Abstract
MicroRNAs (miRNAs) are small silencing RNAs with regulatory roles in gene expression. miRNAs interact with Argonaute (AGO) proteins to form effector complexes that cleave target mRNAs or repress translation. Rice (Oryza sativa) encodes four AGO1 homologs (AGO1a, AGO1b, AGO1c, and AGO1d). We used RNA interference (RNAi) to knock down the four AGO1s. The RNAi lines displayed pleiotropic developmental phenotypes and had increased accumulation of miRNA targets. AGO1a, AGO1b, and AGO1c complexes were purified and further characterized. The three AGO1s all have a strong preference for binding small RNAs (sRNAs) with 5′ U and have Slicer activity. We cataloged the sRNAs in each AGO1 complex by deep sequencing and found that all three AGO1s predominantly bound known miRNAs. Most of the miRNAs were evenly distributed in the three AGO1 complexes, suggesting a redundant role for the AGO1s. Intriguingly, a subset of miRNAs were specifically incorporated into or excluded from one of the AGO1s, suggesting functional specialization among the AGO1s. Furthermore, we identified rice miRNA targets at a global level. The validated targets include transcription factors that control major stages of development and also genes involved in a variety of physiological processes, indicating a broad regulatory role for miRNAs in rice.
INTRODUCTION
In plants, small RNA (sRNA)-based gene silencing systems play important roles in developmental regulation, responses to biotic and abiotic stresses, and epigenetic control of transposable elements (Baulcombe, 2004; Vaucheret, 2006). Central to the sRNA pathways are proteins in the Dicer and Argonaute (AGO) families. Dicer or Dicer-like (DCL) proteins contain two RNaseIII domains and process double-stranded RNAs into ∼21- to 25-nucleotide sRNA duplexes. Upon dicing, a selected strand of the sRNA duplex is bound by an AGO protein to form the core of the effector complex termed RNA-induced silencing complex (RISC). RISC is guided by the sRNA to act on its target (mRNA or chromatin), resulting in mRNA degradation, translational repression, or chromatin modifications (Hannon, 2002; Baulcombe, 2004).
Our understanding of the mechanism and biology of plant sRNA pathways comes mainly from studies using Arabidopsis thaliana and rice (Oryza sativa) as model systems. Thus far, five classes of sRNAs have been discovered in plants: miRNAs and four types of small interfering RNAs (siRNAs), including trans-acting siRNAs (ta-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs), repeat-associated siRNAs (ra-siRNAs), and long siRNA (lsiRNA). Most miRNA loci are encoded by independent transcriptional units in intergenic regions that are transcribed by RNA polymerase II (Xie et al., 2005b). The primary transcript of a miRNA contains a stem-loop structure that can be recognized and processed by DCL1 into a miRNA precursor (pre-miRNA) and further processed by DCL1 into a miRNA/miRNA* duplex (Voinnet, 2009). Rice also encodes natural antisense transcript miRNAs (nat-miRNAs) (Lu et al., 2008). nat-miRNAs are derived from overlapping transcripts antisense to their targets; the maturation of nat-miRNAs resembles that of canonical miRNAs and requires DCL1. Plant miRNAs have near-perfect pairing to their targets and repress target gene expression primarily through mRNA cleavage (Llave et al., 2002). However, there are evidences supporting the notion that miRNA-mediated gene regulation also operates through repression of translation (Aukerman and Sakai, 2003; Chen, 2004; Brodersen et al., 2008; Lanet et al., 2009). ta-siRNAs are a plant-specific class of siRNAs, the production of which is initiated by miRNA-mediated cleavage of noncoding transcripts. The cleavage products serve as substrates for an RNA-dependent RNA polymerase (RDR6 in Arabidopsis) to generate double-stranded RNAs that are further processed by DCL4 into phased siRNAs (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Xie et al., 2005a; Yoshikawa et al., 2005; Axtell et al., 2006). Arabidopsis ta-siRNAs regulate the juvenile-to-adult vegetative phase change (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). nat-siRNAs are derived from natural cis-antisense transcript pairs and are involved in cellular responses to salt stress and pathogen attack (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). lsiRNAs are 30 to ∼40 nucleotides in length and are induced by pathogen infection or under specific growth conditions; the mechanism of lsiRNA biogenesis remains elusive (Katiyar-Agarwal et al., 2007). Plants also contain an abundant class of ∼24-nucleotide ra-siRNAs derived from transposons and repetitive elements. The biogenesis of ra-siRNAs requires activities of DCL3, RDR2, and Pol IV, a plant-specific DNA-dependent RNA polymerase. ra-siRNAs play a role in the methylation and silencing of many transposons and also some genes that are adjacent to repeats (Henderson and Jacobsen, 2007; Zaratiegui et al., 2007).
All types of sRNAs interact with AGO proteins to exert their functions. AGOs contain three characteristic domains: PAZ, MID, and PIWI (Song and Joshua-Tor, 2006). The PAZ domain binds to the 3′ end of sRNAs (Ma et al., 2004), whereas the MID domain provides a binding pocket for the 5′ end (Ma et al., 2005) and is thought to confer the binding specificity of an AGO to an sRNA with a particular 5′ end nucleotide (Mi et al., 2008). PIWI domain adopts the structure of RNase H that contains the catalytic site formed by three residues (Asp, Asp, and His) and provides the Slicer activity that executes the sRNA-guided cleavage of target RNA (Liu et al., 2004; Song et al., 2004; Rivas et al., 2005). Arabidopsis encodes 10 AGO proteins (Morel et al., 2002; Zheng et al., 2007). AGO1 mainly recruits miRNAs that initiate with a 5′ U (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008), whereas AGO4 and AGO6 play partially redundant roles in ra-siRNA–mediated silencing of repeats (Zilberman et al., 2003; Qi et al., 2006; Zheng et al., 2007). AGO7 interacts with miR390 that is required for the biogenesis of ta-siRNAs (Montgomery et al., 2008). AGO10 is involved in the maintenance of the stem cell population in shoot apical meristem (Moussian et al., 1998; Lynn et al., 1999), but its associated sRNAs remain to be identified. We and others have previously shown that AGO2 binds to sRNAs with 5′ A, whereas AGO5 has a binding preference for sRNAs that initiate with C (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008). However, the biological functions of these AGOs remain to be understood.
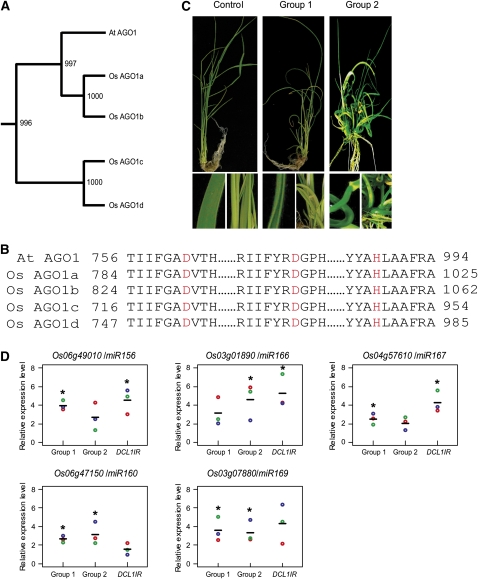
Compared with 10 AGO family members in Arabidopsis, rice has 19 AGO family members that can be phylogenetically divided into four subgroups: MEL1, AGO1, AGO4, and AGO7 (see Supplemental Figure 1 and Supplemental Data Set 1 online). The MEL1 subgroup contains five rice AGOs, including MEL1. MEL1 is specifically expressed in germ cells and involved in the progression of premeiotic mitosis and meiosis during sporogenesis (Nonomura et al., 2007). The MEL1-associated sRNAs remain to be identified, and it is unknown whether the other four members in the subgroup also play a role in sporogenesis. The AGO7 subgroup contains three members, namely AGO2, AGO3, and SHL4. SHL4 has a demonstrated role in the ta-siRNA pathway (Nagasaki et al., 2007). In the AGO4 subgroup, AGO4a and AGO4b share high similarity to Arabidopsis AGO4, whereas AGO16 is closely related to Arabidopsis AGO6. It remains to be tested whether these AGOs play a role in repeat silencing as their counterparts in Arabidopsis. The AGO1 subgroup includes one AGO (PNH1) (Nishimura et al., 2002) that shares high similarity with Arabidopsis AGO10 and four AGO1 homologs (AGO1a, AGO1b, AGO1c, and AGO1d) that are clustered with Arabidopsis AGO1 (Figure 1A; see Supplemental Data Set 2 online). All four rice AGO1 homologs have the catalytic site formed by three residues (Asp, Asp, and His) (Figure 1B).
Figure 1.
Knockdown of Rice AGO1s Results in Increased Accumulation of miRNA Targets and Pleiotropic Developmental Phenotypes.
(A) Phylogenetic relationships of Arabidopsis and rice AGO1 proteins. Alignments of full-length AGO protein sequences were produced by ClustalW and used for phylogenetic analysis. The midpoint-rooted phylogenetic tree was constructed with the MEGA program using the neighbor-joining method with bootstrap values from 1000 trials. The alignment file is included in Supplemental Data Set 2 online.
(B) A partial alignment of the PIWI domains of Arabidopsis and rice AGO1proteins. The residues forming the catalytic DDH motif are shown in red. The starting and ending positions of the sequences are as labeled.
(C) Phenotypes of a weak loss of AGO1 function RNAi line (Group 1), a strong loss of AGO1 function line (Group 2), and a line transformed with an empty vector (Control). Group 1 lines showed mild dwarfism with narrow and rolled leaves and lower seed-setting rate. Group 2 lines displayed pleiotropic developmental phenotypes, including severe dwarfism and tortuous shoot.
(D) Relative expression levels of five miRNA target genes in the leaves of two AGO1 RNAi lines, DCL1IR line, and control plants. The target gene expression levels were normalized using the signal from the GAPDH gene. Values (colored circles) from three biological repeats of quantitative RT-PCR are shown. Horizontal bars represent the average values. Asterisks show where differences between RNAi lines and the control line were significant (P ≤ 0.05 from Student's t test).
To examine the function of rice AGO1s, here, we used a combination of genetic, biochemical, and bioinformatic techniques to examine the phenotypes of AGO1 knockdown lines, to characterize the set of sRNAs present within each AGO1 complex and to identify the target genes of miRNAs. Our analyses demonstrated the importance of AGO1 proteins in rice development and suggested that different AGO1 complexes have both overlapping and nonredundant functions in rice miRNA pathway.
RESULTS
Knockdown of Rice AGO1s Results in Increased Levels of miRNA Targets and Pleiotropic Developmental Phenotypes
To investigate the function of rice AGO1s in the RNAi pathways, especially in the miRNA pathways, we used an RNAi approach to knock down AGO1s. We designed an inverted repeat (IR) construct (AGO1IR) targeting the four rice AGO1 homologs, AGO1a, b, c, and d. Over 30 transgenic lines were regenerated and displayed various degrees of developmental defects. Based upon the severity of developmental defects, these lines could be divided into two groups, designated Groups 1 and 2 (Figure 1C). Weak loss-of-function lines (Group 1) showed mild dwarfism with narrow and rolled leaves and lower seed-setting rate. Strong loss-of-function lines (Group 2) displayed pleiotropic developmental phenotypes, including severe dwarfism and tortuous shoot, and their development was arrested at young seedling stage (Figure 1C). These phenotypes are reminiscent of those caused by loss-of-function of DCL1, which is required for rice miRNA biogenesis (Liu et al., 2005).
To examine whether the observed phenotypes correlated with IR-mediated downregulation of AGO1s, we analyzed the expression levels of the IR-derived sRNAs and AGO1s. RNA gel blot analysis of representative transgenic lines showed that both Groups 1 and 2 expressed sRNAs targeting AGO1s (see Supplemental Figure 2A online). The accumulation levels of all four AGO1s were decreased by ∼35 to 70% in Group 1 and ∼40 to 90% in Group 2, as measured by quantitative RT-PCR (see Supplemental Figure 2B online). As a control, the expression of AGO4a was not significantly changed in the transgenic RNAi lines (see Supplemental Figure 2B online).
In order to determine whether AGO1s are required for miRNA-directed target gene regulation in rice, we used quantitative RT-PCR to examine the accumulation levels of several miRNA target genes in AGO1IR lines. As shown in Figure 1D, the levels of all the target genes in the AGO1IR lines were increased by threefold to fivefold compared with those in control plants and were comparable to those in the DCL1IR line, indicating that rice AGO1s are involved in miRNA pathway, as is their homolog in Arabidopsis.
Purification and Characterization of Rice AGO1s
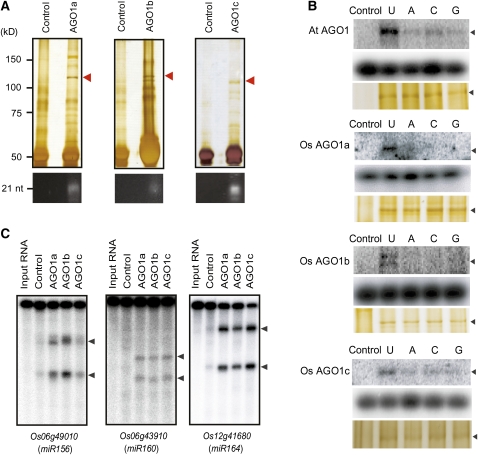
To characterize the rice AGO1s biochemically, we purified rice AGO1a, AGO1b, and AGO1c by immunoprecipitation using antibodies that specifically recognize the N-terminal sequence of each AGO1. As shown on silver-stained gels (Figure 2A), bands of predicted sizes of each AGO1 were recovered from immunoprecipitations using antibodies raised against each AGO1 but not from control experiments, and their identities were further verified by mass spectrometry (see Supplemental Figure 3 online). Immunoblot analysis with each immunoprecipitate showed that there were no obvious cross-reactions between the antibodies (see Supplemental Figure 4 online). Next, we extracted RNAs from the purified AGO1 complexes. The RNAs were resolved on denaturing polyacrymide gels and visualized by SYBR gold staining. We found that all three rice AGO1s bound sRNAs of 21 nucleotides predominantly (Figure 2A).
Figure 2.
Purification and Characterization of Rice AGO1 Complexes.
(A) The AGO1 complexes were immunopurified using peptide-specific antibodies and separated on 10% SDS-PAGE (top panels). Preimmune antisera were used for the control purifications. The proteins were visualized by silver staining. The positions of protein size markers, electrophoresed in parallel, are shown to the left of the gels. The AGO1 protein bands are indicated by red arrowheads. sRNAs were extracted from each AGO1 complex, analyzed by denaturing polyacrymide gels, and visualized by SYBR gold staining (bottom panels). The position of a RNA size marker is shown to the left.
(B) Immunoprecipitates of Arabidopsis AGO1 and rice AGO1a, AGO1b, and AGO1c, as indicated, were incubated with single-stranded 32P-labeled 21-nucleotide siRNAs bearing the indicated 5′ terminal nucleotides. Mixtures were irradiated with UV and resolved by 10% SDS-PAGE. The cross-linked products are indicated by arrowheads. Lower regions of the gels with a shorter exposure (middle panels) are shown to indicate that equal amounts of siRNAs were added into each reaction. Silver-stained gels (bottom panels) are shown as controls for the proteins used in the cross-linking reactions.
(C) Cleavage activity of rice AGO1 complexes was assayed by incubating the indicated immunoprecipitates with uniformly labeled rice miRNA target transcripts. The cleavage products were resolved in 8% denaturing gels. Positions of 5′ and 3′ cleavage products are indicated by the arrowheads.
[See online article for color version of this figure.]
We have previously shown that different Arabidopsis AGOs preferentially recognize and recruit sRNAs that initiate with different 5′-end nucleotides (Mi et al., 2008). To test whether this also holds true for rice AGO1s, we incubated immunopurified AGO1a, AGO1b, and AGO1c with four 21-nucleotide RNA oligos that only differ at their 5′ ends (U, A, C, and G, respectively). The reaction mixtures were then irradiated with UV and resolved by SDS-PAGE. As shown in Figure 2B, all three rice AGO1 exhibited a much higher binding affinity for sRNA beginning with a U than for sRNAs initiating with an A, C, or G, similar to Arabidopsis AGO1. This suggests that the sorting of sRNAs into rice AGOs could also be determined by the 5′ end nucleotide.
An alignment of the PIWI domain of rice AGO1s with Arabidopsis AGO1 revealed that all rice AGO1s possess the residues forming the catalytic triad (Figure 1B), suggesting that rice AGO1s are Slicers. To determine whether rice AGO1s have cleavage activity, we incubated AGO1 immunoprecipitates with in vitro–transcribed target RNAs that are complementary to several miRNAs. All three AGO1 complexes indeed cleaved targets of miR156 (Os06g49010), miR160 (Os06g43910), and miR164 (Os12g41680) (Figure 2C). These data indicated that AGO1 proteins bound to miRNAs in vivo and formed cleavage-competent RISC complexes.
Profiling of sRNAs Associated with Rice AGO1s by Deep Sequencing
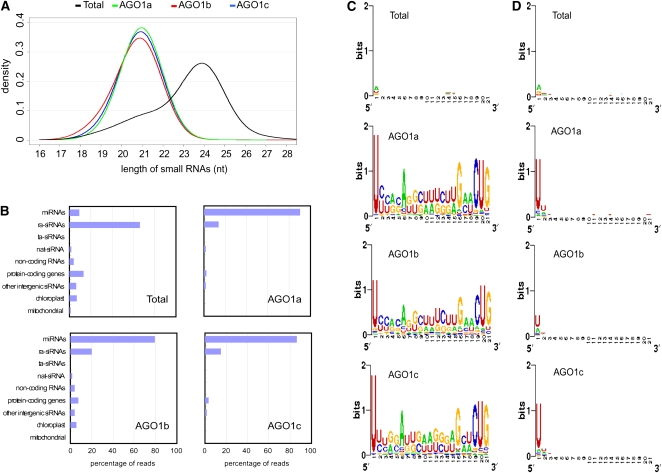
To further understand the biological functions of AGO1s, we determined the sRNA populations that are present in each rice AGO1 complex. sRNAs were purified from the gels, sequentially ligated to RNA adaptors at their 5′ and 3′ ends. The ligation products were converted to cDNAs by RT-PCR and then subjected to Illumina 1G sequencing (Margulies et al., 2005). For comparison, sRNAs isolated from total extract (referred to as Total hereafter) were also sequenced. After removing the adaptor sequences, the sequences were compared with rice nuclear and organellar genomes, and the sequences that perfectly matched to the genomes were used for further analysis. In total, 4,029,462, 3,791,013, 1,230,668, and 3,891,687 sRNA reads were obtained for Total, AGO1a, AGO1b, and AGO1c, respectively (see Supplemental Table 1 online). These reads represent 1,991,942 (Total), 22,588 (AGO1a), 4911 (AGO1b), and 16,024 (AGO1c) unique sRNA sequences (see Supplemental Table 1 online). While sRNAs of 24 nucleotides were the predominant size class in Total, those from AGO1 complexes were mostly 21 nucleotides in length (Figure 3A), which is in full agreement with SYBR gold staining results (Figure 2A).
Figure 3.
Profiling of sRNAs Associated with Rice AGO1 Complexes by Deep Sequencing.
(A) Size distribution of sequenced sRNAs in total RNA (Total) and those bound by AGO1a, AGO1b, and AGO1c complexes.
(B) Bar charts summarizing the annotation of sRNA populations in total RNA (Total) and those bound by the three AGO1 complexes. Some sRNAs can match more than one category of the sequences listed, so the sum of the numbers may be bigger than the input total number.
(C) The relative nucleotide bias at each position of the sRNAs in total RNA (Total) and those bound by AGO1a, AGO1b, and AGO1c complexes.
(D) The relative nucleotide bias at each position of the sRNAs (excluding miRNAs) in total RNA (Total) and those bound by AGO1a, AGO1b, and AGO1c complexes. The graphics in (C) and (D) were made using WebLogo (Crooks et al., 2004). The sequence conservation at each position is indicated by the overall height of the stack of symbols (U, A, C, and G), while the relative frequency of each nucleotide is represented by the height of the corresponding symbol.
sRNAs were categorized based on their genomic locations and functions (Figure 3B; see Supplemental Table 1 online). Sixty-six percent of the Total sRNA reads were matched to repetitive sequences in the genome, whereas 14% of AGO1a, 20% of AGO1b, and 16% of AGO1c were repeat-derived. By contrast, only 9% of the Total were mapped to annotated miRNA precursors, whereas 90% of AGO1a, 79% of AGO1b, and 86% of AGO1c were miRNAs. These data indicated that miRNAs are highly enriched in rice AGO1s and that AGO1s play a major role in the miRNA pathway.
Statistical analysis with the sRNA data sets revealed that rice AGO1s displayed a strong preference for sRNAs initiating with a U (Figure 3C). It was possible that such bias was due to the fact that AGO1s preferentially bind to miRNAs that have propensity to begin with a U. To rule out this possibility, we did the analysis with sRNA data sets excluding miRNAs, and we obtained similar results (Figure 3D). These observations were in agreement with the results obtained from the in vitro sRNA binding experiments (Figure 2B).
Identification of Novel Rice miRNAs
Exhaustive bioinformatic and sRNA sequencing efforts have identified many conserved and nonconserved miRNAs in rice (Wang et al., 2004a; Liu et al., 2005; Sunkar et al., 2005, 2008; Heisel et al., 2008; Lu et al., 2008; Morin et al., 2008; Zhu et al., 2008). However, the limited overlap between the results from these studies suggests that more nonconserved miRNAs remain to be discovered. We applied the criteria that have been recently proposed (Meyers et al., 2008) to identify novel miRNAs in our Total and AGO1 data sets. Twenty-five novel miRNAs that belong to 20 families were identified (Table 1; see Supplemental Data Set 3 online). It is noteworthy that, among these new miRNAs, some are 24 nucleotides in length. Most of these miRNAs are encoded by a single locus, suggesting their recent evolutionary origin. Consistent with this observation, none of these miRNAs have significant conserved counterparts in any other plant species with genomic or EST sequences based on sequence similarity.
Table 1.
Newly Identified miRNAs
| miRNA Readsa |
miR* Readsa |
|||||||
|---|---|---|---|---|---|---|---|---|
| miRNA family | Sequence (5′–3′) | Size | Loci | Total | AGO1s | Total | AGO1s | Precursor Location |
| osa-miR2862 | UCCAACAGCUUAGAUUCGUCC | 21 | 1 | 0 | 165 | 8 | 32 | chr01:27131024..27130804 |
| osa-miR2863a | UUGUCCCAUUCUAGUUUAGCU | 21 | 1 | 6 | 93 | 2 | 45 | chr03:27057512..27057632 |
| osa-miR2863b | UUCGUUUAUUUGGACUAGAGU | 21 | 1 | 7 | 45 | 22 | 0 | chr02:4195762..4195662 |
| osa-miR2864.1 | UUUUGCUGCCCUUGUUUUGCA | 21 | 1 | 18 | 177 | 4 | 0 | chr12:25631216..25631116 |
| osa-miR2864.2 | UUGUUUUGCAUUGUAUAGGUA | 21 | 17 | 128 | 0 | 0 | ||
| osa-miR2865 | CUCAGCAGUCGACUGUACCGUG | 22 | 1 | 2 | 13 | 1 | 0 | chr09:8514793..8514693 |
| osa-miR2866 | UCUAGUUUGUGUUCAGCAUC | 20 | 1 | 0 | 16 | 3 | 0 | chr03:7310813..7310713 |
| osa-miR2867 | UGUGCCAUCCCACACAUCCCGA | 22 | 1 | 0 | 14 | 1 | 0 | chr11:15775943..15776043 |
| osa-miR2868 | UUGGUUUUGUGUAGUAGAAA | 20 | 1 | 0 | 20 | 4 | 0 | chr11:17435548..17435628 |
| osa-miR2869 | UCCCGACAUUAAAUUCUGGGC | 21 | 1 | 29 | 18 | 1 | 0 | chr05:26832219..26832059 |
| osa-miR2870 | UAAUCAGUUUGGGGAGACAAA | 21 | 1 | 11 | 16 | 1 | 0 | chr01:27508880..27508720 |
| osa-miR2871a | UAUUUUAGUUUCUAUGGUCAC | 21 | 2 | 188 | 98 | 10 | 0 | chr05:3855032..3855152 |
| osa-miR2871b | UAUUUUAGUUUCUAUGGUCAC | 21 | 2 | 188 | 98 | 0 | 0 | chr04:8320641..8320781 |
| osa-miR2872 | UGGGGUUCUACAAACCGAACU | 21 | 1 | 60 | 47 | 7 | 9 | chr11:2520898..2521257 |
| osa-miR2873 | AAGUUUGGACUUAAAUUUGGUAAC | 24 | 1 | 208 | 0 | 1 | 0 | chr11:28375679..28375579 |
| osa-miR2874 | AUGUGAACAGUGUCAAACAGUGUC | 24 | 1 | 292 | 0 | 7 | 0 | chr08:15465185..15465285 |
| osa-miR2875 | AUUUACAGUCAUAUACAGUUUAUA | 24 | 1 | 33 | 0 | 14 | 0 | chr08:26959218..26959298 |
| osa-miR2876.1 | UUCCUAUAUGAACACUGUUGC | 21 | 1 | 7 | 102 | 0 | 0 | chr06:17008158..17007978 |
| osa-miR2876.2 | UCCUAUAUGAACACUGUUGCCAGC | 24 | 7 | 0 | 2 | 0 | ||
| osa-miR2877 | UUGCAUCCUCUGCACUUUGGGCCU | 24 | 1 | 18 | 0 | 1 | 0 | chr04:28366463..28366103 |
| osa-miR2878-5p | UACAUGUAUAAAAUUCUGAGGAUG | 24 | 1 | 164 | 0 | 0 | 0 | chr08:26048530..26048630 |
| osa-miR2878-3p | CAGGAUUUUAUACAUGUAAAGAAU | 24 | 175 | 0 | 134 | 0 | ||
| osa-miR2879 | GCCAGAUGUGUUAAAAUAAUGACC | 24 | 1 | 86 | 0 | 1 | 0 | chr02:18890039..18890339 |
| osa-miR2880 | ACGGUAUCCCGUUCGGACAGGAUG | 24 | 1 | 12 | 0 | 1 | 0 | chr03:22696406..22696227 |
| osa-miR2905 | UACAUGUCAGUGACAAAGGCA | 21 | 1 | 20 | 33 | 2 | 0 | chr03:13570806..13570726 |
| osa-miR168a-3p | GAUCCCGCCUUGCACCAAGUGAAU | 24 | 1 | 333 | 0 | 0 | 0 | chr02:1553240..1553154 |
| osa-miR393b-3p | UCAGUGCAAUCCCUUUGGAAU | 21 | 1 | 8251 | 10578 | 3058 | 270 | chr04:34712941..34712810 |
| osa-miR395a.2 | UGAAGUGUUUGGGGGAACUC | 20 | 17 | 165 | 1107 | 0 | 0 | chr04:31586572..31586431 |
| osa-miR396e-3p | AUGGUUCAAGAAAGCCCAUGGAAA | 24 | 9 | 133 | 0 | 0 | 0 | chr04:34217758..34217941 |
| osa-miR396f-3p | AUAGUUCAAGAAAGUCCUUGGAAA | 24 | 9 | 532 | 0 | 0 | 0 | chr02:35630852..35630677 |
| osa-miR396g | UCCACAGGCUUUCUUGAACGG | 21 | 9 | 147 | 1493 | 0 | 0 | chr06:5299811..5299631 |
| osa-miR396 h | UCCACAGGCUUUCUUGAACGG | 21 | 9 | 147 | 1493 | 0 | 0 | chr02:32835792..32835932 |
| osa-miR396i | UCCACAGGCUUUCUUGAACGG | 21 | 9 | 147 | 1493 | 0 | 0 | chr04:30101014..30101134 |
| osa-miR397a.2 | UUGAGUGCAGCGUUGAUGAAC | 21 | 2 | 581 | 18447 | 0 | 0 | chr06:28488787..28488900 |
| osa-miR397b.2 | UUGAGUGCAGCGUUGAUGAAC | 21 | 2 | 581 | 18447 | 5 | 976 | chr02:3280896..3280779 |
| osa-miR437-3p.2 | AAAGUUAGAGAAGUUUGACUUAGG | 24 | 1 | 273 | 0 | 20 | 0 | chr02:17044678..17044466 |
| osa-miR437-3p.3 | AGAAGUUUGACUUAGGACAAAACU | 24 | 560 | 0 | 0 | 0 | ||
| osa-miR820a-5p.2 | UCGGCCUCGUGGAUGGACCAGGAG | 24 | 3 | 205 | 0 | 0 | 0 | chr01:14113150..14112960 |
| osa-miR820b-5p.2 | UCGGCCUCGUGGAUGGACCAGGAG | 24 | 3 | 205 | 0 | 35 | 0 | chr07:13119040..13118840 |
| osa-miR820c-5p.2 | UCGGCCUCGUGGAUGGACCAGGAG | 24 | 3 | 205 | 0 | 26 | 0 | chr10:6693840..6694030 |
| osa-miR1317-3p | GAAAUGAUCUUGGACGUAAUCUAG | 24 | 1 | 589 | 0 | 414 | 0 | chr10:9989677..9989517 |
| osa-miR1317-5p.2 | AGAUUGCUUUCAAGGUCAUUUCUU | 24 | 414 | 0 | 589 | 0 | ||
| osa-miR1320-3p | UGUAAAAUUCAUUCGUUCCAA | 21 | 1 | 63 | 20 | 2 | 0 | chr06:5072603..5072508 |
| osa-miR1423-5p.2 | AGGCAACUACACGUUGGGCGCUCG | 24 | 1 | 76 | 0 | 47 | 0 | chr04:19527720..19527855 |
| osa-miR1423-3p.2 | AGCGCCCAAGCGGUAGUUGUCUCC | 24 | 47 | 0 | 76 | 0 | ||
| osa-miR1429-5p.2 | GUAAUAUACUAAUCCGUGCAUCCA | 24 | 1 | 327 | 0 | 221 | 0 | chr08:2030728..2030605 |
| osa-miR1429-3p.2 | GUUGCACGGGUUUGUAUGUUGCAG | 24 | 221 | 0 | 327 | 0 | ||
| osa-miR1849.2 | AAUGCCCUAUCGUAUCCUAGGUUG | 24 | 1 | 127 | 0 | 32 | 0 | chr04:22005738..22005827 |
| osa-miR1850.2 | GAAGUUGUGUGUGAACUAAACGUG | 24 | 1 | 168 | 0 | 0 | 0 | chr05:26212703..26212571 |
| osa-miR1857-5p.2 | UGGAGCAUGAGGUUAUCUCUC | 21 | 1 | 14 | 118 | 0 | 0 | chr11:2729806..2729935 |
| osa-miR1863b | AGCUCUGAUACCAUGUUAACUGUU | 24 | 2 | 23 | 0 | 1 | 0 | chr12:4866242..4866601 |
| osa-miR1863c | UAGAAACUUGGCUGAUGCAUUACU | 24 | 1 | 7 | 0 | 1 | 0 | chr12:4868192..4868482 |
| osa-miR1868.2 | AAGCGUGCUCACGGAAAACGAGGG | 24 | 1 | 1466 | 0 | 0 | 0 | chr04:19543483..19543651 |
| osa-miR1870-3p | UUUAGGGCUAAUUCAGCAUGAACA | 24 | 1 | 159 | 0 | 0 | 0 | chr06:21163758..21163966 |
| osa-miR1873.2 |
ACCUCAACAUGGUAUCAGAGCUGG |
24 |
1 |
34 |
0 |
0 |
0 |
chr07:12754920..12755116 |
Total reads in Total and AGO1 data sets.
In addition to the novel miRNAs, we also identified additional loci or new variants for a number of previously annotated miRNA families (Table 1; see Supplemental Data Set 3 online). miR396 has three sequence variants that are encoded by six loci (miR396a∼f), these variants differ in one to two nucleotides (see Supplemental Figure 5 online). In our data sets, we found an additional variant; the sequence of this new variant is different from known variants by one to two nucleotides. Intriguingly, this variant originates from the antisense strands of their targets, MADS box genes (see Supplemental Figure 5 online). Thus, we propose that this new miR396 variant is a nat-miRNA. In our data sets, we also detected 24-nucleotide sRNAs produced from the previously annotated miRNA precursors. Most of these 24-nucleotide sRNAs had star sequences and accumulated to equal or higher levels than the annotated 21-nucleotide miRNAs. We annotated these sRNAs as miRNA variants of the corresponding miRNA families (Table 1; see Supplemental Data Set 3 online).
Distribution of sRNAs among Rice AGO1 Complexes
With the findings that all three characterized rice AGO1 homologs predominantly recruit miRNAs and display the same 5′ nucleotide preference, we sought to examine whether rice AGO1s have redundancy or specificity in recruiting sRNAs.
We first examined the sequencing frequency of previously annotated miRNAs and newly identified miRNAs in Total and AGO1 data sets. No sequencing reads were found for miR413-420 and miR426. These miRNAs were identified solely by computational methods (Wang et al., 2004b). For miR439, 441-443, 445, 446, 806-819, 821, 1319, 1436-1442, and 1847, scattered sRNAs were detected across both sense and antisense strands of the precursors, indicating their origins of perfectly double-stranded RNAs formed by sense and antisense transcripts of the loci. Thus, these sRNAs are unlikely bona fide miRNAs and were excluded from further analysis.
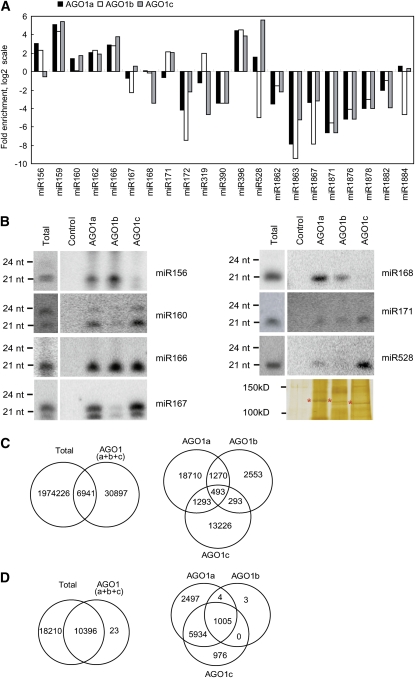
The distribution of the remaining miRNAs within the rice AGO1 complexes was assessed by calculating enrichment or depletion in the AGO1 immunoprecipitates relative to the total extract. Normalized reads in Total and AGO1 data sets were determined for each miRNA family, and enrichment or depletion in each AGO1 complexes was calculated using the reads in each AGO1/the reads in Total ratio (Figure 4A; see Supplemental Table 2 online). The distribution of some of the miRNAs was also confirmed by RNA gel blot analysis (Figure 4B). We found that most of the known miRNAs were evenly distributed in the three rice AGO1s, suggesting a redundant role for AGO1s in recruiting these miRNAs. Intriguingly, we also found that a subset of miRNAs was specifically incorporated into or excluded from one of the AGO1s. For instance, miR156 and 168 were predominantly associated with AGO1a and AGO1b, not with AGO1c; miR160 and 167 were mainly recruited by AGO1c, not by AGO1b; whereas miR528 was overwhelmingly associated with AGO1c. We also found that a subset of miRNAs was underrepresented in all three AGO1s. These include miR172 and 390, both of which are initiated with an A. Intriguingly, we noticed that the 24-nucleotide miRNAs were not present in either AGO1s.
Figure 4.
Distribution of sRNAs among Rice AGO1 Complexes.
(A) The distribution of miRNAs within the rice AGO1 complexes was assessed by calculating enrichment or depletion in the AGO1 immunoprecipitates relative to the total extract. Normalized reads in Total and AGO1 data sets were determined for each miRNA family. Enrichment or depletion in each AGO1 complex was calculated using the reads in each AGO1/the reads in Total ratio and is shown as fold enrichment on a log scale.
(B) Detection of miRNAs in total RNA, AGO1a, AGO1b, AGO1c, and control immunoprecipitates. The RNA gel blots were stripped and reprobed multiple times. A silver-stained gel is shown to indicate that comparable amounts of each AGO complex were used for RNA preparation. The asterisks indicate the bands of AGO proteins. The positions of RNA size markers, electrophoresed in parallel, are shown to the left of the gels.
(C) Comparison between sRNAs in total extract and the AGO1 complexes. The numbers represent the unique sRNA reads that overlap between total extract and the AGO1 complexes.
(D) Comparison between sRNA clusters (excluding miRNA loci) in total extract and the AGO1 complexes. The numbers represent the clusters that overlap between the AGO1 complexes.
[See online article for color version of this figure.]
We also compared the non-miRNA sequences among the data sets. We found that a limited number of sRNAs were shared between Total and AGO1s: <0.4% of unique sRNA sequences from Total were present in AGO1s, and these sequences represent ∼20% of the unique sRNAs in AGO1s. Surprisingly, we found only ∼1.3% of unique sequences were shared among all three AGO1s, and only ∼7.5% between any two AGO1s (Figure 4C). In the data sets of AGO1s, ∼79 to 90% of the reads were miRNAs, but they only represent a minor portion (∼1.2 to 3.3%) of the unique sRNA populations. By contrast, 71 to 75% of the unique sequences were the siRNAs derived from repeats (see Supplemental Table 1 online). Thus, we hypothesized that the limited overlap of sRNA sequences between the data sets was largely the result of heterogeneity of siRNAs that were derived from the same loci, especially from the repeats. To test this hypothesis, we used the following criteria to define sRNA clusters: (1) in one cluster, each sRNA is within 200 nucleotides of a neighboring sRNA; (2) one cluster contains at least 10 unique sRNAs; and (3) one cluster contains at least 50 sRNA reads. A total of 28,629 sRNA clusters were identified. There were 28,606 clusters present in Total and 10,419 in AGO1s. A total of 10,396 clusters were shared between Total and AGO1s, indicating clusters in AGO1s were essentially a subset of those in Total. A total of 1005 clusters (9.6%) were shared among all three AGO1s, and 5938 (57%) between any two of the AGO1s. In AGO1a, AGO1b, and AGO1c, 2497, 3, and 976 clusters were specifically present, respectively (Figure 4D). These data suggested that rice AGO1s function partially redundantly in recruiting sRNAs.
Identification of Rice miRNA Targets
Plant miRNAs usually have extensive complementarity to their target genes and regulate target gene expression predominantly through mRNA cleavage. These two features have allowed fast computational prediction and efficient experimental validation of plant miRNA targets. In Arabidopsis, many miRNA targets have been identified through bioinformatic, genetic, and high-throughput approaches (Jones-Rhoades et al., 2006; Addo-Quaye et al., 2008; German et al., 2008; Voinnet, 2009). However, only a handful of miRNA targets have been studied in rice.
We applied a bioinformatic method (Allen et al., 2005) to predict targets for previously annotated miRNAs and the new miRNAs identified in this study. By applying a cutoff mispairing score of ≤4.0, 311 targets were predicted for 77 miRNA families (see Supplemental Table 3 online).
Next, we employed a high-throughput degradome sequencing approach that has been recently developed to validate the predicted miRNA targets (Addo-Quaye et al., 2008; German et al., 2008). Plant miRNAs mediate the cleavage of their target mRNAs, resulting in 3′ cleavage products with a 5′ monophosphate and a 3′ poly(A) tail. We constructed a cDNA library for transcripts with a 5′ monophosphate and a 3′ poly(A) tail that were isolated from rice seedlings and flowers. The library was subjected to Illumina sequencing. We obtained a total of 12,377,555 reads, representing 3,025,688 unique signatures that perfectly match the sense strand of one or more annotated transcripts. Using previously developed methods (Addo-Quaye et al., 2008; German et al., 2008), we searched our library for cleavage products of predicted miRNA targets. Among 311 predicted targets, 66 had at least one degradome tag with a 5′ end that was precisely opposite the 10th nucleotide of the miRNA, a characteristic of miRNA-mediated cleavage (see Supplemental Table 3 online). To visualize the cleavage events within the target mRNAs, we plotted the abundance of each signature as a function of its position in the target transcript (referred to as t-plots) as described (German et al., 2008) (Figure 5; see Supplemental Figure 6 online). The targets were then classified into three categories (I, II, and III) based on the relative abundance of signatures at the target site and those along the transcript as previously described for Arabidopsis (Addo-Quaye et al., 2008). A total of 19, 35, and 12 targets fall into Categories I, II, and III, respectively (see Supplemental Table 3 online). Most of the identified miRNA targets were conserved between plant species, and a vast majority of the predicted targets of nonconserved miRNAs were not verified in our study, which could be partly explained by the general correlation between expression level and conservation of miRNAs.
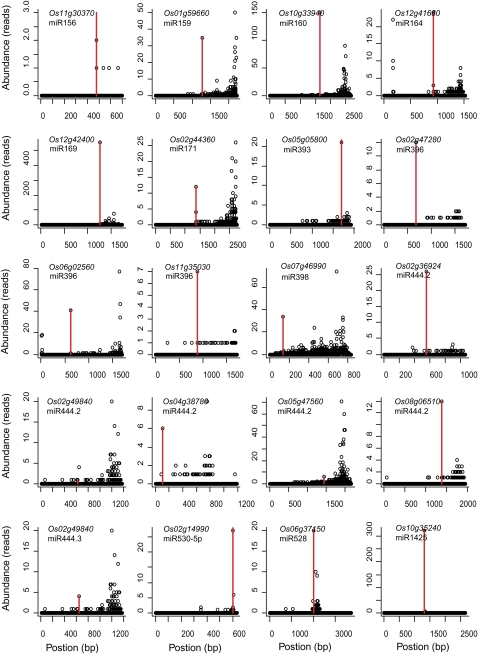
Figure 5.
Target Plots of Validated Rice miRNA Targets.
A degradome cDNA library for transcripts with a 5′ monophosphate and a 3′ poly(A) tail that were isolated from rice seedlings and flowers was constructed and subjected to Illumina sequencing. The abundance of each signature from degradome sequencing is plotted as a function of its position in the transcript. The signatures matching ±1 positions of the expected miRNA cleavage site are combined and shown in red.
DISCUSSION
In contrast with Arabidopsis, which has a single AGO1 predominating in the miRNA pathway, rice has four AGO1 homologs. We showed that knockdown of all four AGO1s in rice resulted in elevated accumulation of miRNA targets and pleiotropic developmental phenotypes (Figure 1), suggesting their roles in miRNA-mediated gene regulation. However, efforts to make RNAi lines that are specific for each of the AGO1s have not yielded any success (data not shown), which makes it difficult to assess the contribution of each AGO1 to the rice miRNA pathway genetically. We took a biochemical approach to purify and characterize three of the AGO1s (AGO1a, b, and c). We found they all had Slicer activity and displayed preference to bind sRNAs with 5′ U as their counterpart in Arabidopsis (Figure 2). Profiling of sRNAs associated with each of the AGO1s by deep sequencing revealed that they all predominantly recruited miRNAs (Figure 3; see Supplemental Table 1 online). These data suggested that in general the rice AGO1s function redundantly in the miRNA pathway. Nevertheless, we also found that some miRNAs were predominantly recruited into one or two rice AGO1s, not into others, suggesting that each AGO1 has also evolved certain specificity in recognizing a subset of miRNAs. Such specificity could be conferred by spatial or temporal expression patterns or by cellular or subcellular compartmentalization of a miRNA and an AGO1 or by yet to be identified sequence or structural features in miRNAs that can be specifically recognized by an AGO1.
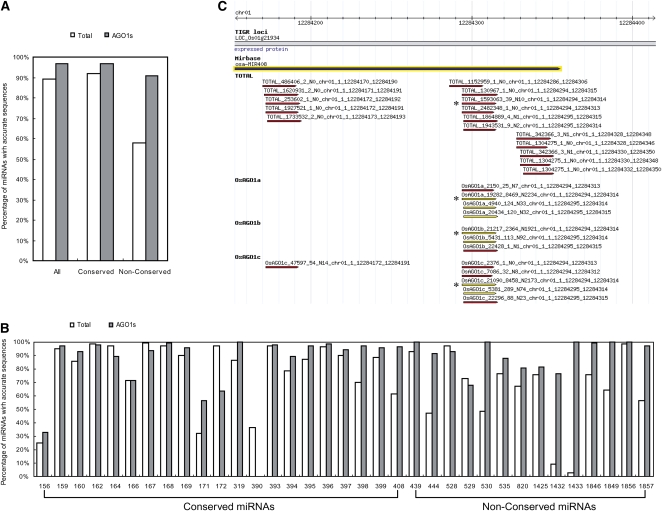
In Drosophila melanogaster, it has been shown that AGO loading could reduce the heterogeneity of the miRNA 5′ ends that is created by the imprecision in pre-miRNA cleavage by Drosha (Seitz et al., 2008). In plants, DCL1 cleavage is not always precise, which could give rise to miRNA variants with 5′ heterogeneity. Moreover, some miRNA precursors, especially the newly evolved ones, produce particularly heterogeneous sRNAs (Rajagopalan et al., 2006). We have previously proposed that the specific recruitment of sRNAs bearing a 5′ U by the Arabidopsis AGO1 complex can help to compensate for inaccuracy of Dicer processing, which could otherwise lead to off-target effects. The availability of sRNA profiles of total extract and AGO1 complexes makes it possible to examine whether miRNAs with accurate sequences are selectively incorporated into effector complexes. We calculated the percentage of miRNA reads in total reads of all pre-miRNA–derived sRNA sequences. As shown in Figures 6A and 6B, the percentage of accurate miRNAs in AGO1 complexes is higher than that in total extract, with the difference being more evident for nonconserved miRNAs. For instance, at the miR408 locus, only 61% of the pre-miRNA–derived reads are miRNAs in total extract, whereas in AGO1 complexes, the percentage increases to 96% (Figures 6B and 6C). These data suggest that AGO1 loading may function as a quality control step to compensate the slippage of Dicer cleavage, which could be especially important for the function of nonconserved miRNAs.
Figure 6.
Loading of miRNAs into AGO1 Complexes Improves the Precision of miRNAs.
(A) The percentages of annotated miRNA sequences in all sRNAs processed from miRNA precursors in total extract and AGO1 complexes.
(B) The percentages of miRNAs with accurate sequences are shown for each miRNA.
(C) A diagram showing the sRNA species (red and yellow) processed from the miR408 precursor (black) and their distribution in total extract and AGO1 complexes. The miRNA strand with accurate sequence is marked by asterisks.
[See online article for color version of this figure.]
Among the identified targets, several miRNA targets are especially noteworthy. Four MADS box genes (Os02g36924, Os02g49840, Os04g38780, and Os08g33488) were predicted targets of three miR444 variants (miR444.1, miR444.2, and miR444.3) (Sunkar et al., 2005; Lu et al., 2008). Our study confirmed that all four genes were indeed cleaved by miR444.2. Os02g49840 was also targeted by miR444.3, indicating its regulation by combinatorial action of two miR444 variants (Figure 5). Surprisingly, no miR444.1-mediated cleavage events were detected in our study (see Supplemental Table 3 online), although miR444.1 is the most abundant variant (see Supplemental Table 2 online), and two of the predicted targets are fully complementary to the miR444.1. In addition to the MADS box genes, two unrelated genes, Os08g06510, encoding a Zn-finger family protein, and Os05g47560, encoding a putative Ser/Thr protein kinase, were validated as targets of miR444.2 (Figure 5; see Supplemental Table 3 online). This indicates that one miRNA can regulate several unrelated genes.
We confirmed Os07g46990 as a miR398 target and Os06g37150 as a miR528 target (Figure 5; see Supplemental Table 3 online). Os07g46990 and Os06g37150 encode a superoxide dismutase and an ascorbate oxidase, respectively, both of which are involved in responses to oxidative stresses. Reactive oxygen species (ROS) are produced in both stressed and unstressed cells. Plants have evolved defense systems against ROS by both limiting the ROS formation as well as instituting its removal. Superoxide dismutases (SODs) constitute the first line of defense against ROS (Alscher et al., 2002). SOD enzymes catalyze the conversion of superoxide to oxygen and peroxide, and the formed peroxide is converted into water through the activity of stromal ascorbate peroxidase. In Arabidopsis, miR398 targets two closely related Cu/Zn SODs (CSD1 and CSD2) (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). It has been shown that the expression of miR398 is downregulated transcriptionally by oxidative stresses; such downregulation increases the CSD1 and CSD2 mRNA accumulation and oxidative stress tolerance (Sunkar et al., 2006). Ascorbate oxidase is a cell wall–localized enzyme that uses oxygen to catalyze the oxidation of ascorbate (AA) to the unstable radical monodehydroascorbate, which rapidly disproportionates to yield dehydroascorbate and AA and thus contributes to the regulation of the AA redox state and oxidative stress response (Fotopoulos et al., 2006). We propose that two unrelated miRNAs, miR398 and miR528, may function in a combinatorial way to promote tolerance to oxidative stress in rice.
In this study, we identified some novel 24-nucleotide miRNAs and 24-nucleotide variants of previously annotated miRNAs (Table 1). Twenty-four-nucleotide miRNAs have been previously reported in rice and Arabidopsis (Vazquez et al., 2008; Zhu et al., 2008). The biogenesis pathway and function of these noncanonical rice miRNAs remain to be determined. Intriguingly, we found that all 24-nucleotide miRNAs were absent in the three characterized AGO1 complexes (Figure 4A; see Supplemental Table 2 online), suggesting that they might enter alternative AGO complexes. We envision that these 24-nucleotide miRNAs might have biological functions different from that of the canonical 21-nucleotide miRNAs associated with AGO1s. It is tempting to propose that these 24-nucleotide miRNAs are recruited by AGO4 to induce cytosine methylation of their precursor sequences in cis or their target genes in trans.
METHODS
Construction of RNAi Vector and Plant Transformation
We designed an IR to target a homologous region of rice AGO1s (see Supplemental Figure 7 online). The fragment was RT-PCR amplified using primers described in Supplemental Table 4 online and cloned into the pUCC-RNAi vector (Liu et al., 2005) to generate an IR. After confirmation by sequencing, the IR was introduced into pCam23ACT:OCS, a binary vector containing the rice (Oryza sativa) Actin1 promoter, resulting in binary AGO1IR RNAi construct. The construct was transformed into rice (japonica cv Nipponbare) essentially as described (Hiei et al., 1994), except that G418 was used for selection.
Generation of Antibodies against Rice AGO1s
Synthetic peptides AGO1aN (KKKTEPRNAGEC), AGO1bN (KKRTGSGSTGEC), and AGO1cN (MASRRPTHRHHTC) were used to raise rabbit polyclonal antibodies against AGO1a, AGO1b, and AGO1c, respectively, essentially as described (Mi et al., 2008). The antisera were affinity purified and used for immunoprecipitation (1:50 dilution) and immunoblots (1:1000 dilution).
Purification of Rice AGO1 Complexes and Associated sRNAs
Rice AGO1 complexes were immunopurified from 3-week-old seedlings as previously described (Qi et al., 2005). The quality of the purifications was examined by SDS-PAGE followed by silver staining, and the bands of expected sizes were confirmed as AGO1 proteins by mass spectrometry and immunoblot analysis.
RNAs were extracted from total extracts and the purified AGO1 complexes by Trizol reagent (Invitrogen), resolved on a 15% denaturing PAGE gel, and visualized by SYBR-gold (Invitrogen) staining. Gel slices within the range of 18 to 28 nucleotides were excised, and the RNAs were eluted and purified for cloning.
sRNA RNA Gel Blot
RNA gel blot analysis with enriched total sRNAs or RNAs prepared from purified AGO complexes was performed as described (Qi et al., 2005). 32P end-labeled oligonucleotide probes complementary to sRNAs were used for the blots. The sequences of the probes are described in Supplemental Table 4 online.
UV Cross-Linking and Slicer Activity Assays
UV cross-linking assays with RNA oligos bearing different 5′ terminal nucleotides were performed as described (Mi et al., 2008). Slicer activity assays were performed essentially as described (Qi et al., 2005) using in vitro–transcribed miRNA target transcripts. The PCR primers used for generating in vitro transcription templates are listed in Supplemental Table 4 online.
Quantitative RT-PCR Analysis
Total RNAs were extracted from 3-week-old rice seedlings with Trizol (Invitrogen). After removal of contaminated DNAs by digestion with RNase-free DNaseI (Promega), the RNAs were reverse-transcribed by M-MLV (Promega) using oligo(dT). The cDNAs were then used as templates for quantitative PCR. Quantitative PCR was performed using SYBR Premix EX Taq (TaKaRa) on Mastercycler ep realplex (Eppendorf). The rice Actin or GAPDH genes were detected in parallel and used as the internal controls. The primers used for PCR are listed in Supplemental Table 4 online.
sRNA Library Preparation and Sequencing
sRNAs cloning for Illumina 1G sequencing was performed essentially as described (Mi et al., 2008). A detailed protocol is presented as a Supplemental Method online.
Bioinformatic Analysis of sRNAs
The adaptor sequences in Illumina 1G sequencing reads were removed using vectorstrip in the EMBOSS package. The sRNA reads with length of 19 to 27 nucleotides were mapped to the O. sativa nuclear, chloroplast, and mitochondrial genomes (http://rice.plantbiology.msu.edu/, version 6.0). The sRNAs with perfect genomic matches were used for further analysis. Annotation of sRNAs was performed using the following databases: miRBase (http://microrna.sanger.ac.uk/sequences, version 12.0) for miRNA annotations, Rfam (http://www.sanger.ac.uk/Software/Rfam/) for noncoding RNAs (rRNAs, tRNAs, snoRNAs, and snRNAs) sequences, the The Institute for Genomic Research rice genome annotation (version 6.0) resource (http://rice.plantbiology.msu.edu/) for protein coding gene and intergenic region sequences, and Repbase (http://www.girinst.org) for transposons and repeats. ta-siRNA sequences were extracted from published results (Zhu et al., 2008) and trans-natural antisense genes were extracted from published results (Osato et al., 2003). The relative frequency of each nucleotide at each position of the sRNAs was calculated and graphically represented using WebLogo (Crooks et al., 2004).
Identification of Novel miRNAs and Prediction of miRNA Targets
The recently proposed criteria for miRNA annotation (Meyers et al., 2008) were applied to identify novel miRNAs. These criteria include the following: (1) miRNA and miRNA* form a duplex with two-nucleotide, 3′ overhang; (2) less than four mismatches in the miRNA/miRNA* duplex; and (3) >75% of the sequenced sRNAs from the precursor are miRNA or miRNA*. Prediction of miRNA targets was performed as described (Zhao et al., 2007). The putative target sites of all miRNA candidates were identified by aligning miRNA sequences to the annotated gene sequences using Perl script. The method enables identification of multiple target sites of a single miRNA on the same target sequence. An initial pool of predicted targets were created, with at most four unpaired nucleotides (including G:U pairs, mismatches, and single-nucleotide bulges) and two single-nucleotide bulges allowed between a miRNA and its targets. A mispair scoring system was then applied to these initial targets. Mismatches and single-nucleotide bulges were each scored as 1, and G:U pairs were each scored as 0.5. The scores were doubled if mismatches, G:U pairs, and bulges were located at positions at 2 to 13 as counted from the 5′-end of a miRNA. Genes with a mispair score ≤4 were selected as putative miRNA targets.
Degradome Library Construction and Data Processing
Degradome libraries were constructed for rice 3-week-old seedling and young panicles as described (Addo-Quaye et al., 2008; German et al., 2008). Sequencing data from the two libraries were combined and processed as described (Addo-Quaye et al., 2008; German et al., 2008).
Accession Number
Databases of sRNAs from rice total extract, AGO1a, AGO1b, and AGO1c complexes and rice degradome sequencing are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE18251.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Relationships of Arabidopsis and Rice AGO Proteins.
Supplemental Figure 2. Molecular Characterization of AGO1 RNAi Lines.
Supplemental Figure 3. Sequencing of Purified AGO1a, AGO1b, and AGO1c by Mass Spectrometry.
Supplemental Figure 4. Immunoblot Analysis with AGO1 Immunoprecipitates.
Supplemental Figure 5. Three Loci Encode a New Variant of miR396.
Supplemental Figure 6. Target Plots of Validated Rice miRNA Targets.
Supplemental Figure 7. An Alignment of cDNA Sequences of Rice AGO1 Homologs.
Supplemental Table 1. Summary of Sequenced Small RNAs from Total Extract and AGO1 Complexes.
Supplemental Table 2. Distribution of miRNAs in Each Rice AGO1 Complex.
Supplemental Table 3. Prediction and Validation of Rice miRNA Targets.
Supplemental Table 4. Primers and Probes.
Supplemental Method. A Protocol for Cloning Small RNAs for Illumina Sequencing.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 1.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Tree in Figure 1A.
Supplemental Data Set 3. Structures of Newly Identified miRNAs.
Supplementary Material
Acknowledgments
We thank Xiaofeng Cao for the DCL1IR line and the National Institute of Biological Sciences Antibody Facility for generating the antisera used in this study. Y.Q. is supported by the Chinese Ministry of Science and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yijun Qi (qiyijun@nibs.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Addo-Quaye, C., Eshoo, T.W., Bartel, D.P., and Axtell, M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Alscher, R.G., Erturk, N., and Heath, L.S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53 1331–1341. [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., and Bartel, D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and Van de Peer, Y. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y.Y., Sieburth, L., and Voinnet, O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Fotopoulos, V., Sanmartin, M., and Kanellis, A.K. (2006). Effect of ascorbate oxidase over-expression on ascorbate recycling gene expression in response to agents imposing oxidative stress. J. Exp. Bot. 57 3933–3943. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- German, M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26 941–946. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. (2002). RNA interference. Nature 418 244–251. [DOI] [PubMed] [Google Scholar]
- Heisel, S.E., Zhang, Y., Allen, E., Guo, L., Reynolds, T.L., Yang, X., Kovalic, D., and Roberts, J.K. (2008). Characterization of unique small RNA populations from rice grain. PLoS One 3 e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., and Jacobsen, S.E. (2007). Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Willmann, M.R., Wu, G., Yoshikawa, M., de la Luz Gutierrez-Nava, M., and Poethig, S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P., and Bartel, B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57 19–53. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Gao, S., Vivian-Smith, A., and Jin, H. (2007). A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21 3123–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., Jr., Zhu, J.K., Staskawicz, B.J., and Jin, H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet, E., Delannoy, E., Sormani, R., Floris, M., Brodersen, P., Crete, P., Voinnet, O., and Robaglia, C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Li, P., Li, X., Liu, C., Cao, S., Chu, C., and Cao, X. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305 1437–1441. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, C., et al. (2008). Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 105 4951–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126 469–481. [DOI] [PubMed] [Google Scholar]
- Ma, J.B., Ye, K., and Patel, D.J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.B., Yuan, Y.R., Meister, G., Pei, Y., Tuschl, T., and Patel, D.J. (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, T.A., Howell, M.D., Cuperus, J.T., Li, D., Hansen, J.E., Alexander, A.L., Chapman, E.J., Fahlgren, N., Allen, E., and Carrington, J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133 128–141. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, R.D., Aksay, G., Dolgosheina, E., Ebhardt, H.A., Magrini, V., Mardis, E.R., Sahinalp, S.C., and Unrau, P.J. (2008). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki, H., Itoh, J., Hayashi, K., Hibara, K., Satoh-Nagasawa, N., Nosaka, M., Mukouhata, M., Ashikari, M., Kitano, H., Matsuoka, M., Nagato, Y., and Sato, Y. (2007). The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 104 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A., Ito, M., Kamiya, N., Sato, Y., and Matsuoka, M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30 189–201. [DOI] [PubMed] [Google Scholar]
- Nonomura, K., Morohoshi, A., Nakano, M., Eiguchi, M., Miyao, A., Hirochika, H., and Kurata, N. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato, N., et al. (2003). Antisense transcripts with rice full-length cDNAs. Genome Biol. 5 R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Qi, Y., He, X., Wang, X.J., Kohany, O., Jurka, J., and Hannon, G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443 1008–1012. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., and Joshua-Tor, L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12 340–349. [DOI] [PubMed] [Google Scholar]
- Seitz, H., Ghildiyal, M., and Zamore, P.D. (2008). Argonaute loading improves the 5′ precision of both microRNAs and their miRNA strands in flies. Curr. Biol. 18 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.J., and Joshua-Tor, L. (2006). Argonaute and RNA–Getting into the groove. Curr. Opin. Struct. Biol. 16 5–11. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Girke, T., Jain, P.K., and Zhu, J.K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Kapoor, A., and Zhu, J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Zhou, X., Zheng, Y., Zhang, W., and Zhu, J.K. (2008). Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 8 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, A., Iwasaki, S., Watanabe, T., Utsumi, M., and Watanabe, Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49 493–500. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Blevins, T., Ailhas, J., Boller, T., and Meins, F., Jr. (2008). Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 36 6429–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136 669–687. [DOI] [PubMed] [Google Scholar]
- Wang, J.F., Zhou, H., Chen, Y.Q., Luo, Q.J., and Qu, L.H. (2004. a). Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 32 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., and Gaasterland, T. (2004. b). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5 R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A., and Carrington, J.C. (2005. b). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005. a). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui, M., Irvine, D.V., and Martienssen, R.A. (2007). Noncoding RNAs and gene silencing. Cell 128 763–776. [DOI] [PubMed] [Google Scholar]
- Zhao, T., Li, G., Mi, S., Li, S., Hannon, G.J., Wang, X.J., and Qi, Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Zhu, J., Kapoor, A., and Zhu, J.K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q.H., Spriggs, A., Matthew, L., Fan, L., Kennedy, G., Gubler, F., and Helliwell, C. (2008). A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 18 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.