the transacting factors NONEXPRESSER OF PR GENES1 (NPR1) and the TGA2 clade of transcription factors, which includes TGA2/5/6 (Cao et al., 1994; Delaney et al., 1995; Zhang et al., 2003; Rochon et al., 2006), in addition to a collection of cis-regulatory elements located in the PR-1 promoter (Lebel et al., 1998). Linker scanning (LS) mutagenesis of the PR-1 promoter revealed that it contained at least three cis-regulatory elements (LS5, LS7, and LS10), displaying positive or negative character (Lebel et al., 1998). LS5 and LS7 both contain the cognate TGA binding sequence, TGACG, and, indeed, TGA2 has been shown to independently bind the LS5 and LS7 promoter elements in vitro (Després et al., 2000). The LS5 element appears to contribute to the negative regulation of PR-1 expression both in the absence and in the presence of SA, while LS7 is required for SA-mediated induction of PR-1 (Lebel et al., 1998).
In unstimulated cells, the endogenous NPR1 protein is localized to both the nucleus and the cytosol (Després et al., 2000). Nuclear localization of NPR1 is critical to PR-1 activation (Kinkema et al., 2000). Chromatin immunoprecipitation experiments revealed that the NPR1 protein is specifically present in the promoter region of the PR-1 gene under both uninduced and SA-stimulated conditions and that its recruitment is independent of the TGA2 clade of transcription factors (Rochon et al., 2006). Similarly, chromatin immunoprecipitation revealed that the recruitment of TGA2 to the PR-1 promoter is both SA and NPR1 independent. Furthermore, plant two-hybrid and protein complementation assays have demonstrated that these two proteins, NPR1 and TGA2, do not interact inside the nucleus until after stimulation with SA (Subramaniam et al., 2001; Rochon et al., 2006). Interestingly, TGA2 is a constitutive repressor, and the TGA2 clade of transcription factors is required for the basal repression of PR-1 (Zhang et al., 2003; Rochon et al., 2006). However, following SA stimulation, TGA2 forms an enhanceosome with NPR1, which provides the transactivation domain. This domain is located in the C-terminal end of NPR1 and contains Cys residues critical to its function (Rochon et al., 2006).
The current model for PR-1 activation proposes that after a rise in SA concentration, nuclear-localized NPR1 interacts with TGA2 to stimulate its DNA binding activity to the SA-responsive promoter element LS7, ultimately resulting in the activation of the gene (Lebel et al., 1998; Després et al., 2000, 2003; Kinkema et al., 2000; Subramaniam et al., 2001; Fan and Dong, 2002; Johnson et al., 2003; Mou et al., 2003; Zhang et al., 2003; Rochon et al., 2006). PR-1 is also negatively regulated by proteins such as SUPPRESSOR OF NPR1 INDUCIBLE1 (Li et al., 1999b) and NIM1-INTERACTING1 (NIMIN1; Weigel et al., 2005), but in the absence of chromatin immunoprecipitation data, one cannot rule out that these proteins act indirectly on PR-1. Although NIMIN1 has been shown to interact with NPR1 in vivo and to form a ternary complex with TGA2 in yeast, these interactions may occur away from the PR-1 promoter. It is thus difficult to include with certainty these proteins in a molecular model of PR-1 activation.
NPR1 contains two identifiable protein–protein interaction motifs: ankyrin repeats (Cao et al., 1997; Ryals et al., 1997; Li et al., 2006) and a BTB/POZ (for Broad-Complex, Tramtrack, and Bric-a-brac/Pox virus and Zinc finger) domain (Aravind and Koonin 1999; Bardwell and Treisman 1994). The ankyrin repeats mediate interactions with TGA factors, and their mutation abolishes NPR1-TGA complex formation, PR gene expression, and SAR (Cao et al., 1997; Ryals et al., 1997; Zhang et al., 1999; Després et al., 2000, 2003). The functional role of the NPR1 BTB/POZ domain in disease resistance is unclear, but this domain is critical to the transactivation function of the TGA2-NPR1 enhanceosome in spite of the fact that it is not an autonomous transactivation domain (Rochon et al., 2006). In mammals, BTB/POZ domains are actively studied because deregulation of proteins bearing this motif often result in disease states such as cancer (Deltour et al., 1999; Collins et al., 2001; Kelly and Daniel, 2006), but not much is known about the function of BTB/POZ domains in plants.
The BTB/POZ domain is an evolutionarily conserved and widely distributed structural motif found in a battery of proteins involved in different biological processes, such as transcriptional regulation, cytoskeletal organization, and formation of voltage-gated channels (Aravind and Koonin, 1999; Collins et al., 2001). This domain has been shown to homodimerize, multimerize, and heterodimerize with other BTB/POZ domains or with proteins devoid of the motif (Bardwell and Treisman, 1994; Aravind and Koonin 1999; Li et al., 1999a; Collins et al., 2001). Although all BTB/POZ domains identified thus far contain a core of ∼90 amino acids, a long form of the BTB/POZ domain also exists that contains an N-terminal extension ∼30 residues in length (Stogios et al., 2005). The structure of residues 7 to 122 of the promyelocytic leukemia zinc finger (PLZF) has been solved and offers a three-dimensional view of the long form of the BTB/POZ domain (Li et al., 1999a), the form to which NPR1 likely belongs.
Here, we identify the repression and oligomerization domains of TGA2 and show that the repression domain dictates the stoichiometry of the TGA2-DNA complex. We establish the stoichiometry of the TGA2-NPR1-DNA complex and solve the molecular function of the NPR1 BTB/POZ domain in the enhanceosome.
RESULTS
The N Terminus of TGA2 Is a Nonautonomous Repression Domain
TGA2 is a constitutive repressor (Rochon et al., 2006), and to identify domains responsible for repression, TGA2 deletions were generated. We focused on the N terminus (a region arbitrarily defined as the sequence located between the first residue and the basic DNA binding domain) since this is where the transactivation domain is located in tobacco (Nicotiana tabacum) TGA1a (Neuhaus et al., 1994). TGA2 and a variant lacking the first 43 amino acids (Δ43) were respectively fused to the DNA binding domain of Gal4 (:DB) and assayed using a transient in vivo plant transcription assay system in Arabidopsis leaves, in which the luciferase reporter gene is first activated by a chimeric LexA:VP16 transcriptional activator (Figure 1B). The baseline level of transcription was determined by transfecting leaves with Gal4 DB (not fused to any other protein or protein domain) along with the reporter construct shown in Figure 1A. To activate the reporter gene, leaves were simultaneously transfected with Gal4 DB and LexA:VP16. In Figure 1B and as reported previously (Rochon et al., 2006), transfection with TGA2:DB and LexA:VP16 resulted in some repression of the reporter gene in untreated (white bars) and SA-treated (gray bars) cells. However, expression of the Δ43 variant of TGA2 (Δ43:DB) along with LexA:VP16 did not result in repression, as values were not below those observed with Gal4 DB + LexA:VP16, and this result was obtained regardless of whether cells were treated with SA. These data indicate that the N terminus of TGA2 is required for the transcriptional repression properties of TGA2.
Figure 1.
The N Terminus of TGA2 Is Not an Autonomous Repression Domain.
(A) Graphic representation of the synthetic 3X Gal4:1X LexA:minimal promoter:Firefly Luciferase reporter gene. The upward arrow indicates the position of the TATA box relative to the RNA start site. The 60bp and 30bp indicate the spacing in base pairs between the most downstream Gal4 element and the LexA element and between the LexA element and the TATA box, respectively. Not shown is an omega translational enhancer in the transcribed region of the Luciferase gene.
(B) Bar graph illustrating the assessment of potential transcriptional repression conferred by TGA2, TGA2 with a 43–amino acid N terminus deletion (Δ43), and the first 47, 101, and 144 amino acids of TGA2 (Nt47, Nt101, and Nt144), all tethered to DNA through the Gal4 DNA binding domain (:DB). Where indicated, the LexA DB fused to the viral particle 16 (LexA:VP16) transactivation domain was also transfected in order to activate the reporter gene. The constructs were transfected along with the 3X UASGAL4:1X LexA DNA element:minimal promoter:Firefly luciferase reporter and the CaMV35S:Renilla luciferase internal standard vectors.
(C) Bar graph illustrating the fact that TGA2, Nt47, Nt101, and Nt144 tethered to DNA through Gal4 DB (TGA2:DB, Nt47:DB, Nt101:DB, and Nt144:DB) do not activate transcription, while Δ43:DB and a chimeric transcription activator composed of the Gal4 DB fused to the transactivation domain of viral particle 16 (Gal4 DB:VP16 TA) do. Gal4 DB represents the baseline level of transcription. The constructs were transfected along with the 5X UASGAL4:Firefly luciferase reporter and the CaMV35S:Renilla luciferase internal standard vectors. In (B) and (C), Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). Data are reported as relative luciferase units. Values consist of n = 25 samples and represent averages ± 1 sd. Every bar represents five bombardments repeated five times (n = 25).
We next addressed whether the N terminus of TGA2 is an autonomous repression domain. The first 47 amino acids of TGA2 were fused to Gal4 DB (Nt47:DB) and coexpressed with LexA:VP16 (Figure 1B). Whether cells were treated or not with SA, this did not result in repression of the activated reporter gene, as values were not below those observed with Gal4 DB + LexA:VP16. Similar findings were observed with longer constructs harboring the first 101 (Nt101:DB) or 144 (Nt144:DB) amino acids of TGA2. These results show that the N terminus of TGA2 is not an autonomous repression domain. Of note, Δ43:DB + LexA:VP16 displayed values that were higher than those observed with Gal4 DB + LexA:VP16 whether cells were treated with SA or not, while Nt47:DB, Nt101:DB, or Nt144:DB + LexA:VP16 were higher than the control in the absence of treatment and not significantly different from it in the presence of SA. In the case of the former, the results suggest that Δ43:DB synergizes with LexA:VP16 for activating the promoter whether cells have received a treatment or not. In the case of Nt47:DB, Nt101:DB, or Nt144:DB, this synergy with LexA:VP16 was abrogated by SA. This would tend to suggest that in this system, Δ43:DB and Nt47:DB/Nt101:DB/Nt144:DB use different mediators to effect gene activation. Furthermore, in the case of Nt47:DB/Nt101:DB/Nt144:DB, these proteins may yet use different mediators depending on whether cells were stimulated with SA or not.
Since the Δ43 variant of TGA2 and the N terminus of TGA2 did not repress transcription of the activated reporter gene, we wanted to determine whether they can activate transcription of a reporter construct consisting of a firefly luciferase gene under the control of five copies of the Gal4 upstream activating sequences (UAS) fused to a minimal promoter (Rochon et al., 2006). Figure 1C shows results from such an experiment. The baseline level of transcription was determined by transfecting leaves with Gal4 DB (not fused to any other protein or protein domain) along with the reporter construct. Transfection with TGA2:DB or the N-terminal constructs Nt47:DB or Nt101:DB did not result in reporter gene activation beyond the baseline level, regardless of whether cells were treated with SA (gray bars) or not (white bars), while construct Nt144:DB showed transcription activation beyond baseline only in the absence of SA. Transfection of the Δ43 variant of TGA2 (Δ43:DB, Figure 1C) led to SA-independent expression of the reporter gene well above the baseline. These data indicate that removal of the N-terminal repression domain of TGA2 leads to the creation of a protein with transcriptional activating properties.
TGA2 Exists as a Higher-Order Complex That Requires the Leucine Zipper
Since the N terminus of TGA2 did not behave as an autonomous repression domain, we hypothesized that it might play a dynamic structural role in TGA2, imposing a stoichiometry observable in gel filtration or discernible in the context of DNA interaction. We first set out to express soluble TGA2 fused to a C-terminal His-tag in Escherichia coli and purify it by affinity chromatography. Purified TGA2 was then analyzed by Sephacryl S300 gel filtration.
The elution profile for TGA2 (Figure 2A) shows that the protein eluted in the void volume and thus formed an oligomer of unknown stoichiometry, but containing 40 or more units of TGA2, based on the S300 theoretical size exclusion of 1.5 MD and the molecular mass of TGA2 (37.51 kD with His-tag) (see Supplemental Figures 1 and 2 online). This void volume entity was not constituted of aggregated, nonfunctional proteins, since it was our source of TGA2 to perform electrophoretic mobility shift assays (EMSAs) and was competent to interact with DNA (Figure 3). An immunoblot analysis using an anti-His-tag antibody confirmed that TGA2 could only be detected in the void volume of the column and could not be found in the included volume (one-column volume) (Figure 2A, inset). Deleting the N-terminal repression domain of TGA2 (Δ43) or the basic DNA binding domain (Δ68) had no effect on its capacity to form a high-order complex as the Δ43 and Δ68 variants also eluted in the void volume (Figures 2B and 2C). This result was again confirmed by immunoblots (Figures 2B and 2C, insets). The Δ43 contained in the void volume is also composed of functional proteins and was our source for EMSAs shown in Figure 4. However, further deleting to Δ93, effectively removing the leucine zipper, abolished the formation of the oligomer and resulted in a species forming mainly a dimer and possibly also a monomer, which can be observed as a slight shoulder around elution volume 70 mL (Figure 2D). The Δ93 elution profile was also confirmed by an immunoblot (Figure 2D, inset). These data show that the leucine zipper is required for the formation of a TGA2 oligomer.
Figure 2.
The Leucine Zipper Is Responsible for TGA2 Oligomerization.
Chromatogram illustrating the elution profile of purified His-tagged TGA2 with N-terminal deletions: TGA2 (A), Δ43 (B), Δ68 (C), and Δ93 (D). The leucine zipper is located between amino acids 72 and 86. In (B) to (D), the TGA2 deletion constructs all contain the AUG initiator codon; therefore, the effective deletion starts at amino acid number 2. In (A), the higher absorbance curve contains 50 nmol of TGA2 in 2 mL (25 μM), while the lower one contains 5 nmol of TGA2 in 2 mL (2.5 μM). The concentrations of Δ43, Δ68, and Δ93 were 15, 3, and 10 μM, respectively. In (A) to (D), insets are immunoblot analysis of pooled protein fractions from the chromatogram using an anti-His antibody. Five fractions for each observed or theoretical peak (peak fraction + the two fractions preceding it + the two fractions following it) were acetone precipitated, and 100% of each precipitate was loaded on SDS-PAGE. The remaining fractions between peaks were pooled and precipitated, and 100% of each precipitate was loaded on SDS-PAGE to assess data points between peaks (V0-T, T-D, V0-D, and D-M). V0-T and V0-D pools ([A] to [D]) started from fraction corresponding to mL 43 and V0-D pools (D) ended at fraction corresponding to mL 60. The apparent mobility of the various TGA2 proteins coincided with the expected mobility, based on the size standards (New England Biolabs Broad Range). In (A), the top and bottom panels represent data from the high and low TGA2 concentration, respectively. Void indicates fractions collected from the void volume, while Tetra, Dimer, and Mono represent pooled fractions from the predicted elution profile of a theoretical TGA2, Δ43, or Δ68 tetramer, dimer, and monomer, respectively. V0-T, T-D, and D-M indicate pooled samples corresponding to fractions located between the void volume and tetramer, between the tetramer and dimer, and between the dimer and monomer, respectively. In (D), V0-D indicates pooled samples corresponding to fractions located between the void volume and dimer.
Figure 3.
The TGA2 Oligomer Can Bind to Its Cognate Sequence LS7 in PR-1.
(A) EMSA using recombinant TGA2 (lanes 2 to 18) together with the LS7 DNA as the probe (all lanes). The numbers indicate the ratio of probe concentration to TGA2 concentration. The black, gray, and white arrows indicate the positions of three distinct complexes. FP stands for free probe and refers to an experiment in which only DNA is present.
(B) Chromatogram based on the elution profile of the LS7 DNA probe derivatized with fluorescein. The profile of free DNA appears as a dashed line, while that of the DNA (5 μM) incubated with 15 or 5 μM TGA2 is represented by a solid or jagged line, respectively. These profiles are from three separate experiments repeated at least three times. The positions of the maxima corresponding to the void volume and to theoretical entities containing four TGA2 and one DNA probes as well as two TGA2 and one DNA probe are indicated.
(C) and (D) Immunoblot analysis of pooled protein fractions from the experiments shown in the chromatogram in (B), using an anti-His antibody. The data are from the high (C) and low (D) TGA2 concentrations. Void indicates fractions collected from the void volume, while Tetramer and Dimer represent pooled fractions from the predicted elution profile of a theoretical TGA2 tetramer bound to two DNA molecules and a TGA2 dimer bound to one DNA molecule. V0-T indicates pooled samples corresponding to fractions located between the void volume and tetramer.
Figure 4.
Binding of the TGA2 Oligomer to DNA Requires the N-Terminal Domain of TGA2.
(A) to (C) EMSA using recombinant Δ43 ([A] and [B]) and TGA2 (C) together with the LS7 DNA probe (A) or the PR-1 probe ([B] and [C]). The numbers indicate the ratio of probe concentration to TGA2 concentration. FP stands for free probe and refers to an experiment in which only DNA is present. In (C), the black, gray, and white arrows indicate the position of three distinct complexes.
(D) Fluorescence polarization experiments using recombinant TGA2 together with the LS5 (jagged line) or LS7 (solid line, which appears under the jagged line) DNA as the probe. Values are reported as millianisotropy units.
(E) qPCR analyses performed with DNA from wild-type plants, tga2/5/6 mutants, and tga2/5/6 mutant transfected with the Δ43 variant of TGA2 treated (gray bars) or not (white bars) with SA. Following sonication, the cross-linked chromatin was separated by gel filtration on S300. The void volume was collected, the cross-linking reversed, and qPCR was performed using PR1 and ubiquitin (UBQ) primer pairs. Data were reported as the ratio of PR-1 over UBQ and represent averages ± 1 sd. Every bar consists of three technical replicates on two biological replicates (n = 6).
The Oligomeric Species of the TGA2 Repressor Binds Its Cognate Sequence in PR-1
To establish the possible biological significance of the higher-order complex form of TGA2, we tested whether the oligomer could bind to cognate DNA elements. To do so, EMSA experiments were performed using the SA response element LS7 (Lebel et al., 1998), which contains a single TGA binding sequence. The EMSA depicted in Figure 3A shows that, at low ratios of protein to probe, the TGA2-DNA complex formed a single retarded band (solid black arrow), while at higher protein-to-probe ratios, two slower mobility bands (solid gray and open arrow) could also be observed. The very slow migrating band (open arrow) likely represents a high-order complex binding to DNA.
As an alternate means to confirm that TGA2 oligomers could bind DNA, we performed gel filtration experiments in which TGA2 was incubated with a fluorescein-derived LS7 probe, and the elution profile was monitored by fluorimetry. In Figure 3B, the dashed line represents the elution profile of the probe alone (free DNA), while the solid black line depicts the profile of the DNA probe incubated with a high concentration of TGA2 (30 nmol in 2 mL). One peak corresponds to the void volume and contains protein-DNA complexes of undetermined stoichiometry, while the other peak, based on the standard curve, corresponds to a complex of four TGA2 and one DNA probe molecule. These results confirm that the TGA2 high-order complex binds to DNA and that a TGA2 tetramer can also bind to DNA. An immunoblot confirmed that the elution fractions displaying fluorescence also contained TGA2 (Figure 3C). TGA2 was also observed in fractions corresponding to the dimer; however, this may have been the trailing of the tetramer peak. To determine whether TGA2 could bind DNA as a dimer, the probe was incubated with a low concentration of TGA2 (10 nmol in 2 mL). This gave rise to the elution profile represented by a jagged line, which suggests that a TGA2 dimer can interact with one DNA probe. Figure 3D confirmed the presence of TGA2 in the fluorescence-containing fraction. Overall, the data presented in Figure 3 suggest that dimers, tetramers, and oligomers of TGA2 can bind cognate DNA.
The N-Terminal Repression Domain of TGA2 Is Mandatory for Binding of the TGA2 Oligomer to DNA in Vitro and in Vivo
Given that the N terminus of TGA2 did not manifest itself in a discernible way on gel filtration (profiles from TGA2 or Δ43 are identical), we performed EMSAs using the SA response element LS7 (Lebel et al., 1998) to determine whether the repression domain would affect the DNA binding behavior of TGA2. The EMSA performed with the Δ43 variant of TGA2 (Δ43TGA2) shows that only a single retarded band was observed regardless of the protein-to-probe ratio (Figure 4A). This band migrates slightly faster than the fastest migrating band in the TGA2 EMSA, suggesting a similar stoichiometry (see Supplemental Figure 3 online).
To study binding in a more relevant context, we also used the LS5 to LS7 region of the PR-1 promoter as probe (PR-1) (Després et al., 2000), which contains two TGA binding elements and compared the binding of Δ43TGA2 (Figure 4B) to that of TGA2 (Figure 4C). As with the LS7 probe (Figure 4A), binding of the Δ43 variant of TGA2 to the PR-1 probe showed preferentially only a single retarded band, despite the presence of two potential binding sites. By contrast, when the EMSA was performed with TGA2 (Figure 4C), three bands could be observed and presented a pattern of migration similar to what was obtained with the LS7 probe (Figure 3A). We had previously demonstrated that TGA2 can bind to LS5 and LS7, separately, in EMSAs (Després et al., 2000). In Figure 4D, we performed fluorescence polarization experiments to compare the relative binding affinity of TGA2 toward LS5 (solid line) and LS7 (jagged line). The two curves being essentially identical points to the fact that, in vitro, TGA2 cannot discriminate between these two elements of the PR-1 promoter. The data of Figures 4A to 4D thus suggest that the N-terminal repression domain of TGA2 controls whether the high-order complex can bind to DNA in vitro.
We next set out to test whether oligomers form on DNA in vivo by combining chromatin cross-linking, gel filtration, and quantitative PCR (qPCR). The rationale was that, if an oligomer forms on the PR-1 promoter in vivo, we should be able to detect the presence of PR-1 by qPCR in the void fraction of an S300 after the chromatin had been cross-linked and sheared by sonication. Figure 4E presents such an experiment and indicates that indeed in wild-type plants, such an oligomer forms on the PR-1 promoter in the absence of SA (open bar), but not after a treatment with SA (gray bar). Next, to demonstrate that this oligomer depends on the presence of TGA2, we repeated the experiment in the tga2/5/6 mutant background (Zhang et al., 2003). Indeed, the data show that binding of the oligomer to PR-1 is TGA2 dependent. Finally, to test whether the N-terminal repression domain of TGA2 was required in vivo for the oligomer to bind DNA, as is the case in vitro, an experiment was performed in the tga2/5/6 mutant background transfected with the Δ43 variant of TGA2. The selected plant showed that the Δ43 variant of TGA2 complements the tga2/5/6 mutation with respect to SA-dependent PR-1 induction (see Supplemental Figure 4 online). These data demonstrate that the oligomer did not form on the DNA when the N terminus of TGA2 is lacking.
The BTB/POZ Domain of NPR1 Negates the Effects of the N-Terminal Repression Domain of TGA2
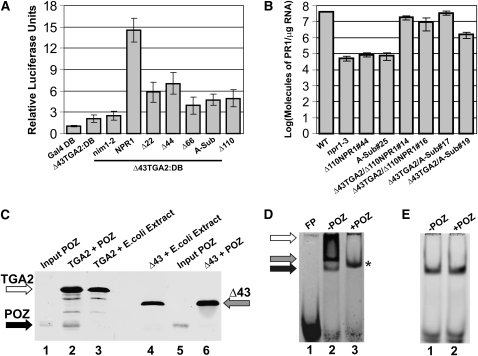
TGA2 interacts with NPR1 to form an enhanceosome with transcriptional activation properties requiring the BTB/POZ domain of NPR1 (Rochon et al., 2006). However, the molecular role of the NPR1 BTB/POZ domain within the context of the enhanceosome remains elusive. Given that TGA2 is a constitutive repressor (Rochon et al., 2006; Figure 1B), a hypothetical scenario is that the NPR1 BTB/POZ domain serves to mask or counteract the function of the TGA2 N-terminal repression domain. To test this hypothesis, we performed in vivo plant transcription assays and assessed the transactivation capacity of the TGA2-NPR1 enhanceosome using the Δ43 variant of TGA2 (Δ43TGA2:DB) that lacks the N-terminal repression domain, in combination with NPR1 variants mutated in the BTB/POZ domain (Figure 5A). The rationale for using these NPR1 mutants has been described in detail (Rochon et al., 2006). Briefly, deletion 22 removes a potential β-strand formed by residues 19 to 22 (FVAT). Deletion 44 removes a β-strand that corresponds to β1 in PLZF, which has been shown to partially destabilize the PLZF dimer (Ahmad et al., 1998). Deletion 66 removes all the structural determinants (β1, α1, and D65) mandatory for BTB/POZ domain homodimerization (Ahmad et al., 1998). Deletion 110 removes what is referred to as the core of the BTB/POZ domain right after β4, but before the next structural element. Finally, the Ala substitution mutant alters the core of the NPR1 BTB/POZ domain, which has been shown in PLZF to result in the disruption of the BTB/POZ fold (Melnick et al., 2000). The nim1-2 is an NPR1 mutant protein that does not interact with TGA2 (Després et al., 2000) and is shown as a negative control. Deleting the first 22, 44, or 66 amino acids, or removing the core of the BTB/POZ domain by deleting to amino acid 110 or by substituting it with Ala (A-sub) did not substantially affect the capacity of NPR1 to form an enhanceosome when complexed with Δ43TGA2:DB (after SA treatment). This is in contrast with the previous observation that the Δ110NPR1 and the A-subNPR1 did not form a transactivating complex with TGA2, despite their interaction (Rochon et al., 2006). The discrepancy between our previous results with TGA2 and the current ones employing Δ43TGA2 highlights the fact that the BTB/POZ domain of NPR1 is only required when the N-terminal repression domain of TGA2 is present in the NPR1-TGA2 enhanceosome.
Figure 5.
The BTB/POZ Domain of NPR1 Interacts with the N terminus of TGA2 and Precludes Binding of the TGA2 Oligomer to DNA.
(A) Bar graph illustrating the transactivation properties of Δ43TGA2 fused to the Gal4 DB in complex with NPR1 and five NPR1 BTB/POZ domain mutants not fused to any domain. Results obtained with Gal4 DB alone (Gal4 DB) and Δ43TGA2:DB alone are also shown. Δ22, Δ44, Δ66, and Δ110 indicate NPR1 variants in which the first 22, 44, 66, or 110 amino acids have been deleted, while A-Sub refers to an NPR1 variant in which the core of the BTB/POZ domain has been substituted with Ala residues (see Rochon et al., 2006 for an in-depth rationale of these mutations). nim1-2 is an NPR1 mutant that does not interact with TGA2. Conditions were identical to those described in Figure 1C. Gray bars indicate a treatment with SA. Data are reported as relative luciferase units. Values consist of n = 25 samples and represent averages ± 1 sd. Every bar represents five bombardments repeated five times (n = 25).
(B) Bar diagram illustrating the abundance of PR-1 transcript present in wild-type, npr1-3, line 44 of an npr1-3 mutant plant expressing a variant of NPR1 lacking the first 110 amino acids (Δ110NPR1#44), and line 25 of an npr1-3 mutant plant expressing a variant of NPR1 mutated by Ala substitutions in the BTB/POZ domain (A-Sub#25). Δ110NPR1#44 was used as parent to express a TGA2 variant lacking the first 43 amino acids (Δ43TGA2). Two independent lines were tested for PR-1 expression (Δ43TGA2/Δ110NPR1#14 and Δ43TGA2/Δ110NPR1#16). These lines express Δ43TGA2 and Δ110NPR1 in the npr1-3 background. Similarly, A-Sub#25 was used as parent to express a TGA2 variant lacking the first 43 amino acids (Δ43TGA2). Two independent lines were tested for PR-1 expression (Δ43TGA2/A-Sub#17 and Δ43TGA2/A-Sub#19). These lines express Δ43TGA2 and a variant of NPR1, mutated by Ala substitutions in the BTB/POZ domain, in the npr1-3 background. Data for each bar represent averages containing two biological replicates, each composed of six plants. Errors are equal to ± 1 se. Note that the scale is logarithmic.
(C) Variation of the pull-down assay in which the solid phase was produced by linking biotinylated LS7 DNA to paramagnetic beads followed by binding of TGA2 (lanes 2 and 3) or Δ43TGA2 (lanes 4 and 6) to the DNA. The NPR1 BTB/POZ domain (lanes 2 and 6) or an E. coli extract (lanes 3 and 4) was incubated with the solid phase. Lanes 1 and 5 contain 20% of the amount of BTB/POZ domain used in lanes 2 and 6. All other lanes contain eluents from the solid phase. Proteins (TGA2, Δ43TGA2, and POZ) were revealed by immunoblots with an anti-His antibody. The black, gray, and white arrows indicate the position of the BTB/POZ domain of NPR1, Δ43TGA2, and TGA2, respectively.
(D) EMSA using recombinant TGA2 (lanes 2 and 3) together with the LS7 DNA as the probe. FP stands for free probe and refers to an experiment in which only DNA was present. POZ indicates that the BTB/POZ domain of NPR1 had been added (+) or not (−) to the EMSA reaction. The black, gray, and white arrows indicate the position of three distinct complexes. An asterisk denotes the position of a TGA2-BTB/POZ complex.
(E) EMSA identical to that in (D) with the exception that Δ43TGA2 replaced TGA2.
These results were also confirmed in transgenic plants, where levels of PR-1 transcripts, after SA treatment, were analyzed by qPCR (Figure 5B). Results show that wild-type plants express higher levels of PR-1 than plants mutated at the npr1 locus (npr1-3) and plants of the same mutant background (npr1-3) expressing a variant of NPR1 lacking the first 110 amino acids (Δ110NPR1#44) or mutated in the core of the BTB/POZ domain (A-Sub#25). These plants, carrying a nonfunctional BTB/POZ domain, had previously been described and analyzed (Rochon et al., 2006). Δ110NPR1#44 and A-Sub#25 were used as parent plants for the introduction of the Δ43 variant of TGA2 (Δ43TGA2), and two independent transgenic lines from each genotype (Δ110NPR1#14 and 16 and A-Sub#17 and 19) were selected for qPCR analyses. Results indicate that activation of PR-1 is restored in these lines expressing NPR1 variants carrying a defective BTB/POZ domain when coexpressed with the Δ43TGA2. When these plants were expressing full-length TGA2, instead of Δ43TGA2, PR-1 activation was not restored (see Supplemental Figure 5 online). The findings of Figures 5A and 5B suggest that the core of the BTB/POZ domain of NPR1 is required for the TGA2 coactivator function of NPR1 only when the repression domain of TGA2 is present in the complex. The corollary is that the BTB/POZ domain of NPR1 negates the function of the N-terminal repression domain of TGA2.
The BTB/POZ Domain of NPR1 Interacts with the N-Terminal Repression Domain of TGA2 to Preclude the Oligomeric Form of TGA2 from Binding to DNA
Since the BTB/POZ domain of NPR1 negates the function of the N-terminal repression domain of TGA2, we hypothesized that these two domains could interact with each other. To test this potential interaction in the context of a cognate DNA element, we performed a variation on the pull-down assay in which TGA2 was first allowed to interact with cognate DNA (unlabeled version of LS7 used in the EMSAs of Figures 3 and 4) before addition of the BTB/POZ domain. Figure 5C demonstrates that the BTB/POZ domain of NPR1 could indeed be recruited by TGA2 bound to DNA (lane 2, black arrow). A pull down performed with an E. coli extract served as a negative control (Figure 5C, lane 3). The same experiment was then performed with Δ43TGA2. This time, the BTB/POZ domain could not be detected in the pull down, indicating that the N-terminal repression domain of TGA2 is essential for recruitment (Figure 5C, lane 6).
Given that Δ43TGA2 cannot bind DNA as an oligomer and that the BTB/POZ domain interacts with the N terminus of TGA2, we envisioned a scenario in which the BTB/POZ domain of NPR1 could possibly prevent the interaction of higher-order forms of TGA2 with DNA. An EMSA was thus performed using the LS7 probe and in which TGA2 was incubated in the presence or absence of the NPR1 BTB/POZ domain. Figure 5D shows that incubation of TGA2 with the probe yielded the fast- (black arrow), intermediate- (gray arrow), and slow-migrating (open arrow) protein species binding to DNA (lane 2). However, upon incubation with the BTB/POZ domain, the only form observed was a migrating species with an intermediate mobility between the fast- and intermediate-migrating bands (asterisk). These results demonstrate that the presence of the BTB/POZ domain prevents the oligomer from binding to DNA. The intermediate mobility of the BTB/POZ-TGA2-DNA complex can be interpreted as a supershift of the TGA2-DNA fast-migrating complex by the BTB/POZ domain, and as such, the BTB/POZ domain would also prevent binding of the intermediate-migrating complex to the probe. A similar experiment was performed with Δ43 (Figure 5E) and shows that the BTB/POZ domain had no effect on the binding of Δ43 to LS7 or the mobility of the Δ43-LS7 complex.
The TGA2-NPR1 Enhanceosome Has a Stoichiometry of Two TGA2 to Two NPR1 to One DNA
Knowing that BTB/POZ domains can homodimerize (Bardwell and Treisman, 1994; Ahmad et al., 1998; Melnick et al., 2000), we set out to test for NPR1 BTB/POZ domain dimerization (Figure 6A) as a first step in establishing the stoichiometry of the TGA2-NPR1 complex. To this end, we employed a plant two-hybrid assay system (Després et al., 2003; Rochon et al., 2006). In this system, reporter gene activation is based on the reconstitution of an active transcription factor. Transfection of the reporter gene and the internal standard along with Gal4 DB served to determine the baseline level of the system. Coexpressing the NPR1 BTB/POZ domain fused to the VP16 transactivation domain (POZ:TA) with Gal4 DB did not lead to expression beyond baseline. Similarly, expressing the domain fused to Gal4 DB (POZ:DB) along with the VP16 TA did not activate the reporter gene. However, cotransfecting POZ:DB and POZ:TA lead to a statistically significant (P < 0.05) increase in normalized luciferase activity, indicating that the NPR1 BTB/POZ domain can self-associate. Association was independent of whether cells were treated (gray bars) or not (white bars) with SA.
Figure 6.
Stoichiometry of the TGA2-NPR1-DNA Enhanceosome.
(A) Bar graph illustrating the in vivo self-association of the NPR1 BTB/POZ domain (POZ) domain. Self-association was monitored in transient plant two-hybrid system through the reconstitution of an active transcription factor, which activated a reporter gene. POZ was fused to either the Gal4 DB (POZ:DB) or VP16 TA (POZ:TA). Results obtained with Gal4 DB alone (Gal4 DB) and POZ:DB coexpressed with VP16 TA are also shown for comparison.
(B) Immunoblot analysis of pooled protein fractions from a Sephacryl S100 chromatogram using an anti-His antibody. Void indicates fractions collected from the void volume, while Dimer and Monomer represent pooled fraction from the predicted elution profile of a theoretical NPR1 BTB/POZ domain dimer and monomer, respectively. V0-D, D-M, and M-Vt indicate pooled samples corresponding to fractions located between the void volume and dimer, between the dimer and monomer, and between the monomer and one column volume, respectively. Crude refers to a crude E. coli extract expressing the NPR1 BTB/POZ domain.
(C) Cross-linking experiment of the NPR1 BTB/POZ domain followed by SDS-PAGE analysis indicating that the domain dimerizes. Extracts in lanes 1 and 2 were cross-linked for 5 and 30 min, respectively. XL indicates cross-linking. Predicted positions of the monomer and dimer, as determined from molecular weight standards, are indicated.
(D) Bar graph illustrating that the in vivo self-association of NPR1 is dependent on the presence of TGA2. Self-association was monitored in a transient plant two-hybrid or three-hybrid system through the reconstitution of an active transcription factor, which activated a reporter gene. NPR1 was fused to either Gal4 DB (NPR1:DB) or VP16 TA (NPR1:TA). Results obtained with Gal4 DB alone (Gal4 DB) and TGA2:DB are shown for comparison. TGA2:DB coexpressed with NPR1:TA and NPR1:DB coexpressed with TGA2:TA are presented to confirm that TGA2 can interact with both types of NPR1 fusion proteins. Black bars indicate experiments performed following SA treatment and in which TGA2 not fused to any domain was also coexpressed. For (A) and (D), conditions were identical to those described in Figure 1C. Gray bars indicate a treatment with SA. White bars indicate no treatment. Data are reported as relative luciferase units. Values consist of n = 25 samples and represent averages ± 1 sd. Every bar represents five bombardments repeated five times (n = 25).
(E) Chromatogram based on the elution profiles of the LS7 DNA probe derivatized with fluorescein. The profile of free DNA appears as a dashed line (the sample also contained NPR1, which does not interact with the DNA), while that of the DNA incubated with 5 μM of TGA2 (which binds as a dimer under these conditions; Figure 3B) is represented by a jagged line. The solid line corresponds to an elution profile in which the sample contained TGA2 (0.5 μM), NPR1 (1 μM), and DNA (0.5 μM). The black arrow corresponds to a theoretical entity containing two TGA2, one NPR1, and one DNA probe, while the gray arrow corresponds to one containing two TGA2, two NPR1, and one DNA probe. The inset is an immunoblot analysis using an anti-NPR1 antibody (Després et al., 2000). Void indicates fractions collected from the void volume. 2NPR1 and 1NPR1 correspond to fractions potentially containing an entity composed of two TGA2, two NPR1, and one DNA probe or two TGA2, one NPR1, and one DNA probe, respectively. V0-2NPR1 indicates pooled samples corresponding to fractions located between the void volume and 2NPR1.
(F) Immunoblot analysis of pooled protein fractions from an S300 elution profile of NPR1 alone using an anti-His antibody. Void indicates fractions collected from the void volume, while Dimer and Monomer represent pooled fractions from the predicted elution profile of a theoretical NPR1 dimer and monomer, respectively. V0-D and D-M indicate pooled samples corresponding to fractions located between the void volume and dimer and between the dimer and monomer, respectively.
The stoichiometry of the BTB/POZ domain self-association was analyzed by size-exclusion chromatography. To do so, soluble BTB/POZ domain fused to a C-terminal His-tag was expressed in E. coli and purified by affinity chromatography followed by Sephacryl S100 gel filtration. Since this domain could not be expressed to levels sufficiently high for monitoring by absorbance, an immunoblot analysis with an anti-His-tag antibody was used to detect the presence of the BTB/POZ domain and confirmed that the domain could self-associate to form a dimer (Figure 6B), based on the theoretical elution profile on an S100 (see Supplemental Figures 1 and 2 online). The dimerization of the domain was also confirmed by cross-linking experiments followed by SDS-PAGE (Figure 6C).
The capacity of the NPR1 BTB/POZ domain to dimerize would suggest that NPR1 itself could be a dimer under certain conditions. We tested this hypothesis using the plant two-hybrid system (Figure 6D), which monitors interaction inside the nucleus. Coexpressing NPR1 fused to the Gal4 DB (NPR1:DB) with NPR1 fused to the VP16 TA (NPR1:TA) did not suggest that NPR1 could self-associate, as values were not significantly different (P > 0.05) from expressing NPR1:DB alone and whether cells were treated (gray bars) or not (white bars) with SA. We then asked whether TGA2 would have an effect on NPR1 self-association by expressing TGA2, not fused to any domain, with the NPR1:DB-NPR1:TA couple. The results (NPR1:DB + NPR1:TA; black bars) show that, indeed, NPR1:DB can interact with NPR1:TA in the context or in the presence of TGA2, since the reporter gene was expressed to higher levels than when NPR1:DB was expressed alone with TGA2 (NPR1:DB; black bars) or when NPR1:DB was coexpressed with NPR1:TA without TGA2 (NPR1:DB + NPR1:TA; gray bars).
We then used gel filtration on Sephacryl S300 to evaluate the stoichiometry of the NPR1-TGA2 complex. The concentration of TGA2 (1 nmol in 2 mL) was set to allow only dimeric species to bind to DNA (Figure 6E; 2TGA2 + 1DNA). At these levels, the elution profile cannot be monitored by absorbance. Since NPR1 can self-associate in the presence of TGA2 (Figure 6D), the premise was that the complex would contain two NPR1 bound to a TGA2 dimer interacting with DNA and, therefore, an equimolar concentration (1 nmol in 2 mL) of NPR1 was required. Unfortunately, we could not produce such an amount of NPR1 in a soluble form. To circumvent this problem, we opted to express a variant of NPR1 mutated in the core of the BTB/POZ domain through Ala substitution (Rochon et al., 2006). Conceding that this is not a perfect solution, since the mutation is known to affect BTB/POZ domain dimerization (Melnick et al., 2000), this variant yielded a sufficiently high concentration of soluble protein to perform the stoichiometry experiment, and a twofold excess of NPR1 (2 nmol in 2 mL) over TGA2 was used. The chromatographic profile suggested that, indeed, the TGA2-NPR1-DNA complex consisted of two TGA2, two NPR1, and one probe. However, we could also detect a complex containing two TGA2, one NPR1, and one probe (see Supplemental Figures 1 and 2 online). The immunoblot (Figure 6E, inset) confirmed the presence of NPR1 in these fractions. However, NPR1 could still be found in the void volume and in fractions located between the void and the TGA2-NPR1-DNA complex, which suggests that NPR1 was only partially redistributed. When analyzed on its own, NPR1 elutes exclusively in the void volume (Figure 6F).
DISCUSSION
Although TGA2 had previously been recognized as a transcriptional repressor through both genetic (Zhang et al., 2003) and biochemical (Rochon et al., 2006) means, its mode of action remained unaddressed. Our study sheds some light on this mode of action by demonstrating that the N-terminal region of TGA2 (amino acids 1 to 43) contains a repression domain that does not act autonomously (Figure 1) but acts by imparting a certain structure on TGA2 (Figure 2). This structure allows TGA2 to bind its cognate DNA sequence as an oligomer of unknown stoichiometry (Figure 3). A repressing mode of action through the formation of a higher-order structure is not typical, since repression domains, just like activation domains, are normally transportable to another protein (Hiratsu et al., 2003; Ikeda and Ohme-Takagi, 2009). Fusions to the Gal4 DB of N-terminal clones of TGA2 of progressive length (Nt47, Nt101, and Nt144) did not repress transcription and therefore seemed to indicate that the repression domain of TGA2 may not portable. To add to the complexity governing the behavior of TGA2, our data show that despite the fact that the N-terminal repression domain of TGA2 precluded TGA2 from interacting with the DNA as an oligomer (Figure 4), it did not seem to play a role in the stoichiometry of the factor in the absence of DNA (deletions up to the first 68 amino acids; Figure 2). This suggests that, in vivo, a protein such as Δ43TGA2 could exist as an oligomer away from the DNA, despite the fact that it would bind DNA strictly as a dimer. This oligomer could potentially play an unidentified role, such as that of a reservoir for dimers to bind to DNA. Obviously, a protein such as Δ43TGA2 may not necessarily exist in a wild-type plant. However, it is conceivable that the masking of the N-terminal repression domain of TGA2 by another protein could lead to a complex behaving just like Δ43TGA2.
Such a protein may indeed exist given that we have shown that the NPR1 BTB/POZ domain interacts with the N terminus of TGA2. Furthermore, since a TGA2-NPR1 enhanceosome, in which the N terminus of TGA2 has been deleted, no longer requires the BTB/POZ domain of NPR1 for its transactivation function (Figure 5), our data support the idea that the BTB/POZ domain, not only interacts with, but indeed masks or negates the function of the TGA2 repression domain. As a result of this masking, the BTB/POZ domain precludes oligomeric TGA2 from binding to its cognate DNA sequence (Figure 5). Whether NPR1 can interact directly with TGA2 oligomers away from the DNA and whether this potential complex plays a biological role remain open questions that are difficult to answer.
Chromatographic separation of cross-linked chromatin suggests that the repressive form of TGA2 might be a higher-order form that exists only prior to SA treatment, while the activating species, that is to say, the one present in the TGA2-NPR1 enhanceosome, may be a low-stoichiometric form. Thus, after SA stimulation, which allows TGA2 to recruit NPR1, the BTB/POZ domain of NPR1 would either act as a crowbar chaperone to disassemble TGA2 oligomers or it would interact with TGA2 dimers and evict oligomers from the DNA. The path leading from the repressing form (TGA2 oligomer) to the activating form (low stoichiometry TGA2-NPR1 complex) of TGA2 is unclear. However, regardless of the mechanism, an enhanceosome would result (Rochon et al., 2006). The presence of a TGA2-dependent oligomer and its disassembly or eviction from PR-1 after SA treatment is supported by in vivo data (Figure 4E).
Having demonstrated that NPR1 can self-associate in the presence of TGA2 and that the BTB/POZ domain can dimerize (Figure 6), one can expect the enhanceosome to have a stoichiometry of two TGA2:2 NPR1. This stoichiometry is supported by gel filtration experiments (Figure 6). Although our data also indicate that a stoichiometry of 2 TGA2:1 NPR1 is also possible, this result is plausible but less convincing since it was obtained with a mutated version of NPR1 compromised in the core of the BTB/POZ domain and, therefore, likely to affect dimerization. This mutation, although necessary to obtain a sufficient amount of soluble protein to perform the gel filtration experiment, may have resulted in a less stable TGA2-NPR1 complex.
In EMSA, the leucine zipper of TGA factors is necessary and sufficient for dimerization, but a DS (dimer stabilization) domain, located C-terminal of the leucine zipper, cooperates with it to stabilize the TGA dimer (Katagiri et al., 1992). This has led several groups to conclude, without verification, that whenever the leucine zipper of a TGA factor is deleted or mutated, dimerization is abolished. However, whether the DS domain can function as an autonomous dimerization interface has not been addressed. Deletion of the first 93 residues in TGA2 (Δ93 constructs in Figure 2) reveals that this construct can dimerize, establishing that the leucine zipper is not required for self-association and suggesting that the DS domain is the primary entity responsible for the dimerization of TGA factors. This result, therefore, opens up the possibility that the leucine zipper may perform tasks other than the well-established dimerization interface. Indeed, progressive deletion of the N terminus of TGA2 (Figure 2) revealed that TGA2 stopped oligomerizing once the leucine zipper was deleted (deletion of the first 93 amino acids), indicating the importance of this domain in higher-order complex formation. Oligomerization through the leucine zipper is well documented in proteins such as FOXP3 and Translin, where it has been shown that mutations in the domain abolish oligomer formation (Aoki et al., 1999; Li et al., 2007). In the case of Translin, the protein forms a ring of 8 to 10 protomers depending on the species of origin (Kasai et al., 1997; Gupta et al., 2008). FOXP3 elutes from gel filtration as a monomer, dimer, tetramer, and oligomer. Similarly, and depending on protein concentration, full-length TGA2 can elute as a dimer, tetramer, or oligomer, but smaller species seem to only exist in the presence of cognate DNA (Figure 2A versus 3B).
Concentration-dependent regulation of gene expression by transcription factors has been known for over two decades. A well-documented case is that of the Krüppel zinc-finger protein, which can act as both an activator and repressor on the same DNA element within a promoter depending on protein stoichiometry (Sauer and Jackle, 1991). At low concentration, Krüppel binds DNA as a monomer and activates transcription, but at higher concentration, it forms a homodimer and acts as a repressor (Sauer and Jackle, 1993). This is reminiscent of TGA2, where the full-length protein, which is capable of binding DNA as an oligomer, represses transcription, while variants lacking this capacity activate transcription. As is the case with Krüppel, TGA2 concentration also dictates which species forms on the DNA. However, even at high concentrations, where oligomers binding to DNA are present, the BTB/POZ domain of NPR1 is capable of either excluding oligomers from forming on cognate DNA or disassembling them altogether (Figure 5C). Thus, the NPR1 BTB/POZ domain's ability to specifically direct the recruitment of only those TGA2 species competent for gene activation presents a novel means of derepression since this motif typically serves to recruit corepressors (Kelly and Daniel, 2006). Furthermore, the NPR1 BTB/POZ domain is also atypical in that it not only functions to dismiss or occlude oligomeric TGA2 species from the DNA, but it accomplishes this feat directly rather than through the recruitment of any cofactors. The NPR1 protein appears to marry the duties of derepressor, contributed by the N-terminal BTB/POZ motif, and coactivator, by way of the C-terminal transactivation domain, reinforcing its role as a key regulator of PR-1 gene induction.
The data presented here and summarized in Figure 7 constitute a significant advancement to the understanding of the mechanisms by which domains of the global regulator NPR1 control PR-1 gene activation in Arabidopsis and contribute to new insights into the function of BTB/POZ domains across kingdoms.
Figure 7.
Working Model for the Regulation of TGA2 by NPR1 and Stoichiometry of the TGA2-NPR1 Enhanceosome.
Left panel: In untreated cells, where NPR1 does not interact with TGA2, TGA2 would form an oligomer capable of binding to its cognate TGACG sequence in the promoter of target genes. This oligomer would repress transcription by a mechanism yet to be identified. Oligomerization of TGA2 on DNA involves the leucine zipper and the N-terminal repression domain. Right panel: After a rise in SA, the NPR1 BTB/POZ domain (POZ) would either assist in disassembling the TGA2 oligomer or assist in recruiting TGA2 dimers to cognate DNA, while excluding TGA2 tetramers and oligomers from binding DNA. The TGA2-NPR1 enhanceosome is likely to have a stoichiometry of 2:2 (TGA2:NPR1). The BTB/POZ domain of NPR1 dimerizes and interacts with the N-terminal repression domain of TGA2 (gray) to mask its capacity to form an oligomer on DNA. The ankyrin repeats (ANK) are the major interfaces stabilizing the TGA2-NPR1 complex, while the C-terminal region of NPR1 contains the transactivation domain (TA) of the enhanceosome.
METHODS
Plant Transcription Assays
All procedures for the plant two-hybrid assays were previously described (Després et al., 2003). All constructs were created by PCR as previously described (Després et al., 2003; Rochon et al., 2006). Arabidopsis thaliana (Columbia) leaves were harvested from 4-week-old plants grown at 21°C (day) and 18°C (night) with a 10-h photoperiod and transferred to Petri dishes containing Murashige and Skoog salts and micronutrients supplemented with B5 vitamins, 1% sucrose, and 0.8% agar at a pH of 5.8. When required, filter-sterilized SA was added to the medium at a final concentration of 1 mM. Coating of the gold particles and general procedures and preparation of the biolistic experiments were as per the manufacturer's instructions (Bio-Rad). After bombardment, leaves were kept in the conditions described above for a period of 24 h before assaying. Assays were performed using the Dual-Luciferase Reporter Assay system (Promega) following the manufacturer's instructions. Luminescence was measured on a Berthold Lumat LB9507 luminometer (Bad Wildbad), and the data obtained represent the value of the reporter gene divided by the value of the internal standard and expressed as relative luciferase units. One microgram of each effector plasmid, 1 μg of the firefly luciferase reporter plasmid, and 0.1 μg of the renilla internal standard plasmid were mixed together and the mixture used to coat beads. This amount of DNA was used to perform five bombardments. Every bar in each graph represents five bombardments repeated five times (n = 25). Arabidopsis ecotype Columbia was used throughout this study.
Chromatography
His-tagged purified proteins were diluted to the required concentrations in a final volume of 2 mL using S300 running buffer (50 mM HEPES, pH 7.4, and 250 mM NaCl) prior to gel filtration analysis on the Sephacryl S100 HR or Sephacryl S300 HR packed in 50-cm-long HR 16 columns (GE Health) and equilibrated with S300 running buffer. Elutions, in 0.5 mL fractions, were performed in the same buffer at a flow rate of 0.8 mL/min. Where indicated, proteins were incubated with DNA probes at room temperature in the dark for 20 min prior to chromatography as described above.
EMSAs
Probes were labeled on the 5′-end of each strand with IRDye-700nm (LI-COR). The probes used were the LS7 probe (5′-TATTTTACTTACGTCATAGATGTGGCGGCA-3′ annealed to 5′-TGCCGCCACATCTATGACGTAAGTAAAATA-3′) or the PR-1 probe (5′-GTTTCTCTACGTCACTATTTTACTTACGTCATAGATGTGG-3′ annealed to 5′-CCACATCTATGACGTAAGTAAAATAGTGACGTAGAGAAAC-3′). Binding reactions were performed in the dark at room temperature for 20 min in 50 μL of EMSA buffer (20 mM HEPES-KOH, pH 7.9, 250 mM NaCl, 2 mM DTT, 20% [v/v] glycerol, and 0.5% [v/v] Tween 20) with 100 fmol of probe prior to loading onto 4% polyacrylamide gels (29.2:0.8 acrylamide-bisacrylamide in 100 mM Tris, 100 mM borate, and 10 mM EDTA) and running at 8 V/cm for 70 min. Gels were then scanned on the Odyssey infrared scanner (LI-COR).
Fluorescence Anisotropy
The probes employed were the LS7 probe (5′-TATTTTACTTACGTCATAGATGTGGCGGCA-3′ annealed to 5′-FAM-TGCCGCCACATCTATGACGTAAGTAAAATA-3′) and LS5 probe (5′-FAM-TGACTGTTTCTCTACGTCACTATTTTACTT-3′ annealed to 5′-AAGTAAAATAGTGACGTAGAGAAACAGTCA-3′). DNA binding reactions were performed in the dark at room temperature for 4 h in 100 μL of S300 running buffer containing 0.1 nM probe prior to measurements to allow the reactions to come to equilibrium. The incubation and analysis were conducted in Costar (Corning) 96-well black nontreated round bottom polypropylene plates. Anisotropy determinations were made with the POLARstar Optima (BMG Labtechnologies), and each reaction was analyzed in quintuplicate.
DNA-Dependent Pull-Down Assays
The biotinylated LS5-LS7 probe (5′-GTTTCTCTACGTCACTATTTTACTTACGTCATAGATGTGG-3′-Biotin annealed to 5′-CCACATCTATGACGTAAGTAAAATAGTGACGTAGAGAAAC-3′) was coupled to Streptavidin iron oxide particles according to the manufacturer's instructions (Sigma-Aldrich). His-tagged purified factor TGA2 or Δ43TGA2 (500 pmol), in 400 μL of binding buffer (200 mM NaCl, 1 mM EDTA, and 20 mM HEPES, pH 7.9), was incubated with the beads for 30 min at room temperature, with continuous inversion. The beads were washed twice with 500 μL of binding buffer and once with binding buffer containing 1% milk. Crude Escherichia coli–produced NPR1 BTB/POZ or a crude E. coli extract in 500 μL of binding buffer, containing 1% (w/v) milk, was added to the beads containing the DNA-TGA2 or DNA-Δ43TGA2 complex and was incubated for 30 min. After this incubation, the beads were washed twice without milk as described above. Proteins were eluted by boiling in 45 μL of SDS-PAGE sample buffer and subjected to immunoblot analysis.
qPCR
Total RNA was extracted from leaves using the RNeasy plant mini kit (Qiagen) according to the supplier's instructions. After treatment with DNase I (Invitrogen), first-strand cDNA synthesis was generated using SuperScript II reverse transcriptase (Invitrogen) and the (dT)17VN oligo in the presence of 0.4 units of RNasin (Fisher Scientific). The newly synthesized cDNA was diluted 1/200 to reflect a concentration of 10 ng μL−1 input total RNA.
qPCR was performed on an MX3000 spectrofluorometric thermal cycler (Stratagene) using a two-temperature cycling regime initiated with a 15-min activation at 95°C, followed by 40 cycles of 2 min of annealing and extension at 66°C and 10 s denaturation at 95°C. Each assay contained 5 ng cDNA, 1× SYBR Green (Quantitech; Qiagen), and 0.5 pmol oligonucleotides (PR1F 5′-GCTCTTGTAGGTGCTCTTGTTCTTCC-3′ and PR1R 5′-AGTCTGCAGTTGCCTCTTAGTTGTTC-3′), prepared as described by Rutledge and Stewart (2008). The fluorescence data collected at the end of each PCR cycle were analyzed by the absolute quantification via Ct method (Rutledge and Stewart, 2008).
Cross-Linked Chromatin Chromatography
Plant treatment, cross-linking, sonication, and cross-linking reversal were performed as we do for chromatin immunoprecipitation (Rochon et al., 2006). Chromatography was as described above. qPCR was performed as we described previously (González -Lamothe et al., 2008), with the exception that PR-1 primers (5′-CGCCACATCTATGACG-3′ and 5′-GATCGGTCACCTAGAGT-3′) and Ubiquitin 5 primers (5′-GACGCTTCATCTCGTCC-3′ and 5′-GTAAACGTAGGTGAGTCCA-3′) were used.
Statistical Methods
All pooled data are expressed as averages, and error bars represent ± 1 sd or se, as specified in the figure legends. When data from two independent populations were compared, statistical significance was assessed using a two-tailed Student's t test.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT1G64280 (NPR1), AT2G14610 (PR1), AT5G06950 (TGA2), and AT3G62250 (Ubiquitin 5).
Supplemental Data
The following materials are available on the online version of this article.
Supplemental Figure 1. Operational Parameters of the S300 and S100 Gel Filtration Columns.
Supplemental Figure 2. Predicted and Observed Elution Volumes Relating to Data from Figures 2D, 3B, 6B, and 6E.
Supplemental Figure 3. Blow-Ups of Selected EMSA Lanes.
Supplemental Figure 4. Quantitation of PR-1 mRNA in the Wild Type and tga2/5/6 Mutant.
Supplemental Figure 5. Expression of TGA2 in the Δ110 and A-Sub Backgrounds Does Not Lead to PR-1 Activation after SA Treatment.
Supplementary Material
Acknowledgments
We thank the DNA Technology Unit of the Plant Biotechnology Institute for oligonucleotide synthesis and DNA sequencing. This research was supported by the Canada Foundation for Innovation (C.D.), the Ontario Innovation Trust (C.D.), the National Research Council Plant Biotechnology Institute core funding (P.R.F.), the National Science and Engineering Research Council (NSERC) discovery grant program (C.D.), and the NSERC graduate scholarship program (P.B. and A.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pierre R. Fobert (pierre.fobert@nrc-cnrc.gc.ca).
Online version contains Web-only data.
References
- Ahmad, K.F., Engel, C.K., and Prive, G.G. (1998). Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Suzuki, K., Ishida, R., and Kasai, M. (1999). The DNA-binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 443 363–366. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285 1353–1361. [DOI] [PubMed] [Google Scholar]
- Bardwell, V.J., and Treisman, R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 8 1664–1677. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Collins, T., Stone, J.R., and Williams, A.J. (2001). All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltour, S., Guerardel, C., and Leprince, D. (1999). Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: The case of HIC-1 and gammaFBP-B. Proc. Natl. Acad. Sci. USA 96 14831–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. [DOI] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe, R., Boyle, P., Dulude, A., Roy, V., Lezin-Doumbou, C., Kaur, G.S., Bouarab, K., Després, C., and Brisson, N. (2008). The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene. Plant Cell 20 3136–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, G.D., Makde, R.D., Rao, B.J., and Kumar, V. (2008). Crystal structures of Drosophila mutant translin and characterization of translin variants reveal the structural plasticity of translin proteins. FEBS J. 275 4235–4249. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50 970–975. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, M., Matsuzaki, T., Katayanagi, K., Omori, A., Maziarz, R.T., Strominger, J.L., Aoki, K., and Suzuki, K. (1997). The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J. Biol. Chem. 272 11402–11407. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., Seipel, K., and Chua, N.H. (1992). Identification of a novel dimer stabilization region in a plant bZIP transcription activator. Mol. Cell. Biol. 12 4809–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, K.F., and Daniel, J.M. (2006). POZ for effect – POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 16 578–587. [DOI] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16 223–233. [DOI] [PubMed] [Google Scholar]
- Li, B., et al. (2007). FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int. Immunol. 19 825–835. [DOI] [PubMed] [Google Scholar]
- Li, J., Mahajan, A., and Tsai, M.D. (2006). Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 45 15168–15178. [DOI] [PubMed] [Google Scholar]
- Li, X., Peng, H., Schultz, D.C., Lopez-Guisa, J.M., Rauscher III, F.J., and Marmorstein, R. (1999. a). Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 59 5275–5282. [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999. b). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329–339. [DOI] [PubMed] [Google Scholar]
- Melnick, A., Ahmad, K.F., Arai, S., Polinger, A., Ball, H., Borden, K.L., Carlile, G.W., Prive, G.G., and Licht, J.D. (2000). In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20 6550–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944. [DOI] [PubMed] [Google Scholar]
- Neuhaus, G., Neuhaus-Url, G., Katagiri, F., Seipel, K., and Chua, N.H. (1994). Tissue-specific expression of as-1 in transgenic tobacco. Plant Cell 6 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M., and Van Loon, L.C. (2004). NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7 456–464. [DOI] [PubMed] [Google Scholar]
- Rochon, A., Boyle, P., Wignes, T., Fobert, P.R., and Despres, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge, R.G., and Stewart, D. (2008). A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 8 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, F., and Jackle, H. (1991). Concentration-dependent transcriptional activation or repression by Kruppel from a single binding site. Nature 353 563–566. [DOI] [PubMed] [Google Scholar]
- Sauer, F., and Jackle, H. (1993). Dimerization and the control of transcription by Kruppel. Nature 364 454–457. [DOI] [PubMed] [Google Scholar]
- Stogios, P.J., Downs, G.S., Jauhal, J.J., Nandra, S.K., and Prive, G.G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6 R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19 769–772. [DOI] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Metraux, J.P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, R.R., Pfitzner, U.M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.