Abstract
Virally induced structures called viral factories form throughout the cytoplasm of cells infected with mammalian orthoreoviruses (MRV). When expressed alone in cells, MRV nonstructural protein μNS forms factory-like structures very similar in appearance to viral factories, suggesting that it is involved in forming the structural matrix of these structures. μNS also associates with MRV core particles; the core proteins μ2, λ1, λ2, λ3, and σ2; and the RNA-binding nonstructural protein σNS. These multiple associations result in the recruitment or retention of these viral proteins or particles at factory-like structures. In this study, we identified the regions of μNS necessary and sufficient for these associations and additionally examined the localization of viral RNA synthesis in infected cells. We found that short regions within the amino-terminal 220 residues of μNS are necessary for associations with core particles and necessary and sufficient for associations with the proteins μ2, λ1, λ2, σ2, and σNS. We also found that only the λ3 protein associates with the carboxyl-terminal one-third of μNS and that viral RNA is synthesized within viral factories. These results suggest that μNS may act as a cytoplasmic scaffolding protein involved in localizing and coordinating viral replication or assembly intermediates for the efficient production of progeny core particles during MRV infection.
Mammalian orthoreoviruses (MRV) are members of the family Reoviridae, which includes important human, animal, and plant pathogens (e.g., rotavirus, bluetongue virus, and rice dwarf virus). All members of the family Reoviridae share a number of characteristics including a similar genome comprised of 9 to 12 segments of double-stranded RNA (dsRNA). These segments are enclosed within a multilayered protein capsid, including one or more inner layers that contact the genome and play roles in viral RNA synthesis and one or more outer layers that mediate virus attachment and entry into the host cell (17, 37, 41). During the entry process, the outer capsid(s) is largely removed from these viruses, and the inner capsid(s) is released into the cytoplasm. Upon release, the genome-enclosing inner capsid(s) serves as the viral transcriptase particle, synthesizing and capping viral plus-strand RNAs, which are then released into the cytoplasm for translation into viral proteins (17, 37, 41).
At early times after entry, distinctive structures, which grow in size over time, appear throughout the cytoplasm of infected cells. These cytoplasmic structures are variously termed viral factories (VF) (MRV), viroplasms (rotavirus and phytoreovirus), or viral inclusion bodies (VIB) (orbivirus). In each case, they contain many viral proteins, particles, and dsRNAs and are thought to be the locations of viral RNA replication and packaging into progeny particles (13, 15, 31, 42, 43, 46).
In previous studies, either one or two nonstructural proteins of each virus were shown to be required for forming the cytoplasmic structures. In MRV and avian orthoreoviruses, the nonstructural protein μNS expressed alone in cells forms factory-like structures (FLS) that appear to be similar by light microscopy to VF formed in infected cells (4, 8, 49). Likewise, orbiviruses such as bluetongue virus and epizootic hemorrhagic disease virus encode a single nonstructural protein, NS2, that forms VIB-like structures when expressed alone in cells (25, 47, 48). In phytoreoviruses such as rice dwarf virus, the nonstructural protein Pns12 expressed alone in cells forms viroplasm-like structures (VLS) (51). Rotaviruses, on the other hand, encode two nonstructural proteins, NSP2 and NSP5, which must be coexpressed under most circumstances to form VLS (16, 18, 34).
In MRV, the μNS sequences required for forming FLS have been carefully examined. The carboxyl-terminal (C-terminal) 250 amino acids (aa) of μNS are sufficient to form FLS, with four distinct regions within this portion of the protein shown to be necessary (5). These regions include two previously predicted coiled-coil domains (30), a linker region containing a putative zinc hook between the coiled coils, and a short C-terminal tail region.
Importantly, the capacity of MRV to form VF is necessary for viral replication. When μNS expression is knocked down by RNA interference, viral growth is severely inhibited (1, 27). Moreover, when wild-type μNS is plasmid expressed in trans, viral growth is rescued (1, 27). Plasmid-expressed μNS with mutations in the putative zinc hook or the C-terminal tail, in contrast, does not rescue viral growth (1, 28). These results strongly suggest that the formation of VF is an important and necessary function for successful MRV multiplication.
Previous studies have shown that μNS associates with six other MRV proteins: five structural proteins that make up the core particle (λ1, λ2, λ3, σ2, and μ2) and the single-stranded RNA (ssRNA) binding nonstructural protein σNS (4, 6, 8, 32, 33). Limited mapping of μNS associations with other viral proteins has shown that μNS aa 14 to 41 are necessary and that aa 1 to 41 are sufficient for the association with the minor core protein μ2 (8). In addition, μNS aa 1 to 13 are necessary for the association with the nonstructural protein σNS (33), and aa 1 to 41 are dispensable for the association with the core surface proteins λ1, λ2, and σ2 (6). μNS additionally associates with MRV core particles in vitro and in cells (6, 7).
In light of these data, we have hypothesized that in addition to its role in forming the structural matrix of VF, a second role for μNS in MRV infection is to act as a type of cytoplasmic scaffolding protein for the coordinated recruitment and assembly of MRV replication complexes within VF. Based on our previously reported data, we have developed a model of MRV VF assembly. In this model, following entry and release into the cytoplasm, the MRV core particle begins transcribing the viral plus-strand RNAs. MRV proteins, including μNS, are synthesized by the cellular translational machinery, either cotranscriptionally or adjacent to the site of core RNA transcription. Following translation, μNS may associate with the adjacent core particle to seed a new VF, or μNS may self-associate first to form a small VF, which then further associates with the adjacent MRV core. Proteins required for the assembly of progeny core particles (λ1, λ2, λ3, μ2, and σ2), as well as σNS, also associate with μNS either by direct association with μNS in the VF matrix or in the cytoplasm with subsequent recruitment to VF by μNS. The VF-localized core particle continues to transcribe the MRV plus-strand RNAs, some of which are now bound by adjacent viral proteins to form replication and assembly complexes for the production of progeny core particles and virions within the VF. The VF is additionally tethered to the cellular cytoskeleton via μNS associations with a microtubule-associated viral protein, μ2. This association allows VF to move toward the perinuclear region of the cell, merging with other VF during the journey, ultimately forming the large perinuclear VF that are characteristic of MRV-infected cells.
We previously developed an assay that exploits the characteristic ability of μNS, as well as the rotavirus protein NSP5 fused at its amino terminus (N terminus) to enhanced green fluorescent protein (EGFP) (34), to form distinctive structures in the cytoplasm in order to identify and map protein-protein interactions (32). In the current study, we modified and made extensive use of this novel assay to explore further the associations between μNS and other MRV proteins. In new experiments, we first defined the region of μNS that is necessary for associations with individual MRV proteins, as well as the MRV core particle, by determining the ability of these proteins to be recruited to FLS formed by a series of μNS deletion mutants. We then constructed a number of plasmids expressing μNS fragments fused to EGFP/NSP5 as a method to map regions of μNS sufficient for associations with each MRV protein. In sum, we found that small, largely nonoverlapping regions of μNS are necessary and sufficient for the association with individual MRV proteins and necessary for the association with core particles. Because the MRV core particle transcribes viral plus-strand RNAs (22), we additionally examined the localization of viral RNA synthesis and found that it too occurs in VF. These results advance our understanding of how a viral nonstructural protein, MRV μNS, has evolved to build discrete regions of cytoplasmic scaffolding within which critical viral activities occur.
MATERIALS AND METHODS
Cells, viruses, antibodies, and other reagents.
CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (HyClone) and 10 μg/ml gentamicin (Invitrogen). MRV strain type 1 Lang was our laboratory stock, originally obtained from B. N. Fields. MRV strain type 3 Dearing was obtained from the laboratory of L. W. Cashdollar (Medical College of Wisconsin). Virus was propagated on L929 cells and plaque purified as previously described (19). Top-component infectious subvirion particles (ISVPs) were made by chymotrypsin digestion (200 μg/ml; Sigma) of purified top-component virions (1 × 1013 particles/ml) for 10 min at 32°C. The reaction was quenched by the addition of 2 mM phenylmethylsulfonyl fluoride (Sigma). Rabbit polyclonal antiserum and purified polyclonal antibodies specific for μNS, μ2, and MRV core particles were described previously (7, 8, 11, 26). Monoclonal antibodies against σNS (3E10) and λ2 (7F4) were also described previously (3, 50). Monoclonal antibody HA.11, specific for the influenza virus hemagglutinin (HA) epitope, was obtained from Covance Research Products. In the indicated experiments, protein A-purified rabbit anti-μNS immunoglobulin G (IgG) was conjugated to Texas Red by using a kit obtained from Molecular Probes. Monoclonal antibody against bromodeoxyuridine was purchased from Sigma. The following secondary antibodies were used as appropriate for different experiments: Alexa 488- or Alexa 594-conjugated goat anti-mouse or anti-rabbit IgG (Molecular Probes) and horseradish peroxidase (HRP)-conjugated donkey anti-mouse or anti-rabbit IgG (Pierce). For microscopy, antibodies were titrated to optimize signal-to-noise ratios. Bromouridine (BrU) was purchased from Sigma. Actinomycin D was purchased from Sigma. All restriction enzymes were obtained from New England Biolabs.
Plasmid constructions.
pCI-μNS, pCI-σNS, pCI-μ2, pCI-λ1, pCI-λ2, and pCI-σ2, expressing μNS, σNS, μ2, λ1, λ2, and σ2, respectively, were described previously but originally named by their genes, pCI-M3, pCI-S3, pCI-M1, pCI-L3, pCI-L2, and pCI-S2, respectively (6, 8, 33, 38). pCI-μNS(14-721), pCI-μNS(41-721), pCI-μNS(173-721), pCI-μNS(221-721), and pCI-μNS(471-721) were described previously but were originally named pCI-M3(14-721), pCI-M3(41-721), pCI-M3(173-721), pCI-M3(221-721), and pCI-M3(471-721), respectively (5, 8, 33). pCI-λ3/HA expressing a C-terminally HA-tagged version of λ3 was described previously (32). pEGFP/μNS(471-721) was described previously but was originally named pEGFP-C1-M3(471-721) (5). pEGFP/NSP5 was obtained from Oscar Burrone and was described previously (16). To make μNS deletion plasmids pCI-μNS(20-721), pCI-μNS(26-721), pCI-μNS(55-721), pCI-μNS(65-721), pCI-μNS(75-721), pCI-μNS(85-721), and pCI-μNS(95-721), PCR was performed by using pCI-μNS as a template. The PCR product was purified, digested with NheI and BlpI or EcoRI and NotI, and ligated into appropriately digested pCI-μNS. To make the μNS fragment fusion plasmids p-μNS(41-221)/EGFP/NSP5, p-μNS(55-221)/EGFP/NSP5, p-μNS(173-221)/EGFP/NSP5, p-μNS(1-20)/EGFP/NSP5, p-μNS(41-173)/EGFP/NSP5, p-μNS(55-173)/EGFP/NSP5, and p-μNS(41-110)/EGFP/NSP5, PCR was performed by using pCI-μNS as a template. The PCR product was purified, digested with AgeI, and ligated into AgeI-digested pEGFP/NSP5. Orientation was determined by restriction digestion. p-μNS(1-227)/EGFP/NSP5 was made in two steps. First, pGEM-M3 was digested with BsaHI to liberate a fragment containing the coding region for μNS aa 1 to 227. This fragment was gel purified, the overhanging ends were filled in with Klenow fragment, and a HindIII digestion was performed on the resulting fragment, which was then ligated into HindIII/SmaI-digested pEGFP-N1 to make p-μNS(1-227)/EGFP. Second, p-μNS(1-227)/EGFP was digested with NdeI and BsrGI and ligated into NdeI/BsrGI-digested EGFP/NSP5 to make p-μNS(1-227)/EGFP/NSP5. To make p-μNS(95-221)/EGFP/NSP5, p-μNS(14-41)/EGFP/NSP5, p-μNS(20-41)/EGFP/NSP5, and p-μNS(95-173)/EGFP/NSP5, PCR was performed by using pCI-μNS as a template. The PCR product was purified, digested with NheI and AgeI, and ligated into NheI/AgeI-digested p-μNS(1-227)/EGFP/NSP5. p-μNS(1-12)/EGFP/NSP5 was made by digesting p-μNS(1-12)/EGFP/μNS(471-721) and EGFP/NSP5 with NdeI and BsrGI followed by ligating the appropriate purified products. p-μNS(1-41)/EGFP/NSP5 was made by digesting p-μNS(1-41)/EGFP/μNS(471-721) and EGFP/NSP5 with NdeI and BsrGI, followed by ligating the appropriate purified products. p-σNS/EGFP was made by PCR using pCI-σNS as a template. The PCR product was purified, digested with EcoRI and SacII, and ligated into EcoRI/SacII-digested pEGFP-N1. p-σNS/EGFP/NSP5 was made by digesting p-σNS/EGFP and pEGFP/NSP5 with NdeI and BsrGI, followed by ligating the appropriate purified products. p-λ3/EGFP/NSP5 was made by PCR using pCI-λ3/HA as a template. The PCR product was purified, digested with AgeI, and ligated into AgeI-digested pEGFP/NSP5. Orientation was confirmed by restriction digestion. All plasmid sequences were confirmed by sequencing. Primer sequences used in PCRs are available upon request.
Transfections and infections.
For transfections, a total of 4 μg plasmid DNA was mixed with 10 μl Lipofectamine (Invitrogen) in Optimem (Invitrogen). After a 20-min incubation, the mixture was added to cells containing DMEM with 10% fetal calf serum (FCS) but lacking antibiotics and incubated for 18 h at 37°C as suggested by the manufacturer. After incubation, cells were processed for immunostaining and microscopy. For other transfections, 8 μg plasmid DNA was mixed with 20 μl Lipofectamine in Optimem. All other steps of the above-described protocol were then followed except that cells were processed for immunoprecipitation and immunoblotting.
For immunostaining studies, 2 × 105 CV-1 cells were seeded the day before transfection in six-well plates (9.6 cm2 per well) containing 18-mm round glass coverslips. For immunoprecipitation studies, 4 × 105 cells were seeded onto 60-mm dishes the day before transfection. Infections were performed by removing media from cells and overlaying cells with virus diluted in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 [pH 7.5]) supplemented with 2 mM MgCl2 for 1 h at room temperature to allow virus to adsorb to cells. Cells were then refed with medium and allowed to incubate further at 37°C. Top-component ISVP infection of cells transfected with μNS expression plasmids was described previously (6).
Immunostaining and microscopy.
Infected or transfected cells were fixed for 20 min with 2% paraformaldehyde in PBS, except for BrU experiments, in which cells were fixed by incubation for 3 min in 100% methanol. Fixed cells were washed three times with PBS and permeabilized for 5 min with 0.2% Triton X-100 in PBS. Cells were again washed three times with PBS and blocked for 5 min with 2% bovine serum albumin in PBS. Primary and secondary antibodies were diluted in 2% bovine serum albumin in PBS. After blocking, cells were incubated for 1 h with primary antibodies, washed three times with PBS, and incubated for 1 h with secondary antibodies. Immunostained cells were washed a final three times with PBS and mounted onto slides with Prolong Plus reagent (Molecular Probes). Immunostained samples were examined by using an Optiphot-2 epifluorescence upright microscope (Nikon) or an Axiovert 200 inverted fluorescence microscope (Zeiss). Images were collected digitally by using either a Photometrics CoolSnapcf camera (Roper Scientific) and MetaVue imaging software (Molecular Devices) or an AxioCam MR color camera and Axiovision AC imaging software (Zeiss). Images were prepared for presentation by using Photoshop and Illustrator (Adobe Systems).
Core immunoprecipitation assay.
Transfected cells were lysed by incubation for 30 min on ice in nondenaturing lysis (Raf) buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% [vol/vol] glycerol, 1% [vol/vol] NP-40) containing protease inhibitors (Roche Biochemicals). 7F4 (λ2-specific) monoclonal antibodies that had been incubated for 2 h with protein A-conjugated magnetic beads (Dynal Biotech) and washed six times with Raf buffer were separated into two aliquots. MRV core particles, prepared as previously described (11), were added to one of the two 7F4-coated bead aliquots at a concentration of 1 × 1012 cores/ml and incubated with rotation for an additional 2 h at 4°C. 7F4/core-coated beads were washed an additional six times with Raf buffer. 7F4-coated or 7F4/core-coated beads were added in equal volumes to the μNS protein-containing cell lysates, which were then rotated overnight at 4°C. Immunoprecipitated proteins were separated from the cell lysate and washed four times with Raf buffer. Sample buffer (125 mM Tris [pH 6.8], 10% [wt/vol] sucrose, 1% [wt/vol] sodium dodecyl sulfate, 0.02% [vol/vol] β-mercaptoethanol, 0.01% bromophenol blue) was added to both the immunoprecipitated proteins and the postbinding supernatant. Samples were then boiled for 3 min and separated on denaturing 10% (wt/vol) polyacrylamide gels containing 0.01% (wt/vol) sodium dodecyl sulfate. Proteins were transferred from the gels onto pieces of nitrocellulose by electroblotting in transfer buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol [pH 8.3]). Nitrocellulose was then blocked by incubation with 5% milk in Tris-buffered saline (20 mM Tris [pH 7.6], 137 mM NaCl) containing 0.5% Tween 20 (Sigma) (TBS-T) for 30 min at room temperature. Primary antibodies were allowed to bind during incubation overnight at room temperature in TBS-T containing 1% milk. HRP-conjugated secondary antibodies were allowed to bind during a 2-h incubation at room temperature in TBS-T containing 1% milk. Before and after the incubation with secondary antibodies, the nitrocellulose was washed three times with TBS-T. The binding of HRP conjugates was detected by incubation with chemiluminescence substrate (Bio-Rad) according to the manufacturer's recommendations, followed by exposure to film (Fuji).
BrU assay.
CV-1 cells were plated onto 8-well chamber slides at a density of 1 × 104 cells per well and then infected with type 1 Lang or type 3 Dearing ISVPs, prepared as previously described (19), at a multiplicity of infection of 100 PFU/cell. Infection was allowed to proceed for 2, 3, 4, or 6 h, at which time cells were treated for 30 min with 10 μg/ml actinomycin D to inhibit cellular RNA polymerase II (52). Cells were then transfected with 10 mM BrU in 3.7 μl Lipofectamine in 30 μl Optimem in the presence or absence of 10 μg/ml actinomycin D and incubated for an additional 60 min before fixation and immunostaining.
RESULTS
μNS and EGFP-fused NSP5 form nonoverlapping cytoplasmic structures.
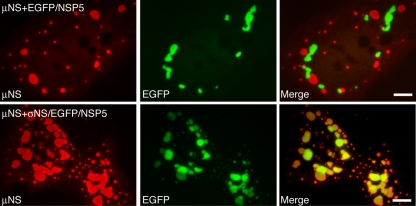
To establish the feasibility of our new approach for defining regions of μNS sufficient for associating with other MRV proteins, we examined its capacity to detect such a known association. A plasmid expressing μNS was cotransfected into cells with a plasmid expressing either EGFP/NSP5 or EGFP/NSP5 fused to the μNS-associating protein σNS. At 18 h posttransfection (p.t.), cells were fixed and stained with antibodies against μNS. The inherent fluorescence of EGFP was used to detect EGFP/NSP5 or σNS/EGFP/NSP5. Interestingly, when the FLS-forming μNS was coexpressed with the VLS-forming EGFP/NSP5, the proteins formed distinctive structures in cells as previously described (8, 32, 34); however, the respective structures did not colocalize, suggesting that the proteins do not associate (Fig. 1, top). In contrast, when μNS was coexpressed with σNS/EGFP/NSP5, the respective structures completely colocalized in cells (Fig. 1, bottom), reflecting the known association of μNS with σNS and validating the use of EGFP/NSP5-formed VLS as a way to identify regions of μNS that are sufficient for associations with other MRV proteins.
FIG. 1.
MRV μNS and EGFP-fused rotavirus NSP5 form nonoverlapping structures that can be made to colocalize through a known protein-protein association. CV-1 cells cotransfected with a plasmid expressing μNS and either EGFP/NSP5 (top) or σNS/EGFP/NSP5 (bottom) were fixed at 18 h p.t. FLS were visualized by staining with μNS-specific polyclonal antibodies followed by Alexa 594-conjugated goat anti-rabbit IgG (left). VLS were visualized by the inherent fluorescence of EGFP (middle). Merged images are also shown (right). Bar, 10 μm.
μNS aa 20 to 25 are necessary and aa 14 to 41 are sufficient for associations with μ2.
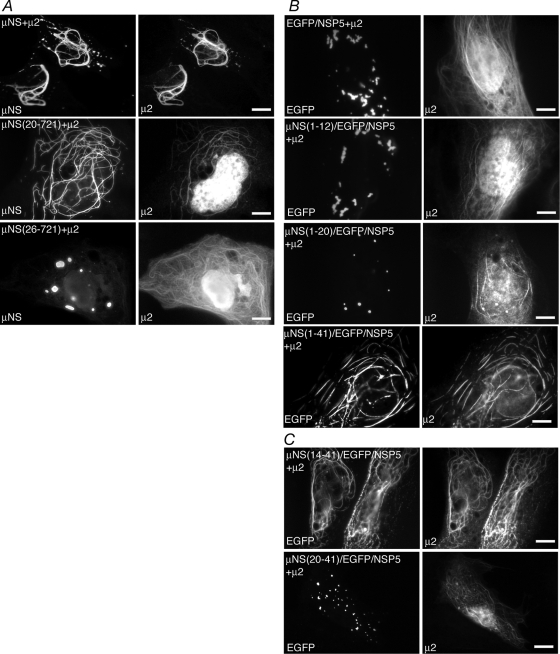
The MRV μ2 protein was previously shown to be a strain-specific microtubule-associated protein (38). When expressed alone in transfected cells, μ2 derived from most MRV strains localizes to cellular microtubules and the nucleus (38). When coexpressed with μNS (Fig. 2A, top) and in infected cells, μ2 and μNS from these strains are associated with microtubules (8, 38). For occasional MRV strains in which μ2 does not associate with microtubules, it colocalizes with μNS in FLS and VF, suggesting that the association between μNS and μ2 is independent of the μ2 microtubule association but necessary for μNS microtubule localization (8). To identify the regions of μNS that are necessary and sufficient for the association with μ2 more precisely, we created a series of μNS deletion mutants and N-terminal fusions to EGFP/NSP5 and examined the ability of μ2 to associate with these proteins following plasmid cotransfections into cells. We defined the region of μNS necessary for the association with μ2 by creating deletions from the N terminus of μNS. We cotransfected cells with plasmids expressing μ2 and the μNS deletion mutants and examined the localization of μ2 and μNS within the cells at 18 h p.t. by immunofluorescence microscopy. In cells coexpressing μ2 and μNS(20-721), μNS colocalized with μ2 on cellular microtubules, suggesting that this deletion did not affect their association (Fig. 2A, middle). In cells coexpressing μ2 and μNS(26-721), while μ2 localized to both the nucleus and cellular microtubules (Fig. 2, bottom right), μNS(26-721) was found in FLS that did not associate with microtubules (Fig. 2A, bottom left), suggesting that μNS aa 20 to 25 are necessary for the μ2 association.
FIG. 2.
μNS aa 20 to 25 are necessary and aa 14 to 41 are sufficient for associations with minor core protein μ2. For each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing μ2 and either μNS (top), μNS(20-721) (middle), or μNS(26-721) (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against μ2 followed by Texas Red-conjugated μNS-specific rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-rabbit IgG to visualize μ2 (right). (B) CV-1 cells were cotransfected with plasmids expressing μ2 and either EGFP/NSP5 (top row), μNS(1-12)/EGFP/NSP5 (second row), μNS(1-20)/EGFP/NSP5 (third row), or μNS(1-41)/EGFP/NSP5 (bottom row). After fixation, cells were stained with rabbit polyclonal antibodies against μ2 followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μ2 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). (C) CV-1 cells were cotransfected with plasmids expressing μ2 and μNS(14-41)/EGFP/NSP5 (top) or μNS(20-41)/EGFP/NSP5 (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against μ2 followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μ2 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). Bar, 10 μm.
To identify regions of μNS sufficient for the association with μ2, we created a series of plasmids expressing fusion proteins connecting fragments of the N-terminal 41 aa of μNS to EGFP/NSP5. We then tested the ability of μ2 to associate with each of these fusion proteins in transfected cells. In each case, protein localization was examined at 18 h p.t. by immunofluorescence microscopy. μ2 did not associate with either μNS(1-12)/EGFP/NSP5 (Fig. 2B, second row) or μNS(1-20)/EGFP/NSP5 (Fig. 2B, third row) but did associate with μNS(1-41)/EGFP/NSP5 (Fig. 2B, bottom row), mirroring our previous findings that these N-terminal amino acids of μNS are not necessary for the μ2 association and that μNS(1-41) is sufficient for the μ2 association (8, 33). Additional N-terminal deletions were made from the μNS(1-41)/EGFP/NSP5 fusion protein to determine the smallest region of μNS sufficient for the association with μ2. When μ2 was coexpressed with μNS(14-41)/EGFP/NSP5, the two proteins completely colocalized on cellular microtubules (Fig. 2C, top); however, μ2 did not efficiently associate with μNS(20-41)/EGFP/NSP5 (Fig. 2C, bottom). Taken together, these data suggest that μNS aa 14 to 41 are sufficient for the association with μ2.
μNS aa 1 to 12 and aa 14 to 41 are sufficient for association with σNS.
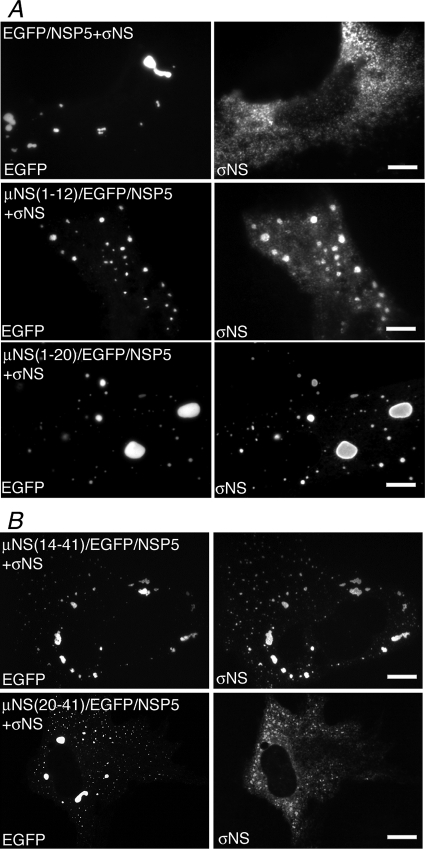
Our previously described deletion of 13 aa from the μNS N terminus (33) disrupted its association with σNS, and because this was already a small region, we did not create additional deletions to dissect it further. These previous findings suggested that σNS might associate with the N-terminal 12 aa of μNS. Indeed, when σNS was coexpressed with μNS(1-12)/EGFP/NSP5 in cells, we found that the two proteins colocalized in VLS, although colocalization was incomplete (Fig. 3A, second row). When an additional 8 aa from μNS were added to the EGFP/NSP5 fusion protein, σNS colocalized with VLS in every cell (Fig. 3A, third row), suggesting that μNS aa 13 to 19 contribute to the σNS association. When the N-terminal 13 aa were deleted from the μNS fusion protein, σNS also completely colocalized with VLS in every cell (Fig. 3B, top). μNS aa 14 to 19 again appear to be important for this putative binding region for σNS, because when deleted in the fusion protein, there was no association between the two proteins (Fig. 3B, bottom). These findings suggest that σNS may bind independently to two different regions within the N-terminal 41 aa of μNS: aa 1 to 12 and aa 14 to 41.
FIG. 3.
μNS aa 1 to 12 or aa 14 to 41 are sufficient for associations with the nonstructural protein σNS. For each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing σNS and either EGFP/NSP5 (top), μNS(1-12)/EGFP/NSP5 (middle), or μNS(1-20)/EGFP/NSP5 (bottom). After fixation, cells were stained with mouse monoclonal antibody 3E10 against σNS followed by Alexa 594-conjugated goat anti-mouse IgG to visualize σNS (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). (B) CV-1 cells were cotransfected with plasmids expressing σNS and either μNS(14-41)/EGFP/NSP5 (top) or μNS(20-41)/EGFP/NSP5 (bottom). After fixation, cells were stained with mouse monoclonal antibody 3E10 against σNS followed by Alexa 594-conjugated goat anti-mouse IgG to visualize σNS (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). Bar, 10 μm.
μNS aa 65 to 74 are necessary and aa 41 to 173 are sufficient for associations with λ1.
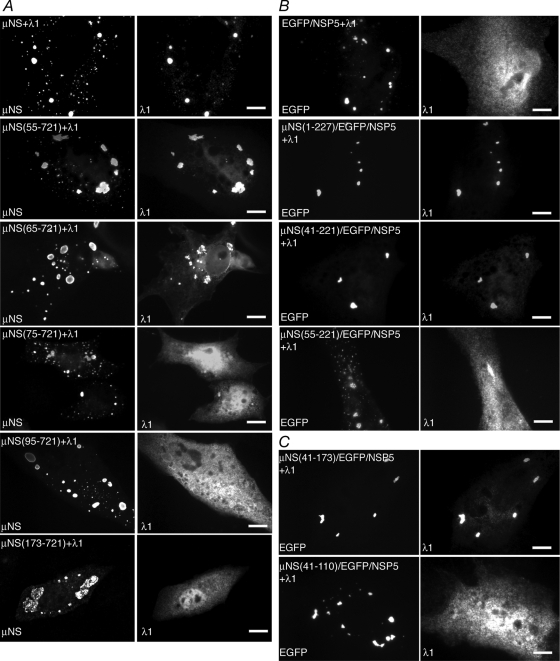
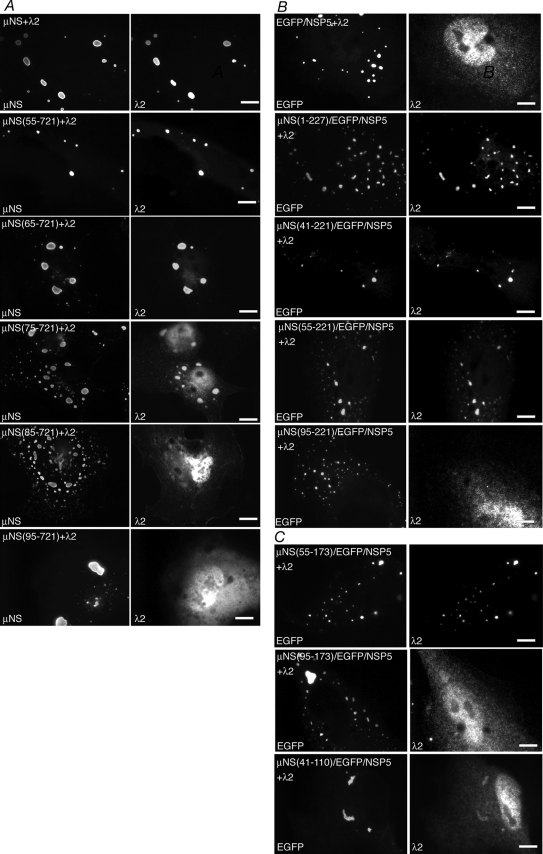
A previous study showed that the deletion of the N-terminal 41 aa of μNS does not disrupt its associations with the viral core surface protein λ1 (6). To identify the region of μNS that associates with λ1 more precisely, we utilized a series of N-terminal deletion mutants of μNS. We cotransfected plasmids expressing each deletion mutant with plasmids expressing λ1 in cells and then visualized the localization of λ1 relative to FLS at 18 h p.t. by immunofluorescence microscopy. We found that the deletion of 55 aa from the N terminus of μNS did not disrupt the association with λ1 (Fig. 4A, second row). The deletion of an additional 10 aa from μNS did not completely abrogate the λ1 association; however, λ1 formed aggregates in these cells, which appeared to localize around FLS, and a portion of λ1 was diffusely distributed in cells (Fig. 4A, third row). The deletion of 74 aa from the μNS N terminus resulted in the loss of an association with λ1 (Fig. 4A, fourth row), suggesting that μNS aa 65 to 74 are necessary for the λ1 association. Further μNS deletions of 94 and 172 aa also resulted in the loss of an association with λ1 (Fig. 4A, fifth and bottom rows).
FIG. 4.
μNS aa 65 to 74 are necessary and aa 41 to 173 are sufficient for associations with the core surface protein λ1. For each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing λ1 and either μNS (top row), μNS(55-721) (second row), μNS(65-721) (third row), μNS(75-721) (fourth row), μNS(95-721) (fifth row), or μNS(173-721) (bottom row). After fixation, cells were stained with rabbit polyclonal antibodies against MRV cores followed by Texas Red-conjugated μNS-specific rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-rabbit IgG to visualize λ1 (right). (B) CV-1 cells were cotransfected with plasmids expressing λ1 and either EGFP/NSP5 (top row), μNS(1-227)/EGFP/NSP5 (second row), μNS(41-221)/EGFP/NSP5 (third row), or μNS(55-221)/EGFP/NSP5 (bottom row). After fixation, cells were stained with rabbit polyclonal antibodies against MRV cores followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize λ1 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). (C) CV-1 cells were cotransfected with plasmids expressing λ1 and either μNS(41-173)/EGFP/NSP5 (top) or μNS(41-110)/EGFP/NSP5 (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against MRV cores followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize λ1 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). Bar, 10 μm.
We were next interested in identifying the μNS regions sufficient for the association with λ1. To identify these regions, we constructed plasmids to express a series of EGFP/NSP5 fusion proteins containing contiguous short regions of μNS. Work with our deletion mutants suggested that the N-terminal 221 aa of μNS are necessary for the association with each of the tested proteins except the viral RNA-dependent RNA polymerase (RdRp) λ3 (see below), and thus, we restricted our μNS/EGFP/NSP5 fusions to the N-terminal 227 aa of μNS. We cotransfected a plasmid expressing each of the fusion proteins with a plasmid expressing λ1 and visualized the protein localizations at 18 h p.t. by immunofluorescence microscopy. We found that λ1 associated with the EGFP/NSP5 fusion containing either μNS(1-227) (Fig. 4B, second row) or μNS(41-221) (Fig. 4B, third row) but not with the fusion containing μNS(55-221) (Fig. 4B, bottom row). In contrast to what was found for the μNS N-terminal deletion mutants, these findings suggest that in the absence of the μNS C terminus, aa 42 to 54 are necessary for the λ1 association. We additionally examined the localization of λ1 when coexpressed with EGFP/NSP5 fusions from which an additional 49 aa and 63 aa were deleted from the C terminus of the μNS fragment. In these experiments, λ1 associated with the fusion containing μNS(41-173) (Fig. 4C, top) but not with that containing μNS(41-110) (Fig. 4C, bottom), suggesting that amino acids in the region of aa 111 to 173 of μNS are important for the λ1 association. These findings suggest that μNS aa 41 to 173 are sufficient for the λ1 association.
μNS aa 75 to 84 are necessary and aa 41 to 173 are sufficient for associations with λ2.
Similar to the case for λ1, we previously found that the N-terminal 41 aa of μNS are not necessary for the association with λ2 (6). We next investigated the region of μNS that was necessary for the association with λ2 by individually cotransfecting a plasmid expressing λ2 with a panel of μNS N-terminal deletion mutants and examining the localization of λ2 relative to FLS by immunofluorescence microscopy. We found that λ2 associated with FLS formed by μNS(55-721) (Fig. 5A, second row) and μNS(65-721) (Fig. 5A, third row) was less completely associated to FLS formed by μNS(75-721) (Fig. 5A, fourth row) and did not colocalize with FLS formed by μNS(85-721) or μNS(95-721) (Fig. 5A, fifth and bottom rows). These findings suggest that μNS aa 75 to 84 are necessary for the λ2 association with μNS.
FIG. 5.
μNS aa 75 to 84 are necessary and aa 55 to 173 are sufficient for associations with the core surface protein λ2. For each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing λ2 and either μNS (top row), μNS(55-721) (second row), μNS(65-721) (third row), μNS(75-721) (fourth row), μNS(85-721) (fifth row), or μNS(95-721) (bottom row). After fixation, cells were stained with rabbit polyclonal antibodies against μNS and mouse monoclonal antibody 7F4 against λ2 followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-mouse IgG to visualize λ2 (right). (B) CV-1 cells were cotransfected with plasmids expressing λ2 and either EGFP/NSP5 (top row), μNS(1-227)/EGFP/NSP5 (second row), μNS(41-221)/EGFP/NSP5 (third row), μNS(55-221)/EGFP/NSP5 (fourth row), or μNS(95-221)/EGFP/NSP5 (bottom row). After fixation, cells were stained with mouse monoclonal antibody 7F4 against λ2 followed by Alexa 594-conjugated goat anti-mouse IgG to visualize λ2 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). (C) CV-1 cells were cotransfected with plasmids expressing λ2 and either μNS(55-173)/EGFP/NSP5 (top), μNS(95-173)/EGFP/NSP5 (middle), or μNS(41-110)/EGFP/NSP5 (bottom). After fixation, cells were stained with mouse monoclonal antibody 7F4 against λ2 followed by Alexa 594-conjugated goat anti-mouse IgG to visualize λ2 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). Bar, 10 μm.
To identify the region of μNS sufficient for the association with λ2, we examined the localization of this protein when coexpressed with our panel of μNS/EGFP/NSP5 fusion proteins. We found that λ2 associated with the EGFP/NSP5 fusion containing μNS(1-227) (Fig. 5B, second row), μNS(41-221) (Fig. 5B, third row), or μNS(55-221) (Fig. 5B, fourth row) but not with that containing μNS(95-221) (Fig. 5B, bottom row). We then tested its association with the fusions from which an additional 48 aa were deleted from the C terminus of the μNS fragment and found that λ2 associated with that containing μNS(55-173) (Fig. 5C, top) but not with that containing μNS(95-173) (Fig. 5C, middle). The deletion of 63 aa more from the C terminus of the μNS fragment abrogated colocalization with λ2 (Fig. 5C, bottom), suggesting that amino acids in the region of aa 110 to 173 of μNS are important for the λ2 association. These findings suggest that μNS aa 55 to 173 are sufficient for the λ2 association.
μNS aa 173 to 220 are necessary and sufficient for associations with σ2.
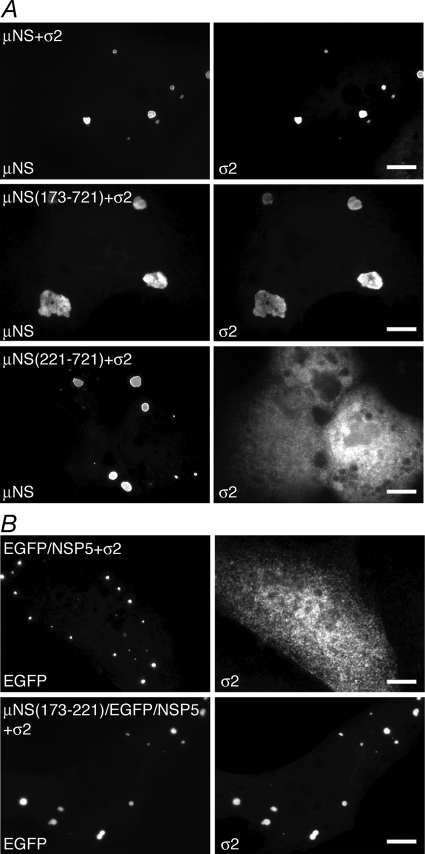
As found for the core surface proteins λ1 and λ2, our previous studies have shown that the N-terminal 41 aa of μNS are not required for its association with the other core surface protein, σ2 (6). To map the region of μNS necessary for the association with σ2 more precisely, we cotransfected a plasmid expressing σ2 with plasmids expressing μNS N-terminal deletion mutants and then examined the localization of σ2 relative to FLS formed by each of the μNS deletions at 18 h p.t. by immunofluorescence microscopy. We found that σ2 colocalized with FLS formed by μNS(173-721) (Fig. 6A, middle) but was diffusely distributed throughout cells expressing μNS(221-721), even though the latter deletion continued to form distinctive FLS (Fig. 6A, bottom). These data suggest that μNS aa 173 to 220 are necessary for the association with σ2.
FIG. 6.
μNS aa 173 to 220 are necessary and aa 173 to 221 are sufficient for associations with the core surface protein σ2. For each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing σ2 and either μNS (top), μNS(173-721) (middle), or μNS(221-721) (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against MRV cores followed by Texas Red-conjugated μNS-specific rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-rabbit IgG to visualize σ2 (right). (B) CV-1 cells were cotransfected with plasmids expressing σ2 and either EGFP/NSP5 (top) or μNS(173-221)/EGFP/NSP5 (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against MRV cores followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize σ2 (right). The inherent fluorescence of EGFP was used to visualize each of the fusion proteins (left). Bar, 10 μm.
To identify the region of μNS sufficient for the association with σ2, we determined the localization of σ2 when coexpressed with plasmids expressing μNS/EGFP/NSP5 fusion proteins. Cells were transfected, and the localization of σ2 relative to VLS formed by the fusion proteins was examined at 18 h p.t. by immunofluorescence microscopy. Consistent with results described above for nonfused μNS deletions, we found that σ2 localized to VLS formed by μNS(173-221)/EGFP/NSP5 (Fig. 6B, bottom), suggesting that μNS aa 173 to 220 are sufficient for the association with σ2.
The C-terminal 250 aa of μNS are sufficient for associations with λ3.
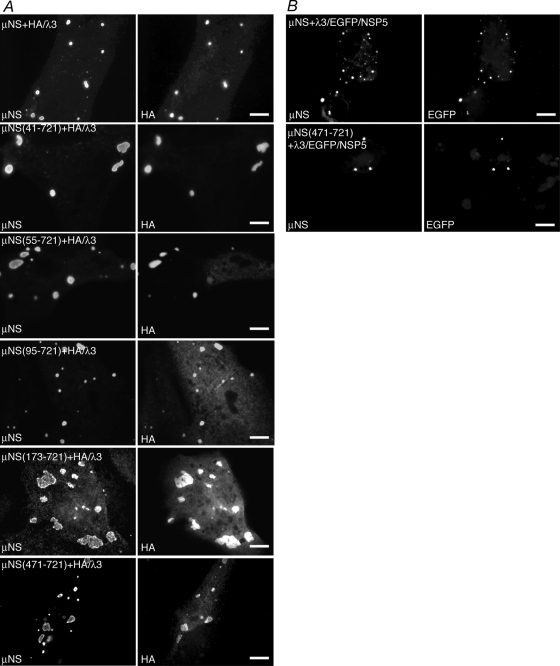
Although our previous studies have shown that full-length μNS associates with the MRV RdRp λ3 (32), this protein was not examined for its ability to associate with μNSC, a second form of μNS expressed in cells that is missing the N-terminal 41 aa (29). We utilized our new panel of μNS deletion mutants in this study to identify the region of μNS necessary for the association with λ3. A plasmid expressing an HA-tagged form of λ3 was cotransfected with plasmids expressing each of the μNS deletions. The localization of HA-tagged λ3 was then examined relative to the FLS formed by each of the μNS deletions at 18 h p.t. by immunofluorescence microscopy. We found that λ3/HA associated completely with μNS(41-721) (Fig. 7A, second row) and μNS(55-721) (Fig. 7A, third row) and continued to colocalize partially with μNS(95-721) (Fig. 7A, fourth row), μNS(173-721) (Fig. 7A, fifth row), or μNS(471-721) (Fig. 7A, bottom row), suggesting that μNS aa 471 to 721 are sufficient for the association with λ3/HA. Since further deletions from the N terminus of μNS result in the loss of FLS formation (8), such mutants could not be examined using this assay.
FIG. 7.
μNS aa 471 to 721 are necessary and sufficient for associations with MRV RdRp λ3. In each experiment, cells were processed for fluorescence microscopy at 18 h p.t. (A) CV-1 cells were cotransfected with plasmids expressing λ3/HA and either μNS (top row), μNS(41-721) (second row), μNS(55-721) (third row), μNS(95-721) (fourth row), μNS(173-721) (fifth row), or μNS(471-721) (bottom row). After fixation, cells were stained with rabbit polyclonal antibodies against μNS and mouse monoclonal antibody against HA followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-mouse IgG to visualize HA-tagged λ3 (right). (B) CV-1 cells were cotransfected with plasmids expressing λ3/EGFP/NSP5 and either μNS (top) or μNS(471-721) (bottom). After fixation, cells were stained with rabbit polyclonal antibodies against μNS followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μNS (left). The inherent fluorescence of EGFP was used to visualize λ3/EGFP/NSP5 (right). Bar, 10 μm.
Because the localization of λ3/HA to FLS appeared to be partially diminished with some of the μNS deletion mutants, we utilized an additional assay to confirm that μNS aa 471 to 721 are sufficient for the association with λ3-HA. Our previous experiments showed that when coexpressed in cells, MRV μNS and rotavirus EGFP/NSP5 do not colocalize and instead form nonoverlapping FLS and VLS, respectively (Fig. 1, top). We therefore cloned the L1 gene, encoding the λ3 protein, upstream of the sequences encoding EGFP/NSP5 to form a plasmid expressing λ3/EGFP/NSP5. This plasmid was then cotransfected with a plasmid expressing either full-length μNS or μNS(471-721), and the localization of λ3/EGFP/NSP5 relative to μNS(1-721) or μNS(471-721) FLS was examined at 18 h p.t. by immunofluorescence microscopy to determine if the addition of λ3 to EGFP/NSP5 would cause a coalescence of FLS and VLS. As expected, λ3/EGFP/NSP5 associated with μNS(1-721) (Fig. 7B, top). Importantly, the λ3/EGFP/NSP5 fusion also completely colocalized with μNS(471-721) (Fig. 7B, bottom), confirming that the C-terminal 250 aa of μNS are sufficient for the association with λ3.
We also attempted to examine the association of λ3/HA with fusion proteins consisting of fragments of μNS fused to EGFP/NSP5 as we had done to define regions of μNS sufficient for associations with other MRV proteins. In this case, however, we found that λ3/HA colocalized with EGFP/NSP5 VLS even in the absence of μNS fusions to the latter protein. This is perhaps not completely unexpected, as the MRV RdRp λ3 shares notable homology with the rotavirus RdRp VP1, a known NSP5-associating protein (2).
Core particles associate with μNS aa 173 to 220.
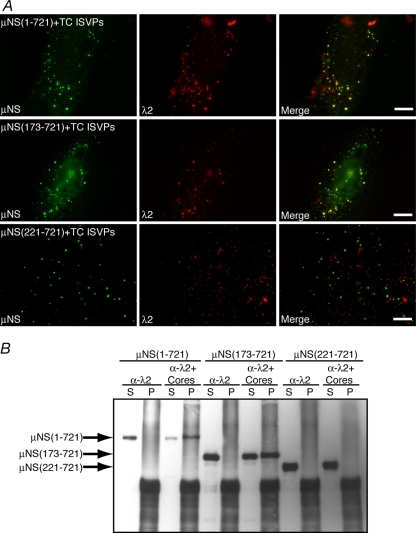
We have previously shown that in addition to recruiting each of the viral core proteins and σNS to VF, parental input core particles are recruited to FLS when cells transfected with a plasmid expressing full-length μNS are infected with top-component (genome-minus) ISVPs of MRV (6). To identify the region of μNS necessary for the recruitment of core particles to FLS, we transfected cells with a plasmid expressing either full-length μNS or the deletion mutant μNS(41-721), μNS(173-721), or μNS(221-721). At 6 h p.t., we infected the transfected cells with top-component ISVPs at 1,000 PFU/cell in the presence of cycloheximide to prevent protein synthesis. At 90 min postinfection (p.i.), cells were fixed and stained with antibodies against μNS and MRV core particles, and the localization of cores relative to FLS was examined by immunofluorescence microscopy. We found that cores localized to FLS in cells expressing both full-length μNS (Fig. 8A, top) (as previously shown [6]) as well as the deletion mutant μNS(173-721) (Fig. 8A, middle). In cells expressing μNS(221-721), however, core particles did not associate with FLS (Fig. 8A, bottom), suggesting that μNS aa 173 to 220 are necessary for core recruitment to FLS.
FIG. 8.
μNS aa 173 to 220 are necessary for MRV core particle localization to FLS and associations with MRV cores. (A) CV-1 cells were transfected with a plasmid expressing either μNS (top row), μNS(173-721) (middle row), or μNS(221-721) (bottom row). At 6 h p.t., 100 μg/ml of cycloheximide (CHX) was added to the cells for 30 min, at which point the cells were incubated (still in the presence of cycloheximide) with type 1 Lang top-component (TC) ISVPs (1,000 PFU/cell) at 4°C for 30 min and then shifted to 37°C for 90 min and fixed. Cells were stained with mouse monoclonal antibody 7F4 against λ2 followed by Texas Red-conjugated μNS-specific rabbit IgG to visualize μNS (left) and Alexa 488-conjugated goat anti-mouse IgG to visualize λ2 (middle). Merged images are also shown (right). Bars, 10 μm. (B) CV-1 cells transfected with a plasmid expressing either μNS (lanes 1 to 4), μNS(173-221) (lanes 5 to 8), or μNS(221-721) (lanes 9 to 12) were lysed at 18 h p.t., and the proteins from the resulting lysates were immunoprecipitated with magnetic beads preincubated with mouse monoclonal antibody 7F4 against λ2 either without (lanes 1, 2, 5, 6, 9, and 10) or with (lanes 3, 4, 7, 8, 11, and 12) an additional incubation with MRV core particles. Postbinding supernatants (S) and immunoprecipitated proteins (P) were then separated by SDS-PAGE, and associated μNS proteins were visualized by immunoblotting using rabbit polyclonal antibodies against μNS followed by HRP-conjugated goat anti-rabbit IgG antibody to visualize μNS. μNS, μNS(173-221), and μNS(221-721) are indicated by arrows.
A second experimental approach was used to confirm the region of μNS necessary for the association with parental core particles. Cells transfected with a plasmid expressing full-length μNS or the deletion mutant μNS(173-721) or μNS(221-721) were incubated for 18 h p.t. and then collected and lysed. Magnetic beads conjugated with protein A were incubated with monoclonal antibody 7F4 against the core surface protein λ2 and then split into two aliquots. One aliquot (bead/λ2) was set aside, and the other aliquot was incubated with purified MRV core particles (bead/λ2/cores). The 7F4-bound beads, bound to cores or not, were then incubated for 4 h with wild-type or mutant μNS-containing lysates, and beads and associated proteins were then separated from the supernatant and washed. Proteins contained in the bead (pellet) fraction as well as in the postbinding supernatant fraction were separated by SDS-PAGE, and any μNS protein associated with the samples was visualized by immunoblotting using μNS antiserum. When bead/7F4 complexes were used to immunoprecipitate associated proteins, full-length μNS was found entirely in the supernatant (Fig. 8B, lanes 1 and 2), reflecting the inability of the λ2-specific antibody to immunoprecipitate μNS from the lysate. However, when bead/7F4/core complexes were incubated with lysate containing full-length μNS, most μNS was found in the pellet (Fig. 8B, lanes 3 and 4), reflecting the specific interaction of μNS with core particles. Similar results were seen when lysate containing μNS(173-721) was used, with μNS found entirely in the supernatant following incubation with bead/7F4 complexes (Fig. 8B, lanes 5 and 6) and in both the supernatant and pellet fraction following incubation with bead/7F4/core complexes (Fig. 8B, lanes 7 and 8). This suggests that the N-terminal 172 aa of μNS are not required for the association with core particles. In the lysate expressing μNS(221-721), in contrast, a different result was seen. When incubated with either bead/7F4 complexes or bead/7F4/core complexes, μNS(221-721) was found entirely in the supernatant fraction of the immunoprecipitation (Fig. 8B, lanes 9 to 12). This finding suggests that μNS(221-721) is not able to bind core particles and concurs with our immunofluorescence findings showing that μNS aa 173 to 220 are necessary for the association with MRV core particles.
Newly synthesized MRV RNA is localized to VF in infected cells.
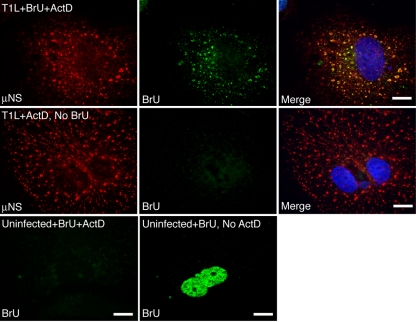
Our previous results showing that parental core particles localize to FLS formed by μNS (6), coupled with the above-described new results defining a region of μNS necessary for this localization, suggest that MRV core particles are embedded in VF in infected cells. Because cores synthesize the plus-strand RNAs of MRV (22), we hypothesized that the localization of cores to VF may result in the production of MRV plus-strand RNAs within VF during infection. To test this hypothesis, we infected cells with ISVPs and at early times p.i. transfected the infected cells with BrU, a uridine analog that is efficiently incorporated into RNA as it is synthesized (24), in the presence or absence of actinomycin D, an inhibitor of cellular RNA polymerase II (52), to prevent cellular transcription. At 60 min p.t., cells were fixed and stained with antibodies against μNS to visualize VF as well as with antibodies against bromodeoxyuridine, which cross-react with BrU (23), to visualize newly synthesized RNA. The localization of newly synthesized RNA relative to VF was visualized by immunofluorescence microscopy. We found that in infected cells pulse-labeled with BrU (Fig. 9, top), the localization of newly synthesized BrU-containing RNA was concentrated in VF, supporting our hypothesis that MRV plus-strand RNAs are synthesized by core particles localized within these structures. The BrU staining in VF was not a result of nonspecific antibody binding, as no staining was seen in the absence of BrU transfection (Fig. 9, middle). The presence of newly transcribed RNA was not seen in actinomycin D-treated, uninfected cells (Fig. 9, bottom left), and an alternative pattern of nuclear BrU staining was seen in uninfected cells without actinomycin D treatment (Fig. 9, bottom right).
FIG. 9.
Newly synthesized MRV RNA is localized to VF in infected cells. CV-1 cells were infected with type 1 Lang (T1L) ISVPs at 100 PFU/cell (top and middle) or not infected (bottom), and at 6 h p.i., cells were treated (+ActD) or not (no ActD), as indicated, with actinomycin D to inhibit transcription by cellular RNA polymerase II. Cells were then transfected (+BrU) or not transfected (no BrU), as indicated, with BrU. At 60 min p.t., cells were fixed and stained with rabbit polyclonal antibodies against μNS and a mouse monoclonal antibody against bromodeoxyuridine, which also binds to BrU, followed by Alexa 594-conjugated goat anti-rabbit IgG to visualize μNS (top and middle, left) and Alexa 488-conjugated goat anti-mouse IgG to visualize BrU (top, middle, and bottom rows, middle column, and bottom row, left column). Merged images of the top and middle panels are also shown (right). Bar, 10 μM.
DISCUSSION
Early studies found that μNS is a major protein component of replication complexes associated with MRV RNAs (35, 36). Subsequent studies have shown that μNS associates with each of the viral core proteins, σNS, and core particles and that these associations result in their recruitment to or retention in FLS (4, 6-8, 32, 33). The consequent hypothesis is that μNS serves as a cytoplasmic scaffolding protein that organizes core proteins, σNS, and core particles such that viral replication and assembly intermediates can efficiently form in VF, which are sites of MRV genome replication and progeny core assembly (14, 40, 44). Our new results, which show that each associating protein or particle utilizes distinct primary sequence regions within μNS, support this hypothesis by providing evidence that a single molecule of μNS may be capable of associating with multiple MRV proteins at the same time (see Fig. 10 for a summary of results). However, additional work is needed to determine if a single molecule of μNS can bind all, a subset, or just one of its identified binding partners or, alternatively, if the binding of one protein interferes with the binding of others. The sequential association of a particular μNS molecule with different proteins over the course of the process of replication and assembly is another interesting possibility.
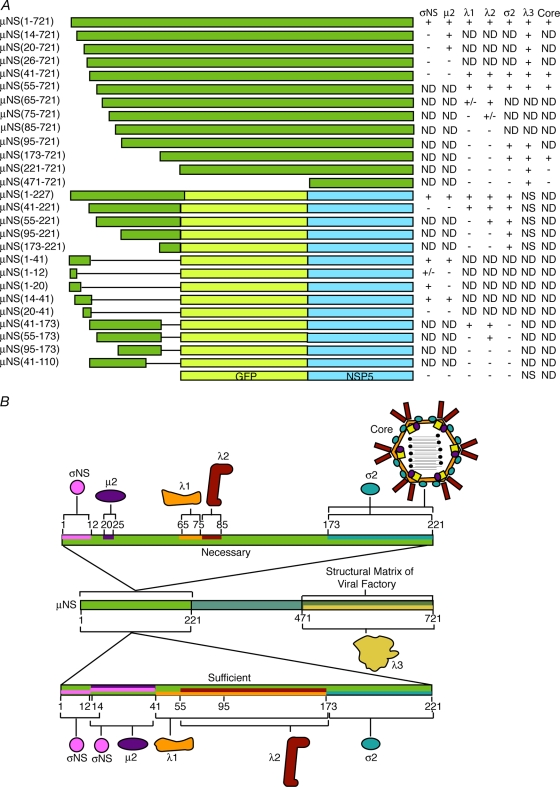
FIG. 10.
Summary of μNS regions necessary and sufficient for associations with σNS, μ2, λ1, λ2, σ2, λ3, and core particles. (A) List of all μNS deletion mutants and EGFP/NSP5 fusion proteins tested in this study and the results of their association with the indicated MRV proteins and MRV cores. ND, not determined; NS, nonspecific association. (B) Schematic representation of μNS showing regions of the protein necessary and sufficient for associations with each MRV protein and core particles.
We previously reported that upon entering cells, MRV core particles are rapidly localized to FLS that have been preformed by plasmid-expressed μNS and thus might be similarly recruited into a newly forming VF during the course of normal infection (6). It was not known, however, if cores are recruited to VF in infected cells. The core serves as the transcriptase particle for the production of MRV plus-strand RNAs (22); thus, if cores localize to VF during infection, it would be expected that newly synthesized MRV RNAs are located within VF. In this study, we found that MRV RNAs are localized to VF from very early times after infection (starting at 3 h p.i.), suggesting that entering cores are recruited to VF as they form at early times in infected cells or, alternatively, that entering cores may seed VF at early times in infection. Together with the localization of all five core structural proteins, as well as the ssRNA-binding nonstructural protein σNS, which is thought to play a role in genome replication and/or packaging (3, 12, 20, 21, 39), this localization of cores to VF may be a particularly efficient way for the virus to ensure that newly transcribed MRV plus-strand RNAs are retained close to the proteins necessary for replication and core assembly. The localization of newly synthesized RNAs at viroplasms was also shown previously for rotavirus- and phytoreovirus-infected cells (45, 51), suggesting that diverse members of the family Reoviridae are similar in this regard.
The region of μNS mapped as necessary for associations with core particles is the same one mapped as both necessary and sufficient for the association with the core surface protein σ2. This suggests that cores may be localized to FLS by the association of μNS with σ2 on the surface of the core particle; however, we have not yet identified the region of σ2 involved in the association with μNS and therefore do not yet know if this region is surface exposed in the context of the assembled core. We have also recently found that μNS associates with a small region of the core surface protein λ2 that is indeed surface exposed in the assembled core (C. L. Miller and M. L. Nibert, unpublished results), therefore raising the possibility that core particles may also associate with μNS via λ2. Furthermore, we have not yet addressed the region of μNS that is sufficient for the association of cores, leaving open the possibility that more than one region of μNS can associate with cores such that they localize to FLS.
Like μNS, orbivirus NS2, phytoreovirus Pns12, and rotavirus NSP2/NSP5 have been shown to be the only viral proteins whose expression is required for forming VIB-like structures or VLS in transfected cells (18, 47, 48, 51). Thus, other, and perhaps all, members of the family Reoviridae encode specific proteins that function in part to form the organizational matrix of these cytoplasmic structures. In addition, there is a good deal of evidence suggesting that these other proteins, like μNS, are involved in recruiting or retaining other viral proteins to their respective structures. For example, most of the proteins that make up the bluetongue virus (orbivirus) core and outer capsid are localized in or near VIBs in infected cells (9). Similarly, several recent studies have shown that in addition to its function in forming the matrix of VIBs, the NS2 protein is involved in recruiting or retaining the bluetongue virus core proteins VP1 (RdRp), VP3 (core shell protein), VP4 (mRNA capping enzyme), and VP6 (dsRNA helicase) to VIBs (25). The remaining core protein (VP7) appears to be recruited to VIBs through its association with VP3 (25). The regions within the NS2 protein necessary and sufficient for each of these functions have not yet been identified, but it would be interesting if, like μNS, NS2 associates with each of the proteins utilizing short, largely nonoverlapping sequences, supporting our hypothesis that these proteins act as cytoplasmic scaffolding for building replication or assembly complexes in cells.
Unlike MRV, orbiviruses, and phytoreoviruses, rotaviruses encode two proteins that must normally associate to form viroplasms in infected cells (18). Whether these two proteins “split” the functional duties of μNS, NS2, or Pns12 is not known. NSP5 and NSP2 have both been reported to associate with the rotavirus RdRp VP1 (2), suggesting that NSP5 and NSP2 may work together to recruit other rotavirus proteins to VLS. A determination of other rotavirus protein associations with VLS, along with the mapping of these associations, will shed light on other potential similarities and differences between μNS and these proteins.
When coexpressed in cells, μNS and EGFP/NSP5 form nonoverlapping structures. Little is known about the cellular proteins that may be involved in forming either FLS or VLS; however, this result suggests that either the two proteins do not require a shared set of cellular proteins to form the structures or any shared cellular proteins that they do require are abundant enough for both structures to form independently in cells. The identity and importance of cellular proteins that associate with μNS are currently under investigation.
One cellular system that does appear to play some role in both VF/FLS and viroplasm/VLS formation is the microtubule network. When cells are treated with the microtubule-depolymerizing drug nocodazole, VF (in infected cells) and FLS (in cells expressing μNS) remain small and diffusely distributed in the cytoplasm (compared to the large, perinuclear structures that are normally seen) (8, 38). This finding suggests that VF movement on microtubules plays a role in VF coalescence. Similarly, a recent study showed that both NSP2 and NSP5 associate with cellular microtubules and that nocodazole treatment of rotavirus-infected cells results in small, diffuse viroplasms that do not develop into larger, more perinuclear structures (10). Nonetheless, while some FLS and VLS localize near each other in the cytoplasm (perhaps suggesting the movement of both of them on the same microtubule track), this association with microtubules does not result in coalescence events between μNS and EGFP/NSP5. Unlike μNS and NSP2/NSP5, it was recently reported that the formation of VIB-like structures by bluetongue virus NS2 is not affected by a disruption of the microtubule network (25), suggesting that these cytoplasmic scaffolding proteins from the different viruses may exploit different cellular mechanisms for cytoplasmic trafficking.
Acknowledgments
We thank Oscar Burrone for the EGFP/NSP5 plasmid, Elaine Freimont for technical assistance, and other members of our laboratories for helpful discussions.
This work was supported by grants F32 AI56939 to C.L.M. and R01 AI47904 and R56 AI067445 to M.L.N. from the U.S. National Institutes of Health. M.M.A. is a Ph.D. graduate of the Harvard Virology Program, which is partially supported by training grant T32 AI07245 from the U.S. National Institutes of Health. In addition, T.J.B. was partially supported by a postdoctoral fellowship from that training grant. Other assistance to C.L.M. was provided by the Office of the Dean, Iowa State University College of Veterinary Medicine.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Arnold, M. M., K. E. Murray, and M. L. Nibert. 2008. Formation of the factory matrix is an important, though not a sufficient function of nonstructural protein μNS during reovirus infection. Virology 375:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnoldi, F., M. Campagna, C. Eichwald, U. Desselberger, and O. R. Burrone. 2007. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J. Virol. 81:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. M., T. R. Peters, and T. S. Dermody. 2003. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 77:5948-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broering, T. J., M. M. Arnold, C. L. Miller, J. A. Hurt, P. L. Joyce, and M. L. Nibert. 2005. Carboxyl-proximal regions of reovirus nonstructural protein μNS necessary and sufficient for forming factory-like inclusions. J. Virol. 79:6194-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broering, T. J., J. Kim, C. L. Miller, C. D. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. L. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brookes, S. M., A. D. Hyatt, and B. T. Eaton. 1993. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J. Gen. Virol. 74:525-530. [DOI] [PubMed] [Google Scholar]
- 10.Cabral-Romero, C., and L. Padilla-Noriega. 2006. Association of rotavirus viroplasms with microtubules through NSP2 and NSP5. Mem. Inst. Oswaldo Cruz 101:603-611. [DOI] [PubMed] [Google Scholar]
- 11.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, R. K., and B. N. Fields. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. Virology 50:799-809. [DOI] [PubMed] [Google Scholar]
- 13.Dales, S. 1965. Replication of animal viruses as studied by electron microscopy. Am. J. Med. 38:699-715. [DOI] [PubMed] [Google Scholar]
- 14.Dales, S., P. Gomatos, and K. C. Hsu. 1965. The uptake and development of reovirus in strain L cells followed with labelled viral ribonucleic acid and ferritin-antibody conjugates. Virology 25:193-211. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, B. T., A. D. Hyatt, and J. R. White. 1987. Association of bluetongue virus with the cytoskeleton. Virology 157:107-116. [DOI] [PubMed] [Google Scholar]
- 16.Eichwald, C., J. F. Rodriguez, and O. R. Burrone. 2004. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 85:625-634. [DOI] [PubMed] [Google Scholar]
- 17.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1786. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 18.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 19.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillian, A. L., and M. L. Nibert. 1998. Amino terminus of reovirus nonstructural protein σNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Gillian, A. L., S. C. Schmechel, J. Livny, L. A. Schiff, and M. L. Nibert. 2000. Reovirus protein σNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J. Virol. 74:5939-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillies, S., S. Bullivant, and A. R. Bellamy. 1971. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science 174:694-696. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, D. A., A. B. Hassan, R. J. Errington, and P. R. Cook. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 12:1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javed, A., S. K. Zaidi, S. E. Gutierrez, C. J. Lengner, K. S. Harrington, H. Hovhannisyan, B. C. Cho, J. Pratap, S. M. Pockwinse, M. Montecino, A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 2004. In situ immunofluorescence analysis: analyzing RNA synthesis by 5-bromouridine-5′-triphosphate labeling. Methods Mol. Biol. 285:29-31. [DOI] [PubMed] [Google Scholar]
- 25.Kar, A. K., B. Bhattacharya, and P. Roy. 2007. Bluetongue virus RNA binding protein NS2 is a modulator of viral replication and assembly. BMC Mol. Biol. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J., X. Zhang, V. E. Centonze, V. D. Bowman, S. Noble, T. S. Baker, and M. L. Nibert. 2002. The hydrophilic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J. Virol. 76:12211-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi, T., J. D. Chappell, P. Danthi, and T. S. Dermody. 2006. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 80:9053-9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, T., L. S. Ooms, J. D. Chappell, and T. S. Dermody. 2009. Identification of functional domains in reovirus replication proteins μNS and μ2. J. Virol. 83:2892-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108:134-146. [DOI] [PubMed] [Google Scholar]
- 30.McCutcheon, A. M., T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus M3 gene sequences and conservation of coiled-coil motifs near the carboxyl terminus of the μNS protein. Virology 264:16-24. [DOI] [PubMed] [Google Scholar]
- 31.McNulty, M. S., W. L. Curran, and J. B. McFerran. 1976. The morphogenesis of a cytopathic bovine rotavirus in Madin-Darby bovine kidney cells. J. Gen. Virol. 33:503-508. [DOI] [PubMed] [Google Scholar]
- 32.Miller, C. L., M. M. Arnold, T. J. Broering, C. Eichwald, J. Kim, J. B. Dinoso, and M. L. Nibert. 2007. Virus-derived platforms for visualizing protein associations inside cells. Mol. Cell. Proteomics 6:1027-1038. [DOI] [PubMed] [Google Scholar]
- 33.Miller, C. L., T. J. Broering, J. S. L. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohan, K. V., J. Muller, I. Som, and C. D. Atreya. 2003. The N- and C-terminal regions of rotavirus NSP5 are the critical determinants for the formation of viroplasm-like structures independent of NSP2. J. Virol. 77:12184-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan, E. M., and H. J. Zweerink. 1975. Characterization of transcriptase and replicase particles isolated from reovirus-infected cells. Virology 68:455-466. [DOI] [PubMed] [Google Scholar]
- 36.Morgan, E. M., and H. J. Zweerink. 1977. Characterization of the double-stranded RNA in replicase particles in reovirus-infected cells. Virology 77:421-423. [DOI] [PubMed] [Google Scholar]
- 37.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 793-842. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 38.Parker, J. S. L., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramig, R. F., R. Ahmed, and B. N. Fields. 1983. A genetic map of reovirus: assignment of the newly defined mutant groups H, I, and J. to genome segments. Virology. 125:299-313. [DOI] [PubMed] [Google Scholar]
- 40.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17:342-355. [DOI] [PubMed] [Google Scholar]
- 41.Roy, P. 2001. Orbiviruses and their replication, p. 1835-1869. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 42.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu, T., M. Yoshii, T. Wei, H. Hirochika, and T. Omura. 2009. Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7:24-32. [DOI] [PubMed] [Google Scholar]
- 44.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41:564-566. [DOI] [PubMed] [Google Scholar]
- 45.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spendlove, R. S., E. H. Lennette, C. O. Knight, and J. H. Chin. 1963. Development of viral antigen and infectious virus on HeLa cells infected with reovirus. J. Immunol. 90:548-553. [PubMed] [Google Scholar]
- 47.Theron, J., H. Huismans, and L. H. Nel. 1996. Site-specific mutations in the NS2 protein of epizootic haemorrhagic disease virus markedly affect the formation of cytoplasmic inclusion bodies. Arch. Virol. 141:1143-1151. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, C. P., T. F. Booth, and P. Roy. 1990. Synthesis of bluetongue virus-encoded phosphoprotein and formation of inclusion bodies by recombinant baculovirus in insect cells: it binds the single-stranded RNA species. J. Gen. Virol. 71:2073-2083. [DOI] [PubMed] [Google Scholar]
- 49.Touris-Otero, F., M. Cortez-San Martin, J. Martinez-Costas, and J. Benavente. 2004. Avian reovirus morphogenesis occurs within viral factories and begins with the selective recruitment of σNS and λA to μNS inclusions. J. Mol. Biol. 341:361-374. [DOI] [PubMed] [Google Scholar]
- 50.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei, T., T. Shimizu, K. Hagiwara, A. Kikuchi, Y. Moriyasu, N. Suzuki, H. Chen, and T. Omura. 2006. Pns12 protein of rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J. Gen. Virol. 87:429-438. [DOI] [PubMed] [Google Scholar]
- 52.Yu, F. L. 1980. Selective inhibition of rat liver nuclear RNA polymerase II by actinomycin D in vivo. Carcinogenesis 1:577-581. [DOI] [PubMed] [Google Scholar]