Abstract
Hepatitis delta antigen (HDAg) is a nuclear protein that is intimately involved in hepatitis delta virus (HDV) RNA replication. HDAg consists of two protein species, the small form (S-HDAg) and the large form (L-HDAg). Previous studies have shown that posttranslational modifications of S-HDAg, such as phosphorylation, acetylation, and methylation, can modulate HDV RNA replication. In this study, we show that S-HDAg is a small ubiquitin-like modifier 1 (SUMO1) target protein. Mapping data showed that multiple lysine residues are SUMO1 acceptors within S-HDAg. Using a genetic fusion strategy, we found that conjugation of SUMO1 to S-HDAg selectively enhanced HDV genomic RNA and mRNA synthesis but not antigenomic RNA synthesis. This result supports our previous proposition that the cellular machinery involved in the synthesis of HDV antigenomic RNA is different from that for genomic RNA synthesis and mRNA transcription, requiring different modified forms of S-HDAg. Sumoylation represents a new type of modification for HDAg.
Hepatitis delta virus (HDV) causes chronic and, occasionally, fulminant hepatitis in humans (16). HDV is a satellite virus which requires hepatitis B virus (HBV) to supply envelope proteins for virus assembly and production (46). It contains a circular RNA genome of 1.7 kb which replicates through a double-rolling-circle mechanism in the nucleus (33). The viral genomic RNA (G-RNA) is first replicated into the full-length antigenomic RNA (AG-RNA) and is also transcribed into a 0.8-kb mRNA, which encodes the only HDV protein, hepatitis delta antigen (HDAg). The AG-RNA, in turn, is replicated into G-RNA by another round of rolling-circle replication. The production of HDAg, which is intimately involved in HDV RNA replication, is a unique feature distinguishing HDV from plant viroids, which do not encode any protein. HDAg consists of two species, the small delta antigen (S-HDAg) (195 amino acids [aa]; 24 kDa) and the large delta antigen (L-HDAg) (214 aa; 27 kDa), which play different roles in HDV replication. S-HDAg is an essential activator for HDV RNA replication (25). In contrast, L-HDAg inhibits certain stages of HDV RNA replication but is required for virion assembly (5, 7, 28, 32). HDAg has been shown to be modified posttranslationally by phosphorylation (6, 9, 40, 42), acetylation (41), methylation (29), and in the case of L-HDAg, isoprenylation (14). Arg-13 methylation, Lys-72 acetylation, and Ser-177 phosphorylation are three major modifications of S-HDAg and are important for the functions of S-HDAg in HDV RNA replication (29, 40, 41, 48). Isoprenylation on Cys-211 of L-HDAg is required for virus assembly (14).
SUMO (small ubiquitin-related modifier) has been identified as a reversible posttranslational protein modifier (34, 35). The human genome encodes four SUMO proteins: SUMO1 to SUMO4 (13, 17). Among them, SUMO1 to SUMO3 are ubiquitously expressed, whereas SUMO4 is expressed mainly in the kidneys, lymph nodes, and spleen (17). Sumoylation is carried out by an E1 activating enzyme (the heterodimer Uba2-Aos1), an E2 conjugating enzyme (Ubc9), and one of several SUMO E3 ligases (38). Although SUMO E1 and E2 are sufficient to modify most substrates in vitro, in vivo sumoylation is usually facilitated by SUMO E3 for substrate selection (13, 24, 44). SUMO modification is a highly dynamic process that can be reversed rapidly by the action of SUMO-specific proteases, which are also involved in the maturation of newly synthesized SUMO proteins (18, 43). Although sumoylated proteins have been found throughout the cell, most of the known SUMO targets are cellular and viral proteins that function in the nucleus (19, 56). The functional outcomes of sumoylation are extremely diverse, including changes in intracellular localization, protein-protein interaction, protein-DNA interaction, and stability and activity of modified proteins (2, 11, 52, 56).
Here we show that HDV S-HDAg is posttranslationally modified by the SUMO pathway both in vivo and in vitro. Using a genetic fusion chimera to mimic sumoylated S-HDAg, we found that SUMO1 conjugation of S-HDAg selectively enhances HDV G-RNA and mRNA synthesis but not AG-RNA synthesis. This result adds to the growing list of different metabolic requirements between HDV G-RNA/mRNA synthesis and AG-RNA synthesis and supports the hypothesis that the cellular machinery involved in the synthesis of HDV AG-RNA is different from that for G-RNA synthesis and mRNA transcription.
MATERIALS AND METHODS
Yeast two-hybrid assay.
Standard techniques were used for the yeast two-hybrid system (8, 12). Briefly, the S-HDAg gene fragment was cloned in frame with the GAL4 DNA binding domain (GAL4-DBD) in the pGBKT7 vector (Clontech) to yield pGBKT7-SHDAg. The human SUMO genes for SUMO1, SUMO2, and SUMO3 and the Ubc9 gene were fused to the GAL4 transcription activation domain (GAL4-AD) by being subcloned into the pACT2 vector (Clontech) to yield pACT2-SUMO1, pACT2-SUMO2, pACT2-SUMO3, and pACT2-Ubc9. Yeast AH109 was cotransformed with the GAL4-DBD plasmid DNA (pGBKT7−SHDAg) and GAL4-AD plasmid DNA (pACT2-SUMO1, pACT2-SUMO2, pACT2-SUMO3, or pACT2-Ubc9). Positive clones were selected based on the ability of cells to grow on Trp, Leu, His, and Ade dropout media supplemented with 5 mM 3-amino-1,2,4-triazole (Sigma) as an indication of interaction.
Plasmid construction and in vitro transcription.
Plasmid pET-Sm, which was used for the expression of S-HDAg in Escherichia coli, has been described elsewhere (47). The expression plasmid pcDNA3.1-SHDAgHA, encoding hemagglutinin (HA)-tagged S-HDAg, was derived from the plasmid pCDNA3.1-WT (29) by introducing an HA tag-encoding sequence into the 3′ end of the S-HDAg coding sequence. Plasmids expressing mutant S-HDAgHAs (containing a lysine-to-arginine mutation and a deletion mutation) were derived from pcDNA3.1-SHDAgHA by PCR-based site-directed mutagenesis or a jumping-PCR method. The expression plasmids pSG5-SUMO1-GG, pcDNA3.1-Ubc9, pcDNA3-SENP1, pcDNA3-SENP2, and pcDNA3-Daxx501-740HA, encoding the mature form of SUMO1 (SUMO1-GG), Ubc9, SENP1, SENP2, and an HA-tagged Daxx(501-740) truncation mutant (30), respectively, were kind gifts from Hsiu-Ming Shih (Institute of Biomedical Science, Academia Sinica). Genomic and antigenomic HDV RNAs (1.9-kb G-RNA and AG-RNA, respectively), which contain the entire HDV genome plus approximately 200 additional nucleotides of HDV sequence, were transcribed from pKS/HDV1.9 (23) by use of MEGAscript (Ambion) after linearization by NotI and SnaBI digestion, respectively. Plasmid PX9-SUMO1, which was used for in vitro transcription of the SUMO1-S-HDAg-encoding mRNA, was derived from plasmid PX9-I/II (39) by introducing the SUMO1-encoding sequence into the 5′ end of the S-HDAg coding sequence by using the jumping-PCR method. Plasmid PX9-I/II-d1nt, which was used for in vitro transcription of an S-HDAg-encoding defective mutant (d1nt) in which the adenosine of the ATG start codon was deleted to abolish S-HDAg translation, has been described elsewhere (48). Capped and poly(A)-tailed S-HDAg-encoding defective mutant (d1nt) mRNA, wild-type S-HDAg-encoding mRNA (wt mRNA), and SUMO-1-S-HDAg-encoding mRNA (SUMO-HDAg mRNA) were transcribed from plasmids PX9-I/II-d1nt, PX9-I/II, and PX9-SUMO1, respectively, by use of T7 mMESSAGE mMACHINE Ultra (Ambion) after linearization by HindIII digestion.
Cell culture and transfection.
The human hepatoma cell line Huh-7 was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and incubated at 37°C in 5% CO2. Plasmid DNA and HDV RNA transfection experiments were performed by use of Lipo2000 and DMRIE-C transfection reagents (Invitrogen), respectively, according to the manufacturer's directions.
In vitro sumoylation.
Bacterially expressed S-HDAg was purified as described previously (47). Glutathione S-transferase (GST) protein, which was used as a negative control for this assay, was expressed in Escherichia coli and purified with glutathione-Sepharose (Pierce) following the manufacturer's protocol. The in vitro sumoylation assay was carried out using a sumoylation kit purchased from Biomol. GST-tagged RanGAP1 (GST-RG1) was provided in this kit as a positive control for this assay. Briefly, 400 ng of purified S-HDAg, GST protein, or GST-RG1 was mixed with 1 μl of 20× SUMO activating enzyme solution (SUMO E1), 1 μl of 20× SUMO conjugating enzyme solution (SUMO E2), and 2 μl of 10× sumoylation buffer, with or without 1 μl of 20× Mg-ATP solution. Water was added to make the final volume 20 μl. The reactions were conducted at 30°C for 1 hour and stopped by adding SDS-PAGE gel loading buffer. Reaction mixtures were then resolved by Western blotting, and S-HDAg and GST/GST-RG1 were detected by a polyclonal antibody to HDAg and a monoclonal antibody to GST, respectively.
In vivo sumoylation.
In Huh-7 cells, pcDNA3-Daxx501-740HA, pcDNA3.1-SHDAgHA wt, or mutant plasmid was cotransfected with SUMO1-GG, Ubc9, SENP1, and/or SENP2 expression plasmid, as indicated. At 48 hours posttransfection, cells were harvested using RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) containing 20 mM N-ethylmaleimide (NEM). The whole-cell extracts were then divided into two aliquots. One aliquot was analyzed by immunoblotting to confirm the expression of transfected proteins. The other aliquot was subjected to immunoprecipitation with a monoclonal antibody to the HA tag (3F10; Roche) and subsequent immunoblotting with the same antibody or a monoclonal antibody to SUMO1 (Santa Cruz) to detect the unmodified protein species and/or sumoylated protein species.
Immunofluorescence microscopy.
Huh-7 cells grown on slide chambers were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. After being blocked with 0.25% bovine serum albumin, the cells were incubated with a mouse monoclonal antibody to HDAg, a rabbit polyclonal antibody to SUMO1 (Biomol), a rabbit polyclonal antibody to HDAg, or a mouse monoclonal antibody to nucleolin (Santa Cruz). After an extensive wash, cells were stained with goat anti-mouse secondary antibody conjugated to Alexa Fluor 488 (green) or goat anti-rabbit secondary antibody conjugated to Alexa Fluor 568 (red) (Molecular Probes). Lastly, images were captured with an inverted fluorescence microscope (Leica DMI6000).
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) procedures, which were used to specifically detect HDV G-RNA, HDV AG-RNA, and HDAg-encoding mRNA by using the G-S, AG-S, and mI-S protocols, respectively, were performed as described previously (48). Briefly, total RNA was extracted from cells by use of TRIzol reagent (Invitrogen) according to the manufacturer's protocol. To detect the HDAg-encoding mRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, 200 ng of total RNA was reverse transcribed in a 20-μl final reaction mixture containing 1× reverse transcriptase buffer, 2.5 μM of oligo(dT)20, a 0.2 mM concentration (each) of dATP, dGTP, dCTP, and dTTP (Roche), 5 mM dithiothreitol, 40 U of RNaseOUT recombinant RNase inhibitor, and 200 U of SuperScript III reverse transcriptase (Invitrogen). The RT reaction was performed at 55°C for 50 min and then inactivated by heating at 95°C for 45 min. For the detection of HDV G-RNA or AG-RNA, 200 ng of total RNA was reverse transcribed in a 20-μl final reaction mixture containing 1× rTth reverse transcriptase buffer, 0.1 μM of tagged strand-specific primer (48), 40 U of RNaseOUT recombinant RNase inhibitor (Invitrogen), 1 mM MnCl2, a 0.2 mM concentration (each) of dATP, dGTP, dCTP, and dTTP, and 5 U of rTth (Applied Biosystems). The RT reaction was performed at 70°C for 15 min and then terminated by adding 2 μl of 10× chelating buffer. Real-time PCR was performed by using a LightCycler FastStart DNA master Sybr green I kit (Roche). Briefly, 5 μl of 10× serially diluted cDNA was mixed with 15 μl of master mix. The reaction consisted of an initiating step of 10 min at 95°C, followed by 45 cycles of amplification, with each cycle consisting of 10 s at 95°C, 3 s at 60°C, and 16 s at 72°C for HDV G-RNA and AG-RNA detection and of 10 s at 95°C, 3 s at 55°C, and 26 s at 72°C for HDV mRNA detection. The input total RNA amount was normalized to the GAPDH mRNA level. The reactions, data acquisition, and analyses were performed using a LightCycler 2.0 system (Roche).
RESULTS
S-HDAg interacts with Ubc9 and SUMO1 in yeast two-hybrid assay.
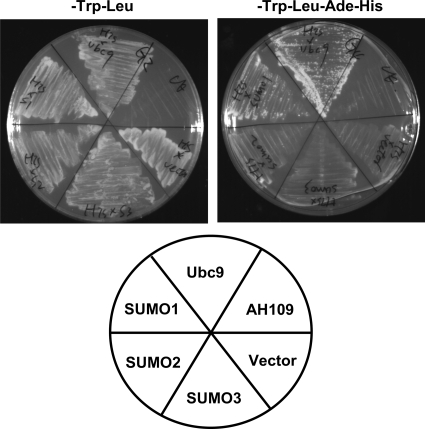
Given that S-HDAg is a nuclear protein intimately involved in HDV RNA replication and that most of the known cellular and viral SUMO targets are transcription factors or proteins that function in the nucleus (2, 50, 52, 56), we set out to investigate whether S-HDAg is a SUMO target protein. Previous studies have indicated that the interaction of a protein with SUMO isoforms or Ubc9 in the yeast two-hybrid assay is often indicative of SUMO conjugation to the interacting protein (1, 10, 15, 36). To investigate whether S-HDAg interacts with Ubc9 or SUMO isoforms, a yeast two-hybrid assay was performed, using S-HDAg as a bait and Ubc9 or SUMO isoforms as preys. As shown in Fig. 1, we observed that S-HDAg interacted with Ubc9 and SUMO1 but not with SUMO2 or SUMO3 in this assay. Although four SUMO isoforms (SUMO1, SUMO2, SUMO3, and SUMO4) have been found in vertebrates, previous studies have shown that the SUMO paralog preferred by viral proteins is SUMO1 (2). This appears to also be the case for HDV utilizing SUMO machinery. Accordingly, we focused on the SUMO1 modification of S-HDAg in the following experiments.
FIG. 1.
S-HDAg interacts with Ubc9 and SUMO1 in yeast two-hybrid assay. The growth of yeast strain AH109 cotransformed with GAL4-DBD plasmid DNA (pGBKT7−S-HDAg) and different GAL4-AD plasmid DNAs (pACT2-SUMO1, pACT2-SUMO2, pACT2-SUMO3, or pACT2-Ubc9) on SD medium is shown. Cotransformants were isolated on SD medium lacking tryptophan and leucine (−Trp−Leu; left plate) or on SD medium lacking tryptophan, leucine, adenine, and histidine (−Trp−Leu−Ade−His; right plate). The lower panel represents the isolated cotransformants in which the GAL4-AD plasmids were used for cotransformation. Vector, pACT2 plasmid without insert DNA; AH109, DNA-free transformation control.
S-HDAg is sumoylated in vitro.
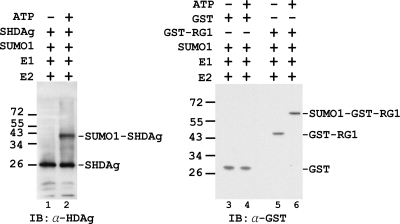
To determine whether S-HDAg can be modified by SUMO1, an in vitro sumoylation assay, using purified S-HDAg, SUMO activating enzyme (E1), Ubc9 (E2), and SUMO1, was performed. After incubation for 1 h at 30°C, proteins in the in vitro sumoylation mixtures were analyzed by immunoblotting with antibody to HDAg. As shown in Fig. 2, in addition to the input S-HDAg, a protein band consistent with the expected size of sumoylated S-HDAg was detected in the reaction mix containing ATP, indicating that S-HDAg was sumoylated by SUMO1 (lane 2). The absence of SUMO1-S-HDAg conjugation in the reaction mix omitting ATP (lane 1) demonstrated that the conjugation is ATP dependent (required for E1 activation) and is thus likely derived from the SUMO cascade. This result revealed that S-HDAg is a SUMO1 target protein in vitro. A parallel reaction showed that GST-RanGAP1 (GST-RG1), the first protein shown to be sumoylated (35), but not the control GST protein, was properly sumoylated, indicating the specificity of this reaction (Fig. 2, lanes 3 to 6).
FIG. 2.
Sumoylation of S-HDAg in vitro. The sumoylation system was reconstituted in vitro with recombinant SUMO E1 (Aos1/Uba2), SUMO E2 (Ubc9), SUMO1, and S-HDAg or the control substrate proteins. GST protein and GST-tagged RanGAP1 (GST-RG1) served as negative and positive control substrate proteins, respectively, for this assay (RG1 was the first protein shown to be posttranslationally modified with SUMO [35]). The reactions were carried out in sumoylation buffer with (+) or without (−) ATP. The S-HDAg substrate and the SUMO1-S-HDAg generated in the reaction were analyzed by immunoblotting (IB) with an antibody (α) to HDAg (lanes 1 and 2). The control substrates and their SUMO1-conjugated forms generated in the reaction were analyzed by immunoblotting with an antibody (α) to GST (lanes 3 to 6). The locations of molecular mass markers are indicated on the left, in kDa, and the positions of substrate proteins and their SUMO1-conjugated forms are marked on the right.
S-HDAg is sumoylated in vivo.
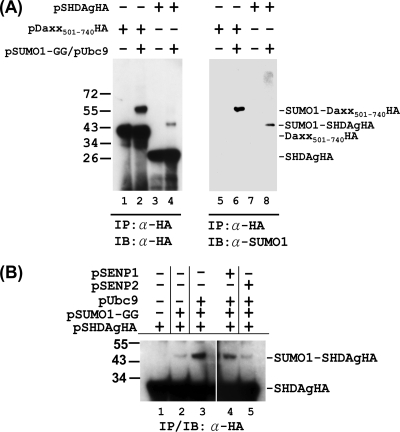
To determine whether S-HDAg is SUMO1 modified in mammalian cells, an in vivo sumoylation assay was performed. We transiently transfected Huh-7 cells with an epitope (HA)-tagged S-HDAg expression plasmid, with or without SUMO1 and Ubc9 expression plasmids. After lysis of cells with RIPA-NEM buffer, proteins were subjected to immunoprecipitation and immunoblotting with antibody to HA tag. When S-HDAgHA was coexpressed with SUMO1 and Ubc9, we observed an S-HDAgHA species of higher molecular weight, in proportion to the size of additional SUMO1 modification (Fig. 3A, lane 4). Sumoylation was further confirmed when immunoprecipitated S-HDAgHA reacted with antibody to SUMO1 (Fig. 3A, lane 8). A control reaction showed that Daxx protein (Fig. 3A, lanes 2 and 6) was properly sumoylated under the same conditions.
FIG. 3.
Sumoylation of S-HDAg in vivo. (A) Detection of SUMO1-conjugated S-HDAg in transfected cells. In Huh-7 cells, the expression plasmid for HA-tagged S-HDAg or the Daxx(501-740) truncation mutant was transfected with or without the expression plasmids for SUMO1-GG (the mature form of SUMO1) and Ubc9. The whole-cell extracts were then prepared and subjected to immunoprecipitation (IP) with an antibody to HA tag followed by immunoblotting (IB) with the same antibody or with an antibody to SUMO1, as indicated. Daxx is a well-documented SUMO target protein (30). Transfection of the expression plasmid for Daxx501-740HA served as a positive control for this assay. (B) Overexpression of SENP2 reduces sumoylation of S-HDAg. The expression plasmid for HA-tagged S-HDAg was transfected into Huh-7 cells with or without the expression plasmids for SUMO1-GG, Ubc9, SENP1, and SENP2, as indicated. The whole-cell extracts were then prepared and subjected to IP/IB with an antibody to the HA tag.
SUMO conjugation is a highly dynamic process that can be reversed rapidly by the action of sentin-specific proteases (SENPs) in mammals. Of the six human proteases (SENP1-3 and SENP5-7), SENP1 and SENP2 have been described to be responsible for the desumoylation of SUMO1-conjugated proteins (18, 43). To examine whether SENP1 and SENP2 are involved in S-HDAg desumoylation, an in vivo sumoylation assay was performed. As shown in Fig. 3B, when SENP2 was coexpressed with SUMO1, Ubc9, and S-HDAgHA, the band intensity of SUMO1-S-HDAgHA was drastically reduced, to the level of SUMO1 and S-HDAgHA coexpression only (compare lane 5 to lanes 2 and 3). As a comparison, coexpression of SENP1 did not show such a reduction (lane 4). This result revealed the reversible nature of the sumoylation of S-HDAg and suggested that SENP2 is the desumoylase involved in the SUMO1 modification of S-HDAg. Collectively, these data further confirmed that S-HDAg is a SUMO1 target protein in mammalian cells.
Detecting sumoylated S-HDAg in HDV RNA-replicating cells.
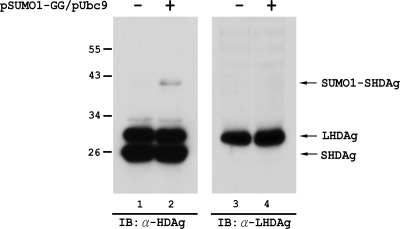
As mentioned above, sumoylation is a highly dynamic process. Most SUMO target proteins undergo rapid cycles of sumoylation and desumoylation; only a few are stably conjugated with SUMO. Thus, most targets appear to be modified to only a small percentage at steady state (13, 38). Therefore, in order to detect the in vivo sumoylated form of a target protein, it is always necessary to overexpress SUMO isoforms and/or Ubc9. Thus, to examine whether HDAg is sumoylated in HDV RNA-replicating cells, SUMO1 and Ubc9 were overexpressed in Huh-7 cells in which HDV RNA replication was established. As shown in Fig. 4, two HDAg species, representing S- and L-HDAg, and an HDAg species of higher molecular weight, corresponding to the size of SUMO1 conjugation of S-HDAg, were detected with a mouse monoclonal antibody against HDAg (lane 2). When the same sample was subjected to immunoblotting with a rabbit antibody (LP3; a kind gift from Stan Lemon) specific for L-HDAg, only L-HDAg of the original size, with no proteins of higher molecular weight corresponding to the size of SUMO1-conjugated L-HDAg, was detected (lane 4). These results suggested that S-HDAg, but not L-HDAg, is a SUMO1 target in the HDV life cycle.
FIG. 4.
S-HDAg produced from HDV RNA-replicating cells is a SUMO1 target protein. In Huh-7 cells, HDV genomic RNA was transfected together with the S-HDAg-encoding mRNA to establish HDV RNA replication. Four days after RNA transfection, the cells were then transfected again, with or without the SUMO1-GG and Ubc9 expression plasmids. Two days later, the whole-cell extracts were prepared and then subjected to immunoblotting with a mouse monoclonal antibody against HDAg (lanes 1 and 2) or with a rabbit polyclonal antibody specific for L-HDAg (LP3) (49) (lanes 3 and 4).
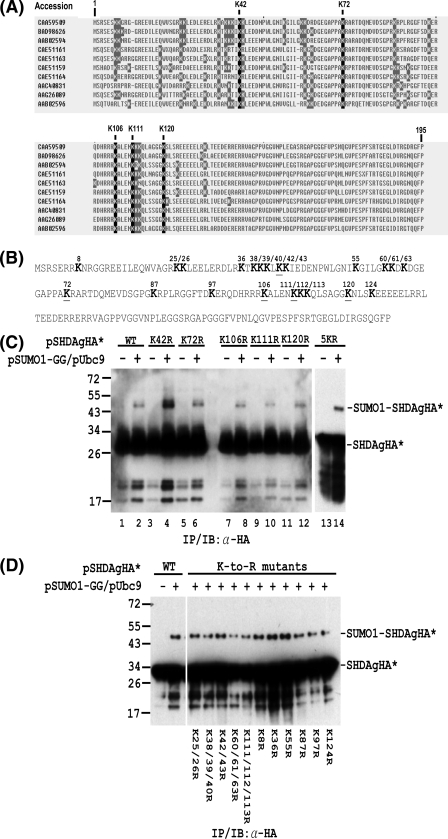
Multiple lysine residues are SUMO1 acceptors within S-HDAg.
Having determined that S-HDAg is sumoylated, we next sought to identify the S-HDAg residues that are modified by SUMO1. Many SUMO acceptor lysines follow the consensus motif ψKXE (in which ψ is a hydrophobic residue and X is any amino acid) (13). However, an increasing number of proteins have turned out to be sumoylated on nonconsensus sites (20, 30, 45, 53). Inspection of the amino acid sequence reveals that none of the lysine residues within S-HDAg match the SUMO consensus. To identify the SUMO acceptor lysine(s) of S-HDAg, we therefore mutated all of the lysine residues that are conserved among different HDV isolates (Fig. 5A) to arginines by site-directed mutagenesis. The S-HDAgHA expression plasmids, which contain individual lysine-to-arginine mutations at the conserved positions 42, 72, 106, 111, and 120 (Fig. 5B), were subsequently used for in vivo sumoylation assay. As shown in Fig. 5C, all five mutants could be sumoylated in the presence of SUMO1 and Ubc9 coexpression (lanes 4, 6, 8, 10, and 12). To examine whether these five conserved lysines are exclusive for S-HDAg sumoylation, an S-HDAgHA expression plasmid containing all five lysine-to-arginine mutations (5KR mutant) was also constructed and used for in vivo sumoylation assay. The 5KR mutant protein synthesized in transfected Huh-7 cells was also sumoylated (Fig. 5C, lane 14), indicating that the SUMO acceptor lysines within S-HDAg are not limited to the five conserved lysines.
FIG. 5.
Mapping of lysine residues in S-HDAg that are conjugated by SUMO1. (A) Alignment of S-HDAg amino acid sequences from different HDV isolates. The nonconserved and conserved lysine (K) residues are shaded in gray and black, respectively. (B) Amino acid sequence of the Italian HDV isolate used in the in vivo sumoylation assay. The lysine (K) residues are shown in bold. The underlined K residues denote the lysine residues which are conserved among different HDV isolates. (C) The SUMO acceptor lysines within S-HDAg are not limited to the five conserved lysines. In Huh-7 cells, expression plasmids encoding wild-type S-HDAgHA or its mutants containing K-to-R substitutions at the conserved lysine(s) were transfected with or without the SUMO1-GG and Ubc9 expression plasmids. The whole-cell extracts were then prepared and subjected to IP/IB with an antibody to the HA tag. (D) Multiple lysine residues are SUMO1 acceptors within S-HDAg. Expression plasmids encoding wild-type S-HDAgHA or its mutants containing K-to-R substitution(s) were used for in vivo sumoylation assay as in panel C. The positions of the S-HDAgHA species (denoted as SHDAgHA*) and the SUMO1-conjugated form (denoted as SUMO1-SHDAgHA*) are marked on the right. Expression plasmids encoding wild-type S-HDAgHA or its mutants are indicated at the top or bottom.
S-HDAg is a lysine-rich protein, especially in its N-terminal two-thirds. For the particular HDV isolate used in the in vivo sumoylation assay (Fig. 5B), about 17% of the amino acids are lysines in this region. Some of these lysines form clusters, as revealed by amino acid sequence alignment between different HDV isolates (Fig. 5A). To avoid missing any potential SUMO acceptor lysine(s), we scanned all the other lysines which are not conserved among different HDV isolates either by mutating individual lysines or by mutating combinations of two or three lysines that are neighbored. The in vivo sumoylation data showed that all of these mutants can be sumoylated (Fig. 5D). Taken together, these results suggest that multiple lysine residues are SUMO1 acceptors within S-HDAg.
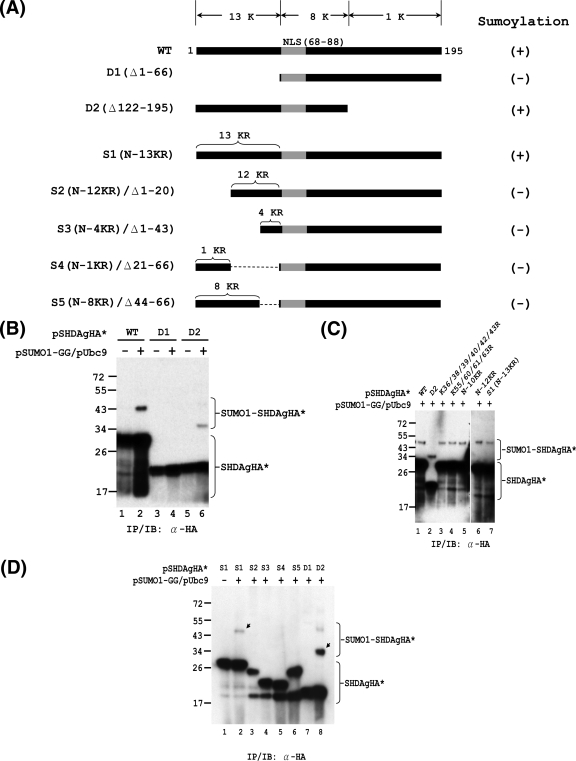
The N-terminal 66 aa of S-HDAg are required for SUMO modification.
The above site-directed mutagenesis data showed that multiple lysine residues are SUMO1 acceptors within S-HDAg. To further examine whether there is a particular region of S-HDAg in which the SUMO acceptor lysines may be located, an N-terminally truncated mutant (D1; deletion of aa 1 to 66 of S-HDAg) and a C-terminally truncated mutant (D2; deletion of aa 122 to 195 of S-HDAg) were generated to test their ability to be sumoylated (Fig. 6A). Given that nuclear localization signals (NLS) are important for the sumoylation of certain proteins (53), the NLS of S-HDAg was preserved in both D1 and D2. The in vivo sumoylation assay showed that D2 was sumoylated (Fig. 6B, lane 6), whereas the N-terminally truncated D1 mutant lost its ability to be sumoylated (Fig. 6B, lane 4). This result suggested two possibilities: either the SUMO acceptor lysines of S-HDAg reside exclusively in the N-terminal 66-aa region or the N-terminal 66-aa region of S-HDAg contains a functional domain that is necessary for SUMO modification of S-HDAg. The N-terminal 66-aa region of S-HDAg contains 13 lysines (Fig. 6A). To test whether these 13 lysines are exclusively the SUMO acceptors of S-HDAg, we sought to generate an S-HDAg mutant in which all 13 N-terminal lysines were replaced by arginines. The intermediate constructs (K36/38/39/40/42R, K55/60/61/63R, N-10KR, and N-12KR) and the final construct, which contains the 13 N-terminal lysine-to-arginine (K-to-R) substitutions (S1 [N-13KR]), were used for in vivo sumoylation assays. As shown in Fig. 6C, all of these N-terminal K-to-R mutants, including S1 (N-13KR), were sumoylated, indicating that the SUMO acceptor lysines of S-HDAg are not limited to the N-terminal 13 lysines.
FIG. 6.
Mapping of functional domain required for SUMO modification of S-HDAg. In vivo sumoylation assays were performed with S-HDAg and its mutants. (A) Schematic representation of S-HDAg and its deletion and K-to-R substitution derivatives. The total numbers of lysine (K) residues within the various regions of S-HDAg are indicated at the top. The nuclear localization signal (NLS; amino acids 66 to 88) is shown in gray. The numbers of K-to-R substitutions within the N-terminal region of S1 and its derivatives are indicated on top of these constructs. The ability of the S-HDAg derivatives to undergo sumoylation is summarized on the right. (B) The N-terminally truncated form of S-HDAg loses its ability to be sumoylated. Expression plasmids encoding the wild-type S-HDAgHA (WT) or its deletion mutants, D1 and D2, were used for in vivo sumoylation and subsequent IP/IB assay with anti-HA antibody. (C) The N-terminal 13 lysines are not the exclusive SUMO acceptor lysines of S-HDAg. Expression plasmids encoding wild-type S-HDAgHA and its derivatives (as indicated) were used for in vivo sumoylation and subsequent IP/IB assay with anti-HA antibody. (D) The N-terminal 66 aa of S-HDAg are required for its SUMO modification. The indicated expression plasmids were used for in vivo sumoylation assay as mentioned above. The arrows indicate SUMO1-modified S-HDAgHA* species.
The above data suggest the possibility that the N-terminal 66-aa region of S-HDAg contains a functional domain that is necessary for SUMO conjugation of S-HDAg. To define which subregion within the N-terminal 66-aa region of S-HDAg is required for its SUMO1 conjugation, we next generated S-HDAg truncation mutants derived from S1. These mutants contain different deletions in subregions within the N-terminal 66-aa region of S1 (S2 to S5) (Fig. 6A). After subjecting these deletion mutants to in vivo sumoylation assay, we observed that all of these deletion mutants lost the ability to be sumoylated (Fig. 6D). This result suggested that the integrity of the N-terminal 66-aa region of S-HDAg is necessary for SUMO conjugation of S-HDAg. In sum, these results showed that multiple lysines are SUMO acceptors throughout the whole sequence of S-HDAg and that the N-terminal 66 aa of S-HDAg are required for its SUMO modification.
SUMO1 conjugation of S-HDAg enhances HDV G-RNA and mRNA synthesis but not AG-RNA synthesis.
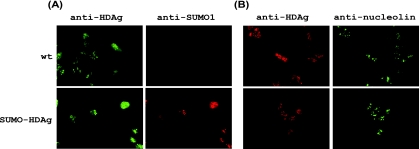
To address the functional consequences of S-HDAg sumoylation, we exploited a fusion strategy in which SUMO1 is fused to S-HDAg to mimic sumoylated S-HDAg. Given that all of the lysine residues are located in the N-terminal two-thirds of S-HDAg, the SUMO1-encoding sequence was fused to the 5′ end of the S-HDAg coding sequence; this mRNA (SUMO-HDAg) encodes a SUMO1-S-HDAg fusion protein. Immunofluorescence staining showed that the subcellular localizations of wild-type S-HDAg and the SUMO1-S-HDAg fusion protein were very similar. Similar to wild-type S-HDAg, the SUMO1-S-HDAg fusion protein was observed in both nucleoli and nucleoplasm, suggesting that SUMO1 modification does not alter the subcellular distribution of S-HDAg (Fig. 7).
FIG. 7.
The SUMO1-S-HDAg fusion protein displays a subcellular localization pattern similar to that of wild-type S-HDAg. The mRNA encoding wild-type S-HDAg (wt) or mutant SUMO1-S-HDAg (SUMO-HDAg) was transfected into Huh-7 cells. At day 1 posttransfection, cells were fixed and prepared for immunofluorescence microscopic examination (as described in Materials and Methods). (A) A mouse anti-HDAg monoclonal antibody or a rabbit anti-SUMO1 polyclonal antibody was used as the first antibody. (B) A rabbit anti-HDAg polyclonal antibody or a mouse anti-nucleolin monoclonal antibody was used as the first antibody.
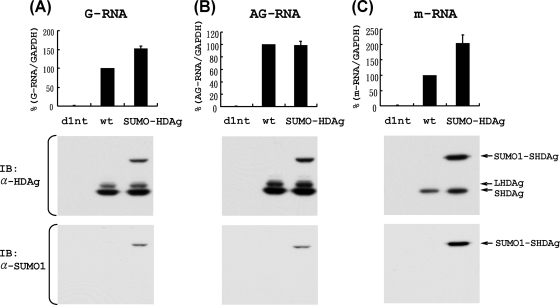
We next examined the ability of the SUMO1-S-HDAg fusion protein to promote the synthesis of HDV G-RNA, AG-RNA, and mRNA by using the established HDV cDNA-free RNA transfection method and qRT-PCR procedure (39, 48). For this purpose, HDV G-RNA (for AG-RNA and mRNA detection) or AG-RNA (for G-RNA detection) was cotransfected with the mRNA encoding either wild-type S-HDAg or SUMO1-S-HDAg fusion protein. This approach allows the study of various HDV RNA species separately (39, 48). As a negative control, HDV G-RNA or AG-RNA was also transfected together with an mRNA encoding a defective S-HDAg mutant (d1nt), in which the adenosine of the ATG start codon was deleted to abolish S-HDAg translation. As shown in Fig. 8, no nascent HDV G-RNA (A), AG-RNA (B), or mRNA (C) was detected when d1nt was cotransfected, demonstrating the specificity of this assay. We found that cotransfection of the mRNA encoding the SUMO1-S-HDAg fusion protein led to significantly larger amounts of HDV mRNA (C) and G-RNA (A) than those produced by cotransfection with the mRNA encoding wild-type S-HDAg. In contrast, HDV AG-RNA synthesis was not affected by the sumoylation state of S-HDAg (B). Besides the transfected SUMO1-S-HDAg fusion protein, the normally sized S-HDAg and some L-HDAg were also detected, indicating that HDV RNA indeed replicated in the cells (Fig. 8). This result suggested that sumoylation of S-HDAg selectively enhances HDV G-RNA and mRNA synthesis but not AG-RNA synthesis.
FIG. 8.
Effects of SUMO1 conjugation of S-HDAg on HDV G-RNA, AG-RNA, and mRNA synthesis. HDV AG-RNA (A) or HDV G-RNA (B and C) was cotransfected with an mRNA encoding wild-type S-HDAg (wt), SUMO1-S-HDAg (SUMO-HDAg), or defective S-HDAg (d1nt) into cells to establish HDV RNA replication. At day 4 (A and B) or day 1 (C) posttransfection, total RNAs were extracted, and HDV G-RNA (A), AG-RNA (B), and mRNA (C) were detected by qRT-PCR, using the G-S, AG-S, and mI-S protocols, respectively (48) (upper panels). The data were normalized relative to the values obtained for the wild-type S-HDAg-encoding mRNA cotransfection. Error bars represent standard deviations from the means for three independent experiments. To demonstrate the expression of the expected forms of HDAg (as indicated by arrows), the whole-cell extracts from one of the qRT-PCR assays were prepared and subjected to immunoblotting with a mouse monoclonal antibody to HDAg or SUMO1, as indicated (lower panels).
DISCUSSION
In recent years, plenty of data have accumulated concerning various viruses and their interaction with the SUMO machinery (2, 50, 52). Consistent with the nuclear predominance of SUMO modification in cells (13, 56), most of the viral SUMO substrates are nuclear proteins of DNA viruses or retroviruses (2, 56). Although HDV belongs to the RNA viruses, HDV is a rare case in which the viral RNA genome is replicated in the nuclei of host cells. This observation prompted us to investigate whether HDV S-HDAg, which is a nuclear protein intimately involved in HDV RNA replication, is a SUMO target protein. In the present study, by using a yeast two-hybrid assay (Fig. 1) and in vitro and in vivo sumoylation assays (Fig. 2 and 3), we demonstrated that S-HDAg is a SUMO1 target protein. Consistent with the reversible nature of the sumoylation process, we also found that S-HDAg sumoylation can be drastically reversed by the action of a specific isopeptidase, SENP2 (Fig. 3B, lane 5). This result suggested that SENP2 is responsible for the desumoylation of S-HDAg. Nevertheless, since a weak reduction of S-HDAg sumoylation was also observed when overexpression of SENP1 was conducted (Fig. 3B, lane 4), it cannot be ruled out that SENP1 is also involved in the desumoylation of S-HDAg. Finally, we showed that S-HDAg, but not L-HDAg, produced in HDV RNA-replicating cells is sumoylated (Fig. 4). This observation suggested that S-HDAg is a SUMO1 target in the HDV life cycle. During HDV replication, L-HDAg is produced in the late stage of the viral replication cycle as a result of a specific RNA-editing event (51). L-HDAg contains all of the functional domains of S-HDAg, with an additional 19 amino acids at the C terminus. By using the in vivo sumoylation assay, we found that, similar to overexpressed S-HDAg, overexpressed L-HDAg can also be sumoylated (data not shown). The underlying mechanism for the selective sumoylation of S-HDAg but not L-HDAg is not clear.
To address the functional consequence of S-HDAg sumoylation in HDV replication, we searched for its SUMO acceptor lysine by mutagenesis and subsequent in vivo sumoylation. However, no individual lysine residue of S-HDAg could be identified as the major SUMO acceptor site, suggesting that multiple lysine residues are SUMO acceptors within S-HDAg (Fig. 5). This is also the case for sumoylation of the cellular transcriptional repressor Daxx (30). An explanation for this kind of sumoylation is that the primary SUMO acceptor site is changed when the preferred lysine is mutated (30, 37).
S-HDAg is a highly lysine-rich protein, and some of these lysines form clusters, as examined by amino acid sequence alignment between different HDV isolates (Fig. 5A). In search of the possible lysine cluster which is responsible for the sumoylation of S-HDAg, we generated an N-terminally truncated mutant (D1), a C-terminally truncated mutant (D2), and a series of N-terminal K-to-R mutants (Fig. 6A). Subsequent in vivo sumoylation assays using these mutants further confirmed that multiple lysine residues throughout the whole sequence of S-HDAg are SUMO acceptors and suggested that the N-terminal 66-aa region is necessary for S-HDAg sumoylation (Fig. 6). Furthermore, studies using N-terminal subregion truncation mutants derived from S1, which contains all 13 N-terminal K-to-R mutations, revealed that the integrity of the N terminus of S-HDAg is necessary for its sumoylation (Fig. 6D).
Since multiple lysine residues are SUMO acceptors within S-HDAg, it is not possible to investigate the functional consequences of S-HDAg sumoylation by using an S-HDAg mutant in which the specific SUMO acceptor lysine is replaced. We therefore utilized a genetic fusion strategy which has been used to study the functional consequences of SUMO modification (3, 4, 27, 54, 55) to investigate the effect of sumoylation of S-HDAg. We found that a SUMO1-S-HDAg fusion protein selectively enhances HDV G-RNA and mRNA synthesis but not AG-RNA synthesis (Fig. 8), suggesting that the sumoylation of S-HDAg is involved in modulation of HDV RNA synthesis. The reported functional outcomes of protein sumoylation are extremely diverse, ranging from changes in intracellular localization to altered activity of the modified protein (2, 11, 52, 56). Immunofluorescence examination showed that SUMO1 conjugation did not alter the subcellular localization of S-HDAg (Fig. 7), suggesting that the effect of sumoylation of S-HDAg does not occur through changes in subcellular targeting. The exact mechanism by which sumoylation of S-HDAg regulates HDV RNA synthesis will require further investigation.
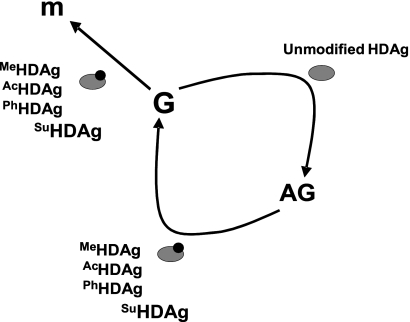
Without encoding its own polymerase, it is intuitive that HDV has to replicate its RNA genome by using cellular enzymes. To date, the detailed mechanism by which the cellular RNA polymerases are redirected for HDV genome replication and mRNA transcription is still unclear. Since it is an essential component of the HDV RNP complex, S-HDAg has many features reminiscent of a transcription factor and is indispensable for HDV replication (21, 26, 31). Besides, S-HDAg binds to HDV RNA and serves as an RNA chaperon to alter RNA conformation (22). Accordingly, S-HDAg must play a very important role in the redirection of the cellular RNA polymerases for HDV replication. Multisite protein modifications have been suggested to play regulatory functions in cells. This is also the case for S-HDAg in regulating HDV RNA replication. Previously, S-HDAg has been shown to be subjected to R-13 methylation, K-72 acetylation, and S-177 phosphorylation, and these posttranslational modifications are important for HDV G-RNA synthesis and mRNA transcription but are dispensable for AG-RNA synthesis (29, 40, 41, 48). In the present study, we show that S-HDAg is also subjected to posttranslational modification by the SUMO pathway. The consequence of SUMO conjugation of S-HDAg is very similar to those of the above-mentioned posttranslational modifications (Fig. 9), providing further support for our previous proposition that the cellular machinery involved in the synthesis of HDV AG-RNA is different from that for G-RNA synthesis and mRNA transcription (26). Whether S-HDAg can be changed posttranslationally by these modifications sequentially and/or synergistically for its optimal functions and precisely how these modifications affect the synthesis of the various HDV RNA species will require further studies.
FIG. 9.
Model of HDAg requirement for synthesis of the three HDV RNA species. MeHDAg, AcHDAg, PhHDAg, and SuHDAg represent methylated, acetylated, phosphorylated, and sumoylated HDAg, respectively. m, HDAg-encoding mRNA; G, HDV genomic RNA; AG, HDV antigenomic RNA.
Acknowledgments
We thank Hsiu-Ming Shih for generous gifts of plasmids, Wei-Che Hsu for assistance with sequence alignment, Pei-Jer Chen for the generous gift of the mouse monoclonal antibody to HDAg, and Shin-Ru Shih and Yi-Ling Lin for helpful discussions and advice.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 2.Boggio, R., and S. Chiocca. 2006. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 9:430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossis, G., C. E. Malnou, R. Farras, E. Andermarcher, R. Hipskind, M. Rodriguez, D. Schmidt, S. Muller, I. Jariel-Encontre, and M. Piechaczyk. 2005. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell. Biol. 25:6964-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, S., and K. H. Vousden. 2008. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle 7:2519-2528. [DOI] [PubMed] [Google Scholar]
- 5.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, M. F., S. C. Baker, L. H. Soe, T. Kamahora, J. G. Keck, S. Makino, S. Govindarajan, and M. M. Lai. 1988. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 62:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. H., K. J. Park, and S. B. Hwang. 2002. Large hepatitis delta antigen is phosphorylated at multiple sites and phosphorylation is associated with protein conformational change. Intervirology 45:142-149. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 11.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 12.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 13.Geiss-Friedlander, R., and F. Melchior. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 8:947-956. [DOI] [PubMed] [Google Scholar]
- 14.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 256:1331-1333. [DOI] [PubMed] [Google Scholar]
- 15.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govindarajan, S., K. P. Chin, A. G. Redeker, and R. L. Peters. 1984. Fulminant B viral hepatitis: role of delta agent. Gastroenterology 86:1417-1420. [PubMed] [Google Scholar]
- 17.Guo, D., M. Li, Y. Zhang, P. Yang, S. Eckenrode, D. Hopkins, W. Zheng, S. Purohit, R. H. Podolsky, A. Muir, J. Wang, Z. Dong, T. Brusko, M. Atkinson, P. Pozzilli, A. Zeidler, L. J. Raffel, C. O. Jacob, Y. Park, M. Serrano-Rios, M. T. Larrad, Z. Zhang, H. J. Garchon, J. F. Bach, J. I. Rotter, J. X. She, and C. Y. Wang. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36:837-841. [DOI] [PubMed] [Google Scholar]
- 18.Hay, R. T. 2007. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17:370-376. [DOI] [PubMed] [Google Scholar]
- 19.Heun, P. 2007. SUMOrganization of the nucleus. Curr. Opin. Cell Biol. 19:350-355. [DOI] [PubMed] [Google Scholar]
- 20.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 21.Huang, W. H., C. W. Chen, H. L. Wu, and P. J. Chen. 2006. Post-translational modification of delta antigen of hepatitis D virus. Curr. Top. Microbiol. Immunol. 307:91-112. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Z. S., and H. N. Wu. 1998. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 273:26455-26461. [DOI] [PubMed] [Google Scholar]
- 23.Jeng, K. S., A. Daniel, and M. M. Lai. 1996. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J. Virol. 70:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 25.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 79:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamsoul, I., J. Lodewick, S. Lebrun, R. Brasseur, A. Burny, R. B. Gaynor, and F. Bex. 2005. Exclusive ubiquitination and sumoylation on overlapping lysine residues mediate NF-kappaB activation by the human T-cell leukemia virus Tax oncoprotein. Mol. Cell. Biol. 25:10391-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, C. Z., P. J. Chen, and D. S. Chen. 1995. Large hepatitis delta antigen in packaging and replication inhibition: role of the carboxyl-terminal 19 amino acids and amino-terminal sequences. J. Virol. 69:5332-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, D. Y., Y. S. Huang, J. C. Jeng, H. Y. Kuo, C. C. Chang, T. T. Chao, C. C. Ho, Y. C. Chen, T. P. Lin, H. I. Fang, C. C. Hung, C. S. Suen, M. J. Hwang, K. S. Chang, G. G. Maul, and H. M. Shih. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24:341-354. [DOI] [PubMed] [Google Scholar]
- 31.Macnaughton, T. B., and M. M. Lai. 2006. HDV RNA replication: ancient relic or primer? Curr. Top. Microbiol. Immunol. 307:25-45. [DOI] [PubMed] [Google Scholar]
- 32.Macnaughton, T. B., and M. M. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 35.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 37.Meulmeester, E., M. Kunze, H. H. Hsiao, H. Urlaub, and F. Melchior. 2008. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30:610-619. [DOI] [PubMed] [Google Scholar]
- 38.Meulmeester, E., and F. Melchior. 2008. Cell biology: SUMO. Nature 452:709-711. [DOI] [PubMed] [Google Scholar]
- 39.Modahl, L. E., and M. M. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu, J. J., D. S. Chen, and P. J. Chen. 2001. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 75:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu, J. J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 319:60-70. [DOI] [PubMed] [Google Scholar]
- 42.Mu, J. J., H. L. Wu, B. L. Chiang, R. P. Chang, D. S. Chen, and P. J. Chen. 1999. Characterization of the phosphorylated forms and the phosphorylated residues of hepatitis delta virus delta antigens. J. Virol. 73:10540-10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhopadhyay, D., and M. Dasso. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286-295. [DOI] [PubMed] [Google Scholar]
- 44.Muller, S., A. Ledl, and D. Schmidt. 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23:1998-2008. [DOI] [PubMed] [Google Scholar]
- 45.Pichler, A., P. Knipscheer, E. Oberhofer, W. J. van Dijk, R. Korner, J. V. Olsen, S. Jentsch, F. Melchior, and T. K. Sixma. 2005. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12:264-269. [DOI] [PubMed] [Google Scholar]
- 46.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheu, G. T., and M. M. Lai. 2000. Recombinant hepatitis delta antigen from E. coli promotes hepatitis delta virus RNA replication only from the genomic strand but not the antigenomic strand. Virology 278:578-586. [DOI] [PubMed] [Google Scholar]
- 48.Tseng, C. H., K. S. Jeng, and M. M. Lai. 2008. Transcription of subgenomic mRNA of hepatitis delta virus requires a modified hepatitis delta antigen that is distinct from antigenomic RNA synthesis. J. Virol. 82:9409-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J. G., J. Cullen, and S. M. Lemon. 1992. Immunoblot analysis demonstrates that the large and small forms of hepatitis delta virus antigen have different C-terminal amino acid sequences. J. Gen. Virol. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, V. G., and D. Rangasamy. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17-27. [DOI] [PubMed] [Google Scholar]
- 51.Wong, S. K., and D. W. Lazinski. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 99:15118-15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, Y. C., A. F. Deyrieux, and V. G. Wilson. 2007. Papillomaviruses and the host SUMOylation system. Biochem. Soc. Trans. 35:1433-1435. [DOI] [PubMed] [Google Scholar]
- 53.Xue, Y., F. Zhou, C. Fu, Y. Xu, and X. Yao. 2006. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 34:254-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, C. K., J. H. Kim, D. K. Ann, and M. R. Stallcup. 2008. Differential regulation of the two transcriptional activation domains of the coiled-coil coactivator CoCoA by sumoylation. BMC Mol. Biol. 9:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yurchenko, V., Z. Xue, and M. J. Sadofsky. 2006. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell. Biol. 26:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, J. 2007. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 64:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]