Abstract
HIV-1 possesses an exquisite ability to infect cells independently from their cycling status by undergoing an active phase of nuclear import through the nuclear pore. This property has been ascribed to the presence of karyophilic elements present in viral nucleoprotein complexes, such as the matrix protein (MA); Vpr; the integrase (IN); and a cis-acting structure present in the newly synthesized DNA, the DNA flap. However, their role in nuclear import remains controversial at best. In the present study, we carried out a comprehensive analysis of the role of these elements in nuclear import in a comparison between several primary cell types, including stimulated lymphocytes, macrophages, and dendritic cells. We show that despite the fact that none of these elements is absolutely required for nuclear import, disruption of the central polypurine tract-central termination sequence (cPPT-CTS) clearly affects the kinetics of viral DNA entry into the nucleus. This effect is independent of the cell cycle status of the target cells and is observed in cycling as well as in nondividing primary cells, suggesting that nuclear import of viral DNA may occur similarly under both conditions. Nonetheless, this study indicates that other components are utilized along with the cPPT-CTS for an efficient entry of viral DNA into the nucleus.
Lentiviruses display an exquisite ability to infect dividing and nondividing cells alike that is unequalled among Retroviridae. This property is thought to be due to the particular behavior or composition of the viral nucleoprotein complexes (NPCs) that are liberated into the cytoplasm of target cells upon virus-to-cell membrane fusion and that allow lentiviruses to traverse an intact nuclear membrane (17, 28, 29, 39, 52, 55, 67, 79). In the case of the human immunodeficiency type I virus (HIV-1), several studies over the years identified viral components of such structures with intrinsic karyophilic properties and thus perfect candidates for mediation of the passage of viral DNA (vDNA) through the nuclear pore: the matrix protein (MA); Vpr; the integrase (IN); and a three-stranded DNA flap, a structure present in neo-synthesized viral DNA, specified by the central polypurine tract-central termination sequence (cPPT-CTS). It is clear that these elements may mediate nuclear import directly or via the recruitment of the host's proteins, and indeed, several cellular proteins have been found to influence HIV-1 infection during nuclear import, like the karyopherin α2 Rch1 (38); importin 7 (3, 30, 93); the transportin SR-2 (13, 20); or the nucleoporins Nup98 (27), Nup358/RANBP2, and Nup153 (13, 56).
More recently, the capsid protein (CA), the main structural component of viral nucleoprotein complexes at least upon their cytoplasmic entry, has also been suggested to be involved in nuclear import or in postnuclear entry steps (14, 25, 74, 90, 92). Whether this is due to a role for CA in the shaping of viral nucleoprotein complexes or to a direct interaction between CA and proteins involved in nuclear import remains at present unknown.
Despite a large number of reports, no single viral or cellular element has been described as absolutely necessary or sufficient to mediate lentiviral nuclear import, and important controversies as to the experimental evidences linking these elements to this step exist. For example, MA was among the first viral protein of HIV-1 described to be involved in nuclear import, and 2 transferable nuclear localization signals (NLSs) have been described to occur at its N and C termini (40). However, despite the fact that early studies indicated that the mutation of these NLSs perturbed HIV-1 nuclear import and infection specifically in nondividing cells, such as macrophages (86), these findings failed to be confirmed in more-recent studies (23, 33, 34, 57, 65, 75).
Similarly, Vpr has been implicated by several studies of the nuclear import of HIV-1 DNA (1, 10, 21, 43, 45, 47, 64, 69, 72, 73, 85). Vpr does not possess classical NLSs, yet it displays a transferable nucleophilic activity when fused to heterologous proteins (49-51, 53, 77, 81) and has been shown to line onto the nuclear envelope (32, 36, 47, 51, 58), where it can truly facilitate the passage of the viral genome into the nucleus. However, the role of Vpr in this step remains controversial, as in some instances Vpr is not even required for viral replication in nondividing cells (1, 59).
Conflicting results concerning the role of IN during HIV-1 nuclear import also exist. Indeed, several transferable NLSs have been described to occur in the catalytic core and the C-terminal DNA binding domains of IN, but for some of these, initial reports of nuclear entry defects (2, 9, 22, 46, 71) were later shown to result from defects at steps other than nuclear import (60, 62, 70, 83). These reports do not exclude a role for the remaining NLSs in IN during nuclear import, and they do not exclude the possibility that IN may mediate this step by associating with components of the cellular nuclear import machinery, such as importin alpha and beta (41), importin 7 (3, 30, 93, 98), and, more recently, transportin-SR2 (20).
The central DNA flap, a structure present in lentiviruses and in at least 1 yeast retroelement (44), but not in other orthoretroviruses, has also been involved in the nuclear import of viral DNA (4, 6, 7, 31, 78, 84, 95, 96), and more recently, it has been proposed to provide a signal for viral nucleoprotein complexes uncoating in the proximity of the nuclear pore, with the consequence of providing a signal for import (8). However, various studies showed an absence or weakness of nuclear entry defects in viruses devoid of the DNA flap (24, 26, 44, 61).
Overall, the importance of viral factors in HIV-1 nuclear import is still unclear. The discrepancies concerning the role of MA, IN, Vpr, and cPPT-CTS in HIV-1 nuclear import could in part be explained by their possible redundancy. To date, only one comprehensive study analyzed the role of these four viral potentially karyophilic elements together (91). This study showed that an HIV-1 chimera where these elements were either deleted or replaced by their murine leukemia virus (MLV) counterparts was, in spite of an important infectivity defect, still able to infect cycling and cell cycle-arrested cell lines to similar efficiencies. If this result indicated that the examined viral elements of HIV-1 were dispensable for the cell cycle independence of HIV, as infections proceeded equally in cycling and arrested cells, they did not prove that they were not required in nuclear import, because chimeras displayed a severe infectivity defect that precluded their comparison with the wild type (WT).
Nuclear import and cell cycle independence may not be as simply linked as previously thought. On the one hand, there has been no formal demonstration that the passage through the nuclear pore, and thus nuclear import, is restricted to nondividing cells, and for what we know, this passage may be an obligatory step in HIV infection in all cells, irrespective of their cycling status. In support of this possibility, certain mutations in viral elements of HIV affect nuclear import in dividing as well as in nondividing cells (4, 6, 7, 31, 84, 95). On the other hand, cell cycle-independent infection may be a complex phenomenon that is made possible not only by the ability of viral DNA to traverse the nuclear membrane but also by its ability to cope with pre- and postnuclear entry events, as suggested by the phenotypes of certain CA mutants (74, 92).
Given that the cellular environment plays an important role during the early steps of viral infection, we chose to analyze the role of the four karyophilic viral elements of HIV-1 during infection either alone or combined in a wide comparison between cells highly susceptible to infection and more-restrictive primary cell targets of HIV-1 in vivo, such as primary blood lymphocytes (PBLs), monocyte-derived macrophages (MDM), and dendritic cells (DCs).
In this study, we show that an HIV-1-derived virus in which the 2 NLSs of MA are mutated and the IN, Vpr, and cPPT-CTS elements are removed displays no detectable nuclear import defect in HeLa cells independently of their cycling status. However, this mutant virus is partially impaired for nuclear entry in primary cells and more specifically in DCs and PBLs. We found that this partial defect is specified by the cPPT-CTS, while the 3 remaining elements seem to play no role in nuclear import. Thus, our study indicates that the central DNA flap specifies the most important role among the viral elements involved thus far in nuclear import. However, it also clearly indicates that the role played by the central DNA flap is not absolute and that its importance varies depending on the cell type, independently from the dividing status of the cell.
MATERIALS AND METHODS
Cells.
293T cells and HelaP4 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS). HeLaP4, referred to as HeLa cells throughout the text, express the CD4 and CXCR4 receptors, bear the HIV long terminal repeat (LTR)-ß-galactosidase reporter cassette, and were obtained from Pierre Charneau, Pasteur Institute, Paris, France. When indicated, HeLa cells were arrested in G1/S phase by treatment with 10 μg/ml aphidicolin (Sigma) for 24 h prior to infection. This treatment dramatically impaired infection with the MLV (not shown). Human primary cells were obtained from peripheral blood samples of healthy donors at the Etablissement Français du Sang de Lyon. Peripheral blood mononuclear cells were obtained after Ficoll and Percoll separation. Primary monocytes were subsequently purified by negative selection with a cocktail of hapten CD3, CD7, CD19, CD45RA, and CD56 anti-immunoglobulin E antibodies coupled to MACS microbeads (Miltenyi Biotec, France) to more than 90% purity (5). Monocytes were differentiated into macrophages by 3 h of serum starvation followed by culture for 4 days in 100 ng/ml of granulocyte-macrophage colony-stimulating factor (GM-CSF; AbCys, Paris, France) (48). Dendritic cells were differentiated from monocytes by culture for 4 days in 100 ng/ml of GM-CSF and interleukin 4 (IL-4; AbCys, Paris, France) (76). PBLs were stimulated with 1 μg/ml phytohemagglutinin (PHA; Sigma) and 150 U/ml interleukin 2 (IL-2; AIDS Reagent and Reference Program of the NIH) for 24 h prior to infection. All primary cells were maintained in RPMI 1640 medium supplemented with 10% FCS (5% for DCs).
Retroviral vectors.
HIV-1 vectors were produced upon transfection of 293T cells with the HIV-1 packaging construct 8.91-coding gag-pro-pol, tat, and rev genes but no other accessory genes (obtained from Didier Trono, Lausanne, Switzerland); the transfer vector coding for a viral minigenome-expressing green fluorescent protein (GFP) under the control of a cytomegalovirus (CMV) promoter (68); either the plasmid MD.G, coding for the vesicular stomatitis virus G (VSVg) envelope protein, or a plasmid coding for the HIV-1 NL4-3 envelope (obtained from Michael Malim, London, United Kingdom); and, when indicated, a Vpr-coding plasmid, at a ratio of 8:8:4:4. Mutations were introduced by standard molecular biology techniques into the packaging construct 8.91 to generate MA NLS−, containing mutations in both putative NLSs of the matrix protein (K26K27K28Y29K30L31K32/A26A27A28A29K30L31K32 and K110S111K112K113K114A115Q116/K110S111A112A113A114A115Q116; amino acid residue positions in MA are marked here) (16, 40); IN 1-50, lacking the catalytic core domain and DNA binding domain of the integrase (residues 51 to 288); and IN D116A, a class I integrase mutant containing a substitution in its catalytic site (80). The wild-type viral minigenome contains a cPPT-CTS cassette that has been removed in the ΔcPPT-CTS mutant genome upon ClaI/HpaI digestion.
Viral particles were produced by calcium phosphate transfection of 293T cells. Supernatants were collected 2 days after transfection and purified by ultracentrifugation through a 25% (wt/vol) sucrose cushion. Virions were resuspended in RPMI medium supplemented with 0.1 mM deoxynucleoside triphosphates (dNTPs), 10 mM MgCl2, and 6 mM CaCl2 and treated twice with DNase (RQ1 DNase, 33 u/ml; Promega) for 45 min at 37°C in order to remove plasmid DNA contaminations. The infectious titers of wild-type HIV-1 viral preparations were determined with HeLa cells. Wild-type and mutant HIV-1 viral preparations were normalized according to their p24CA contents by an enzyme-linked immunosorbent assay (ELISA).
Infections.
Cells were infected at a multiplicity of infection of 1 (HeLa cells) or 5 (PBLs, macrophages, and DCs), as estimated by comparison with WT virus. Virus was removed 2 h after infection, and cells were replenished with fresh medium. The percentage of infected GFP-positive cells was assessed by flow cytometry analysis 3 to 5 days postinfection. For PCR, cells were harvested at 24 to 48 h postinfection and treated with pronase for 10 min at room temperature. Pronase was inactivated with serum-rich medium, and cells were washed 2 times with phosphate-buffered saline (PBS) prior to lysis in 0.25% NP-40, 0.25% Tween 20, 2.5 mM MgCl2, 25 mM KCl, 5 mM Tris, pH 8.3. DNA from the cell lysates was then purified by phenol-chloroform extraction and was finally resuspended in water.
PCR analysis.
Semiquantitative PCRs were performed on serial threefold dilutions of cellular lysates DNA extracted as described above (routinely at least 5 dilutions per sample). PCR products were run on agarose gel and quantified based on the number of dilutions amplified for each sample. Cell lysates infected with WT HIV-1 diluted similarly served as a positive standard curve based on which mutants were quantified (values are thus expressed as percentages of the WT level). Various sets of primers were used to amplify either HIV-1 full-length (FL) DNA (GenBank accession no. M38432) (AC37 [CACTCCCAACGAAGACAAG] and AC 38 [CAGCAAGCCGAGTCCTGCGT]), HIV-1 2-LTR circles (AC 34 [TCCCAGGCTCAGATCTGGTCTAAC] and AC 35 [GCCTCAATAAAGCTTGCCTTG]), or HIV-1 1-LTR circles (CG130 [AATCCAGCGGACCTTCCTTCCCGCGGCCTGCTGCCGGC] and AC252 [CGCGTCTAGACTTTCGCTTTCAAGTCCCTGTTCG]). Values obtained here were normalized to those obtained with similar amplification of sample DNAs for actin DNA (ActinUp [CGAGAAGATGACCCAGGTG] and ActinDown [TGCCGCCAGACAGCACTGT]) or mitochondrial DNA (AC 391 [CTAAAGTGTGTTAATTAATTAATG] and AC 392 [CTAAGCGTTTTGAGCTGCATTGCTGCG]). With the primers used here in the case of 1-LTR amplification, it remains formally possible that single-stranded DNA molecules extended from the ends of linear vDNA in one PCR cycle anneal and are extended subsequently by virtue of their homology in the LTR. This may yield apparent 1-LTR circle amplification from what is instead linear vDNA. However, given that we have observed this phenomenon with vDNA inputs 100- to 1,000-fold higher than what routinely used here, the products obtained here represent bona fide 1-LTR circles. In each experiment and for each mutant, infections were carried out in parallel in the absence or presence of the reverse transcriptase (RT) inhibitors zidovudine (AZT; 10 μM) and dideoxyinosine (ddI; 20 μM) to determine the extent of plasmid DNA carryover in PCR assays, as exemplified in Fig. 2B. For each mutant, the values obtained in the presence of RT inhibitors were subtracted from those obtained in its absence. Statistical relevance was determined using a Student t test.
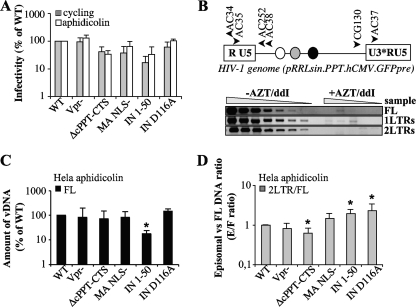
FIG. 2.
Characterization of the infectivity and nuclear import abilities of mutant viruses in HeLa cells. CA-normalized quantities of virion particles were used to infect cycling and arrested HeLa cells that had been treated with aphidicolin for 24 h prior to infection. (A) The quantity of infected GFP-positive cells was assessed at 3 days postinfection by flow cytometry (values are expressed as percentages of the WT level). For PCR, cells were lysed at 24 h postinfection, and the accumulation of reverse transcription products was analyzed by semiquantitative PCR. (B) Schematic representations of the HIV-1 genome used here and of the primers used to amplify the various viral DNA forms. For simplicity, the Rev responsive element, the cPPT-CTS, and the CMV-GFP-Wpre expression cassette are presented as white, gray, and black circles, respectively. Shown are typical results for PCRs performed with the various viral DNA forms, using threefold dilutions of cell lysates infected with WT HIV-1 in the presence or absence of RT inhibitors to ensure that quantification occurred within the linear range of the assay. Full-length (FL) products of reverse transcription (C) are expressed as percentages of the wild-type level. (D) The quantity of 2-LTR circles was normalized to the quantity of FL products for each mutant, and the 2-LTR-circle/FL-DNA ratio for the wild type was set to 1. “E/F ratio” represents the ratio of episomal to full-length products. Graphs are representative of 5 to 8 experiments. Values that are significantly different from those observed for the wild type are indicated by an asterisk (Student test; P < 0.05).
Western blot analysis.
Western blot analysis was carried out on normalized quantities of purified virions by using standard protocols. Antibodies were obtained from the AIDS reagent repository of the NIH, with the exception of the anti-IN antibody, which was a kind gift from J. F. Mouscadet (ENS, Cachan, France).
RESULTS
None of the known viral karyophilic elements alone is required for nuclear import of HIV-1 in G1/S-arrested HeLa cells.
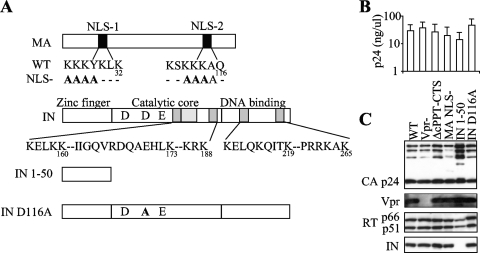
To determine the relative contribution of karyophilic elements in HIV-1 nuclear import, we mutated each of them individually in the context of HIV-1-derived lentiviral vectors (Fig. 1A). Vpr and the cPPT-CTS sequence were simply removed, the first by using a minimal version of an HIV-1 packaging vector devoid of vpr and the second by deletion on the viral minigenome. A large deletion was introduced in IN to remove all but its N-terminal 50 amino acids (IN 1-50) and thus all its NLSs described so far (9, 12, 37). Although large deletions at the 3′ end of polymerase (Pol) have been reported to negatively affect viral assembly (15), we found that this deletion allowed the production of viral particles. A similar large deletion that removed the globular head of MA did result in a dramatic decrease in particle production, contrary to what was described in a previous report (75). Thus, point mutations were introduced into the two previously described NLSs of MA (K26K27K28Y29K30L31K32/A26A27A28A29K30L31K32 and K110S111K112K113K114A115Q116/K110S111A112A113A114A115Q116) (16, 40).
FIG. 1.
Production of HIV-1 vectors mutated in all putative karyophilic elements. (A) Schematic representation of the HIV-1 packaging constructs used here. For simplicity, the ΔcPPT-CTS mutant is not represented here. (B) Virions were produced by transfection of 293T cells by using the indicated packaging construct, a GFP-coding viral minigenome, and the pantropic VSVg envelope protein. The quantity of viral particles released in the supernatant was determined by p24CA ELISA. For infections and Western blot analysis, viral particles were purified from the supernatant by ultracentrifugation through a 25% sucrose cushion and then resuspended and quantified by p24CA ELISA. (C) Normalized quantities of viral particles were analyzed by Western blotting using the indicated antibodies.
All mutant viruses were produced at levels comparable to those observed for the wild type upon DNA transfection of 293T cells (Fig. 1B and C). When analyzed by Western blotting, virions displayed no major processing defect, with the exception of the IN 1-50 mutant, which contained slightly reduced amounts of RT and a greater degree of accumulation of partially processed forms of Gag.
A virus containing a single point mutation in the catalytic site of IN (IN D116A) was also included in our analysis. This mutant is a class I IN integration-defective mutant and has been described before (11).
Quantities of mutant viruses were normalized by a p24CA ELISA and then used to infect cycling versus aphidicolin-treated G1/S-arrested HeLa cells (Fig. 2A). The percentage of GFP-positive cells was determined at 3 days postinfection by flow cytometry. Several mutants displayed a detectable infectivity defect in comparison to the WT (ΔcPPT-CTS, MA NLS−, and IN 1-50). Yet, despite these variations, all mutants behaved similarly on cycling and G1/S-arrested HeLa cells, and actually, a slight increase in the infectivity of arrested versus cycling cells was observed for most of the mutants. Integration-deficient mutant viruses (IN D116A and IN 1-50) yielded GFP-positive cells, despite a 3- to 6-fold infectivity defect in the latter. In the cases of these viruses, GFP expression is driven from extrachromosomal 1- and 2-LTR circles and is lost upon cell division (reference 11 and data not shown).
In order to analyze infection at a molecular level, the accumulation of viral DNA products in aphidicolin-treated HeLa cells was analyzed by PCR at 24 h postinfection by using primers that specifically amplified full-length (FL) viral DNA or 2-LTR circles (Fig. 2C and D). To ensure that amplification occurred within the linear range of the assay, multiple dilutions of sample DNAs were amplified and quantified over a standard curve obtained after amplification of DNA derived from cells infected with WT HIV-1 (Fig. 2B). A defect in 2-LTR circles can be consequent to a general defect in viral DNA accumulation or to a specific nuclear import defect. To this end, we chose to present first the amount of FL DNA synthesized upon infection (Fig. 2C) and then to normalize 2-LTR circles for variations in the amount of FL DNA (Fig. 2D). Under these conditions, a 2-LTR-circle/FL-DNA ratio below 1 indicates a nuclear import defect, while a ratio above 1 indicates increased accumulation of 2-LTR circles with respect to the WT level (as often observed with integrase-defective mutants).
FL DNA was detected at levels comparable to those observed for the WT for all mutants, with the exception of the IN 1-50 mutant, which displayed a 5-fold defect with respect to the WT level. This defect may be directly linked to the smaller amount of processed RT in viral particles (Fig. 1C). The ratio of 2-LTR circles versus FL DNA showed no significant decrease between the WT and most mutants, suggesting no specific nuclear import defect in these cells. However, a small but significant defect in the accumulation of 2-LTR circles was noticed for the cPPT-CTS mutant (1.6-fold), while, on the contrary, the two IN mutants displayed slight increases (2-fold) in 2-LTR-circle/FL-DNA ratio, in line with previous reports indicating an increased accumulation of 2-LTR circles in integration-defective IN mutants.
The cPPT-CTS plays a partial role in nuclear import of HIV-1 DNA that is stronger in primary cells, independent of their cell cycling status.
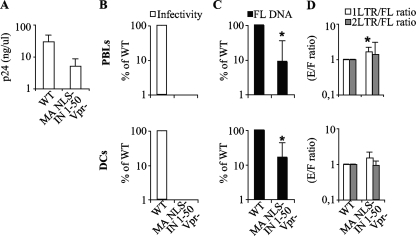
The intracellular milieu is known to influence retroviral replication, and this influence can be exerted in multiple manners: the presence or absence of the nuclear envelope, the presence of cell-type-specific positive or negative factors, the activation status of target cells, the size of the deoxynucleotide pool, and so forth. Thus, it is possible that the role of a given viral protein is mostly apparent in a specific cellular context. To explore this possibility, single mutants were tested on cycling, PHA-IL-2-stimulated peripheral blood lymphocytes (PBLs) as well as on nondividing macrophages (MDM) and dendritic cells (DCs), both differentiated from circulating blood monocytes upon incubation with granulocyte-macrophage colony stimulating factor (GM-CSF), alone or together with interleukin-4 (IL-4).
On these cells, Vpr did not exert a major effect on infectivity (Fig. 3A). The ΔcPPT-CTS deletion resulted in a 2- to 3-fold defect in all cell types and, among the cells tested here, was more pronounced in PBLs (3-fold). Similarly, the infectivity of the MA NLS− mutant was diminished 3- to 7-fold, and this defect was more apparent in PBLs.
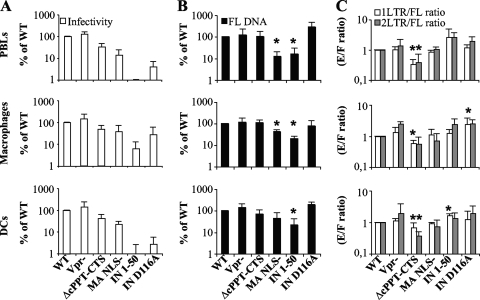
FIG. 3.
Characterization of the infectivity and nuclear import abilities of mutant viruses in primary cells. Normalized quantities of virions were used to infect PHA-IL-2-stimulated PBLs, monocyte-derived macrophages, and DCs. (A) The quantities of infected cells were assessed at 3 days (for PBLs and MDM) or 5 days (for DCs) postinfection by flow cytometry and are presented here normalized to the wild-type level. For PCR analysis, cells were lysed at 24 h (for PBLs and MDM) or 48 h (for DCs) postinfection, and the accumulation of reverse transcription products was analyzed with primers specific for full-length viral DNA (B) and 1- and 2-LTR circles (C). The quantity of viral DNA circles presented is normalized to the amount of FL DNA, thus taking into account possible differences at steps prior to nuclear import. Values that are significantly lower than those observed for the wild type are indicated by an asterisk (Student test; P < 0.05). The graphs present values obtained in 3 to 5 independent experiments.
The infectivities of IN mutants were considerably reduced compared to the WT level on PBLs and DCs and to a lesser extent on MDM. This defect was more dramatic for the IN 1-50 mutant (more that 100-fold on PBLs and DCs and 15-fold on MDM) than for the D116A single point mutant (30-fold in PBL and DCs and 3-fold in MDM), possibly revealing the multiple defects of the IN 1-50 mutant in addition to integration. The reason for the more drastic defect of this mutant in primary cells than in HeLa cells is presently unknown. However, we believe it may be due to smaller amounts of viral DNA present in infected primary cells, lower levels of transcriptional activity from unintegrated viral DNA in primary cells, or both.
At the molecular level, FL DNA accumulations were decreased significantly only in the cases of the MA NLS− and IN 1-50 mutants (Fig. 3B). These decreases were 5-fold in all cell types analyzed for the IN 1-50 mutant, while they varied according to cell type for the MA NLS− mutant (5-fold in PBLs and 2.5-fold in macrophages; the difference was not statistically significant in DCs, according to the Student test [P > 0.05]). Of note, no such defect has previously been observed for an MA NLS mutant during the early steps of viral infection. Next, nuclear import was analyzed by determining the ratio of 2- and 1-LTR circles to FL DNA with respect to the WT level (Fig. 3C). One-LTR circles are generated upon recombination by the cellular machinery of the 5′ and 3′ viral ends and were included in the analysis as a second testimony of viral DNA entry into the nucleus.
No significant differences were observed for viruses lacking Vpr or for viruses mutated in the NLSs of MA. Similarly, no nuclear import defect was observed for the IN 1-50 and D116A mutants, which, on the contrary, displayed increased quantities of extrachromosomal viral DNA circles. This is not unexpected, as increased viral DNA circle formation has been well described for integration-deficient viruses. However, a significant difference was observed after infection with the ΔcPPT-CTS mutant. Accumulation of 2- and 1-LTR circles was impaired 3-fold in PBLs and 2-fold in DCs and macrophages.
A virus lacking all known karyophilic viral elements is still able to enter the nuclei of nondividing cells.
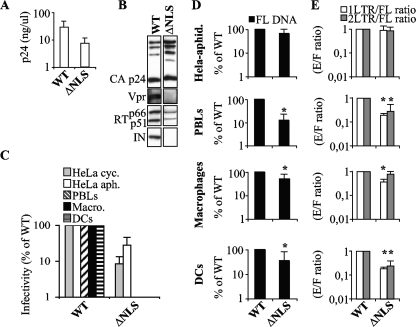
Next, a mutant virus that contained all mutations simultaneously (ΔNLS) was engineered. This mutant was produced with only a 3- to 10-fold defect with respect to the WT upon transfection of 293T cells and, as expected by the absence of IN, contained less RT than the WT (Fig. 4A and B). When normalized quantities of virions were used to infect the various cell types, the ΔNLS mutant retained a substantial infectivity on both cycling and G1/S-arrested HeLa cells (Fig. 4C) (4- to 12-fold lower than the WT level) but was totally noninfectious in PBLs, macrophages, and DCs. When FL DNA accumulation was analyzed, the ΔNLS mutant was found to accumulate less FL DNA in primary but not HeLa cells (2- to 8-fold in primary cells and 1.5-fold in HeLa cells [Fig. 4D]; only aphidicolin-treated cells are shown here). The 2-LTR-circle/FL-DNA and 1-LTR-circle/FL-DNA ratios did not diminish in HeLa cells, but they diminished 3-fold in PBLs and DCs and 1.3-fold in macrophages (Fig. 4E).
FIG. 4.
Characterization of the infectivity and nuclear import abilities of mutant virus lacking all potential karyophilic elements. (A) Quantities of wild type and ΔNLS (MA NLS−, IN 1-50, Vpr−, and ΔcPPT-CTS) mutant viral particles released in the supernatant were determined by p24CA ELISA after purification. (B) Normalized quantities of particles were analyzed by Western blotting using the indicated antibodies. (C) Virions were used to infect cycling and aphidicolin-treated HeLa cells, PHA-IL-2-stimulated PBLs, macrophages, and DCs. The percentages of infected GFP-positive cells were assessed 3 to 5 days following infection by flow cytometry and are presented here normalized to the WT level. (D and E) PCR analysis was carried out as described for Fig. 2, and the relative quantities of FL products of reverse transcription (black bars) and the 1-LTR-circle/FL-DNA (white bars) and 2-LTR-circle/FL-DNA (gray bars) ratios were determined by PCR. The results presented for viral DNA circle species are normalized to the amount of FL DNA synthesized for each mutant, as indicated. Each graph presents values obtained in 2 to 4 experiments. Values that are significantly lower than those observed for the wild type are indicated by an asterisk (Student test; P < 0.05).
The cPPT-CTS is the major determinant of the nuclear import defect of the ΔNLS mutant.
Given that the major defect observed in the accumulation of viral DNA nuclear forms seemed due to the absence of the cPPT-CTS sequence, this sequence was added back in the context of the ΔNLS mutant (thus mutated for IN, Vpr, and MA). The virus displayed only a minor production defect (Fig. 5A) but was noninfectious on PBLs and DCs (Fig. 5B). Despite a defect in FL DNA accumulation, no specific nuclear import defect could be observed, indicating that addition of the cPPT-CTS rescued the nuclear import defect observed in the NLS− mutant virus (Fig. 5C and D).
FIG. 5.
The cPPT-CTS is the major determinant of the nuclear import defect of the ΔNLS mutant. (A) The quantity of viral particles released in the supernatant following transfection of 293T cells with plasmid coding for the wild type and for a mutant virus mutated in its karyophilic elements, with the exception of the cPPT-CTS mutant (MA NLS−, IN 1-50, and Vpr), was determined by p24CA ELISA after purification. (B) Virions were used to infect PHA-IL-2-stimulated PBLs or DCs, and cells were analyzed by flow cytometry 3 to 5 days later. The graph presents data obtained with 3 to 4 independent experiments. For simplicity, data obtained with the ΔNLS mutant are omitted here and below. (C and D) PCR analysis was carried out as described in the legend to Fig. 2, and the relative quantities of FL products of reverse transcription (black bars) and the 1-LTR-circle/FL-DNA (white bars) and 2-LTR-circle/FL-DNA (gray bars) ratios were determined. Graphs present values obtained in 3 to 5 independent experiments. Values that are significantly lower than those observed for the wild type are indicated by an asterisk (Student test; P < 0.05).
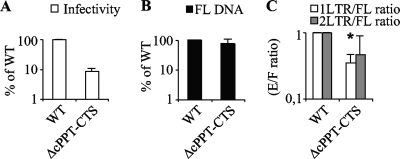
The cPPT-CTS exerts its role on nuclear import also upon HIV-1 X4 Env pseudotyping.
VSVg pseudotyping provides a useful tool when infection of different cell types is examined, as it allows infection independently from cell-specific receptor variations. Yet, as differences between entry pathways have been reported, it was important to perform similar experiments using a WT envelope protein. To this end, the WT and the ΔcPPT-CTS mutant were produced by transfection of 293T cells together with an X4 HIV-1 Env protein, normalized by protein content and tested on target PBLs. Similarly to what described previously for VSVg pseudotypes, an infectivity defect was observed for X4 pseudotyped particles. The defect was more pronounced than in the case of VSVg (10-fold) (Fig. 6A). No defect could be measured in FL-DNA accumulation (Fig. 6B), but clear defects in the 1- and 2-LTR-circle/FL-DNA ratios were again present (Fig. 6C).
FIG. 6.
Characterization of the infectivity and nuclear import abilities of the ΔcPPT-CTS mutant upon HIV-1 X4 Env pseudotyping. CA-normalized quantities of wild-type and ΔcPPT-CTS virions were pseudotyped with the HIV-1NL4-3 X4 envelope by transfection with appropriate coding DNAs, and purified virions were used to infect PHA-IL-2-stimulated PBLs. (A) The quantity of infected cells was assessed at 3 days postinfection by flow cytometry and is expressed as a percentage of the wild-type level. (B and C) PCR analysis was carried out as described for Fig. 3, and the relative quantities of FL products of reverse transcription (black bars) and the 1-LTR-circle/FL-DNA (white bars) and 2-LTR-circle/FL-DNA (gray bars) ratios were determined. Graphs present values obtained in 3 to 5 independent experiments. The asterisk indicates statistically significant differences from the WT level (Student test; P < 0.05).
The cPPT-CTS speeds the kinetics of viral DNA entry into the nucleus.
A defect in the accumulation of viral DNA forms into the nucleus can be due to several causes, such as nonintegrity of viral DNA ends or a delayed kinetics of entry into the nucleus. The lack of significant differences in joining of DNA in 2-LTR circles issued from the WT and the cPPT-CTS mutant (data not shown), as well as the similar defect in nuclear accumulation of 1-LTR circles (formed by recombination and thus less sensitive to viral end integrity), suggested that the latter possibility could be true. To test this hypothesis, PBLs were infected with the WT and the ΔcPPT-CTS mutant and analyzed up to 8 days postinfection (Fig. 7).
FIG. 7.
Kinetic analysis of the infection of primary PBLs by the ΔcPPT-CTS mutant. Virions were produced as described above and used to infect PHA-IL-2-stimulated PBLs. Cells were analyzed at the indicated time postinfection by flow cytometry or lysed and analyzed by PCR, as indicated. Graphs present values obtained in 3 independent experiments. The asterisk indicates statistically significant differences from the WT level (Student test; P < 0.05).
Under these conditions, the infectivity defect of the cPPT-CTS mutant was maintained at each time point analyzed. Similarly, the accumulation of FL DNA for this mutant continued to remain slightly impaired with respect to the WT level. However, the defect in the accumulation of 1- and 2-LTR circles subdued over time and became marginal by day 4 postinfection, suggesting that deletion of the cPPT-CTS element causes a delay in the nuclear import of viral DNA.
DISCUSSION
In the present study, we have investigated the interplay between the intracellular environment and nucleophilic elements of HIV-1 by analyzing the abilities of several mutants to accumulate episomal DNA following infection of various cell types. When it was feasible to do so, the viral elements were deleted, as in the cases of Vpr, the cPPT-CTS, and most of IN, but when the deletion impaired virion assembly, as in the case of MA, previously described NLSs were mutated individually (35, 43). This approach was used to generate mutant viruses of sufficient infectivity to allow nuclear import in a variety of cell types to be investigated by measuring the formation of viral DNA circles. As in most studies concerning HIV-1 nuclear import, we have used the measure of extrachromosomal viral DNA circles as a marker for nuclear import. The possibility that a decrease in 2-LTR circles results from circularization defects (for example, consequent to inefficient completion of viral DNA ends) rather than from nuclear import defects has recently been raised (65). To ensure that this was not the case here, we have analyzed both 1- and 2-LTR circles, formed via recombination and viral DNA end ligation, respectively. If the possibility that incomplete ends affected 2-LTR circle formation exists, this occurrence is unlikely in the case of 1-LTR circles, because recombination does not require intact ends. To further support this claim, we have failed to reveal differences in 2-LTR junctions formed upon infection with the WT and the cPPT-CTS mutant.
Our analysis revealed that among the elements examined here, the cPPT-CTS is the sole affecting nuclear entry. The requirement for this element during nuclear import is moderate and is observed in cycling as well as noncycling cells.
Our results differ somewhat from the sole report that analyzed all 4 of the nucleophilic elements of HIV-1 together. In this report, elements were either deleted or swapped for their MLV counterparts to give raise to chimeric MLV/HIV viruses (Vpr with cPPT-CTS and MA with IN, respectively) equally infectious on cycling and arrested HeLa cells (91). This result led to the conclusion that none of the examined elements were required for cell cycle-independent infection of HIV-1. However, the extremely low level of infectivity of these chimeric viruses precluded an in depth analysis on cells other than HeLa cells. Given that the intracellular environment influences most steps of the viral life cycle, we wanted to explore in more detail the behavior of mutant viruses in various cell types. Indeed, we have previously shown that even the MLV gammaretrovirus undergoes nuclear import in certain nondividing cell types but not in others (48) (in primary macrophages but not in dendritic cells, for instance). Besides, certain viral mutants display a cell-type-dependent phenotype at steps other than nuclear entry in nondividing cells (92), suggesting that cell cycle independence may be the sum of multiple viral abilities, in addition to the ability to traverse the nuclear membrane.
In this study, we have engineered mutants of sufficient infectivity to allow the analysis of nuclear import in a wide comparison among primary cells that were either cycling or growth arrested. Under these conditions, MA, IN, and Vpr played no detectable role in nuclear import, alone or combined. However, a clear, albeit moderate, effect was observed for the cPPT-CTS. This finding is in agreement with previous observations on the temperate (24, 26, 61, 66), rather than extreme (4, 6, 7, 31, 78, 84, 95), role of this cis element in nuclear import. Contrary to other reports, however, these defects were more apparent in primary cells than in HeLa cells, independent of their cycling status (4, 6, 7, 84). We believe that this may depend on their peculiar susceptibility to viral infection in comparison to that observed for primary cell lines.
Interestingly, the cPPT-CTS has been suggested to promote faster kinetics of viral DNA entry into the nucleus by overcoming a saturation block in nuclear entry (87), and more recently, the cPPT-CTS has been proposed to signal the end of reverse transcription and the start of uncoating (8). While we have not obtained evidence of a saturation block in our assays (as similar results were obtained for different viral inputs [not shown]), our results indicate that the cPPT-CTS could play a kinetic role in facilitating faster transition from the cytoplasm to the nucleus. This may in turn protect viral DNA once in the nucleus, explaining why, even when the nuclear import defect decreases over time, the infectivity defect of cPPT-CTS-deleted viruses persists.
The fact that nuclear entry defects were observed in viruses devoid of the same element in cycling and arrested cells may suggest that an active nuclear import through the NPC is required even in cycling cells, as suggested by others (8, 54). This hypothesis is interesting because it argues that passage through the nuclear pore serves a purpose in subsequent postnuclear events, as it occurs for other viruses (19, 63, 89), and that the requirement for this step is common to all cell types.
Although our results indicate a role for the cPPT-CTS in nuclear entry of viral DNA, they nonetheless argue for the presence of other elements, given that even in the absence of this element, viral entry into the nucleus does occur. Recent evidence indicates that CA may be involved in nuclear import (14, 25, 90). CA plays a pleiotropic role during the early steps of viral infection, and mutations in CA can affect reverse transcription, uncoating, and postnuclear entry events (74, 87, 88, 92). However, it is true that one of the major differences between complex and simple retroviruses is the relative paucity of CA in viral nucleoprotein complexes derived from the former. Since most of the CA seems to be lost during the uncoating of HIV-1 (18, 28, 29, 67), it seems unlikely that CA is involved directly in nuclear entry. Thus, CA may influence nuclear import indirectly, either by unmasking nuclear entry signals in other components of viral cores (as for RT or NC) or by leaving a place for recruitment of cellular proteins that piggyback viral DNA complexes into the nucleus. This possibility has not been explored fully, but a number of DNA binding proteins, such as BAF, HMGA1, and LEDGF, are associated with viral nucleoprotein complexes. These proteins normally migrate into the nucleus upon translation and may thus associate directly with viral DNA in the cytoplasm and transport it along into the nucleus. Furthermore, viral complexes of HIV-1 have been shown to use modified tRNAs to attain the nucleus (94), a finding that suggests the possibility that cellular factors intervene in the nuclear import of HIV-1.
Lastly, we have shown that a mutant in which all but the N-terminal zinc binding domain of IN is removed assembles into virions that are infectious at least in permissive HeLa cells. The N-terminal 50 residues of IN seem to provide a nonspecific binding site for DNA. This fragment can be substituted for heterologous zinc finger domains of similar lengths with no apparent phenotypic differences (not shown), indicating that IN can be entirely removed from virion particles. This observation is interesting because it suggests that IN is not an obligatory structural component of viral nucleoprotein complexes, given that infectious viral particles can be obtained in its absence. However, the infectivity of this mutant is drastically diminished in most primary cells, except in macrophages, where this defect is contained within 15 folds. This defect seems more profound than the reverse transcription problem displayed by this mutant. We believe that this may result from a combination of two defects, related to viral DNA synthesis and variability of expression of nonintegrated viral DNA in these cells. Indeed, the strong GFP expression of the IN D116A mutant observed in HeLa cells is drastically diminished in PBLs, DCs, and, to a lesser extent, macrophages. The IN 1-50 mutant displays a clear reverse transcription defect that could be due to the lower observed rates of incorporation of mature RT into virion particles (and thus to a Gag-Pro-Pol processing defect), to a direct effect of IN on reverse transcription, or to both. An interaction between RT and IN in vitro has been reported (42, 82), and certain mutations in IN affect reverse transcription (97).
However, this chimera does not display a defect in nuclear entry in any of the cell types examined. This finding argues against a major functional role for cellular partners of IN during the phase of nuclear import per se (3, 20, 41) and suggests that these cofactors may intervene in the optimization of events that immediately precede or succeed nuclear entry of viral DNA.
Acknowledgments
We thank Jeanine Bernaud and Dominique Rigal for help with blood sample collection. We thank the AIDS Research and Reference Reagent Program of the NIH and J.-F. Mouscadet for material used in this study. We are also indebted to Loraine Jarrosson-Wuilleme for initial engineering of some of the mutants used here.
A.C. acknowledges the support of the CNRS. This work received the support of Sidaction and the ANRS.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Agostini, I., S. Popov, J. Li, L. Dubrovsky, T. Hao, and M. Bukrinsky. 2000. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp. Cell Res. 259:398-403. [DOI] [PubMed] [Google Scholar]
- 2.Ao, Z., K. R. Fowke, E. A. Cohen, and X. Yao. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ao, Z., G. Huang, H. Yao, Z. Xu, M. Labine, A. W. Cochrane, and X. Yao. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282:13456-13467. [DOI] [PubMed] [Google Scholar]
- 4.Ao, Z., X. Yao, and E. A. Cohen. 2004. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J. Virol. 78:3170-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arfi, V., L. Riviere, L. Jarrosson-Wuilleme, C. Goujon, D. Rigal, J. L. Darlix, and A. Cimarelli. 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 82:6557-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arhel, N., S. Munier, P. Souque, K. Mollier, and P. Charneau. 2006. Nuclear import defect of human immunodeficiency virus type 1 DNA flap mutants is not dependent on the viral strain or target cell type. J. Virol. 80:10262-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arhel, N. J., S. Souquere-Besse, and P. Charneau. 2006. Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: intracellular visualization at ultrastructural resolution levels. Retrovirology 3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arhel, N. J., S. Souquere-Besse, S. Munier, P. Souque, S. Guadagnini, S. Rutherford, M. C. Prevost, T. D. Allen, and P. Charneau. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 26:3025-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armon-Omer, A., A. Graessmann, and A. Loyter. 2004. A synthetic peptide bearing the HIV-1 integrase 161-173 amino acid residues mediates active nuclear import and binding to importin alpha: characterization of a functional nuclear localization signal. J. Mol. Biol. 336:1117-1128. [DOI] [PubMed] [Google Scholar]
- 10.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. [DOI] [PubMed] [Google Scholar]
- 11.Berger, G., C. Goujon, J. L. Darlix, and A. Cimarelli. 2009. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1-derived vectors. Gene Ther. 16:159-163. [DOI] [PubMed] [Google Scholar]
- 12.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 13.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 14.Brun, S., M. Solignat, B. Gay, E. Bernard, L. Chaloin, D. Fenard, C. Devaux, N. Chazal, and L. Briant. 2008. VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. U. S. A. 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U. S. A. 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell, E. M., and T. J. Hope. 2005. Gene therapy progress and prospects: viral trafficking during infection. Gene Ther. 12:1353-1359. [DOI] [PubMed] [Google Scholar]
- 20.Christ, F., W. Thys, J. De Rijck, R. Gijsbers, A. Albanese, D. Arosio, S. Emiliani, J. C. Rain, R. Benarous, A. Cereseto, and Z. Debyser. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192-1202. [DOI] [PubMed] [Google Scholar]
- 21.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 22.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 23.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 24.De Rijck, J., and Z. Debyser. 2006. The central DNA flap of the human immunodeficiency virus type 1 is important for viral replication. Biochem. Biophys. Res. Commun. 349:1100-1110. [DOI] [PubMed] [Google Scholar]
- 25.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebina, H., J. Aoki, S. Hatta, T. Yoshida, and Y. Koyanagi. 2004. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 6:715-724. [DOI] [PubMed] [Google Scholar]
- 28.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 32.Fouchier, R. A., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 33.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed, E. O., and M. A. Martin. 1994. HIV-1 infection of non-dividing cells. Nature 369:107-108. [DOI] [PubMed] [Google Scholar]
- 36.Fritz, J. V., P. Didier, J. P. Clamme, E. Schaub, D. Muriaux, C. Cabanne, N. Morellet, S. Bouaziz, J. L. Darlix, Y. Mely, and H. de Rocquigny. 2008. Direct Vpr-Vpr interaction in cells monitored by two photon fluorescence correlation spectroscopy and fluorescence lifetime imaging. Retrovirology 5:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulizia, J., M. P. Dempsey, N. Sharova, M. I. Bukrinsky, L. Spitz, D. Goldfarb, and M. Stevenson. 1994. Reduced nuclear import of human immunodeficiency virus type 1 preintegration complexes in the presence of a prototypic nuclear targeting signal. J. Virol. 68:2021-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 41.Hearps, A. C., and D. A. Jans. 2006. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 398:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. U. S. A. 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyman, T., M. Wilhelm, and F. X. Wilhelm. 2003. The central PPT of the yeast retrotransposon Ty1 is not essential for transposition. J. Mol. Biol. 331:315-320. [DOI] [PubMed] [Google Scholar]
- 45.Iijima, S., Y. Nitahara-Kasahara, K. Kimata, W. Zhong Zhuang, M. Kamata, M. Isogai, M. Miwa, Y. Tsunetsugu-Yokota, and Y. Aida. 2004. Nuclear localization of Vpr is crucial for the efficient replication of HIV-1 in primary CD4+ T cells. Virology 327:249-261. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda, T., H. Nishitsuji, X. Zhou, N. Nara, T. Ohashi, M. Kannagi, and T. Masuda. 2004. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 78:11563-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacquot, G., E. Le Rouzic, A. David, J. Mazzolini, J. Bouchet, S. Bouaziz, F. Niedergang, G. Pancino, and S. Benichou. 2007. Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology 4:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarrosson-Wuilleme, L., C. Goujon, J. Bernaud, D. Rigal, J. L. Darlix, and A. Cimarelli. 2006. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 80:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkins, Y., M. McEntee, K. Weis, and W. C. Greene. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamata, M., and Y. Aida. 2000. Two putative alpha-helical domains of human immunodeficiency virus type 1 Vpr mediate nuclear localization by at least two mechanisms. J. Virol. 74:7179-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamata, M., Y. Nitahara-Kasahara, Y. Miyamoto, Y. Yoneda, and Y. Aida. 2005. Importin-alpha promotes passage through the nuclear pore complex of human immunodeficiency virus type 1 Vpr. J. Virol. 79:3557-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karageorgos, L., P. Li, and C. Burrell. 1993. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retroviruses 9:817-823. [DOI] [PubMed] [Google Scholar]
- 53.Karni, O., A. Friedler, N. Zakai, C. Gilon, and A. Loyter. 1998. A peptide derived from the N-terminal region of HIV-1 Vpr promotes nuclear import in permeabilized cells: elucidation of the NLS region of the Vpr. FEBS Lett. 429:421-425. [DOI] [PubMed] [Google Scholar]
- 54.Katz, R. A., J. G. Greger, P. Boimel, and A. M. Skalka. 2003. Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J. Virol. 77:13412-13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khiytani, D. K., and N. J. Dimmock. 2002. Characterization of a human immunodeficiency virus type 1 pre-integration complex in which the majority of the cDNA is resistant to DNase I digestion. J. Gen. Virol. 83:2523-2532. [DOI] [PubMed] [Google Scholar]
- 56.Konig, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kootstra, N. A., and H. Schuitemaker. 1999. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology 253:170-180. [DOI] [PubMed] [Google Scholar]
- 58.Le Rouzic, E., A. Mousnier, C. Rustum, F. Stutz, E. Hallberg, C. Dargemont, and S. Benichou. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 277:45091-45098. [DOI] [PubMed] [Google Scholar]
- 59.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Limón, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Limón, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu, R., A. Limon, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lux, K., N. Goerlitz, S. Schlemminger, L. Perabo, D. Goldnau, J. Endell, K. Leike, D. M. Kofler, S. Finke, M. Hallek, and H. Buning. 2005. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J. Virol. 79:11776-11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahalingam, S., V. Ayyavoo, M. Patel, T. Kieber-Emmons, and D. B. Weiner. 1997. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J. Virol. 71:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mannioui, A., E. Nelson, C. Schiffer, N. Felix, E. Le Rouzic, S. Benichou, J. C. Gluckman, and B. Canque. 2005. Human immunodeficiency virus type 1 KK26-27 matrix mutants display impaired infectivity, circularization and integration but not nuclear import. Virology 339:21-30. [DOI] [PubMed] [Google Scholar]
- 66.Marsden, M. D., and J. A. Zack. 2007. Human immunodeficiency virus bearing a disrupted central DNA flap is pathogenic in vivo. J. Virol. 81:6146-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 69.Nitahara-Kasahara, Y., M. Kamata, T. Yamamoto, X. Zhang, Y. Miyamoto, K. Muneta, S. Iijima, Y. Yoneda, Y. Tsunetsugu-Yokota, and Y. Aida. 2007. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J. Virol. 81:5284-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 72.Popov, S., M. Rexach, L. Ratner, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347-13352. [DOI] [PubMed] [Google Scholar]
- 73.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi, M., R. Yang, and C. Aiken. 2008. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J. Virol. 82:12001-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romani, N., D. Reider, M. Heuer, S. Ebner, E. Kampgen, B. Eibl, D. Niederwieser, and G. Schuler. 1996. Generation of mature dendritic cells from human blood. An improved method with special regard to clinical applicability. J. Immunol. Methods 196:137-151. [DOI] [PubMed] [Google Scholar]
- 77.Sherman, M. P., C. M. de Noronha, M. I. Heusch, S. Greene, and W. C. Greene. 2001. Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol. 75:1522-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sirven, A., F. Pflumio, V. Zennou, M. Titeux, W. Vainchenker, L. Coulombel, A. Dubart-Kupperschmitt, and P. Charneau. 2000. The human immunodeficiency virus type-1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood 96:4103-4110. [PubMed] [Google Scholar]
- 79.Suzuki, Y., and R. Craigie. 2007. The road to chromatin—nuclear entry of retroviruses. Nat. Rev. Microbiol. 5:187-196. [DOI] [PubMed] [Google Scholar]
- 80.Taddeo, B., W. A. Haseltine, and C. M. Farnet. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taguchi, T., M. Shimura, Y. Osawa, Y. Suzuki, I. Mizoguchi, K. Niino, F. Takaku, and Y. Ishizaka. 2004. Nuclear trafficking of macromolecules by an oligopeptide derived from Vpr of human immunodeficiency virus type-1. Biochem. Biophys. Res. Commun. 320:18-26. [DOI] [PubMed] [Google Scholar]
- 82.Tasara, T., G. Maga, M. O. Hottiger, and U. Hubscher. 2001. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 507:39-44. [DOI] [PubMed] [Google Scholar]
- 83.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.von Schwedler, U., R. S. Kornbluth, and D. Trono. 1994. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 91:6992-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wacharapornin, P., D. Lauhakirti, and P. Auewarakul. 2007. The effect of capsid mutations on HIV-1 uncoating. Virology 358:48-54. [DOI] [PubMed] [Google Scholar]
- 89.Whittaker, G. R. 2003. Virus nuclear import. Adv. Drug Deliv. Rev. 55:733-747. [DOI] [PubMed] [Google Scholar]
- 90.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamashita, M., and M. Emerman. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamashita, M., O. Perez, T. J. Hope, and M. Emerman. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 3:1502-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zaitseva, L., P. Cherepanov, L. Leyens, S. J. Wilson, J. Rasaiyaah, and A. Fassati. 2009. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zaitseva, L., R. Myers, and A. Fassati. 2006. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 4:e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 96.Zennou, V., C. Serguera, C. Sarkis, P. Colin, E. Perret, J. Mallet, and P. Charneau. 2001. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19:446-450. [DOI] [PubMed] [Google Scholar]
- 97.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zielske, S. P., and M. Stevenson. 2005. Importin 7 may be dispensable for human immunodeficiency virus type 1 and simian immunodeficiency virus infection of primary macrophages. J. Virol. 79:11541-11546. [DOI] [PMC free article] [PubMed] [Google Scholar]