Abstract
Nitrogen (N) and nitrate (NO3−) per se regulate many aspects of plant metabolism, growth, and development. N/NO3− also suppresses parts of secondary metabolism, including anthocyanin synthesis. Molecular components for this repression are unknown. We report that three N/NO3−-induced members of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) gene family of transcription factors (LBD37, LBD38, and LBD39) act as negative regulators of anthocyanin biosynthesis in Arabidopsis thaliana. Overexpression of each of the three genes in the absence of N/NO3− strongly suppresses the key regulators of anthocyanin synthesis PAP1 and PAP2, genes in the anthocyanin-specific part of flavonoid synthesis, as well as cyanidin- but not quercetin- or kaempferol-glycoside production. Conversely, lbd37, lbd38, or lbd39 mutants accumulate anthocyanins when grown in N/NO3−-sufficient conditions and show constitutive expression of anthocyanin biosynthetic genes. The LBD genes also repress many other known N-responsive genes, including key genes required for NO3− uptake and assimilation, resulting in altered NO3− content, nitrate reductase activity/activation, protein, amino acid, and starch levels, and N-related growth phenotypes. The results identify LBD37 and its two close homologs as novel repressors of anthocyanin biosynthesis and N availability signals in general. They also show that, besides being developmental regulators, LBD genes fulfill roles in metabolic regulation.
INTRODUCTION
Nitrogen (N) and nitrate (NO3−) itself regulate many aspects of plant metabolism, growth, and development. These include NO3− uptake and assimilation (Crawford, 1995; Lejay et al., 1999), starch and organic acid metabolism (Scheible et al., 1997a), secondary metabolism (e.g., anthocyanin production) (Scheible et al., 2004; Fritz et al., 2006), germination (Alboresi et al., 2005), root architecture (Zhang et al., 1999), root and shoot development (Scheible et al., 1997b), leaf expansion (Walch-Liu et al., 2000), stomatal opening (Guo et al., 2003), flowering (Bernier et al., 1993), seed set, and senescence (Crawford, 1995; Stitt, 1999; Crawford and Forde, 2002). Some molecular components involved in regulation of these N/NO3− responses in Arabidopsis thaliana have been described. The MADS box transcription factor (TF) ARABIDOPSIS NITRATE REGULATED1 (ANR1) was found to act as regulator of systemic NO3− repression and localized NO3− stimulation of lateral root growth (Zhang and Forde, 1998). NITRATE TRANSPORTER1.1 (NRT1.1) was subsequently shown to act upstream of ANR1 in the signaling pathway triggering lateral root growth and root colonization of NO3−-rich patches (Remans et al., 2006) and to regulate expression of the high-affinity NO3− transporter NRT2.1 (Muños et al., 2004). Also, NRT2.1 is suspected to repress lateral root initiation in response to nutritional cues by acting either as a NO3− sensor or signal transducer (Little et al., 2005). Recently, a CBL-interacting protein kinase, CIPK8, was shown to regulate parts of the primary NO3− response, including NO3− transporter and assimilation genes (Hu et al., 2009). Another recent report pinpoints the TF NIN-LIKE PROTEIN7 (NLP7) as an important element of NO3− signal transduction and regulator for N assimilation (Castaings et al., 2009). Furthermore, two bZIP TFs, ELONGATED HYPOCOTYL5 (HY5) and HY5-HOMOLOG (HYH), were suggested to be positive regulators of NITRATE REDUCTASE2 (NIA2) and negative regulators of NRT1.1 in Arabidopsis (Jonassen et al., 2009). Moreover, NRT1.1 was shown to regulate expression of genes involved in NO3− assimilation, energy metabolism, and the pentose-phosphate pathway, further supporting the view that NRT1.1 acts as a NO3− sensor in Arabidopsis (Wang et al., 2009). Despite these advances, a lot remains to be discovered about N/NO3− signal transduction.
Flavonoids are major secondary metabolites and widely distributed in plants. They give color to flowers and fruits (Winkel-Shirley, 2001) and are important antioxidants (Gould et al., 2002). Their antioxidant properties have nutritional value for humans and help to protect plants from oxidative damage (Nagata et al., 2003), allowing nutrient recovery in aging and senescing leaves (Hoch et al., 2003). Synthesis of flavonoids and especially anthocyanins, which are responsible for purple coloration of leaves, is stimulated by abiotic and biotic stresses (Dixon and Paiva, 1995), including cold, high irradiance, excess sugar (Tsukaya et al., 1991), or limitation of inorganic macronutrients like phosphorous (P) and N (Scheible et al., 2004; Morcuende et al., 2007). There is also clear evidence that NO3− per se regulates secondary metabolic pathways, including parts of phenylpropanoid, flavonoid, and anthocyanin metabolism. NO3− addition to N-depleted Arabidopsis seedlings leads to a rapid repression of a set of Phe and flavonoid biosynthetic genes and decrease of Phe (Scheible et al., 2004), which are among the first recorded responses in the seedlings. These changes occur much earlier than any increase in organic N status, indicating that NO3− reduction is not required for these changes. Moreover, tobacco (Nicotiana tabacum) genotypes with low nitrate reductase (NR) activity accumulate NO3− but have a low organic N status comparable to the one of an N-limited wild type (Scheible et al., 1997a, 1997b, 1997c); however, unlike N-limited wild type, they display suppression of parts of secondary metabolism (Fritz et al., 2006).
In Arabidopsis, a number of MYB- and bHLH-type TFs are known that regulate sectors of flavonoid biosynthesis (Nesi et al., 2000, 2001; Mehrtens et al., 2005; Stracke et al., 2007; Dubos et al., 2008, Matsui et al., 2008). The Arabidopsis MYB TF PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1/MYB75) is a key regulator of flavonoid/anthocyanin synthesis genes, including those encoding chalcone synthase (CHS), dihydroflavonol 4-reductase (DFR), and glutathione-S-transferase (GST) (Borevitz et al., 2000; Tohge et al., 2005; Wangwattana et al., 2008). The transcript levels of PAP1 and its close homolog PAP2/MYB90 are strongly induced during N or P limitation (Scheible et al., 2004; Morcuende et al., 2007) and quickly repressed after NO3− addition to nitrogen-depleted Arabidopsis seedlings (Scheible et al., 2004). The E3 ligase NITROGEN LIMITATION ADAPTATION (NLA) is an important player in a potential signaling pathway that leads to induction of anthocyanin biosynthesis during N limitation (Peng et al., 2007a, 2008). When grown in N-limiting conditions, nla mutants fail to induce anthocyanin biosynthetic genes (Peng et al., 2007b) and hence do not produce anthocyanins, but display early senescence. Interestingly, nla mutants produce normal levels of anthocyanins during P limitation or combined P and N limitation, indicating that an NLA-independent P-specific pathway exists. Players in this latter pathway might include the TFs PHOSPHATE STARVATION RESPONSE1 (Nilsson et al., 2007) and bHLH32 (Chen et al., 2007) and an interacting F-box protein (Chen et al., 2008). PAP1 and PAP2 were also recently described as being targets of trans-acting-small interfering RNAs derived from the TAS4 locus (Addo-Quaye et al., 2008). Whether and how the action of trans-acting-small interfering RNAs relates to environmental stresses is unclear. Also, the signaling pathway underlying the N/NO3− repression is unknown.
With the advent of ATH1 GeneChips and high-throughput, quantitative real-time PCR (qRT-PCR), it became possible to obtain a near genome-wide catalog of potentially regulatory genes with N/NO3−-dependent transcript abundances (Wang et al., 2003; Scheible et al., 2004), making them prime candidates for N regulators. Among these are three NO3−-responsive members of the plant-specific ASYMMETRIC LEAVES2 (AS2)-LIKE (ASL)/LATERAL ORGAN BOUNDARY (LOB) DOMAIN (LBD) gene family (Scheible et al., 2004) that consists of 43 members in Arabidopsis (Shuai et al., 2002). ASL/LBD genes encode a new family of zinc-finger DNA binding TFs, and the founder gene LOB was shown to recognize a 6-bp GCGGCG consensus motif and to interact with a specific bHLH protein (Husbands et al., 2007). Phylogenetic analyses subdivide the family into two classes: class I, which comprises LOB and LBD1 through LBD36, and class II, consisting of LBD37 through LBD42. Several Arabidopsis class I members have been implicated in plant development; LBD6/AS2 functions in the specification of adaxial/abaxial organ polarity and negatively regulates expression of KNOX TFs in lateral organs (Ori et al., 2000; Semiarti et al., 2001; Byrne et al., 2002; Lin et al., 2003), LBD36 was shown to regulate proximal-distal patterning in Arabidopsis petals (Chalfun-Junior et al., 2005), LBD16 and LBD29 are required for auxin-dependent lateral root formation (Okushima et al., 2007), LBD18 and LBD30 were shown to regulate tracheary element differentiation in Arabidopsis (Soyano et al., 2008), and LBD30/JAGGED LATERAL ORGANS is essential for Arabidopsis embryo development (Borghi et al., 2007). Developmental functions have also been described for a few class I LBD genes in maize (Zea mays; Bortiri et al., 2006; Evans, 2007; Taramino et al., 2007). However, no biological function was so far reported for a class II LBD gene.
Here, we present a reverse genetic characterization of class II LBD37, LBD38, and LBD39 and show that LBD37 and the two homologs mediate the repressive effect of N/NO3− on anthocyanin biosynthesis and further affect N-responsive genes and N metabolism, thereby revealing a novel function for LBD proteins in the regulation of plant metabolism.
RESULTS
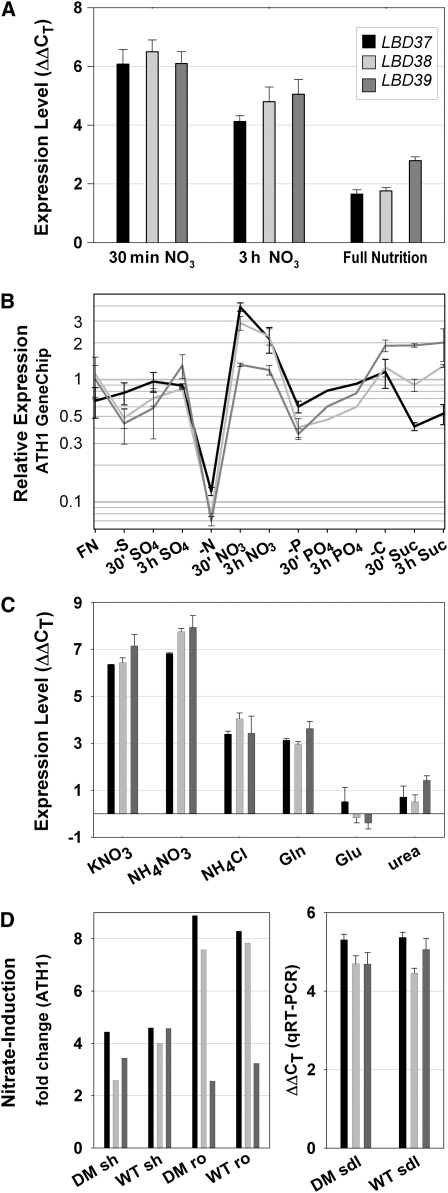
LBD37, 38, and 39 Transcripts Are Strongly Induced by Nitrate and More Moderately by Ammonium and Gln
LBD37, LBD38, and LBD39 are closely related and arose by two segmental duplications of the Arabidopsis genome (http://www.tigr.org/). Their transcripts increase rapidly (within minutes) and strongly (LBD37 and LBD38 >20-fold) after readdition of 3 mM KNO3, but not after readdition of 3 mM KCl, to N-depleted seedlings. The transcript levels also remain 4- to 10-fold higher in seedlings grown in full nutrient conditions compared with N-deprived seedlings (see Supplemental Figure 1 online; Scheible et al., 2004). Other LBD gene transcripts are unaffected by N status and NO3− readdition (see Supplemental Figure 1 online). Rapid and strong NO3− induction was confirmed by qRT-PCR (Figure 1A). The response of the three LBD transcripts was specific for N/NO3−, since limitation and subsequent readdition of sugar, phosphate, or sulfate did not lead to noteworthy transcript changes (Figure 1B; see Supplemental Figure 1 online). Other important N sources, such as ammonium (NH4+) and Gln, also induce the three genes, although to a weaker extent (Figure 1C); for example, induction of LBD37 30 min after readdition of 3 mM NH4Cl to N-limited seedlings was seven times weaker compared with 30 min readdition of NO3−. Induction by NH4NO3 was slightly higher than by KNO3 alone, indicating that the effects of NO3− and NH4+ are somewhat additive. Glu or urea had no effect on the three LBD transcripts (Figure 1C). The induction of the three LBD genes was also detectable after KNO3 readdition to NR-deficient Arabidopsis nia1 nia2 double mutants grown in NO3−-free medium (ATH1 data from Wang et al. [2004] and confirmed by qRT-PCR; Figure 1D), indicating that a signal derived from NO3− per se already triggers the increase of the three LBD transcripts. Sixfold to eightfold induction of the three LBD transcripts was also recorded after addition of micromolar concentrations of nitrite to Arabidopsis roots (Wang et al., 2007).
Figure 1.
Nutrient-Dependent Expression of LBD37, LBD38, and LBD39.
(A) Relative qRT-PCR expression levels after 30 min or 3 h NO3− readdition to N-limited wild-type seedlings or in N-replete relative to N-limited wild-type seedlings. Data are depicted as ΔΔCT; i.e., a logarithmic scale; e.g., ΔΔCT = 6 equals a 26- = 64-fold induction if primer efficiency is 100% (see Supplemental Table 1 online). Gray shading is defined in the key. Results represent mean values ± sd from three independent biological replicates with two technical replicates for each.
(B) Relative expression levels derived from ATH1 GeneChip data for various nutrient stress and nutrient readdition time courses (FN, full nutrition; −S, sulfur limitation; SO4, 3 mM potassium sulfate readdition [W.-R. Scheible and R. Morcuende, unpublished data]; −N, nitrogen limitation; NO3, 3 mM potassium nitrate readdition [Scheible et al., 2004]; −P, phosphorous limitation; PO4, 3 mM potassium phosphate readdition [Morcuende et al., 2007]; −C, carbon limitation; Suc, 15 mM sucrose readdition [Osuna et al., 2007]). The average expression level of each gene in all conditions was set to 1. Data represent mean values ± sd from two independent biological replicates. Gray shading is explained in (A).
(C) Effect of readdition of various N sources (KNO3, 3 mM potassium nitrate; NH4NO3, 1.5 mM ammonium nitrate; NH4Cl, 3 mM ammonium chloride; Gln, 1.5 mM l-Gln; Glu, 3 mM l-Glu; 1.5 mM urea) on LBD transcript levels. Readditions were done for 30 min on N-limited wild-type seedlings. Results represent mean values ± sd from three independent biological replicates with two technical replicates for each. Gray shading is as in (A).
(D) Nitrate induction of LBD37, LBD38, and LBD39 in nitrate reductase-deficient nia1 nia2 double mutants (DM). The left panel depicts public ATH1 GeneChip results from shoots (sh) and roots (ro) (Wang et al., 2004). Addition of 5 mM KNO3 for 2 h; averaged signal ratios between nitrate-treated and chloride-treated plants are shown as reported in the supplemental data sets from Wang et al. (2004). The right panel shows qRT-PCR results (given as ΔΔCT) from N-starved seedlings (sdl) that were resupplied with 3 mM KNO3 during 30 min prior to harvest. Gray shading is as in (A).
LBD37, LBD38, or LBD39 Overexpressing and Mutant Lines
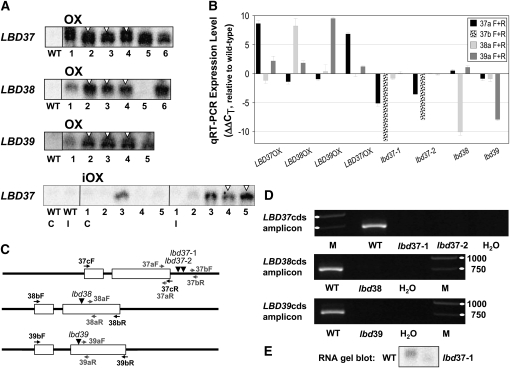
Constitutive and ethanol-inducible overexpression (OX and iOX, respectively) lines were produced and homozygous T-DNA insertion mutants isolated from the SALK and Gabi-Kat collections (Alonso et al., 2003; Rosso et al., 2003). Transgenic lines were characterized for LBD transcript levels by RNA gel blot and qRT-PCR (Figure 2). qRT-PCR analysis revealed that the induction of LBD in the N-limited overexpressing lines (Figure 2B) was comparable to the strong induction in N-limited wild type after NO3− addition (cf. Figures 1A and 1C). By contrast, the respective LBD transcript was strongly reduced (ΔΔCT = −6 to −10) in lbd37, lbd38, and lbd39 T-DNA insertion lines when primers downstream of the insertion site were used (Figures 2B and 2C). In the case of the lbd37 mutant alleles (lbd37-1 and lbd37-2), the insertion sites are both located in the 3′-untranslated region of the transcript (Figure 2C). Still, this led to (1) considerably reduced qRT-PCR amplification of an LBD37 PCR product upstream of the insertion sites (Figure 2B), (2) strongly reduced amplification of another LBD37 PCR product downstream of the insertion sites, (3) no detectable amplification of RT-PCR products spanning the entire LBD37 coding sequence (Figure 2D), and (4) an aberrant and reduced RNA gel blot signal for LBD37 (Figure 2E). We conclude that all analyzed lbd mutants are strong reduction-of-function or null alleles.
Figure 2.
Molecular Characterization of LBD Overexpressor and T-DNA Insertion Lines.
(A) RNA gel blot analysis of LBD expression in five to six constitutive overexpressor lines (OX) or inducible overexpresser lines (iOX). IOX lines were tested in N-limited, noninduced control conditions (C), and after 3 h of induction (I) with 0.2% (v/v) ethanol. Note the constitutive induction in LBD37iOX line 3. White triangles indicate lines that were chosen for subsequent experiments.
(B) Quantitative analysis of LBD expression by qRT-PCR in N-limited OX and iOX lines relative to N-limited wild type and in previously N-starved T-DNA insertion lines after 30 min resupply of 3 mM KNO3 relative to equally treated wild type. Note the logarithmic nature of the y axis (cf. legend to Figure 1A). Primers used are depicted in (C) and are given in Supplemental Table 1 online. Results represent mean values ± sd from three biological replicates/independent lines with two technical replicates for each.
(C) Graphical depiction of LBD gene structures, T-DNA insertion sites, and primer annealing sites. White boxes indicate exons and black lines untranslated regions or introns. T-DNA insertions are represented by black triangles and primers by arrows. Primers shown in gray were used for the analysis presented in (B), whereas those shown in black were used for the analysis shown in (D).
(D) Amplification of PCR products comprising the entire LBD gene coding sequence (LBDcds) from cDNA samples of wild-type and lbd T-DNA insertion mutants. N-starved seedlings were supplied with 3 mM KNO3 for 30 min prior to harvest. Amplification was performed for 25 cycles. A negative control (water) and a DNA marker (M) are shown for comparison. The 1000- and 750-bp marker DNA bands are indicated by white ellipses. Each LBDcds amplicon is expected to be ∼750 bp long. cDNA preparations used were of comparable concentrations as determined by qRT-PCR (CT UBQ10 = 16.7 to 17.1; Czechowski et al., 2005). Representative results for each PCR amplification are shown.
(E) RNA gel blot analysis of LBD37 expression in the wild type and lbd37-1 mutant after 30 min nitrate readdition to previously N-starved seedlings. The signal in the wild type corresponds to a size of 1.05 to 1.1 kb being very similar to the sizes of LBD37 full-length RNAs deposited in GenBank (e.g., BX830145 or AF447894). The signal in lbd37-1 is much weaker and of aberrant size.
Altered Anthocyanin Pigmentation in LBD37, LBD38, or LBD39 Overexpressers and Mutants
OX lines were investigated in N-limited conditions when the three LBD genes display very low expression (Figure 1B). When grown on agar plates or in liquid cultures (Scheible et al., 2004), wild-type seedlings turn reddish within 2 d after the N source (i.e., NO3−) is removed (Figure 3A), due to accumulation of anthocyanins. By contrast, the OX lines of each of the three LBD genes remained green (Figure 3B; see Supplemental Figure 2A online). Homozygous T-DNA insertion mutants for the three LBD genes were grown with sufficient N/NO3−, conditions in which the genes show high expression in the wild type. The lbd37, lbd38, and lbd39 mutants accumulated anthocyanin in the leaves (Figures 3C and 3D; see Supplemental Figure 2B online), whereas the wild type did not (Figure 3E). The suppression of anthocyanin synthesis in LBD37, LBD38, or LBD39 OX lines was robust. Other stress conditions (e.g., cold, high concentrations of sugar, and P limitation) that stimulate anthocyanin production were unable to override the effect of LBD OX in seedlings (Figures 3F and 3G; see Supplemental Figure 2C online).
Figure 3.
Anthocyanin Pigmentation Phenotypes.
(A) and (B) Nine-day-old N-limited wild-type (A) and N-limited LBD37 OX seedlings (B). For additional photographs, see Supplemental Figure 2 online.
(C) to (E) Nine-day-old nitrate-replete lbd37-1 T-DNA mutant seedlings ([C] and [D]) and a NO3−-replete wild-type seedling (E). For additional photomicrographs, see Supplemental Figure 2 online.
(F) and (G) Nine-day-old wild-type seedling (F) and a LBD37 overexpresser seedling (G) grown in medium containing 6% sucrose.
(H) Adult wild-type, LBD37, LBD38, and LBD39 OX plants (from left to right) grown in high illumination (600 μE).
(I) Flowers of wild-type, LBD37, LBD38, and LBD39 OX plants (from left to right) grown in –N soil.
(J) to (M) Adaxial (left) and abaxial (right) surface of mature rosette leaves from wild-type (J), LBD37 (K), LBD38 (L), and LBD39 (M) OX plants grown in –N soil.
(N) Seeds of wild-type, LBD37, and LBD38 OX plants (from left to right, two seeds each) from plants as shown in (H).
Bars = 4 mm in (A), (B), (F), (G), and (I), 2 mm in (C) and (E), 0.4 mm in (D), 2 cm in (H), 1 cm in (J) to (M), and 0.5 mm in (N).
The pigmentation phenotype of the OX lines was present throughout development. When grown in N-limiting conditions or under high irradiance (Figure 3H), the wild type accumulated anthocyanins in leaves, stems, flowers, and siliques, but the OX plants did not. However, some anthocyanin was noticeable in mature and senescing leaves of the OX plants (Figure 3H). Mature OX leaves had anthocyanin on the lower (abaxial) leaf surface and in the leaf petiole, but not the upper (adaxial) leaf surface (Figures 3J to 3M). This indicates that LBD37, LBD38, or LBD39 lose their strong negative effect in leaves with increasing age. Consistent with this observation, transcript levels of the three genes drop sharply with age in wild-type rosette leaves (data from AtGenExpress; Schmid et al., 2005). LBD OX seeds also gave rise to a slight transparent testa phenotype (Figure 3N), indicating reduced proanthocyanin and condensed tannin synthesis in the Arabidopsis seed coat. Consistently, the three genes are not expressed in wild-type seeds (data from AtGenExpress; Schmid et al., 2005). In addition to their striking pigmentation phenotypes, the soil-grown mature LBD OX plants displayed a reduction in shoot growth during reproductive stages (Figure 3H, cf. ultimate Results section).
LBD37 Localizes to the Nucleus
LBD genes are thought to encode zinc-finger TFs. Some class I family members have previously been shown to localize to the nucleus (Husbands et al., 2007; Okushima et al., 2007). The subcellular localization for class II LBDs has not yet been confirmed. We created a fusion of LBD37 to the C terminus of green fluorescent protein (GFP) expressed under control of the cauliflower mosaic virus 35S promoter (P35S). The P35S:GFP-LBD37 construct was transformed into wild-type plants, and transgenic lines with high GFP fluorescence were selected. When grown in N-limiting conditions, the GFP-expressing transgenic lines failed to produce anthocyanin (see Supplemental Figures 3A and 3B online), as did constitutive LBD37 OX plants. Thus, the GFP-LBD37 fusion protein is functional. GFP fluorescence was then examined in the leaves of seedlings and found to localize to nuclei of P35S:GFP-LBD37 plants (see Supplemental Figures 3C to 3G online). These results indicate that GFP-LBD37 fusion protein was targeted to the nucleus, consistent with its predicted role as TF.
Flavonoid Profiling Reveals Specific Changes of Cyanidin Glycosides in lbd Mutants and LBD OX Lines
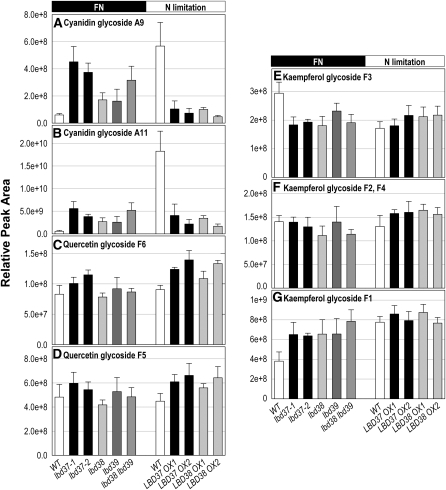
To analyze whether the LBDs affect flavonoid metabolism in general or only in specific sectors, we next analyzed the flavonoid metabolite profiles in shoot material of (1) 9-d-old N-limited LBD37 OX, LBD38 OX, and wild-type seedlings and (2) 9-d-old N-replete lbd37, lbd38, lbd39, and lbd38 lbd39 mutants and wild-type seedlings (Figure 4; see Supplemental Figure 4 online). Compared with N-limited wild types, independent N-limited LBD37 or LBD38 OX lines showed an 80 to 90% lower content of two cyanidin derivatives (Figures 4A and 4B), for example, cyanidin 3-O-[2-O-(2-O-(sinapoyl) xylosyl) 6-O-(p-O-coumaroyl) glucoside] 5-O-[6-O-(malonyl) glucoside] (m/z = 1343; A11; Tohge et al., 2005) and cyanidin 3-O-[2-O-(6-O-(sinapoyl) xylosyl) 6-O-(p-O-(glucosyl)-p-coumaroyl) glucoside] 5-O-(6-O-malonyl) glucoside (m/z = 1181; A9; Tohge et al., 2005). The former is the most abundant and major anthocyanin species that accumulates in PAP1 OX plants (Tohge et al., 2005). The levels of these cyanidin derivatives in N-limited LBD37 or LBD38 OX lines were as low as in NO3−-replete wild type (Figures 4A and 4B; see Supplemental Figure 4 online), indicating that LBD OX fully phenocopies the reduction of these two metabolites that occurs in NO3−-fed wild-type seedlings. By contrast, the two lbd37 T-DNA mutants showed an approximately sixfold increased level of these cyanidin glycosides in comparison to the wild type when grown in NO3−-sufficient conditions (Figures 4A and 4B). Similarly, cyanidin glycoside A9 and A11 levels were also increased in lbd38 and lbd39 mutants, and the effect was even slightly more pronounced in an lbd38 lbd39 double mutant. Other cyanidin derivatives were not reliably detected in the samples.
Figure 4.
Targeted Flavonoid Profiling.
Contents of seven detected cyanidin and flavonol glycoside species in shoot material from NO3−-fed (FN, black horizontal bar) and N-limited (N limitation, white horizontal bar) wild type, lbd mutant lines, two independent LBD37 OX, and two LBD38 OX lines. Results are mean values ± sd from three biological replicates for each genotype. Cyanidin derivatives and flavonol glycosides are named according to Tohge et al. (2005).
(A) Cyanidin 3-O-[2"-O-(2"'-O-(sinapoyl) xylosyl) 6"-O-(p-O-coumaroyl) glucoside] 5-O-[6""-O-(malonyl) glucoside], A9 (m/z = 1181).
(B) Cyanidin 3-O-[2"-O-(6"'-O-(sinapoyl) xylosyl) 6"-O-(p-O-(glucosyl)-p-coumaroyl) glucoside] 5-O-(6""-O-malonyl) glucoside, A11 (m/z = 1343).
(C) Quercetin 3-O-[2"-O-(rhamnosyl) glucoside] 7-O-rhamnoside, F6 (m/z = 757).
(D) Quercetin 3-O-glucoside 7-O-rhamnoside, F5 (m/z = 611).
(E) Kaempferol 3-O-[2"-O-(rhamnosyl) glucoside] 7-O-rhamnoside, F3 (m/z = 741).
(F) Kaempferol 3-O-glucoside 7-O-rhamnoside or quercetin 3-O-rhamnoside 7-O-rhamnoside, F2 or F4 (m/z = 595).
(G) Kaempferol 3-O-rhamnoside 7-O-rhamnoside, F1 (m/z = 579).
Flavonol glycosides (named F1 to F6 according to Tohge et al., 2005) derived from quercetin or kaempferol were largely unchanged in the OX or mutant seedlings irrespective of NO3− supply (Figures 4C to 4G; see Supplemental Figure 4 online). The OX and mutant lines also produce normal levels of lignins as detected by phloroglucinol staining of stem sections (see Supplemental Figure 4H online). Together these results suggest (1) a specific effect of the LBDs on anthocyanin metabolism downstream of dihydroquercetin and (2) that LBD37 or LBD38 OX mimics the effect of N/NO3− supply on cyanidin glycoside levels.
LBD OX Phenocopies the Transcriptional Changes Found in the Cyanidin Pathway after NO3− Addition to N-Limited Wild-Type Seedlings and Results in a Mirror Image of the Changes Observed in PAP1 Overexpressers
N/NO3− deficiency or PAP1 OX strongly induce a shared set of genes involved in late flavonoid synthesis in Arabidopsis (Lillo et al., 2008). N/NO3− deficiency also leads to strong transcriptional induction of PAP1 and PAP2 (Scheible et al., 2004). To further test and cement the emerging role of the LBD genes in N/NO3−-dependent repression of anthocyanin biosynthesis, we used qRT-PCR to analyze the expression of structural or regulatory genes known to be involved in early or late steps of flavonoid biosynthesis. Expression was compared in (1) N-limited LBD OX seedlings versus N-limited wild-type seedlings, (2) NO3−-fed (i.e., N-replete) versus N-limited wild-type seedlings, (3) NO3−-fed pap-1D seedlings (Borevitz et al., 2000) versus NO3−-fed wild-type seedlings, and (4) NO3−-fed lbd37, lbd38, and lbd39 mutant seedlings versus NO3−-fed wild-type seedlings (Figure 5; see Supplemental Figures 5 to 7 online).
Figure 5.
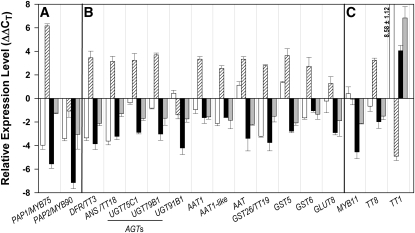
Expression Levels of Structural and Regulatory Anthocyanin Synthesis Genes.
(A) Regulatory genes PAP1 and PAP2.
(B) Anthocyanin synthesis genes.
(C) Regulatory genes MYB11, TT8, and TT1.
Shown are relative expression changes in NO3−-fed versus N-limited wild types (white bars), in the NO3−-fed pap1-D activation tagging line versus NO3−-fed wild-types (hatched bars), and in N-limited LBD37 and LBD38 OXs versus N-limited wild-types (black and gray bars, respectively). Results represent mean values ± sd from three independent biological replicates/lines with two technical replicates for each. Shown are genes with |ΔΔCT| ≥ 1.5 in LBD37 or 38 OX lines. Note the logarithmic character of the y axis (cf. legend to Figure 1A).
The analyses revealed that independent LBD37 OX or LBD38 OX lines had decreased expression of PAP1 and PAP2, resembling the decrease found in NO3−-fed wild-type seedlings or in wild-type seedlings after short-term NO3− readdition (Scheible et al., 2004) (Figure 5). Extension of the comparison to flavonoid biosynthetic genes showed that (1) LBD37 or 38 OX strongly (often 5- to 10-fold) represses late genes encoding pathway steps downstream of dihydroquercetin, such as cyanidin biosynthesis (i.e., DFR, ANTHOCYANIDIN SYNTHASE, ANTHOCYANIN GLYCOSYLTRANSFERASE, ANTHOCYANIN ACYLTRANSFERASE, and GST), including also a gene (GLUT8) encoding a putative carbohydrate transmembrane transporter upregulated by PAP1 that might be involved in vacuolar transport of cyanidin glycosides (Tohge et al., 2005; cf. Figures 5 and 6). Moreover, analysis of the same set of genes in lbd37, lbd38, and lbd39 mutants revealed that the genes downregulated in the OX lines remain induced in NO3−-replete lbd37 lbd38 and lbd39 mutants (see Supplemental Figure 5 online), which is consistent with the observed anthocyanin accumulation (Figures 3C and 3D; see Supplemental Figure 2B online).
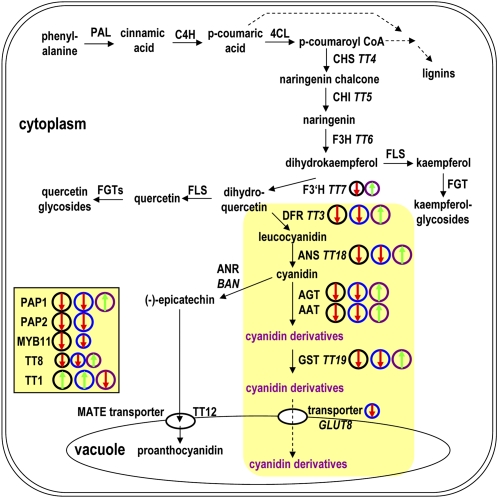
Figure 6.
Summary of the Effects of Nitrate, LBD37, 38, or 39, and PAP1 Overexpression on the Anthocyanin Biosynthesis Pathway.
Anthocyanin-specific pathway steps in flavonoid metabolism are shown in a yellow background. Regulatory genes that were found to be considerably misexpressed in LBD OXs are shown in the yellow shaded box to the left. Black, blue, and purple circles mark the pathway steps/genes affected by NO3−, LBD37 OX, or PAP1 OX, respectively. Red arrows indicate repression and green arrows induction. The size of the circles gives an indication of the strength of the changes. Contents of cyanidin derivatives are low in N/NO3−-fed wild-type plants and N-limited LBD37, LBD38, and LBD39 OX lines and accumulate in N/NO3−-fed lbd37, lbd38, and lbd39 mutants and PAP1OXs (Borevitz et al., 2000; Tohge et al., 2005). Major quercetin- and kaempferol-glycosides were unchanged. PAL, phenylalanine ammonium lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHI, chalcone isomerase; FLS, flavonol synthase; FGT, flavonol glycosyltransferase; BAN, anthocyanidin reductase BANYULS; ANS, anthocyanidin synthase; AGT, anthocyanin glycosyltransferase; AAT, anthocyanin acyltransferase. Pathways are depicted according to Tohge et al. (2005).
Genes encoding steps in phenylpropanoid metabolism (e.g., PHENYLALANINE AMMONIUM LYASE [PAL], CINNAMATE-4-HYDROXYLASE, or 4-COUMARATE:COA LIGASE) or early steps in flavonoid synthesis (CHS, CHALCONE ISOMERASE [CHI], or FLAVANONE 3-HYDROXYLASE [F3H]), as well as genes encoding flavonol glycosyltransferases not specific for anthocyanins or steps in proanthocyanin biosynthesis (e.g., BANYULS), were not or only very weakly (less than twofold) affected by LBD OX (see Supplemental Figure 6A online), showing that LBD OX in seedlings specifically represses anthocyanin synthetic genes. N/NO3− also did not affect these genes much; PAL3 and UDP-GLUCOSYL TRANSFERASE 78D1 (UGT78D1) were approximately twofold induced, and CHS, FLAVONOID 3′ HYDROXYLASE (F3′H), and UGT71C1 were approximately twofold to threefold repressed in the comparison of N/NO3−-fed versus N-limited wild types. The same was also observed for PAP1 OX seedlings: PAL genes, CHS, CHI, and F3H were largely unaffected; only F3′H and UGT78D2 showed a slight/moderate induction (see Supplemental Figure 6A online). These results with seedlings are in slight contrast with previous studies with leaves of rosette stage pap1-D plants (Borevitz et al., 2000; Tohge et al., 2005), where some phenylpropanoid metabolism genes and early genes in flavonoid biosynthesis were moderately induced.
Besides PAP1 and PAP2, we analyzed the expression of additional regulatory genes involved in the regulation of anthocyanin, procyanidin, flavonoid, or phenylpropanoid biosynthesis, including several MYB family TF genes (MYB3, MYB4, MYB11, MYB12, MYB32, MYB66/WEREWOLF, MYB111, MYB123/TRANSPARENT TESTA2 [TT2], MYB0/GLABRA1 [GL1]), bHLH family TF genes (GL3, ENHANCER OF GLABRA1 [EGL1], and TT8), WD40 protein (TRANSPARENT TESTA GLABRA1 [TTG1]), and others (TRANSPARENT TESTA1, HY5, HYH, ANTHOCYANINLESS2, and TTG2) (Figure 5C; see Supplemental Figure 6B online). The analysis showed that three TF genes had an appreciably changed transcript level in LBD OX; TT1 was strongly induced, while MYB11 and TT8 were threefold to fourfold repressed (Figure 5C). PAP1 OX again resulted in a phenotype opposite to that of LBD37/38 OX with respect to TT8 and TT1. N/NO3− had a wider effect; in addition to a strong inductive effect on TT1, it also led to a clear induction of MYB111, EGL1, and GL1 and a repression of MYB123/TT2 (see Supplemental Figure 6B online). The effects on MYB11 and TT8 transcript levels were weak (Figure 5C).
The set of anthocyanin synthesis genes repressed in constitutive LBD OX lines was subsequently also analyzed in inducible LBD37 OX (iOX) lines (see Supplemental Figure 7 online; Figure 2). The results showed that iOX leads to the same general changes as observed before in the constitutive OX lines, except that the decrease of the PAP1, PAP2, and anthocyanin pathway transcripts found after 3 or 6 h induction was less pronounced. Possibly, the endogenous PAP1 and PAP2 transcripts and PAP1 and PAP2 proteins were stable enough to prevent a more dramatic decrease. The result indicates that the changes detected in constitutive OX lines are specific and not merely an unrelated phenotype.
In summary, transcript changes found in LBD37/38 OX lines and in the PAP1 OX line reveal a broad overlap and also surprisingly well reflect the changes found in the comparison of N/NO3−-fed and N-limited wild-type seedlings (Figures 5 and 6). Together, the results pinpoint the LBD37 TF and its close homologs as novel molecular components involved in the transmission of an N/NO3−-derived signal that suppresses anthocyanin synthesis via repression of PAP1 and PAP2.
Transcriptome Profiling Reveals a More Profound Role of the LBDs in N Signaling
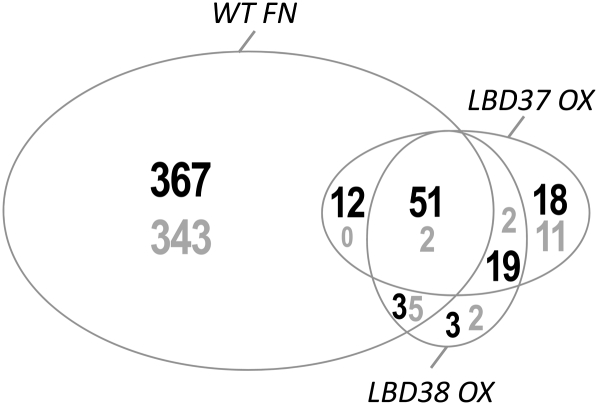
The analysis presented so far was focused on flavonoid/anthocyanin biosynthesis. To test whether LBD37, 38, and 39 have only this specific function or whether they may have a more widespread role in NO3−/N regulation, we investigated the global transcript profiles of N-limited LBD37 and LBD38 OX lines, as well as N-limited and N/NO3−-replete wild-type seedlings with Affymetrix ATH1 GeneChips. N/NO3− availability in the wild type led to a ≥3-fold change of 783 gene transcripts (Figure 7; see Supplemental Data Set 1 online), resembling results of earlier studies (Wang et al., 2003; Scheible et al., 2004; 581 of these transcripts were also ≥2-fold changed in the former study). Interestingly, LBD37 or LBD38 OX led to a ≥3-fold repression of 100 or 76 gene transcripts, respectively, while only very few gene transcripts were induced (Figure 7), revealing that the LBDs mainly function as transcriptional repressors, as noted before. Remarkably, 63 to 71% of the gene transcripts that were ≥3-fold repressed in LBD37 or LBD38 OX seedlings were also ≥3-fold repressed by N/NO3− availability in wild-type seedlings, suggesting that the LBDs play a role in the regulation of transcriptional N responses. This percentage even rises to 69 to 80% when gene transcripts with ≥2-fold repression are considered (see Supplemental Data Set 1 online).
Figure 7.
Shared Transcriptional Changes in Response to Nitrogen Status and LBD37 or LBD38 Overexpression.
The Venn diagram indicates the number of gene transcripts that are ≥3-fold decreased (black numbers) or increased (gray numbers) in either N-limited LBD37 or LBD38 overexpressing seedlings or N-replete wild-type (WT FN) seedlings in comparison to N-limited wild-type seedlings. Data were calculated from the average of three biological replicates for each condition (see Supplemental Data Sets 1 and 2 online). The size of the ellipses is roughly proportional to the number of gene transcripts affected (for more detailed information, see Supplemental Figure 8 online).
Among the N/NO3−-repressed genes with reduced transcript levels in LBD37 or LBD38 OX seedlings were PAP1 and PAP2, several genes encoding glutaredoxin, senescence-associated genes, or the nitrate transporter gene NRT2.5 (see Supplemental Figures 8A and 8B and Supplemental Data Set 1 online). Besides, several additional gene transcripts involved in NO3− uptake and assimilation were altered in LBD OX, including NIA1, which encodes NR (see Supplemental Figure 8C online) and to some lesser extent also NIA2, the NO3− transporter genes NRT1.1, NRT2.1, NRT2.2, and NRT3.1, as well as the Gln synthetase gene GLN1.4 (see Supplemental Figures 8C, 8G, and 8H online). In addition, a number of N-responsive gene transcripts encoding regulatory components were altered in LBD OXs. These include TFs (WRKY38, NF-YA10, and NAC55) and several (receptor) protein kinases or the Glu-receptor protein GLR1.2 (see Supplemental Figures 8A, 8B, and 8I online). Closer inspection of the ATH1 GeneChip results also confirmed the repression of anthocyanin synthetic genes (see Supplemental Figure 8J online) in LBD OX seedlings, although the individual transcript responses were generally less pronounced than observed with qRT-PCR (Figure 5).
We also compared lbd37 and lbd38 seedlings grown in NO3−-replete conditions with NO3−-replete wild-type seedlings. There were only very few (≥3-fold) changes (1 or 12 transcripts induced in lbd37 or lbd38, respectively; no genes ≥3-fold repressed in both mutants) with the most noteworthy being the 3.3-fold induction of NRT2.1 in lbd38 (see Supplemental Data Set 2 online).
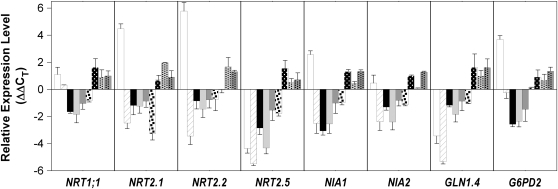
LBD37, 38, and 39 Repress Genes Involved in NO3− Uptake/Assimilation
We aimed to confirm the results observed for genes involved in NO3− uptake and reduction (see Supplemental Figure 8H online), including NR genes NIA1, NIA2, NO3− transporter genes NRT1.1, NRT2.1, NRT2.1, and NRT2.5, as well as GLN1.4 and G6PD2, the latter being involved in the provision of carbon skeletons required for N assimilation. Expression analysis by qRT-PCR confirmed that NRT2.1, NRT2.2, NIA1, and G6PD2 are induced 3 h (and already 30 min) after readdition of NO3− to N-limited wild type (Figure 8; see Supplemental Figure 9 online; Scheible et al., 2004). This induction is abolished or reversed in N-replete wild types, which is attributable to organic nitrogen compounds like Gln (Krapp et al., 1998; Lejay et al., 1999; Nazoa et al., 2003). By contrast, NRT2.5 and GLN1.4 are repressed 3 h (but not 30 min) after NO3− readdition (Scheible et al., 2004; Figure 8), suggesting repression is linked to NO3− reduction. Consequently and similar to NIA1, these two genes are repressed after Gln readdition to N-starved wild type (ΔΔCT = −6.91, −1.82, and −2.25 for NIA1, NRT2.5, and GLN1.4, respectively, after 3 h).
Figure 8.
qRT-PCR Analysis of Gene Transcripts Involved in Nitrate Uptake and Assimilation.
Relative expression changes in (1) N-limited wild type after 3 h NO3− readdition versus N-limited wild type (white bar, 1st in each group), (2) NO3−-fed versus N-limited wild types (hatched bar, 2nd in each group), (3) N-limited overexpressers of LBD37, LBD38, and LBD39 versus N-limited wild types (black, light-gray, and medium-gray bars, respectively; 3rd to 5th in each group), (4) inducible LBD37 overexpressers (iOX) after 3 h induction versus noninduced LBD37 iOX (black dotted white bar, 6th in each group), and (5) N-replete lbd37-1, lbd38, and lbd39 mutants versus N-replete wild type (white dotted black bar, black dotted light-gray bar, black dotted medium-gray bar, 7th to 9th in each group). Results represent mean values ± sd from three independent biological replicates/lines with two technical replicates for each. Note the logarithmic character of the data (cf. legend to Figure 1A).
qRT-PCR analysis further confirmed that expression of NIA genes was 3- to 6-fold reduced, and expression of NRT1.1, NRT2.1, NRT2.2, and NRT2.5 was 2- to 10-fold lower in the OX lines compared with the wild type (Figure 8; cf. Supplemental Figure 8H online). In addition, expression of GLN1.4 and G6PD2 was significantly decreased in the OX lines. These results are further corroborated by similar changes in LBD37 iOX lines and reciprocal, yet weaker changes in lbd37, lbd38, and lbd39 mutants (Figure 8; see Supplemental Figure 9A online) and suggest that LBD37/38/39 expression mimics the effect of organic N compounds on expression of NIA1, NIA2, NRT2.1, NRT2.5, or GLN1.4.
LBD37, 38, and 39 Affect N Status, Growth, and N-Dependent Shoot Branching
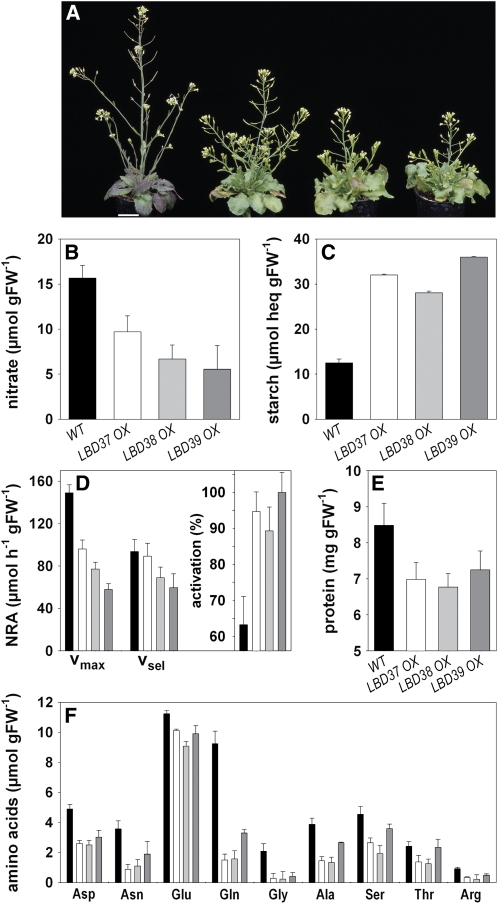
Reduced expression of genes required for NO3− uptake and reduction caused by mimicry of N availability in LBD OX lines should eventually lead to symptoms of N limitation, reduced plant N status, and ultimately growth. The smaller stature of mature LBD OX plants (Figures 3H and 9A) is in agreement with this. To further test the hypothesis, we performed a biochemical analysis of mature wild-type and LBD OX lines (Figures 9B to 9F). This revealed that LBD OX lines contain 40 to 65% less NO3− (Figure 9B) and have 100 to 150% higher starch levels (Figure 9C) than wild-type plants. Moreover, the OX lines displayed a 35 to 60% reduction in maximal NR activity, while NR activation increased from ∼60% in the wild type to 90 to 100% in OX lines (Figure 9D). Furthermore, protein content was decreased by up to 20% (Figure 9E), and total amino acid content dropped in the OX lines. This was mostly due to a fourfold to fivefold decrease of Gln but was also due to substantial decreases in other amino acids, such as Asp, Asn, Arg, or Gly (Figure 9F). The strong decrease of Gln and Asn in the OX lines is further confirmed by a reciprocal increase of Gln and Asn in lbd mutants (see Supplemental Figure 10 online). The analyzed molecular, physiological, and biochemical parameters all show that LBD37, 38, or 39 have more widespread effects and negatively affect N assimilation and, as a consequence, plant N status and growth. Besides, LBD37, 38, or 39 OX also alters N-dependent basal shoot branching (see Supplemental Figure 11 online). Arabidopsis wild-type plants produce no basal rosette branches when grown in N-limited conditions. In comparison, LBD OX plants grow as many basal shoot branches in N-limited conditions as they do or the wild type does in N-replete conditions. This result is again consistent with the idea that these LBD proteins represent important molecular components to signal N/NO3− availability.
Figure 9.
Altered N Status in LBD Overexpressors.
(A) Plant phenotype of the analyzed wild type, LBD37, LBD38, and LBD39 OXs (from left to right). Bar = 2 cm.
(B) NO3− content.
(C) Starch content.
(D) Maximal and selective nitrate reductase activity, NRA, and NR activation (i.e., Vsel/Vmax).
(E) Protein content.
(F) Amino acid contents.
Results are from shoots of the plants shown in (A). Wild type (black bars), LBD37 (white bars), LBD38 (light-gray bar), and LBD39 OXs (medium-gray bars) were grown in high light conditions (cf. Figure 3H). Results are the average ± sd from three independent lines each. For each sample (i.e., line), materials from five individual plants were combined.
DISCUSSION
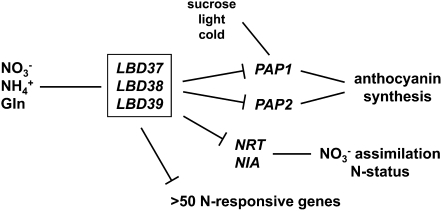
Nitrogen and NO3− (as the major form of N available to many plants) are macronutrients and signals for plant growth and metabolism, but still little is known about the molecular components that constitute or mediate the N/NO3− signal in higher plants. We successfully used a reverse genetic approach to investigate the potential functions of three strongly N/NO3−-inducible members of the novel LBD gene family of TFs (Shuai et al., 2002; Scheible et al., 2004; Husbands et al., 2007). The three genes are highly related and most probably evolved by two subsequent segmental duplications of a chromosome 5 area comprising LBD37 (http://www.tigr.org). Members of the plant-specific LBD TF gene family have been implicated in developmental processes during recent years (see Introduction), but for many LBD genes, functions are still unknown. Our results expand the knowledge about the LBD gene family by showing that some members are involved in regulation of metabolism. They mediate the N/NO3−-dependent regulation of anthocyanin synthesis and also exert wider effects by repressing a larger number of N-responsive gene transcripts and key transcripts for NO3− assimilation, with subsequent alterations of N status and N-dependent growth (Figure 10). An important conclusion from our results is that expression of LBD37, LBD38, or LBD39 constitutes an N availability signal in Arabidopsis.
Figure 10.
Model of LBD37, 38, and 39 Functions.
Major N sources induce LBD37, 38, and 39. Expression of these LBDs represses PAP1 and PAP2, several important NRT and NIA genes, as well as a larger number of additional N-responsive genes, thereby decreasing anthocyanin synthesis, nitrate assimilation, and N status.
LBD37, 38, and 39 Repress Anthocyanin Biosynthesis Upstream of PAP1 and PAP2
The visual pigmentation phenotypes observed in LBD37, LBD38, and LBD39 OX plants and the lbd37, lbd38, and lbd39 mutants prompted us to perform a targeted analysis of flavonoid metabolism at the metabolite and transcript levels. All the results gathered suggest that these LBDs regulate the late anthocyanin-specific steps of flavonoid biosynthesis, thereby giving a striking phenocopy of the N/NO3− effect and a reverse phenocopy of PAP1 OX on this part of secondary metabolism (Figure 6). PAP1 and especially PAP2 are strongly repressed in continuously N/NO3−-fed Arabidopsis wild-type seedlings and in previously N-limited wild-type seedlings that were re-supplied with 3 mM KNO3 for 3 h (Scheible et al., 2004). The same strong repression of PAP1 and especially PAP2 is observed in LBD37 or LBD38 OXs, suggesting that these LBDs are N/NO3−-induced repressors acting upstream of PAP1 and PAP2 (Figure 10). Whether the LBDs can directly bind to PAP1 and PAP2 promoter regions remains to be investigated.
It is noteworthy that LBD37 OX seedlings do not produce anthocyanins in leaves even when challenged with P limitation, high irradiation, and cold or high concentrations of sucrose (Figure 3G; see Supplemental Figure 2C online). In these conditions, PAP2 remains strongly repressed, while PAP1 is derepressed to the level present in the wild type (see Supplemental Figure 12 online). Thus, P deprivation, sucrose, light, and cold can antagonize the LBD-mediated N/NO3− repression of PAP1 but not the repression of PAP2 (Figure 10). Consistent with this, PAP1 was proposed to have a key role in light and sucrose induction of anthocyanin biosynthesis in Arabidopsis (Teng et al., 2005; Solfanelli et al., 2006; Cominelli et al., 2008), whereas PAP2 appears to be more important in the N/NO3− regulation of anthocyanin biosynthesis (Lea et al., 2007). The fact that PAP1 expression in sugar- or P-stressed LBD37 OX seedlings is still ∼10-fold lower than in PAP1-overexpressing pap1-D lines (see Supplemental Figure 12 online) might explain their inability to produce high levels of anthocyanin.
OX of each LBD gene resulted in the same visual, physiological, and overall molecular phenotypes. Likewise, loss of function of each of the three LBD genes resulted in the reciprocal phenotypes. This shows that these putative paralogs have the same biological function and that reduced anthocyanins in LBD OX lines is not the result of ectopic expression. Also, analyses with ATTED II (Obayashi et al., 2007) or Gene-CAT (Mutwil et al., 2008) reveal coexpression of the three LBD genes, suggesting their involvement in the same biological process or possibly even the same molecular heteromeric complex, as exemplified for cellulose synthase isoforms (Taylor et al., 2003; Persson et al., 2007). The latter possibility is supported by the comparable phenotypes of each of the lbd single mutants and would suggest that LBD37, LBD38, and LBD39, although involved in the same biological process, have distinct or at least not fully redundant biochemical functions.
The data discussed so far provide evidence that the three LBDs regulate anthocyanin synthesis via repression of PAP1 and PAP2. A number of other genes encoding MYB and bHLH TFs are known to be involved in regulating anthocyanin biosynthesis in seeds and vegetative tissues. The MYB and bHLH proteins form complexes with the TTG1 WD40-repeat protein in Arabidopsis and are able to regulate several epidermal gene expression programs in Arabidopsis (Broun, 2005), including the regulation of anthocyanin and proanthocyanin biosynthesis as well as mucilage synthesis in the seed coat or trichome and root hair organogenesis. Therefore, expression of additional MYB and bHLH TF genes, the TTG1 gene, and a few other TF genes (e.g., TT1 and TTG2) known to be involved in anthocyanin, procyanidin, flavonoid, or phenylpropanoid (e.g., MYB4) metabolism was analyzed. bHLH TT8 interacts with PAP1/PAP2 (Zimmermann et al., 2004). TT8 was found to be repressed by LBD37 and LBD38 OX and induced in lbd37, lbd38, and lbd39 mutants and by PAP1 OX. This illustrates coregulation of components of the same transcriptional complex and suggests TT8 is not a direct target of LBD but that its expression rather depends on PAP1/PAP2. The closely related MYB11, MYB12, and MYB111 TFs are redundant positive regulators and affect flavonol accumulation in different parts of the Arabidopsis seedling by regulating several genes of flavonoid biosynthesis, including CHS, CHI, F3H, FLS1, and UGT91A1 and UGT84A1 (Stracke et al., 2007). Seedlings of the triple mutant myb11 myb12 myb111 do not form flavonols, but accumulation of anthocyanins is unaffected in these seedlings (Stracke et al., 2007). In LBD OX seedlings, MYB11 was repressed, but MYB12 and MYB111 were unchanged. Consistently, flavonols were also not affected. The small MYB protein Arabidopsis MYBL2 acts as a negative regulator (Dubos et al., 2008; Matsui et al., 2008). Loss of MYBL2 activity leads to strong anthocyanin accumulation and induction of late flavonoid biosynthesis genes as well as TT8 and PAP1, while MYBL2 overexpression suppressed these genes and inhibited the biosynthesis of proanthocyanidins in seeds. These results are analogous to what we observed with LBD OXs and mutants. It will be important to clarify the relationship and interaction between MYBL2 and the LBDs. The TT1 gene encodes a nuclear WIP-domain zinc-finger protein that is involved in seed coat development and pigmentation (Sagasser et al., 2002). LBD OX lines and NO3−-fed wild-types displayed strong induction of TT1, while it was strongly repressed by PAP1 OX. The expression pattern of TT1 is thus opposite to that of PAP1, PAP2, or the late flavonoid biosynthesis genes. Nothing is known about the function of this gene in vegetative tissue. There is no expression history available as it is not represented on ATH1 GeneChips and requires more sensitive methods, such as qRT-PCR, for detection in leaves. In conclusion, it is clear that the expression of some of these regulatory genes is modified in LBD OX lines, in PAP1 OX, or by N/NO3−. However, it will require more work to determine whether these changes are direct or indirect and to finally clarify how the genes and their products interact.
The SPX-domain E3 ligase NLA was described to be required for anthocyanin production and other N limitation responses in Arabidopsis (Peng et al., 2007a, 2007b, 2008), as N-limited nla mutants no longer accumulate anthocyanin. Therefore, it is of interest to compare nla mutants and LBD OXs in more detail. In both genotypes, anthocyanin synthetic genes are repressed, but nla mutants do not show repression of PAP1 or PAP2 transcripts (Peng et al., 2007b), while this is a dominant transcriptional change in LBD OXs. The inability to produce anthocyanins also seems to occur only in mature nla mutant plants, as N-limited nla mutant seedlings display a wild-type-like anthocyanin pigmentation (see Supplemental Figure 2D online), while the three LBDs suppress anthocyanin production at the seedling stage already. Furthermore, the array of transcriptional changes observed in N-limited nla mutants and LBD OX lines is quite different, and neither is NLA transcript abundance affected in LBD OX lines, nor are LBD37, 38, or 39 transcript levels altered in nla (see Supplemental Data Set 1 online and Peng et al., 2007b). These observations lead us to conclude that E3 ligase NLA and the LBDs affect anthocyanin production via different signaling pathways. LBDs appear to repress PAP1/PAP2 transcription, whereas NLA possibly influences PAP1/PAP2 at a posttranslational level.
LBD37, 38, and 39 Expression in N-Limited Plants Mimics N Availability and Affects N-Dependent Traits
Inhibition of sectors of secondary metabolism is part of the primary response to NO3−/N (see Introduction). Repression of LBD37, 38, and 39 during N limitation and rapid strong induction by NO3−, NH4+, or Gln as well as the fact that LBD OX inhibits anthocyanin synthesis and strongly represses PAP1 and PAP2 leads to the conclusion that the LBDs represent a primary component in a signaling pathway that represses anthocyanin production when NO3−/N is available (Figure 10). However, the range of traits altered by LBD expression is not limited to anthocyanins, as transcriptome studies revealed that a large majority of genes repressed in N-limited LBD OX lines are also repressed by NO3−/N availability. The LBDs influence the abundance of >50 or ∼15% of the N-repressed transcripts in Arabidopsis, giving them broader importance in N signaling. An important role of these LBDs in N signaling and adaptation to changing NO3−/N availability can also be inferred from the observation that their transcripts are among the few that decrease when Arabidopsis plants are subjected to mild N limitation (A. Schlereth and M. Stitt, personal communication; see Tschoep et al. [2009] for the definition of mild N limitation).
Among the genes affected by the LBDs are the ones encoding nitrate reductase (NIA) and several NO3− transporters (NRT). NIA and some NRT2 genes are induced as part of the primary response to NO3− (Redinbaugh and Campbell, 1991) and repressed by organic nitrogen sources such as Gln (Krapp et al., 1998; Lejay et al., 1999; Nazoa et al., 2003). LBD OX did not induce genes for NO3− uptake and assimilation; instead, OX of each of the three LBD genes led to a robust repression of NIA and several NRT2 genes (Figure 8). The same was seen with LBD iOX lines, in which NIA and NRT2 genes were repressed after ethanol induction (Figure 8; see Supplemental Figure 9A online). This indicates that, in contrast with NLP7, HY5, or NRT1.1 (see Introduction), the LBDs are not involved in the primary induction of N uptake and assimilation by NO3−. Fast and strong induction of the LBDs by NO3− also implies that the LBD proteins are synthesized, but de novo protein synthesis per se is not required for NO3− induction of NIA or NRT2 genes (Redinbaugh and Campbell, 1991; Gowri et al., 1992). NIA or NRT2 genes are also equally induced in N-limited wild-type and lbd mutants after NO3− readdition (see Supplemental Figure 9B online), providing yet more evidence that the LBDs are not involved in the primary induction of N uptake/assimilation by NO3−.
The best explanation for the observed repression of the genes involved in NO3− uptake and assimilation is that LBD37-39 signal N availability to the plant system, leading to repression of specific N deficiency responses (like anthocyanin synthesis) and feedback repression of NIA or NRT2 genes. The alternative view that LBD37-39 expression could alter sugar signaling leading to repression of NIA or NRT2 genes, which were described as being sugar inducible (Cheng et al., 1992; Lejay et al., 2003), seems unlikely as many of the gene transcripts repressed in LBD OX seedlings and by N availability (e.g., PAP2, NIA1, NRT2.1, NRT2.2, At1g74810, At1g22160, At5g02580, At5g02230, At3g62960, and At1g76960; see Supplemental Data Set 1 online; Scheible et al., 2004) are much less affected or not affected by sugars (cf. data from Osuna et al., 2007). Conversely, strongly sugar-responsive genes in Arabidopsis seedlings (Osuna et al., 2007) are hardly or not affected in LBD OX seedlings.
Mimicry of N availability by LBD expression, the resulting repression of NIA and NRT2 genes, and reduction of NO3− assimilation will eventually affect plant N status. Consistently, mature soil-grown LBD OX plants have a low NO3− content and a shortage of organic N compounds (most notably a reduction of Gln, but also protein content). Other typical indicators of low N status observed in the LBD OX plants are high activation (90 to 100%) of NR (Scheible et al., 1997b, 1997c) and starch accumulation (Scheible et al., 1997a; Peng et al., 2007a). Mimicry of N availability by LBD expression can also be deduced from the alteration of basal shoot branching, an N-dependent growth phenotype (see Supplemental Figure 11 online; Crawford and Forde, 2002; Médiène et al., 2002). Shoot branching has long been known to be influenced by hormones such as cytokinins and auxin (for review, see Ongaro and Leyser, 2008) and recently also by strigolactones (Gomez-Roldan et al., 2008; Umehara et al., 2008). It is also known that cytokinins are produced in response to NO3− addition in Arabidopsis roots (Takei et al., 2002) and move acropetally and promote shoot branching. It will be interesting to establish whether the LBDs affect the levels or the transport of these hormones too.
In conclusion, the identification of the three LBDs as regulators of anthocyanin synthesis and repressors of a larger number of N-repressed gene transcripts pinpoints these LBD genes as important molecular components in plant NO3−/N signaling.
METHODS
Plant Genotypes
Arabidopsis thaliana wild-type Columbia-0 seeds were obtained from an MPI-MP in-house collection. LBD gene T-DNA insertion lines (lbd37-1, SALK_097991; lbd37-2, SALK_057939; lbd39, SALK_049910) and the activation-tagging mutant pap1-D (Borevitz et al., 2000) were obtained through the Nottingham Arabidopsis Stock Centre (NASC). The T-DNA insertion line lbd38 (GABI_049C12) was generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (MPI for Plant Breeding Research, Cologne, Germany). Seeds of nia1nia2 double mutants (Wang et al., 2004) were provided by Marc Lepetit (Institute for Integrative Plant Biology, Montpellier, France).
Homozygous T-DNA insertion lines were identified by standard PCR procedures with wild-type and T-DNA–specific primer pairs (lbd37-1: FwdP 5′-TCGTCCAATATACTGGTTGGAATTT-3′ and RevP 5′-TCCAGCAAAATGATTCCACCG-3′; lbd37-2: FwdP 5′-TTGGTCCAACCACATGCTTA-3′ and RevP 5′-TCTCCAGCAAAATGATTCCAC-3′; lbd38: FwdP 5′-GGAGATAGAACCATGAGTTGCAATGGTTGTCGA-3′ and RevP 5′-CAAGAAAGCTGGGTCTCAAGCGAAGAGATTGAGCAA-3′; lbd39: FwdP 5′-ATCTTTGAGTGTTTTGCGTCG and RevP 5′-TTCGGAAAAGAAATGAGTTGC-3′). To amplify T-DNA–specific products, RevP primers were combined with LBb2 (5′-GCGTGGACCGCTTGCTGCAACT-3′) for lbd37_1 and lbd37_2 or with LBc1 (5′-CCGCAATGTGTTATTAAGTTG-3′) for lbd39. The lbd38 primer, RevP2 (5′-AAGAGATTGAGCAACTTTGTCGTT-3′), was combined with the GabiKat-LB-primer (5′-CCCATTTGGACGTGAATGTAGACAC-3′).
For production of constitutive and inducible LBD OX and GFP-LBD fusion constructs, PCR fragments containing the entire annotated LBD coding sequences were amplified from cDNA pools, directionally cloned into GATEWAY entry vector pENTRY/SD/D-TOPO (Invitrogen), transferred by homologous recombination into destination vector pMDC32 (Curtis and Grossniklaus, 2003), pSRN (Junker et al., 2003), or pK7FWG2 (Karimi et al., 2002), and finally transformed into wild-type Columba-0 via Agrobacterium tumefaciens GV3101 (Clough and Bent, 1998).
Plant Growth Conditions
Wild-type, T-DNA mutant, and OX lines were grown on GS90 soil (100% N; 0.3 g NH4NO3 L−1) (Gebrüder Patzer), 0% N soil (GS90 N-free), or 50% N soil (1:1 mixture of the two soil types) in Percival AR-36L growth chambers (Percival Scientific) in a 16-h-light (∼150 μE, 20°C)/8-h-dark (18°C) or 24-h-light cycle at ∼70% relative humidity. For high-light conditions, the wild type and OXs were grown on GS90 soil for 4 weeks and then transferred to a Percival chamber with 8 h light (600 μE, 20°C)/16 h dark (18°C) at ∼70% relative humidity. Samples were taken in the middle of the light period.
Arabidopsis seedlings were grown in sterile liquid culture (Scheible et al., 2004) either for 9 d in N-replete conditions (FN, 4 mM KNO3) or first for 7 d in FN and then for 2 d in N deprivation conditions (no N added; −N). On day 9, some batches of N-starved seedlings were resupplied with 3 mM KNO3 for 30 min or 3 h (wild type and mutants) or treated with 0.2% ethanol for 3 and 6 h (iOX). For additional stress conditions, seedlings were grown for 7 d in FN and another 3 d in N or P deprivation conditions (Morcuende et al., 2007), high concentrations of sucrose (6% w/v), or transferred to the cold (4°C). For that purpose, seedlings were pregrown in 24 h light (150 to 180 μE, 20°C) on sterile 0.5% (w/v) agar plates (Falcon Integrid 100 mm square) containing 27 mL FN medium, and after 7 d, seedlings were transferred individually to 24-well culture plates (Greiner Bio-one Cellstar 662160) containing 1 mL of medium per well.
Quantitative Real-time PCR
All steps including RNA isolation, cDNA synthesis, quality control, primer design, real-time PCR, and data analysis were performed as described (Czechowski et al., 2004, 2005; Udvardi et al., 2008). Sequences and amplification efficiencies (E) of qRT-PCR primers are listed in Supplemental Table 1 online.
Affymetrix ATH1 GeneChip Experiments
Total RNA was prepared separately from three biological replicates of N-limited LBD37OX, LBD38OX, and wild-type seedlings and N-replete lbd37-1 mutant, lbd38 mutant, and wild-type seedlings (i.e., 18 samples) using a Qiagen RNeasy plant mini kit. RNA concentration was measured with a NanoDrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technologies). Subsequent procedures, including (1) quality controls of RNA, (2) preparation and biotin labeling of cRNA, (3) ATH1 GeneChip hybridizations, washing, staining, and scanning, were performed by ATLAS Biolabs. Processing of ATH1 GeneChip raw data (CEL files) was performed with open source ROBIN software (http://mapman.gabipd.org), which provides a user-friendly interface to powerful microarray processing. First, the CEL files were processed with RMA (log-scale robust multiarray analysis) based on Quantile normalization (Irizarry et al., 2003). Subsequently, a linear model was fitted using the R limma package (Smyth, 2004) and the contrasts (1) wild-type FN versus wild-type –N wild-type, (2) LBD37 OX –N versus wild-type –N, (3) LBD38 OX –N versus wild-type –N, as well as (4 and 5) lbd37 FN and lbd38 FN versus wild-type FN were extracted. P values were estimated using the empirical Bayes procedure implemented in limma. Differential expression across different comparisons was assessed by identifying genes having a false discovery rate below 5% using multiple testing adjustment (Benjamini and Hochberg, 1995) and identifying significant contrasts using the nestedF procedure in limma. The RMA-normalized log2 signal intensities for all 22,750 ATH1 probe sets from 18 arrays, the log2 fold changes, and adjusted P values for probe sets with ≥2-fold differential signal intensity are compiled in Supplemental Data Sets 1 and 2 online. Data and Affymetrix CEL-files are also accessible at the National Center for Biotechnology Information Gene Expression Omnibus database (Edgar et al., 2002) under accession number GSE18818.
Nonradioactive RNA Gel Blot Analysis
Total RNA was isolated from constitutive and inducible LBD OX lines or lbd37-1 mutants, and 10-μg aliquots were loaded on a formaldehyde-containing 1.5% (w/v) agarose gel and electrophoretically separated. For hybridization, digoxigenin-labeled PCR products (encoding gene-specific regions of each LBD gene) were used and detected by chemiluminescence using CDPStar reagent (New England Biolabs). Details are described by Peterhaensel et al. (1998).
Targeted Flavonoid Profiling
Shoot material from three biological replicates of freeze-dried 9-d-old seedlings of N-limited and N-replete LBD37OX, LBD38OX (two independent lines each), and wild-type seedlings, and N-sufficient lbd37-1, lbd37-2, lbd38, lbd39, and lbd38 lbd39 mutants and wild-type seedlings (∼20 mg fresh weight) were homogenized in five volumes of 50% methanol. Two-microliter aliquots were subjected to liquid chromatography–electrospray ionization–mass spectrometry analysis. Extraction and analysis of flavonoid pathway intermediates was performed as described previously (Tohge et al., 2005; Matsuda et al., 2009).
Lignin Staining
Inflorescence stems of 4-week-old plants grown on FN agar (0.5% w/v) plates were harvested, and 50-μm cross sections were cut from stems pieces of equal developmental age. Sections were put on a microscope slide and treated with one to two drops of freshly prepared phloroglucinol solution (25 mg phloroglucinol dissolved in 25 mL 100% ethanol and 25 mL 37% hydrochloric acid). After incubation for 5 min, the sections were observed using a Leica MZ 12.5 stereomicroscope, and photomicrographs were taken with a mounted digital camera system.
Determination of Nitrate, Nitrate Reductase Activity, Amino Acids, Protein, and Starch
Metabolites were extracted twice with 80% ethanol and once with 50% ethanol from shoot tissue of three biological replicates each pooled from five individual LBD37OX, LBD38OX, LBD39OX, and wild-type plants grown in high light conditions (600 μE) or 9-d-old N-sufficient lbd mutant and wild-type seedlings. The same ethanolic extracts were used for determination of nitrate, nitrate reductase activity, amino acids, protein, and starch. Details are described by Cross et al. (2006), Gibon et al. (2004), and Hendriks et al. (2003).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under AT5G67420 (LBD37), AT3G49940 (LBD38), AT4G37540 (LBD39), and the accession numbers compiled in Supplemental Table 1 online.
Author Contributions
G.R. and W.-R.S. designed the research and analyzed data. G.R. performed the research. T.T. and F.M. measured flavonoids and analyzed flavonoid profiling data. F.M. and K.S. contributed tools and equipment for flavonoid analysis. W.-R.S. wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. N/NO3−-Specific, Strong Induction of LBD37, LBD38, and LBD39.
Supplemental Figure 2. Anthocyanin Pigmentation Phenotypes.
Supplemental Figure 3. Nuclear Localization of LBD37-GFP Fusion Protein.
Supplemental Figure 4. Flavonoid Levels and Lignin Staining.
Supplemental Figure 5. Expression of Late Flavonoid Pathway Genes and Regulators in lbd37, lbd38, and lbd39 Mutants.
Supplemental Figure 6. qRT-PCR Transcript Profiling of Early Flavonoid Pathway Genes and Regulators.
Supplemental Figure 7. Expression of Flavonoid Pathway Genes and Regulators in Inducible LBD37 Overexpressor Lines.
Supplemental Figure 8. Comparison of Transcriptome Changes in Response to Nitrogen Status and LBD37 or LBD38 Overexpression.
Supplemental Figure 9. Expression of NRT2.1 and NIA Genes in LBD37 iOX Lines and lbd Mutant Seedlings.
Supplemental Figure 10. Amino Acid Levels in lbd Mutant Seedlings.
Supplemental Figure 11. Nitrogen-Dependent Basal Shoot Branching Phenotype.
Supplemental Figure 12. Expression of PAP1 and PAP2 in LBD37 Overexpressers during Environmental Stress Conditions.
Supplemental Table 1. Accession Numbers and Primer Sequences for (Semi-) Quantitative RT-PCR.
Supplemental Data Set 1. ATH1 GeneChip Results for Wild-Type and LBD Overexpressers.
Supplemental Data Set 2. ATH1 GeneChip Results for Wild-Type and lbd Mutants.
Supplementary Material
Acknowledgments
The work was supported by the Max-Planck Society. We thank our former and present colleagues Tomasz Czechowski, Jens-Holger Dieterich, Regina Feil, Yves Gibon, John Lunn, Rosa Morcuende, Magdalena Musialak-Lange, Armin Schlereth, Mark Stitt, Michael Udvardi, and Pia Walch-Liu for valuable input, help, discussions, and suggestions on the manuscript. Thanks also to Björn Usadel and Marc Lohse for implementing ROBIN, the software for processing and statistical treatment of the ATH1 GeneChip data. We also thank Josef Bergstein and Eugenia Maximova (Max-Planck Institute for Molecular Plant Physiology) for providing photographical and microscopy expertise. The SALK and Gabi-Kat consortia and ABRC/NASC are acknowledged for generating and providing T-DNA insertion lines. Finally, we also express our gratitude to Marc Lepetit (Institute for Integrative Plant Biology, Montpellier, France) and Steven J. Rothstein (University of Guelph, Ontario, Canada) for providing nia1 nia2 and nla mutant seeds, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wolf-Rüdiger Scheible (scheible@mpimp-golm.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Addo-Quaye, C., Eshoo, T.W., Bartel, D.P., and Axtell, M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi, A., Gestin, C., Leydecker, M.T., Bedu, M., Meyer, C., and Truong, H.N. (2005). Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 28 500–512. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., Gadrinab, C., and Heller, C. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57 289–300. [Google Scholar]
- Bernier, G., Havelange, A., Houssa, C., Petitjean, A., and Lejeune, P. (1993). Physiological signals that induce flowering. Plant Cell 5 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi, L., Bureau, M., and Simon, R. (2007). Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8 272–279. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., Taconnat, L., Renou, J.P., Daniel-Vedele, F., Fernandez, E., Meyer, C., and Krapp, A. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57 426–435. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior, A., Franken, J., Mes, J.J., Marsch-Martinez, N., Pereira, A., and Angenent, G.C. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 57 559–575. [DOI] [PubMed] [Google Scholar]
- Chen, Z.H., Jenkins, G.I., and Nimmo, H.G. (2008). Identification of an F-box protein that negatively regulates P(i) starvation responses. Plant Cell Physiol. 49 1902–1906. [DOI] [PubMed] [Google Scholar]
- Chen, Z.H., Nimmo, G.A., Jenkins, G.I., and Nimmo, H.G. (2007). BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 405 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.L., Acedo, G.N., Christinsin, M., and Conkling, M. (1992). Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc. Natl. Acad. Sci. USA 89 1861–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cominelli, E., Gusmaroli, G., Allegra, D., Galbiati, M., Wade, H.K., Jenkins, G.I., and Tonelli, C. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165 886–894. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M. (1995). Nitrate: Nutrient and signal for plant growth. Plant Cell 7 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M., and Forde, B.G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Cross, J.M., von Korff, M., Altmann, T., Bartzetko, L., Sulpice, R., Gibon, Y., Palacios, N., and Stitt, M. (2006). Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 142 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Bari, R.P., Stitt, M., Scheible, W.-R., and Udvardi, M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38 366–379. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., Routaboul, J.M., Alboresi, A., Weisshaar, B., and Lepiniec, L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55 940–953. [DOI] [PubMed] [Google Scholar]
- Edgar, R., Domrachev, M., and Lash, A.E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M.M. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, C., Palacios-Rojas, N., Feil, R., and Stitt, M. (2006). Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: Nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 46 533–548. [DOI] [PubMed] [Google Scholar]
- Gibon, Y., Blaesing, O.E., Hannemann, J., Carillo, P., Höhne, M., Hendriks, J.H., Palacios, N., Cross, J., Selbig, J., and Stitt, M. (2004). A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan, V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 189–194. [DOI] [PubMed] [Google Scholar]
- Gould, K.S., McKelvie, J., and Markham, K.R. (2002). Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ. 25 1261–1269. [Google Scholar]
- Gowri, G., Kenis, J.D., Ingemarsson, B., Redinbaugh, M.G., and Campbell, W.H. (1992). Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol. Biol. 18 55–64. [DOI] [PubMed] [Google Scholar]
- Guo, F.Q., Young, J., and Crawford, N.M. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks, J.H., Kolbe, A., Gibon, Y., Stitt, M., and Geigenberger, P. (2003). ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 133 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch, W.A., Singsaas, E.L., and McCown, B.H. (2003). Resorption protection. Anthocyanins facilitate nutrient recovery in autumn by shielding leaves from potentially damaging light levels. Plant Physiol. 133 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H.C., Wang, Y.Y., and Tsay, Y.F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57 264–278. [DOI] [PubMed] [Google Scholar]
- Husbands, A., Bell, E.M., Shuai, B., Smith, H.M., and Springer, P.S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Jonassen, E.M., Sévin, D.C., and Lillo, C. (June 17, 2009). The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J. Plant Physiol. 166 2071–2076. [DOI] [PubMed] [Google Scholar]
- Junker, B.H., Chu, C., Sonnewald, U., Willmitzer, L., and Fernie, A.R. (2003). In plants the alc gene expression system responds more rapidly following induction with acetaldehyde than with ethanol. FEBS Lett. 535 136–140. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Krapp, A., Fraisier, V., Scheible, W.R., Quesada, A., Gojon, A., Stitt, M., Caboche, M., and Daniel-Vedele, F. (1998). Expression studies of Nrt2:1Np, a putative high-affinity nitrate transporter: evidence for its role in nitrate uptake. Plant J. 14 723–731. [Google Scholar]
- Lea, U., Slimestad, R., Smedvig, P., and Lillo, C. (2007). Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225 1245–1253. [DOI] [PubMed] [Google Scholar]
- Lejay, L., Gansel, X., Cerezo, M., Tillard, P., Müller, C., Krapp, A., von Wirén, N., Daniel-Vedele, F., and Gojon, A. (2003). Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 15 2218–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay, L., Tillard, P., Lepetit, M., Olive, F., Filleur, S., Daniel-Vedele, F., and Gojon, A. (1999). Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18 509–519. [DOI] [PubMed] [Google Scholar]
- Lillo, C., Lea, U.S., and Ruoff, P. (2008). Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 31 587–601. [DOI] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, D.Y., Rao, H., Oliva, S., Daniel-Vedele, F., Krapp, A., and Malamy, J.E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 102 13693–13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, F., Yonekura-Sakakibara, K., Niida, R., Kuromori, T., Shinozaki, K., and Saito, K. (2009). MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 57 555–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55 954–967. [DOI] [PubMed] [Google Scholar]
- Médiène, S., Pagès, L., Jordan, M.-O., Le Bot, J., and Adamowicz, S. (2002). Influence of nitrogen availability on shoot development in young peach trees Prunus persica (L.) Batsch. Trends Ecol. Evol. 16 547–554. [Google Scholar]
- Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende, R., Bari, R., Gibon, Y., Zheng, W., Pant, B.D., Blasing, O., Usadel, B., Czechowski, T., Udvardi, M.K., Stitt, M., and Scheible, W.-R. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30 85–112. [DOI] [PubMed] [Google Scholar]
- Muños, S., Cazettes, C., Fizames, C., Gaymard, F., Tillard, P., Lepetit, M., Lejay, L., and Gojon, A. (2004). Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16 2433–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil, M., Obro, J., Willats, W.G., and Persson, S. (2008). GeneCAT – Novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36 W320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Todoriki, S., Masumizu, T., Suda, I., Furuta, S., Du, Z., and Kikuchi, S. (2003). Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis. J. Agric. Food Chem. 51 2992–2999. [DOI] [PubMed] [Google Scholar]
- Nazoa, P., Vidmar, J.J., Tranbarger, T.J., Mouline, K., Damiani, I., Tillard, P., Zhuo, D., Glass, A.D.M., and Touraine, B. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52 689–703. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, L., Müller, R., and Nielsen, T.H. (2007). Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30 1499–1512. [DOI] [PubMed] [Google Scholar]
- Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K., and Ohta, H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro, V., and Leyser, O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59 67–74. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Osuna, D., Usadel, B., Morcuende, R., Gibon, Y., Bläsing, O.E., Hohne, M., Gunter, M., Kamlage, B., Trethewey, R., Scheible, W.R., and Stitt, M. (2007). Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49 463–491. [DOI] [PubMed] [Google Scholar]
- Peng, M., Bi, Y.M., Zhu, T., and Rothstein, S.J. (2007. b). Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase NLA. Plant Mol. Biol. 65 775–797. [DOI] [PubMed] [Google Scholar]
- Peng, M., Hannam, C., Gu, H., Bi, Y.M., and Rothstein, S.J. (2007. a). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50 320–337. [DOI] [PubMed] [Google Scholar]
- Peng, M., Hudson, D., Schofield, A., Tsao, R., Yang, R., Gu, H., Bi, Y.M., and Rothstein, S.J. (2008). Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 59 2933–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., Khitrov, N., Auer, M., and Somerville, C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhaensel, C., Obermaier, I., and Rueger, B. (1998). Nonradioactive Northern blot analysis of plant RNA and the application of different haptens for reprobing. Anal. Biochem. 264 279–283. [DOI] [PubMed] [Google Scholar]
- Redinbaugh, M.G., and Campbell, W.H. (1991). Higher plant responses to environmental nitrate. Physiol. Plant. 82 640–650. [Google Scholar]
- Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., Tillard, P., Forde, B.G., and Gojon, A. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 103 19206–19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sagasser, M., Lu, G.-H., Hahlbrock, K., and Weisshaar, B. (2002). A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.-R., Gonzalez-Fontes, A., Lauerer, M., Muller-Rober, B., Caboche, M., and Stitt, M. (1997. a). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.-R., González-Fontes, A., Morcuende, R., Lauerer, M., Geiger, M., Glaab, J., Gojon, A., Schulze, E.-D., and Stitt, M. (1997. c). Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203 304–319. [DOI] [PubMed] [Google Scholar]
- Scheible, W.-R., Lauerer, M., Schulze, E.D., Caboche, M., and Stitt, M. (1997. b). Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J. 11 671–691. [Google Scholar]
- Scheible, W.-R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., Schindelasch, D., Thimm, O., Udvardi, M.K., and Stitt, M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article 3. [DOI] [PubMed]
- Solfanelli, C., Poggi, A., Loreti, E., Alpi, A., and Perata, P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano, T., Thitamadee, S., Machida, Y., and Chua, N.H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20 3359–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, M. (1999). Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2 178–186. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., and Weisshaar, B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei, K., Takahashi, T., Sugiyama, T., Yamaya, T., and Sakakibara, H. (2002). Multiple routes communicating nitrogen availability from roots to shoots: A signal transduction pathway mediated by cytokinin. J. Exp. Bot. 53 971–977. [DOI] [PubMed] [Google Scholar]
- Taramino, G., Sauer, M., Stauffer, J.L., Jr., Multani, D., Niu, X., Sakai, H., and Hochholdinger, F. (2007). The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50 649–659. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Tschoep, H., Gibon, Y., Carillo, P., Armengaud, P., Szecowka, M., Nunes-Nesi, A., Fernie, A.R., Koehl, K., and Stitt, M. (2009). Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ. 32 300–318. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., Ohshima, T., Naito, S., Chino, M., and Komeda, Y. (1991). Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol. 97 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi, M.K., Czechowski, T., and Scheible, W.-R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., Magome, H., Kamiya, Y., Shirasu, K., Yoneyama, K., Kyozuka, J., and Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200. [DOI] [PubMed] [Google Scholar]
- Walch-Liu, P., Neumann, G., Bangerth, F., and Engels, C. (2000). Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J. Exp. Bot. 51 227–237. [DOI] [PubMed] [Google Scholar]
- Wang, R., Okamoto, M., Xing, X., and Crawford, N.M. (2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Tischner, R., Gutierrez, R.A., Hoffman, M., Xing, X., Chen, M., Coruzzi, G., and Crawford, N.M. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Xing, X., and Crawford, N.M. (2007). Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 145 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Xing, X., Wang, Y., Tran, A., and Crawford, N.M. (2009). A genetic screen for nitrate-regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 135 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangwattana, B., Koyama, Y., Nishiyama, Y., Kitayama, M., Yamazaki, M., and Saito, K. (2008). Characterization of PAP1-upregulated glutathione S-transferase genes in Arabidopsis thaliana. Plant Biotechnol. 25 191–196. [Google Scholar]
- Winkel-Shirley, B. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127 1399–1404. [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Jennings, A., Barlow, P.W., and Forde, B.G. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 96 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.