Abstract
The immune correlates of human/simian immunodeficiency virus control remain elusive. While CD8+ T lymphocytes likely play a major role in reducing peak viremia and maintaining viral control in the chronic phase, the relative antiviral efficacy of individual virus-specific effector populations is unknown. Conventional assays measure cytokine secretion of virus-specific CD8+ T cells after cognate peptide recognition. Cytokine secretion, however, does not always directly translate into antiviral efficacy. Recently developed suppression assays assess the efficiency of virus-specific CD8+ T cells to control viral replication, but these assays often use cell lines or clones. We therefore designed a novel virus production assay to test the ability of freshly ex vivo-sorted simian immunodeficiency virus (SIV)-specific CD8+ T cells to suppress viral replication from SIVmac239-infected CD4+ T cells. Using this assay, we established an antiviral hierarchy when we compared CD8+ T cells specific for 12 different epitopes. Antiviral efficacy was unrelated to the disease status of each animal, the protein from which the tested epitopes were derived, or the major histocompatibility complex (MHC) class I restriction of the tested epitopes. Additionally, there was no correlation with the ability to suppress viral replication and epitope avidity, epitope affinity, CD8+ T-cell cytokine multifunctionality, the percentage of central and effector memory cell populations, or the expression of PD-1. The ability of virus-specific CD8+ T cells to suppress viral replication therefore cannot be determined using conventional assays. Our results suggest that a single definitive correlate of immune control may not exist; rather, a successful CD8+ T-cell response may be comprised of several factors.
CD8+ T cells may play a critical role in blunting peak viremia and controlling human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) replication. The transient depletion of CD8+ cells in SIV-infected macaques results in increased viral replication (26, 31, 51, 70). The emergence of virus-specific CD8+ T cells coincides with the reduction of peak viremia (12, 39, 42, 63), and CD8+ T-cell pressure selects for escape mutants (6, 9, 13, 28, 29, 38, 60, 61, 85). Furthermore, particular major histocompatibility complex (MHC) class I alleles are overrepresented in SIV- and HIV-infected elite controllers (15, 29, 33, 34, 46, 56, 88).
Because it has been difficult to induce broadly neutralizing antibodies (Abs), the AIDS vaccine field is currently focused on developing a vaccine designed to elicit HIV-specific CD8+ T cells (8, 52, 53, 82). Investigators have tried to define the immune correlates of HIV control. Neither the magnitude nor the breadth of epitopes recognized by virus-specific CD8+ T-cell responses correlates with the control of viral replication (1). The quality of the immune response may, however, contribute to the antiviral efficacy of the effector cells. It has been suggested that the number of cytokines that virus-specific CD8+ T cells secrete may correlate with viral control, since HIV-infected nonprogressors appear to maintain CD8+ T cells that secrete several cytokines, compared to HIV-infected progressors (11, 27). An increased amount of perforin secretion may also be related to the proliferation of HIV-specific CD8+ T cells in HIV-infected nonprogressors (55). While those studies offer insight into the different immune systems of progressors and nonprogressors, they did not address the mechanism of viral control. Previously, we found no association between the ability of SIV-specific CD8+ T-cell clones to suppress viral replication in vitro and their ability to secrete gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), or interleukin-2 (IL-2) (18).
Evidence suggests that some HIV/SIV proteins may be better vaccine targets than others. CD8+ T cells recognize epitopes derived from Gag as early as 2 h postinfection, whereas CD8+ T cells specific for epitopes in Env recognize infected cells only at 18 h postinfection (68). Additionally, a previously reported study of HIV-infected individuals showed that an increased breadth of Gag-specific responses was associated with lower viral loads (35, 59, 65, 66). CD8+ T-cell responses specific for Env, Rev, Tat, Vif, Vpr, Vpu, and Nef were associated with higher viral loads, with increased breadth of Env in particular being significantly associated with a higher chronic-phase viral set point.
None of the many sophisticated methods employed for analyzing the characteristics of HIV- or SIV-specific immune responses clearly demarcate the critical qualities of an effective antiviral response. In an attempt to address these questions, we developed a new assay to measure the antiviral efficacy of individual SIV-specific CD8+ T-cell responses sorted directly from fresh peripheral blood mononuclear cells (PBMC). Using MHC class I tetramers specific for the epitope of interest, we sorted freshly isolated virus-specific CD8+ T cells and determined their ability to suppress virus production from SIV-infected CD4+ T cells. We then looked for a common characteristic of efficacious epitope-specific CD8+ T cells using traditional methods.
MATERIALS AND METHODS
SIVmac239 virus stocks.
SIVmac239 (GenBank accession no. M33262) was generated as previously described (23). Briefly, Vero cells were transfected with plasmid DNA encoding the SIV proviral sequences. One day after transfection, CEMx174 cells were added to the Vero cultures. Virus was expanded on CEMx174 cells, and cell-free supernatant was collected 2 days after peak syncytium formation. Harvested virus was analyzed by Gag p27 enzyme-linked immunosorbent assay (ZeptoMetrix Corporation) and quantitative reverse transcription (RT)-PCR prior to use in ex vivo studies.
Animals and viral load analysis.
Indian rhesus macaques (Macaca mulatta) from the Wisconsin National Primate Research Center were cared for according to the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee. Animals were typed for the MHC class I alleles Mamu-A*01, Mamu-A*02, and Mamu-B*08 by sequence-specific PCR (32, 46). Viral RNA (vRNA) was extracted using guanidine hydrochloride as previously described (19, 23, 24) for EDTA-anticoagulated plasma and using an M48 virus minikit (Qiagen) for virus production assay samples. Viral RNA from both plasma and virus production assay supernatant was quantified with forward primer SIV1552 (5′-GTCTGCGTCATCTGGTGCATTC-3′), reverse primer SIV1635 (5′-CACTAGCTGTCTCTGCACTATGTGTTTTG-3′), and probe 5′-6-carboxyfluorescein-CTTCCTCAGTGTGTTTCACTTTCTCTTCTGCG-6-carboxytetramethylrhodamine-3′ using the SuperScript III Platinum one-step quantitative RT-PCR kit (Invitrogen) as previously described (26).
Isolating ex vivo epitope-specific CD8+ T cells.
We obtained large blood draws from infected animals with known CD8+ tetramer-positive (tetramer+) responses. We depleted isolated PBMC of CD14+ cells using nonhuman primate CD14 microbeads and LS columns (Miltenyi Biotec). CD14− PBMC were resuspended at 40 × 106 cells/ml and stained with 25 μg/ml of either phycoerythrin (PE)- or allophycocyanin (APC)-conjugated tetramer for 1 h at 37°C. Cells were washed once and stained with either anti-PE or anti-APC (Miltenyi Biotec) according to the manufacturer's instructions. Cells were washed again and run over Miltenyi Biotec LS magnetic columns. The bound cells were flushed out, resuspended in 1 ml of complete RPMI medium (RPMI 1640 medium supplemented with 15% fetal bovine serum, 2 mM l-glutamine, and 50 μg/ml antimycotic/antibiotic [all purchased from HyClone Laboratories, Inc.]) supplemented with 100 U/ml of IL-2 (NIH AIDS Research and Reagent Program) for counting, and incubated overnight at 37°C in a 48-well plate. We used epitope-specific CD8+ T cells that were sorted to at least 50% specificity as determined by surface staining postsorting. The average percent specificity was 67%, with a range from 50.2% to 92.7%. There was no correlation between the percent specificity of each sort and viral suppression.
CD4+ T-cell targets and infection.
CD4+ T-cell targets were generated, as previously described (68), from SIV-naïve animals expressing Mamu-A*01, Mamu-A*02, Mamu-B*08, or none of these alleles. Briefly, nonhuman primate CD4 microbeads (Miltenyi Biotec) were used to positively select CD4+ T cells. Isolated cells were stimulated overnight with 2.5 μg/ml staphylococcal enterotoxin B (SEB; Sigma), 2.5 μg/ml CD28 (clone L293; BD Biosciences), 2.5 μg/ml CD49d (clone 9F10; BD Biosciences), and 2.5 μg/ml CD3 (clone SP34; BD Biosciences) antibodies for 24 h and cultured in complete RPMI medium supplemented with 100 U/ml of IL-2 until infected. The SIVmac239 clone used throughout this study was competent at downregulating MHC class I as observed by CD4 and Gag p27 staining (Fig. 1e and data not shown). SIVmac239 was purified over a 20% sucrose cushion and magnetized with ViroMag beads (OZ Biosciences) as previously described (67, 68). ViroMag-bound virus was layered over activated CD4+ T cells and placed onto a magnetic plate for 15 min. With this technique, CD4+ T cells were synchronously infected with a multiplicity of infection of between 0.1 and 1. Cells were harvested and washed three times in complete RPMI medium to remove unbound virus and resuspended at 1 × 106 cells/ml in complete RPMI medium supplemented with 100 U/ml of IL-2. Cells treated with tenofovir (NIH AIDS Research and Reference Reagent Program) were pretreated with 400 μM tenofovir for 2 h prior to infection and throughout the experiment.
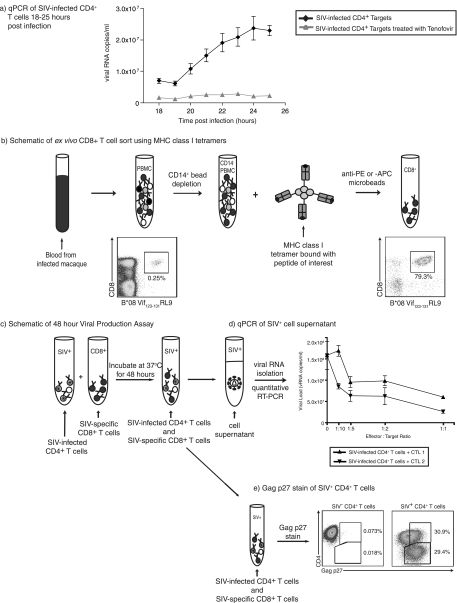
FIG. 1.
Viral life cycle timing and viral production assay schematic. (a) The length of the SIV life cycle was measured by analyzing viral RNA copies per milliliter of cell supernatant each hour between 18 and 25 h postinfection. Each assay for each time point was performed in triplicate. CD4+ T-cell targets were used from at least four animals and repeated in three different experiments. Cells pretreated with tenofovir for 2 h prior to infection and throughout the assay served as controls. (b) Effector cells were generated using MHC class I tetramers specific for the epitope of interest. Cells were incubated with either anti-PE or anti-APC Miltenyi microbeads depending on the tetramer fluorochrome used and passed through a Miltenyi magnetic column. (c) Activated, SIV-infected CD4+ T cells were incubated with ex vivo-sorted, unstimulated, SIV-specific CD8+ T cells for 48 h. (d and e) Supernatant was then removed for analysis of viral RNA content by quantitative RT-PCR (d), and the remaining cells were stained for CD4, CD8, and Gag p27 (e). qPCR, quantitative PCR.
Virus production assay.
We added 2.5 × 104 activated, SIV-infected CD4+ T cells per test. Ex vivo tetramer-sorted CD8+ T cells were added at effector-to-target ratios of 1:1, 1:2, 1:5, and 1:10. Complete RPMI medium with 100 U/ml of IL-2 was added to bring the final volume to 400 μl per test. Assays were set up in duplicate or triplicate when cell numbers allowed. Cells were incubated at 37°C in a 5% CO2 incubator for approximately 48 h. At that time, cells were pelleted, and 300 μl of supernatant was removed and frozen for viral RNA isolation and quantitative RT-PCR analysis as previously described (26). The remaining cells were stained for CD8 peridinin chlorophyll protein (PerCP) (clone SK1; BD Biosciences) and CD4 APC (clone L200; BD Biosciences) surface expression and intracellularly stained with fluorescein isothiocyanate (FITC)-conjugated anti-Gag p27 Ab (clone 55-2F12 from the NIH AIDS Research and Reference Reagent Program) as previously described (68). The percent maximum viral suppression for each epitope-specific CD8+ T-cell population was calculated according to the following equation: (vRNA copies/ml without cytotoxic T lymphocytes [CTL] − vRNA copies/ml with CTL)/vRNA copies/ml without CTL × 100 = percent maximum viral suppression. Assays were performed in duplicate or triplicate with two or more different CD4+ T-cell target populations from MHC class I-matched animals and repeated at least once with CD8+ T cells sorted in a second independent experiment at least 1 month apart. Independent experiments had an average standard deviation of less than 11% maximum suppression. Incubation of epitope-specific CD8+ T cells with MHC class I-mismatched SIV-infected CD4+ T cells consistently showed no suppression of virus production (data not shown), indicating that the suppression of matched cells is MHC class I restricted.
Immunological assays.
Peptide avidity assays were performed using freshly isolated PBMC in an IFN-γ enzyme-linked immunospot (ELISPOT) assay (Mabtech) as previously described (77). Briefly, 1 × 105 PBMC were stimulated with 10-fold-diluted amounts of peptide ranging from 10 pM to 10 μM and incubated at 37°C in a 5% CO2 incubator overnight. Plates were imaged using an ELISPOT reader (Autoimmun-Diagnostika), counted with an ELISPOT Reader, version 4.0 (Autoimmun-Diagnostika), and analyzed as previously described (84). Functional avidity was calculated as the peptide concentration required to elicit 50% of the maximal IFN-γ response. Multifunctional intracellular cytokine staining was performed as previously described (11, 73, 84). Briefly, freshly isolated PBMC were incubated with 5 μM peptide and anti-CD28 (clone L293; BD Biosciences) and anti-CD49d (clone 9g; Pharmingen) antibodies for 1.5 h at 37°C before the addition of 10 μg of brefeldin A per test. Cells were incubated a further 5 h before being stained for the surface expression of CD3-Alexa 700 (clone SP34; BD Biosciences), CD4-PerCP-Cy5.5 (L200; BD Biosciences), and CD8-Pacific blue (clone RPA-T8; BD Biosciences) and fixed overnight with 1% paraformaldehyde. The following day, cells were permeabilized with 0.1% saponin buffer; intracellularly stained for CD69-PE-Texas Red (clone TP1.55.3; Beckman Coulter), IFN-γ-FITC (clone 4S.B3; BD Biosciences), TNF-α-PE (clone MAb11; BD Biosciences), and IL-2-APC (clone MQ1-17H12; BD Biosciences); and fixed with 1% paraformaldehyde. Programmed death 1 (PD-1) (clone EH12.2H7; Biolegend) and central and effector memory stainings were performed with freshly isolated PBMC. Cells were stained for the surface expression of the MHC class I tetramer of interest-PE and CD3-Alexa 700, CD8-Pacific blue, CD28-PE-Cy5 (clone CD28.2; BD Biosciences), and PD-1-APC antibodies.
Statistical analysis.
Statistical correlations were determined by Mann-Whitney tests using GraphPad Prism 4 for Macintosh (San Diego, CA).
RESULTS
Conventional assays fail to evaluate the antiviral efficacy of epitope-specific CD8+ T cells. To directly test and compare the ability of epitope-specific CD8+ T cells to suppress viral replication, we developed a novel ex vivo viral suppression assay using freshly sorted CD8+ T-cell populations. We then sought to find a correlate of the ability of these different cells to suppress viral replication.
SIV-infected CD4+ T cells begin to release virus at 20 h postinfection.
Before analyzing the antiviral efficacy of individual virus-specific CD8+ T-cell populations, we wanted to determine the length of the viral life cycle. To do this, we infected activated CD4+ T cells from four SIV-naïve animals. Tests were set up in triplicate using 1.0 × 105 SIV-infected cells, and these cells were incubated at 37°C for up to 25 h. Supernatant was removed every hour from 18 to 25 h postinfection and analyzed for viral RNA content by quantitative RT-PCR. Viral RNA copies increased between 19 and 20 h postinfection, indicating that virus was being released from the cells at this time (Fig. 1a). Cells treated with tenofovir showed no increase in levels of viral release by quantitative RT-PCR (Fig. 1a) or Gag p27 staining (data not shown).
An antiviral hierarchy exists among 12 SIV epitopes.
To quantify the antiviral efficacy of SIV-specific CD8+ T cells, we isolated epitope-specific cells using MHC class I tetramers (Fig. 1b). We depleted PBMC of CD14+ cells prior to tetramer sorting since monocytes and macrophages were stained by the tetramer and would therefore be sorted with the CD8+ tetramer+ cells. These ex vivo-sorted CD8+ T cells were incubated with activated SIV-infected CD4+ T cells at effector-to-target ratios ranging from 1:1 to 1:10 (Fig. 1c). Due to the modest number of effector cells sorted, we used 2.5 × 104 SIV-infected CD4+ T cells per test throughout our viral production assay. We incubated the effector and target cells for 48 h to allow epitope-specific CD8+ T cells that recognize their cognate epitope late in the viral life cycle an opportunity to recognize virus-infected cells. Also, incubating cells for 48 h rather than 24 h resulted in greater differences between wells that contained epitope-specific CD8+ T cells and the control wells that did not have any CD8+ T cells (Fig. 2a). We therefore removed supernatant at 48 h postinfection and assayed viral RNA concentrations (Fig. 1d). We stained the remaining cells for the presence of intracellular Gag p27 (Fig. 1e). We selected animals with a wide range of viral loads and MHC class I alleles for these studies (Table 1). We sorted CD8+ T cells that were specific for 12 different MHC class I SIV epitopes restricted by Mamu-A*01, -A*02, and -B*08 and tested them for their antiviral efficacies (Table 2).
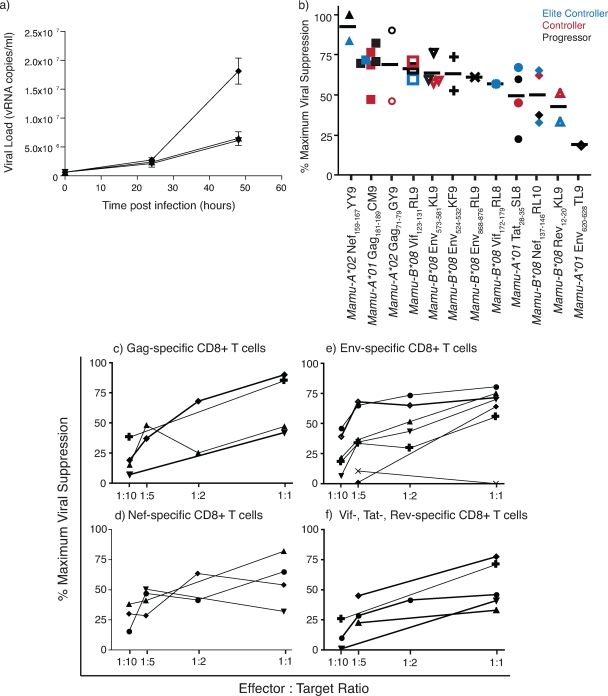
FIG. 2.
Inhibition of viral production by ex vivo-sorted, unstimulated, SIV-specific CD8+ T cells. (a) Viral RNA copies per milliliter of supernatant from cultures containing SIV-infected CD4+ T cells only (⧫), SIV-infected CD4+ T cells and Gag181-189CM9-specific CD8+ T cells (▾), and SIV-infected CD4+ T cells and Env573-581KL9-specific CD8+ T cells (▴). Supernatant was removed at 0, 24, and 48 h postinfection in duplicate or triplicate. (b) Antiviral hierarchy of SIV-specific CD8+ T cells specific for 12 epitopes. All assays were performed at an effector-to-target cell ratio of 1:1. Each data point represents the average percent maximum suppression capacity of one CD8+ T-cell specificity from one animal, with the black bar indicating the average percent maximum suppression of each specificity. Elite controllers (blue) have viral loads of <10e3 viral copies/ml, controllers (red) have viral loads between 10e3 and 2 × 10e4 viral copies/ml, and progressors (black) have viral loads of >2 × 10e4 viral copies/ml at the date of assay. (c to f) Percent maximum suppression capacity of SIV-specific CD8+ T cells at various effector-to-target cells ratios. Each data set is the average maximum suppression capacity of one CD8+ T-cell specificity from one animal. (c) SIV-specific CD8+ T cells directed to Gag. ⧫, Gag71-79GY9; +, Gag181-189CM9; ▴, Gag71-79GY9; ▾, Gag181-189CM9. (d) SIV-specific CD8+ T cells directed to Nef. ▴, Nef159-167YY9; •, Nef137−146RL10; ⧫, Nef137-146Rl10; ▾, Nef137-146RL10. (e) SIV-specific CD8+ T cells directed to Env. •, Env573-581KL9; ▴, Env524-532KF9; ◊, Env573-581KL9; ▾, Env868-876RL9; ⧫, Env573-581KL9; +, Env524-532KF9; ×, Env620-628TL9. (f) SIV-specific CD8+ T cells directed toward Vif, Tat, or Rev. ⧫, Vif123-131RL9; +, Tat28-35SL8; •, Vif172-179RL8; ▾, Tat28-35SL8; ▴, Rev12-20KL9.
TABLE 1.
MHC class I genotypes and SIV infection details for rhesus macaques used in this study
| Animal | MHC class I genotype(s) | Vaccine | Infection strain | Wk p.i. at date of assayf | Avg viral load set point (vRNA copies/ml)g |
|---|---|---|---|---|---|
| rh2029 | A*01, A*08 | None | 239B*08-8xd | <16 | 8.59 × 105 |
| rh2062 | A*02 | None | 239B*08-6xe | 5 | 2.85 × 106 |
| r90149 | A*02, B*01 | Delta nefa | 239 | >81 | 1.09 × 103 |
| r95061 | A*01, A*02, B*17, B*29 | DNA/MVA CM9b | 239 | >433 | <30 |
| r95071 | A*02, B*17, B*29 | None | 239 | 173 | 7.64 × 104 |
| r96009 | B*08 | None | 239B*08-8xd | >28 | 3.02 × 106 |
| r96067 | A*01, B*08 | None | 239 | >30 | 1.16 × 102 |
| r96141 | A*01, A*11 | None | 239B*08-8xd | <16 | 7.16 × 102 |
| r97042 | B*08 | None | E660 | 53 | 9.47 × 106 |
| r97113 | A*01 | DNA/Ad5 GRNTc | 239 | >170 | 1.65 × 105 |
| r98016 | A*02, B*08, B*17, B*29 | None | 239 | >270 | 3.97 × 102 |
| r98031 | A*01, B*01 | None | 239 | <9 | 3.87 × 106 |
| r98037 | A*01, B*01 | Delta nefa | 239 | >53 | 2.69 × 102 |
| r99006 | A*01, B*08, B*17, B*29 | None | 3× 239h | >286 | <30 |
| r99039 | A*01 | None | 239 | <9 | 5.64 × 103 |
| r00032 | A*02, B*08 | None | 239 | >75 | 9.16 × 103 |
| r00047 | A*02 | Delta nefa | 239 | >90 | 2.20 × 104 |
| r00078 | A*08, B*08, B*29 | None | 239 | >230 | 9.79 × 105 |
| r01027 | A*01, B*08 | None | 239 | >80 | 3.95 × 104 |
| r01080 | A*01 | DNA/Ad5 GRNTc | 239 | >160 | 1.94 × 103 |
| r02019 | A*08, B*08 | None | 239 | >80 | <30 |
| r02120 | A*02, B*01, B*08 | None | 239B*08-8xd | >23 | 2.22 × 106 |
| r02129 | A*01, B*01 | None | E660 | 66 | 8.21 × 105 |
See reference 64.
Vaccinated with DNA/MVA encoding Gag CM9 only (4).
Vaccinated with DNA/Ad5 encoding Gag, Rev, Nef, and Tat (84).
SIVmac239 containing mutations in 8 Mamu-B*08 epitopes (L. Valentine et al., unpublished data).
SIVmac239 containing mutations in Mamu-B*08 Vif123-131RL9, Vif172-179RL8, Nef8-16RL9, Nef137-146RL10, and Nef246-254RL9.
p.i., postinfection.
Average chronic vRNA copies/ml from each assay time point.
See reference 25.
TABLE 2.
Ex vivo SIV-specific CD8+ T cells used in the 48-h virus suppression assay
| Epitope | Protein | Amino acid positions | Sequence | MHC restriction | IC50 (nM) |
|---|---|---|---|---|---|
| SL8 | Tat | 28-35 | STPESANL | A*01 | 43a |
| RL10 | Nef | 137-146 | RRHRILDIYL | B*08 | 11b |
| YY9 | Nef | 159-167 | YTSGPGIRY | A*02 | 2.7c |
| KL9 | Rev | 12-20 | KRLRLIHLL | B*08 | 3.2b |
| KF9 | Env gp41 | 524-532 | KRGVFVLGF | B*08 | 7.2b |
| KL9 | Env gp41 | 573-581 | KRQQELLRL | B*08 | 12b |
| TL9 | Env gp41 | 620-628 | TVPWPNASL | A*01 | 10d |
| RL9 | Env gp41 | 868-876 | RRIRQGLEL | B*08 | 5.5b |
| GY9 | Gag p17 matrix | 71-79 | GSENLKSLY | A*02 | 4.9c |
| CM9 | Gag p27 capsid | 181-189 | CTPYDINQM | A*01 | 22a |
| RL9 | Vif | 123-131 | RRAIRGEQL | B*08 | 7.5b |
| RL8 | Vif | 172-179 | RRDNRRGL | B*08 | 152b |
We previously used suppression assays to establish an antiviral hierarchy of SIV-specific CD8+ T cells (18, 44). However, these assays used cell lines (44) and clones (18) that were cultured for long periods of time in IL-2 and required frequent restimulations with irradiated B-lymphoblastoid cell lines pulsed with high concentrations of peptide. For a more physiologically relevant analysis of effector cell function, we used freshly isolated ex vivo cells. We calculated the virus-suppressive ability of each tetramer-sorted epitope-specific CD8+ T-cell population using the number of copies of viral RNA per milliliter of supernatant at 48 h with and without effector cells: (vRNA copies/ml without CTL − vRNA copies/ml with CTL)/vRNA copies/ml without CTL × 100. Viral suppression was calculated with tetramer-isolated CD8+ T cells sorted to greater than 50% epitope specificity as measured by postsort tetramer stains. Some freshly isolated CD8+ T-cell populations were better than others at suppressing viral replication at an effector-to-target cell ratio of 1:1 (Fig. 2b). CD8+ T cells recognizing Mamu-A*02-restricted Nef159-167YY9 suppressed viral replication by more than 80%. As expected, Gag-specific CD8+ T cells effectively suppressed viral replication. Even though Env-specific CD8+ T-cell responses were previously associated with higher HIV viral loads (35), CD8+ T cells recognizing three Mamu-B*08-restricted Env-specific epitopes suppressed viral replication by >50%. The inhibition of viral production from infected CD4+ T cells by epitope-specific CD8+ T cells was unrelated to the viral load of each animal at the time of the assay (Fig. 2b and data not shown). Epitope-specific CD8+ T cells from elite controllers (viral load of <10e3 copies/ml plasma) did not always control viral replication, while epitope-specific CD8+ T cells from progressors (viral load of >2 × 10e4 copies/ml plasma) often suppressed viral replication as well as, if not better than, cells of the same specificity from elite controllers or controllers. CD8+ T cells directed against all tested proteins still suppressed SIV-infected CD4+ T cells at low effector-to-target cell ratios (Fig. 2c to f). Nef-specific CD8+ T cells, in particular, suppressed virus replication at effector-to-target cell ratios as low as 1:10.
Commonly used assays do not predict whether a particular epitope-specific CD8+ T cell will be efficacious.
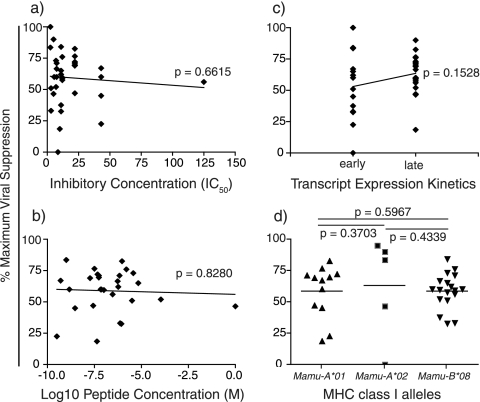
After determining the percent maximum viral suppression for each epitope-specific CD8+ T-cell population tested, we investigated whether efficacious CD8+ T cells had a common characteristic. We first looked at each epitope's affinity for its respective restricted MHC class I allele. The 50% inhibitory concentration (IC50) of each of the 12 epitopes studied (Table 2) (5, 23, 48, 49) was compared to the antiviral efficacy of epitope-specific CD8+ T cells (Fig. 3a). We found no evidence for a correlation between the affinity with which an MHC class I molecule bound its cognate epitope and viral suppression. We next looked at each epitope's ability to trigger cytokine secretion from CD8+ T cells. High-avidity epitope-specific CD8+ T cells require fewer MHC-epitope complexes on the surface of a target cell to induce cytokine secretion. Previously reported data suggested that antiviral efficacy can be correlated with epitope avidity (7, 10, 54, 74). High-avidity CD8+ T cells may therefore have superior antiviral efficacy compared with low-avidity CD8+ T cells, perhaps due to more efficient epitope processing, epitope transport to the cell surface, delayed epitope-MHC complex decay kinetics (58), or the ability to induce killing with fewer MHC-epitope complexes on the surface of infected cells. Additionally, Gag-specific CD8+ T-cell lines were previously shown to be more avid than Env-specific CD8+ T-cell lines (17). We used IFN-γ ELISPOT with PBMC responding to 10-fold dilutions of peptide to calculate the 50% effective concentration (EC50) needed to induce half the maximal cytokine secretion. Interestingly, there was no correlation between T-cell avidity for its cognate epitope and the ability of ex vivo-sorted epitope-specific CD8+ T cells to suppress viral replication (Fig. 3b).
FIG. 3.
Correlation between percent maximum viral suppression and epitope affinity, epitope avidity, mRNA expression kinetics, and MHC class I-restricting alleles. Each data point represents the average percent maximum suppression capacity of one CD8+ T-cell specificity at an effector-to-target cell ratio of 1:1 from one animal. (a) Peptide binding to the restricting MHC molecule (50% inhibitory concentration [49]) was determined as described previously (5, 23, 48, 49). (b) The effective concentration (EC50) to induce half the maximal TCR activation and secrete IFN-γ in response to 10-fold dilutions of cognate peptide was calculated using an IFN-γ ELISPOT assay with fresh PBMC prior to tetramer sorting. (c) Early viral transcripts include Tat, Rev, and Nef, while the viral transcripts expressed late in the SIV viral life cycle are Gag, Pol, Env, Vpr, Vpx, and Vif. (d) The MHC class I-restricting alleles for each epitope studied were defined previously (5, 45, 49). Statistical correlations were made using the Mann-Whitney test.
It was previously suggested that proteins expressed early in the HIV/SIV life cycle may be the most efficacious vaccine immunogens (3, 68). CD8+ T cells specific for epitopes derived from the proteins translated early, Tat, Rev, and Nef, should then suppress viral replication better than those CD8+ T cells specific for epitopes derived from the later proteins Gag, Pol, Env, Vpr, Vpx, and Vif (3, 20, 36, 37, 62, 72, 78, 79). Surprisingly, however, CD8+ T cells specific for epitopes derived from proteins whose viral transcripts are expressed late in the viral life cycle still inhibited virus production (Fig. 3c). Interestingly, however, recently reported data have shown that CD8+ T cells specific for Gag and Pol recognize infected cells prior to proviral integration (68, 69). Our data suggest that CD8+ T cells recognizing infected cells early in the viral life cycle, Gag and Pol, are not superior in their antiviral function to CD8+ T cells recognizing infected cells late (Fig. 2a and data not shown). Additionally, viral suppression does not correlate with the MHC class I-restricting allele of each epitope (Fig. 3d). Little difference in antiviral efficacy was seen among CD8+ T cells recognizing epitopes bound by Mamu-A*01, Mamu-A*02, or Mamu-B*08.
Cytokine secretion does not correlate with SIV-specific CD8+ T-cell antiviral efficacy.
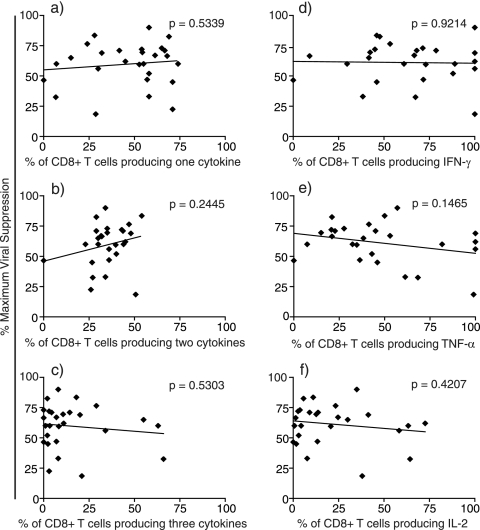
PBMC from elite controllers maintained functional CD8+ T cells capable of degranulation and cytokine and chemokine production compared with CD8+ T cells from HIV progressors (11, 27). We therefore explored whether ex vivo, tetramer-sorted SIV-specific CD8+ T cells that suppress viral replication have a high percentage of CD8+ T cells that secrete a variety of cytokines. We stimulated fresh PBMC prior to tetramer sorting with their cognate peptide and analyzed their ability to secrete IFN-γ, TNF-α, and IL-2 simultaneously by intracellular cytokine staining. Surprisingly, CD8+ T cells that produced only one cytokine suppressed viral production effectively (Fig. 4a). In contrast, epitope-specific CD8+ T cells that produced all three cytokines were not the most effective suppressor cells (Fig. 4c). In almost all cases, 25 to 50% of activated, epitope-specific CD8+ T cells secreted two of the three tested cytokines; however, no significant correlation existed between the suppressive efficacy of the various CD8+ T-cell populations and their secretion of two of three cytokines (Fig. 4b). We also analyzed the quantity of activated, epitope-specific CD8+ T cells secreting each cytokine individually in response to cognate peptide. All epitope-specific CD8+ T cells secreted each cytokine tested to some extent except for one Mamu-A*02-restricted Gag71-79GY9-specific and one Mamu-B*08-restricted Env524-532KF9-specific CD8+ T-cell population. The highest proportion of activated CD8+ T cells secreted IFN-γ, followed by TNF-α (Fig. 4d and e). Far fewer activated CD8+ T cells secreted IL-2 (Fig. 4f). Secretion of any of the three tested cytokines by activated CD8+ T cells did not correlate with ex vivo epitope-specific CD8+ T-cell antiviral efficacy (Fig. 4d to f). Interestingly, therefore, cells can suppress virus replication with a limited cytokine repertoire. Indeed, multifunctional cells secreting high percentages of all three cytokines simultaneously were not associated with strong suppression.
FIG. 4.
Correlation between percent maximum viral suppression and cytokine secretion. Each data point represents the average percent maximum suppression capacity of one CD8+ T-cell specificity at an effector-to-target cell ratio of 1:1 from one animal. On the same day as the ex vivo SIV-specific CD8+ T-cell sort, PBMC were stained intracellularly for their ability to secrete IFN-γ, TNF-α, and IL-2 in response to peptide stimulation. (a) Activated CD8+ T cells producing only one of the three cytokines following peptide stimulation. (b) Activated CD8+ T cells producing two of the three cytokines following peptide stimulation. (c) Activated CD8+ T cells producing all three cytokines following peptide stimulation. (d to f) Total percentages of activated CD8+ T cells producing IFN-γ (d), TNF-α (e), or IL-2 (f) following peptide stimulation. Statistical correlations were made using the Mann-Whitney test.
PD-1 expression on CD28+ and CD28− memory CD8+ T-cell populations failed to correlate with viral suppression.
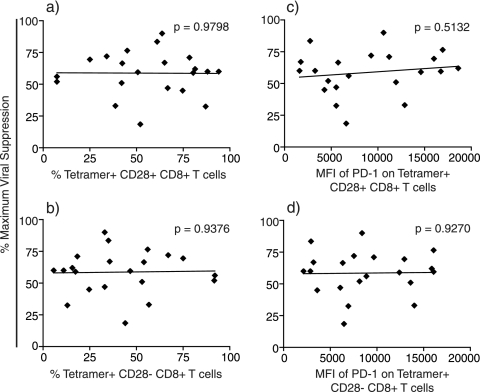
Recent data have shown that HIV-infected patients with low viral set points have a higher percentage of central memory CD8+ T cells than patients with high viral set points. Patients with high viral set points, on the other hand, tend to have higher frequencies of effector memory CD8+ T cells than those with lower viral set points (14, 83). We therefore determined the percentages of central (CD28+) and effector (CD28−) memory CD8+ T-cell populations from PBMC from the approximate date of the viral production assays. CD8+ tetramer+ T cells had a higher percentage of effector memory CD8+ T cells than central memory CD8+ T cells; however, the balance between central and effector memory CD8+ tetramer+ T cells did not predict antiviral efficacy (Fig. 5a and b). Those CD8+ tetramer+ T cells with a greater proportion of central memory CD8+ T cells did not suppress viral replication more efficiently than CD8+ tetramer+ T cells with a greater proportion of effector memory CD8+ T cells.
FIG. 5.
Correlation between percent maximum viral suppression and central/effector memory CD8+ T-cell populations and their expression of PD-1. Each data point represents the average percent maximum suppression capacity of one CD8+ T-cell specificity at an effector-to-target cell ratio of 1:1 from one animal. (a and b) Percentage of CD3+ CD8+ tetramer+ T cells that are central memory (CD28+) (a) or effector memory (CD28−) (b) cells. (c) Mean fluorescence intensity (MFI) of PD-1 on CD3+ CD8+ tetramer+ CD28+ central memory T cells. (d) Mean fluorescence intensity of PD-1 on CD3+ CD8+ tetramer+ CD28− effector memory T cells. Statistical correlations were made using the Mann-Whitney test.
Programmed death 1 (PD-1) is upregulated on exhausted virus-specific CD8+ T cells during chronic HIV infection. Upregulation of PD-1 correlates with impaired effector function and increased disease progression in both HIV-infected humans and SIV-infected macaques (21, 80, 89). Moreover, blocking the engagement of PD-1 to its ligand restored virus-specific CD8+ T-cell function, survival, and proliferation; reduced viral loads; and increased rates of animal survival (81). We therefore examined whether expression levels of PD-1 on CD8+ tetramer+ T cells correlated with suppressive ability. Unexpectedly, PD-1 expression on both CD28+ and CD28− memory CD8+ T cells did not correlate with viral suppression (Fig. 5c and d). CD8+ tetramer+ cells with high mean fluorescence intensities of PD-1 on their cell surface suppressed virus production by more than 50%. There was also no correlation between viral suppression and PD-1 expression on all CD8+ tetramer+ T cells irrespective of central or effector memory differentiation (data not shown).
DISCUSSION
We developed a novel viral suppression assay to test the antiviral efficacy of ex vivo, tetramer-sorted, SIV-specific CD8+ T cells. Using this assay, we established an antiviral hierarchy among 12 epitope-specific CD8+ T cells. Suppression did not correlate with disease status, the protein from which the epitopes were derived, or the MHC class I allele restricting the epitopes. Epitope avidity, epitope affinity, viral transcript expression kinetics, CD8+ T-cell recognition kinetics, CD8+ T-cell cytokine secretion, the balance of central and effector memory cells, or PD-1 expression on the surface of memory epitope-specific CD8+ T cells showed no correlation with viral suppression. Standard laboratory assays such as IFN-γ ELISPOT assay and intracellular cytokine staining, therefore, failed to predict whether epitope-specific CD8+ T cells might control viral replication.
Conventional assays do not directly measure the antiviral efficacy of epitope-specific CD8+ T cells. IFN-γ ELISPOT assays and intracellular cytokine staining measure the magnitude of cytokine secretion by peptide-stimulated epitope-specific CD8+ T cells, while tetramer staining assays simply count epitope-specific CD8+ T cells. Proliferation assays test the ability of CD8+ T cells to proliferate in the presence of peptide or infected cells. Finally, using B cells and large amounts of peptide, chromium release assays indirectly measure the killing ability of either bulk PBMC or epitope-specific cell lines or clones. None of these assays directly measures the antiviral efficacy of epitope-specific CD8+ T cells. Previously developed viral suppression assays to test the antiviral efficacies of epitope-specific CD8+ T cells were performed with CD8+ T-cell lines or clones (17, 18, 44, 47, 57, 75, 76, 86, 87). Both of these cell types require culturing cells in IL-2 and frequent restimulation using B-lymphoblastoid cell lines pulsed with high concentrations of peptide. This may result in less physiologically relevant cell populations.
We therefore developed a novel assay to isolate ex vivo epitope-specific CD8+ T cells from PBMC and test their antiviral function in a more physiologically relevant setting. These cells represent a heterogeneous, realistic population of circulating cells in various stages of activation, proliferation, and differentiation. In addition to using ex vivo-sorted epitope-specific CD8+ T cells, we shortened the viral suppression assay to 48 h. Limited cell growth occurs in 48 h, removing the possibility of one cell type outgrowing another. Longer assays may skew the effector-to-target cell ratio and, potentially, the suppression capacity of the SIV-specific CD8+ T cells. A 48-h assay is advantageous over a 24-h assay, as it allows those SIV-specific CD8+ T cells that recognize cognate epitopes late in infection to make up for any initial time lag. Additionally, we saw little difference in viral RNA content in the supernatant containing epitope-specific CD8+ T cells and the control lacking any CD8+ T cells in a 24-h assay. All ex vivo CD8+ T-cell populations in this study were sorted to at least 50% specificity as measured by postsort tetramer staining. Although CD8+ T cells were sorted to less purity than cell lines or clones, the antiviral hierarchy and lack of correlation still held true when cells of only >80% purity were analyzed (data not shown). Finally, we detected little residual tetramer staining of the sorted CD8+ T cells after they were rested overnight (data not shown). It is therefore unlikely that the tetramers interfered with T-cell receptor (TCR) interactions with SIV-infected CD4+ T cells.
Although a hierarchy is seen among the 12 epitopes tested, our new assay clearly shows that almost all epitope-specific CD8+ T cells have some capacity to suppress viral replication. Data from previous studies suggested that Mamu-A*02-restricted Nef159-167YY9-specific CD8+ T cells were largely ineffective at suppressing viral replication from SIV-infected activated CD4+ T cells (18, 44); however, our new assay revealed that ex vivo-harvested Nef159-167YY9-specific CD8+ T cells suppressed viral replication efficiently. Additionally, we previously observed that Mamu-A*01-restricted Tat28-35SL8 CD8+ T-cell lines had enhanced antiviral efficacy compared to that of Mamu-A*01-restricted Gag181-189CM9 CD8+ T-cell lines (18, 47). Ex vivo viral suppression assays, however, revealed a more comparable antiviral profile for these two particular epitope-specific CD8+ T-cell populations. Long-term culture of the CD8+ T-cell lines could account for this difference, as they may become exhausted or undergo phenotypic changes in vitro after multiple stimulations and may therefore exhibit varied antiviral efficacies.
Gag-specific CD8+ T cells have been implicated in the control of viral replication (30, 35, 68, 71). Indeed, the vaccination of rhesus macaques with Gag-encoded DNA and Ad5 vectors afforded a short-term reduction of viral loads after SIVmac239 infection (16, 43). The addition of Tat, Rev, and Nef to this vaccine regimen further augmented the observed viral control (84). Additionally, increased breadth of Gag-specific CD8+ T-cell responses was significantly associated with lower viral loads in a cohort of 578 HIV-infected individuals (35), and the frequency of Gag-specific CD8+ T-cell responses inversely correlated with viral load in HIV-infected children (30). In this study, the two tested Gag-specific CD8+ T-cell responses suppressed viral replication ex vivo by >70% on average.
Env-specific CD8+ T cells suppressed virus production similarly to Gag-specific CD8+ T cells, indicating that these cells may also contribute to the control of viral replication. Although increased breadth of Env-specific CD8+ T-cell responses correlated with increased viral loads in HIV-infected patients (35), Env-specific CD8+ T cells can suppress viral replication ex vivo. Sequence diversity and rapid viral escape in Env might reduce antiviral function in vivo. Our data suggest that effective antiviral CD8+ T cells can recognize epitopes from several SIV proteins including Env, Tat, Nef, Rev, and Vif.
Control of SIV infection by elite controller rhesus macaques is not mediated by Gag-specific CD8+ T-cell responses alone. Nearly all SIV-infected rhesus macaque elite controllers express Mamu-B*17 and/or Mamu-B*08 (46). Of the five common Mamu-B*17-restricted CD8+ T-cell responses and 14 Mamu-B*08-restricted CD8+ T-cell responses discovered thus far, only one infrequently observed response is directed against Gag (45, 48, 50). Additionally, reduction of recrudescent virus after CD8+ depletion in elite controller rhesus macaques was mediated by subdominant CD8+ T-cell responses, few of which were Gag specific (26). Although Gag-specific CD8+ T cells suppressed viral replication ex vivo and are important in the control of HIV infection, control of SIV infection in rhesus macaques may be mediated by CD8+ T cells directed against a variety of epitopes derived from multiple proteins.
The efficacy with which an epitope-specific CD8+ T cell kills an SIV-infected CD4+ T cell does not depend on how tightly the processed epitope binds to its respective MHC class I molecule. Additionally, the amount of cognate exogenous peptide required on the surface of presenting cells to induce TCR activation is not correlated with viral suppression. Some groups have shown a relationship between peptide avidity and antiviral efficacy (7, 54, 74). Using CD8+ T-cell clones specific for HLA-B*27-Gag KK10, Almeida et al. previously found that the peptide avidity of each clone correlated with the patient's viral load (7). This finding corroborates earlier data illustrating that high-avidity CD8+ T cells not only have superior antiviral efficacy compared with low-avidity CD8+ T cells but also lyse target cells more rapidly (2, 22, 74). It was also shown that although Gag-specific CD8+ T cells were significantly more avid than Env-specific CD8+ T cells, there was no correlation between the ability to inhibit virus and epitope avidity (17). Cell lines or clones were used in those studies rather than fresh PBMC. The avidity of cultured cells can, however, be affected by the quantity of peptide used to restimulate cell lines (2, 40, 41). Additionally, measuring epitope avidity with high concentrations of exogenous peptide bypasses viral protein expression, epitope processing in the proteasome, and epitope transport to the cell surface, all important events within an infected cell (10). Determining the functional avidity of epitopes using virally infected cells may therefore be a better way to look for a correlation between avidity and CD8+ T-cell suppression. The lack of any significant correlation between peptide avidity and CD8+ T-cell antiviral efficacy in our studies suggests that low-avidity CD8+ T cells can still inhibit viral release. Even though low-avidity CD8+ T cells may require more peptide antigen on the surface of the target cells to trigger cytokine secretion, this lag in time appears to have little effect on the suppressive capacities of these cells.
The multifunctionality of epitope-specific CD8+ T cells may be a key contributor to CD8+ T-cell antiviral efficacy. Elite controllers maintain highly functional CD8+ T cells capable of concurrently secreting IFN-γ, TNF-α, IL-2, and macrophage inflammatory protein 1β (MIP-1β) and cell surface expression of CD107a compared with CD8+ T cells from progressors (11, 27). This implies that a successful CD8+ T-cell response consists of antiviral cells that can secrete several cytokines. Analysis of the cytokine profile of CD8+ T cells revealed that ex vivo, CD8+ T cells do not need to secrete several cytokines simultaneously in order to suppress viral production from SIV-infected CD4+ T cells. Additionally, secretion of a particular cytokine did not correlate with suppression. When greater than 50% of the epitope-specific CD8+ T cells secreted a single cytokine, that cell population still maintained suppressive function. While epitope-specific CD8+ T-cell multifunctionality may improve the overall anti-HIV immune response, it may not necessarily correlate directly with reduction of viral replication from virus-infected CD4+ T cells.
Several studies implicated the upregulation of PD-1 on the surface of CD8+ T cells as an indicator for poor effector function and increased viral loads (21, 80, 89). Our data showed no correlation between the mean fluorescence intensity of PD-1 on epitope-specific memory CD8+ T cells and antiviral efficacy. This is congruent with a previous study looking at mRNA levels of PD-1 in Gag- and Env-specific CD8+ T-cell lines (17). Although PD-1 expression on HIV-specific CD8+ T cells and the total CD8+ T-cell population significantly correlates with viral load and inversely correlates with the CD4 count of HIV-infected patients (21), it had little effect on the epitope-specific CD8+ T cell's ability to suppress viral replication in our 48-h assay.
Although we established an antiviral hierarchy among 12 epitope-specific CD8+ T-cell populations using a novel viral suppression assay with freshly sorted cells, we found no clear correlate of immune control. Viral suppression could not be predicted by any of the conventional assays, including epitope avidity, expression or recognition kinetics, and cytokine secretion. CD8+ T-cell proliferation and perforin and granzyme secretion may correlate with viral suppression and will be the focus of future studies. In vivo, epitope-specific CD8+ T cells may work in concert with each other and with other arms of the immune system to control viral replication. The focus to find one particular correlate of immune protection may oversimplify the complex interplay of the anti-HIV immune response.
Acknowledgments
This research was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease grants R01 AI076114, R01 AI049120, R24 RR015371, and R24 RR016038; in part by grant R51 RR000167 from the National Center for Research Resources (NCRR), awarded to the WNPRC, University of Wisconsin—Madison; and by grant RR000168, awarded to the New England Primate Research Center.
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: human IL-2 (item 136) from Hoffman-La Roche, tenofovir disoproxil fumarate (item 10198), and SIVmac p27 hybridoma (55-2F12, item 1547) from Niels Pedersen. We acknowledge Enrique Leon, Caitlin McNair, and Thomas Friedrich for production of high-titer SIV. We also thank Chrystal Glidden, Gretta Borchardt, and Debra Fisk for MHC typing of animals.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander-Miller, M. A., G. R. Leggatt, and J. A. Berzofsky. 1996. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc. Natl. Acad. Sci. U. S. A. 93:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, A., R. Lubong, H. Ng, D. G. Brooks, J. A. Zack, and O. O. Yang. 2004. Impacts of epitope expression kinetics and class I downregulation on the antiviral activity of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes. J. Virol. 78:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, T. M., P. Jing, B. Calore, H. Horton, D. H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B. R. Mothé, C. Emerson, N. Wilson, J. D. Lifson, I. M. Belyakov, J. A. Berzofsky, C. Wang, D. B. Allison, D. C. Montefiori, R. C. Desrosiers, S. Wolinsky, K. J. Kunstman, J. D. Altman, A. Sette, A. J. McMichael, and D. I. Watkins. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, T. M., B. R. Mothé, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothé, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 7.Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appay, V., D. C. Douek, and D. A. Price. 2008. CD8+ T cell efficacy in vaccination and disease. Nat. Med. 14:623-628. [DOI] [PubMed] [Google Scholar]
- 9.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 10.Bennett, M. S., H. L. Ng, M. Dagarag, A. Ali, and O. O. Yang. 2007. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J. Virol. 81:4973-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 14.Burgers, W. A., C. Riou, M. Mlotshwa, P. Maenetje, D. de Assis Rosa, J. Brenchley, K. Mlisana, D. C. Douek, R. Koup, M. Roederer, G. de Bruyn, S. A. Karim, C. Williamson, C. M. Gray, and the CAPRISA 002 Acute Infection Study Team. 2009. Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J. Immunol. 182:4751-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 16.Casimiro, D. R., F. Wang, W. A. Schleif, X. Liang, Z. Q. Zhang, T. W. Tobery, M. E. Davies, A. B. McDermott, D. H. O'Connor, A. Fridman, A. Bagchi, L. G. Tussey, A. J. Bett, A. C. Finnefrock, T. M. Fu, A. Tang, K. A. Wilson, M. Chen, H. C. Perry, G. J. Heidecker, D. C. Freed, A. Carella, K. S. Punt, K. J. Sykes, L. Huang, V. I. Ausensi, M. Bachinsky, U. Sadasivan-Nair, D. I. Watkins, E. A. Emini, and J. W. Shiver. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 79:15547-15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, H., A. Piechocka-Trocha, T. Miura, M. A. Brockman, B. D. Julg, B. M. Baker, A. C. Rothchild, B. L. Block, A. Schneidewind, T. Koibuchi, F. Pereyra, T. M. Allen, and B. D. Walker. 2009. Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes. J. Virol. 83:3138-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung, C., W. Lee, J. T. Loffredo, B. Burwitz, T. C. Friedrich, J. P. Giraldo Vela, G. Napoé, E. G. Rakasz, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2007. Not all cytokine-producing CD8+ T cells suppress simian immunodeficiency virus replication. J. Virol. 81:1517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cline, A. N., J. W. Bess, M. Piatak, and J. D. Lifson. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303-312. [DOI] [PubMed] [Google Scholar]
- 20.Cullen, B. R. 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249:203-210. [DOI] [PubMed] [Google Scholar]
- 21.Day, C. L., D. E. Kaufmann, P. Kiepiela, J. A. Brown, E. S. Moodley, S. Reddy, E. W. Mackey, J. D. Miller, A. J. Leslie, C. DePierres, Z. Mncube, J. Duraiswamy, B. Zhu, Q. Eichbaum, M. Altfeld, E. J. Wherry, H. M. Coovadia, P. J. Goulder, P. Klenerman, R. Ahmed, G. J. Freeman, and B. D. Walker. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350-354. [DOI] [PubMed] [Google Scholar]
- 22.Derby, M., M. Alexander-Miller, R. Tse, and J. Berzofsky. 2001. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J. Immunol. 166:1690-1697. [DOI] [PubMed] [Google Scholar]
- 23.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothé, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 24.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich, T. C., A. B. McDermott, M. R. Reynolds, S. Piaskowski, S. Fuenger, I. P. De Souza, R. Rudersdorf, C. Cullen, L. J. Yant, L. Vojnov, J. Stephany, S. Martin, D. H. O'Connor, N. Wilson, and D. I. Watkins. 2004. Consequences of cytotoxic T-lymphocyte escape: common escape mutations in simian immunodeficiency virus are poorly recognized in naive hosts. J. Virol. 78:10064-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. León, T. Soma, G. Napoé, S. V. Capuano, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genescà, M., T. Rourke, J. Li, K. Bost, B. Chohan, M. B. McChesney, and C. J. Miller. 2007. Live attenuated lentivirus infection elicits polyfunctional simian immunodeficiency virus Gag-specific CD8+ T cells with reduced apoptotic susceptibility in rhesus macaques that control virus replication after challenge with pathogenic SIVmac239. J. Immunol. 179:4732-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 29.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 30.Huang, S., J. Dunkley-Thompson, Y. Tang, E. A. Macklin, J. Steel-Duncan, I. Singh-Minott, E. G. Ryland, M. Smikle, B. D. Walker, C. D. Christie, and M. E. Feeney. 2008. Deficiency of HIV-Gag-specific T cells in early childhood correlates with poor viral containment. J. Immunol. 181:8103-8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59:693-703. [DOI] [PubMed] [Google Scholar]
- 33.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Muñoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. R. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 34.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 35.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klotman, M. E., S. Kim, A. Buchbinder, A. DeRossi, D. Baltimore, and F. Wong-Staal. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. U. S. A. 88:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, and C. Carter. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 39.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroger, C. J., and M. A. Alexander-Miller. 2007. Cutting edge: CD8+ T cell clones possess the potential to differentiate into both high- and low-avidity effector cells. J. Immunol. 179:748-751. [DOI] [PubMed] [Google Scholar]
- 41.Kroger, C. J., and M. A. Alexander-Miller. 2007. Dose-dependent modulation of CD8 and functional avidity as a result of peptide encounter. Immunology 122:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 43.Liu, J., K. L. O'Brien, D. M. Lynch, N. L. Simmons, A. La Porte, A. M. Riggs, P. Abbink, R. T. Coffey, L. E. Grandpre, M. S. Seaman, G. Landucci, D. N. Forthal, D. C. Montefiori, A. Carville, K. G. Mansfield, M. J. Havenga, M. G. Pau, J. Goudsmit, and D. H. Barouch. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loffredo, J. T., B. J. Burwitz, E. G. Rakasz, S. P. Spencer, J. J. Stephany, J. P. Vela, S. R. Martin, J. Reed, S. M. Piaskowski, J. Furlott, K. L. Weisgrau, D. S. Rodrigues, T. Soma, G. Napoé, T. C. Friedrich, N. A. Wilson, E. G. Kallas, and D. I. Watkins. 2007. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 81:2624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loffredo, J. T., T. C. Friedrich, E. J. León, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8 T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One 2:e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81:8827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loffredo, J. T., E. G. Rakasz, J. P. Giraldo, S. P. Spencer, K. K. Grafton, S. R. Martin, G. Napoé, L. J. Yant, N. A. Wilson, and D. I. Watkins. 2005. Tat(28-35)SL8-specific CD8+ T lymphocytes are more effective than Gag(181-189)CM9-specific CD8+ T lymphocytes at suppressing simian immunodeficiency virus replication in a functional in vitro assay. J. Virol. 79:14986-14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loffredo, J. T., J. Sidney, A. T. Bean, D. R. Beal, W. Bardet, A. Wahl, O. E. Hawkins, S. Piaskowski, N. A. Wilson, W. H. Hildebrand, D. I. Watkins, and A. Sette. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 182:7763-7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loffredo, J. T., J. Sidney, C. Wojewoda, E. Dodds, M. R. Reynolds, G. Napoé, B. R. Mothé, D. H. O'Connor, N. A. Wilson, D. I. Watkins, and A. Sette. 2004. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J. Immunol. 173:5064-5076. [DOI] [PubMed] [Google Scholar]
- 50.Maness, N., L. Yant, C. Chung, J. Loffredo, T. Friedrich, S. Piaskowski, J. Furlott, G. May, T. Soma, E. Leon, N. Wilson, H. Piontkivska, A. Hughes, J. Sidney, A. Sette, and D. Watkins. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J. Virol. 82:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 53.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 54.Messaoudi, I., J. A. Guevara Patiño, R. Dyall, J. LeMaoult, and J. Nikolich-Zugich. 2002. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298:1797-1800. [DOI] [PubMed] [Google Scholar]
- 55.Migueles, S. A., A. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. Mclaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 56.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minang, J. T., E. V. Barsov, F. Yuan, M. T. Trivett, M. Piatak, J. D. Lifson, D. E. Ott, and C. Ohlen. 2008. Efficient inhibition of SIV replication in rhesus CD4+ T-cell clones by autologous immortalized SIV-specific CD8+ T-cell clones. Virology 372:430-441. [DOI] [PubMed] [Google Scholar]
- 58.Motozono, C., S. Yanaka, K. Tsumoto, M. Takiguchi, and T. Ueno. 2009. Impact of intrinsic cooperative thermodynamics of peptide-MHC complexes on antiviral activity of HIV-specific CTL. J. Immunol. 182:5528-5536. [DOI] [PubMed] [Google Scholar]
- 59.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothé, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 61.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, and C. R. Rizza. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 62.Ranki, A., A. Lagerstedt, V. Ovod, E. Aavik, and K. J. Krohn. 1994. Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch. Virol. 139:365-378. [DOI] [PubMed] [Google Scholar]
- 63.Reimann, K. A., K. Tenner-Racz, P. Racz, D. C. Montefiori, Y. Yasutomi, W. Lin, B. J. Ransil, and N. L. Letvin. 1994. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J. Virol. 68:2362-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. León, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivière, Y., M. B. McChesney, F. Porrot, F. Tanneau-Salvadori, P. Sansonetti, O. Lopez, G. Pialoux, V. Feuillie, M. Mollereau, and S. Chamaret. 1995. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res. Hum. Retroviruses 11:903-907. [DOI] [PubMed] [Google Scholar]
- 66.Rolland, M., D. Heckerman, W. Deng, C. M. Rousseau, H. Coovadia, K. Bishop, P. J. Goulder, B. D. Walker, C. Brander, and J. I. Mullins. 2008. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One 3:e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 68.Sacha, J. B., C. Chung, E. G. Rakasz, S. P. Spencer, A. K. Jonas, A. T. Bean, W. Lee, B. J. Burwitz, J. J. Stephany, J. T. Loffredo, D. B. Allison, S. Adnan, A. Hoji, N. A. Wilson, T. C. Friedrich, J. D. Lifson, O. O. Yang, and D. I. Watkins. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sacha, J. B., C. Chung, J. Reed, A. K. Jonas, A. T. Bean, S. P. Spencer, W. Lee, L. Vojnov, R. Rudersdorf, T. C. Friedrich, N. A. Wilson, J. D. Lifson, and D. I. Watkins. 2007. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J. Virol. 81:11703-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 71.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartz, O., V. Maréchal, O. Danos, and J. M. Heard. 1995. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J. Virol. 69:4053-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seder, R. A., P. A. Darrah, and M. Roederer. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247-258. [DOI] [PubMed] [Google Scholar]
- 74.Sedlik, C., G. Dadaglio, M. F. Saron, E. Deriaud, M. Rojas, S. I. Casal, and C. Leclerc. 2000. In vivo induction of a high-avidity, high-frequency cytotoxic T-lymphocyte response is associated with antiviral protective immunity. J. Virol. 74:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Severino, M. E., N. V. Sipsas, P. T. Nguyen, S. A. Kalams, B. D. Walker, R. P. Johnson, and O. O. Yang. 2000. Inhibition of human immunodeficiency virus type 1 replication in primary CD4(+) T lymphocytes, monocytes, and dendritic cells by cytotoxic T lymphocytes. J. Virol. 74:6695-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomiyama, H., M. Fujiwara, S. Oka, and M. Takiguchi. 2005. Epitope-dependent effect of Nef-mediated HLA class I down-regulation on ability of HIV-1-specific CTLs to suppress HIV-1 replication. J. Immunol. 174:36-40. [DOI] [PubMed] [Google Scholar]
- 77.Valentine, L. E., S. M. Piaskowski, E. G. Rakasz, N. L. Henry, N. A. Wilson, and D. I. Watkins. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82:575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Baalen, C. A., C. Guillon, M. van Baalen, E. J. Verschuren, P. H. Boers, A. D. Osterhaus, and R. A. Gruters. 2002. Impact of antigen expression kinetics on the effectiveness of HIV-specific cytotoxic T lymphocytes. Eur. J. Immunol. 32:2644-2652. [DOI] [PubMed] [Google Scholar]
- 79.Van Baalen, C. A., M. Schutten, R. C. Huisman, P. H. Boers, R. A. Gruters, and A. D. Osterhaus. 1998. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J. Virol. 72:6851-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Velu, V., S. Kannanganat, C. Ibegbu, L. Chennareddi, F. Villinger, G. J. Freeman, R. Ahmed, and R. R. Amara. 2007. Elevated expression levels of inhibitory receptor programmed death 1 on simian immunodeficiency virus-specific CD8 T cells during chronic infection but not after vaccination. J. Virol. 81:5819-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velu, V., K. Titanji, B. Zhu, S. Husain, A. Pladevega, L. Lai, T. H. Vanderford, L. Chennareddi, G. Silvestri, G. J. Freeman, R. Ahmed, and R. R. Amara. 2008. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature 458:206-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 83.Wherry, E. J., V. Teichgräber, T. C. Becker, D. Masopust, S. M. Kaech, R. Antia, U. H. von Andrian, and R. Ahmed. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225-234. [DOI] [PubMed] [Google Scholar]
- 84.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 86.Yang, O. O., S. A. Kalams, M. Rosenzweig, A. Trocha, N. Jones, M. Koziel, B. D. Walker, and R. P. Johnson. 1996. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J. Virol. 70:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang, O. O., S. A. Kalams, A. Trocha, H. Cao, A. Luster, R. P. Johnson, and B. D. Walker. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J. Virol. 71:3120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, J., Z. Zhang, X. Wang, J. Fu, J. Yao, Y. Jiao, L. Chen, H. Zhang, J. Wei, L. Jin, M. Shi, G. Gao, H. Wu, and F. Wang. 2007. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood 109:4671-4678. [DOI] [PubMed] [Google Scholar]