Abstract
Moloney murine leukemia virus (MoMLV) Gag utilizes its late (L) domain motif PPPY to bind members of the Nedd4-like ubiquitin ligase family. These interactions recruit components of the cell's budding machinery that are critical for virus release. MoMLV Gag contains two additional L domains, PSAP and LYPAL, that are believed to drive residual MoMLV release via interactions with cellular proteins Tsg101 and Alix, respectively. We found that overexpression of Tsg101 or Alix failed to rescue the release of PPPY-deficient MoMLV via these other L domains. However, low-level expression of the ubiquitin ligase Itch potently rescued the release and infectivity of MoMLV lacking PPPY function. In contrast, other ubiquitin ligases such as WWP1, Nedd4.1, Nedd4.2, and Nedd4.2s did not rescue this release-deficient virus. Efficient rescue required the ubiquitin ligase activity of Itch and an intact C2 domain but not presence of the endophilin-binding site. Additionally, we found Itch to immunoprecipitate with MoMLV Gag lacking the PPPY motif and to be incorporated into rescued MoMLV particles. The PSAP and LYPAL motifs were dispensable for Itch-mediated virus rescue, and their absence did not affect the incorporation of Itch into the rescued particles. Itch-mediated rescue of release-defective MoMLV was sensitive to inhibition by dominant-negative versions of ESCRT-III components and the VPS4 AAA ATPase, indicating that Itch-mediated correction of MoMLV release defects requires the integrity of the host vacuolar sorting protein pathway. RNA interference knockdown of Itch suppressed the residual release of the MoMLV lacking the PPPY motif. Interestingly, Itch stimulation of the PPPY-deficient MoMLV release was accompanied by the enhancement of Gag ubiquitination and the appearance of new ubiquitinated Gag proteins in virions. Together, these results suggest that Itch can facilitate MoMLV release in an L domain-independent manner via a mechanism that requires the host budding machinery and involves Gag ubiquitination.
Retroviruses require access to the host budding machinery to exit the cell (5, 13, 40). To this end, retroviral Gag polyproteins use short sequences called late (L) domains to promote virus release by recruiting members of the host vacuolar protein sorting (vps) machinery. In the cell, vps proteins are involved in membrane dynamics that facilitate the separation of daughter cells at the completion of cytokinesis (9, 39) and the budding of vesicles into endosomal compartments or multivesicular bodies (MVB) (2, 23), a process topologically similar to virus budding (57). Class E vps proteins are organized into three heteromeric endosomal complexes (called endosomal sorting complexes) required for transport, namely, ESCRT-I, -II, and -III (2). In the current model for budding, sequential recruitment of ESCRT components on the cytoplasmic face of the membrane facilitates vesicle invagination into MVB compartments and viral egress from the cell (2). The disassembly of ESCRT-III components is catalyzed by the activity of VPS4 AAA-type ATPase, which in turn is presumed to trigger membrane fission events (3, 50). Any disruption in this sequence, such as mutations in L domain motifs or dominant-negative interference with the function of ESCRT-III members or the VPS4 ATPase, adversely affects virus release. This indicates that Gag interactions with the ESCRT machinery are necessary for virus budding and separation from the cell (19, 21, 34, 49, 57).
Currently, three types of L domain motifs have been identified: PT/SAP, LYPXnL, and PPPY. All retroviral Gag molecules contain at least one of these motifs, as multiple L domains are believed to synergistically function to ensure efficient viral release. Moloney murine leukemia virus (MoMLV) Gag carries all three L domain motifs, PSAP, LYPAL, and PPPY, which bind the vps protein Tsg101, the ESCRT-associated protein Alix (46), and members of the Nedd4-ubiquitin ligase family (33), respectively. In HIV-1, the PTAP motif in the p6 region of Gag binds Tsg101 (16, 56), which functions in viral budding (16, 35) as a member of ESCRT-I (16, 36, 57). The LYPXnL motif is also located in p6 and is the binding site for Alix (49, 57), a protein that also interacts with the nucleocapsid domain of HIV-1 Gag (14, 43) and links Gag to components of ESCRT-III (14). Similarly, the human T-cell leukemia virus (HTLV-I) Gag carries PPPY and PTAP L domains, which both contribute to efficient HTLV-1 release (6, 7, 21). The PPPY L domain motif, which is found in numerous retroviral Gag polyproteins (6, 7, 19, 21, 27, 28, 61, 62), plays a critical role in MoMLV release, as mutations disrupting its sequence lead to significant decreases in virus budding and release (33, 62). PSAP and LYPAL, the additional L domain motifs, are believed to serve little to no role in the release of MoMLV Gag virus-like particles (45, 46).
The role of Nedd4-like ubiquitin ligases in budding events was initially established by data obtained with the yeast Nedd4-like ligase Rsp5, an enzyme that ubiquitinates surface proteins, thus signaling their incorporation into the MVB pathway (26). From retroviral budding studies, multiple findings support the notion that Nedd4-like ubiquitin ligases link PPPY-containing Gag proteins to the host ESCRT machinery. For example, mutations in the PPPY motif or expression of dominant-negative versions of Nedd4-like ligases resulted in budding defects similar to those seen upon interference with the function of ESCRT-III members (7, 21, 27, 28, 33, 62). Overexpression of Nedd4-like ligases WWP1 and Itch corrected the budding defects of a MoMLV PPPY mutant that retained residual binding to both ligases (33). Also, when transplanted to a heterologous retroviral Gag, the PPPY L domain creates a requirement for Nedd4-like ubiqutin ligase activity to facilitate viral release that is dependent on the presence of a functional ESCRT pathway (63). Collectively, these observations support the notion that Nedd4-like ubiquitin ligases link retroviral Gag polyproteins to components of the ESCRT pathway necessary for budding.
Both endosomal and viral budding require the ubiquitin conjugation properties of Nedd4-like ligases, indicating that ubiquitin transfer to a key protein(s) is necessary to promote budding. A role for Gag ubiquitination in viral budding has been suggested (8, 20, 22, 48). In fact, ubiquitin attachment to equine infectious anemia virus (EIAV) Gag can substitute for the lack of L domains and rescue viral budding (25), suggesting that ubiquitin molecules conjugated to Gag can signal the recruitment of the host ESCRT machinery. For feline immunodeficiency virus, efficient budding seems to require L domain-dependent ubiquitination of Gag proteins (8) that is independent of the L domain ability to directly recruit Nedd4-like ubiquitin ligases (i.e., by means of the PT/SAP L domain motif) (8). Similarly, ubiquitination of HTLV-1 Gag was also shown to play a significant role in viral release (22). Conversely, data arguing in favor of a role for the ubiquitination of transacting factors, but not Gag, in the facilitation of viral budding have also been reported (10, 63). Thus Gag polyproteins recruit, in a PPPY-dependent or -independent manner, enzymatically active Nedd4-like ubiquitin ligases that conjugate ubiquitin molecules to Gag or to Gag-binding host factors. Such interactions, whether direct or indirect, are believed to link the viral protein to the host ESCRT pathway and facilitate release.
In addition to the well-characterized cellular proteins that bind primary L domain motifs, retroviral Gag can recruit other host factors, either via secondary L domains or independently of L domains (10, 24, 29, 55, 59). These cellular factors are believed to promote virus production by facilitating Gag protein trafficking to the plasma membrane and/or providing additional L domain-independent links to the host vps pathway. Examples of these parallel pathways are illustrated in the rescue of a budding-defective HIV-1 lacking the PTAP domain by overexpression of Alix (15, 54) and in the remarkably potent rescue of HIV-1 lacking all known L domains by the overexpression of Nedd4.2s, a Nedd4.2 isoform that belongs to the Nedd4-like ubiquitin ligase family (10, 55). In this study, we sought to identify host cell factors that rescue budding defects of the MoMLV mutant lacking the PPPY motif (MoMLV AAAY mutant). Our studies provide evidence that Itch overexpression rescued budding and infectivity defects of the MoMLV AAAY mutant virus, indicating that Gag can recruit the ubiquitin ligase Itch in an L domain-independent manner to facilitate MoMLV release via a mechanism that involves Gag ubiquitination.
MATERIALS AND METHODS
Proviral constructs.
In this study, we used the proviral clone pNCA, which contains an infectious copy of MoMLV and was a generous gift from Steve Goff (11). Mutations in pNCA were introduced using the QuikChange kit (Stratagene) and the following primers: for the AAAY mutation, 5′-CTACTTACAGAAGACGCCGCGGCTTATAGGGACCCAAGACCACCC-3′; for the ASAA mutation, 5′-CCGCCTCCTCTTCCTGCATCCGCCGCGTCTCTCCCCCTTGAACC-3′; and for the AAPAL mutation, 5′-CCGCCTCGATCCTCCGCTGCTCCAGCCCTCACTCCTTCTCTAGGC-3′.
Protein expression plasmids.
Nedd4.1 cDNA (gene name, KIAA0093; Swiss-Prot accession no. P46934) was obtained from the Kazusa DNA Institute, Chiba, Japan. Nedd4.1 was amplified by PCR and cloned into pHM6 (Roche) between the HindIII and KpnI sites. Nedd4.2 (KIAA0439) cDNA was purchased from Open Biosystems and cloned into pHM6 between HindIII and KpnI sites. WWP1 (accession number NM_007013), a gift from the Miyazano Laboratory (Fred Hutchinson Cancer Research Institute), and Itch (accession number NM_031483), a gift from Annie Angers (University of Montreal), were cloned between the NotI and KpnI sites of the pHM6 plasmid. The Alix/AIP-1 and Nedd4.2s expression vectors were described by Dussupt et al. (14). The Tsg101 expression plasmid was a gift from Limin Li and Stanley Cohen (Stanford University) and was previously described by Li et al. (30). The expression vector for the VPS4aK173Q mutant was generously provided by Wes Sundquist (University of Utah) and was previously described by von Schwedler et al. (57). Endophilin expression vector was a generous gift from Steve Goff (Columbia University) and was previously described by Wang et al. (59).
Virus release analysis.
293T cells or HeLa cells (2 to 3 million) were seeded in T25 flasks and transfected the following day by using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. At 32 h after transfection, cell culture medium was filtered through a 0.45-μm-pore-size filter and pelleted in an SW41 rotor at 151,000 × g for 1 h 15 min on a 20% sucrose (wt/vol)-phosphate-buffered saline (PBS) cushion. After centrifugation, the supernatant was removed and the virus pellet was resuspended in cell lysis buffer (1% [vol/vol] NP-40, 50 mM Tris [pH 8.0], 150 mM NaCl, and a protease inhibitor cocktail [Roche]) and boiled in 1× Laemmli buffer. Cells were scraped in cold PBS and pelleted at 1,000 × g; then, cell pellets were lysed in 250 μl of the aforementioned lysis buffer for 20 min on ice. Cell lysates were cleared by centrifugation, and the supernatants were collected and boiled in 1× Laemmli sample buffer. Proteins in virions and cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using a goat anti-MoMLV CA antibody. Protein expression was detected using monoclonal anti-hemagglutinin (anti-HA) or anti-Flag antibodies (Sigma).
MoMLV infectivity assay.
Virus particles were produced using transient transfection of 293T cells and Lipofectamine 2000 according to the manufacturer's instructions. Infectivity was then measured by infecting NIH 3T3 cells. The XC overlay test was used to assess susceptibility to virus infection. Subconfluent cultures of NIH 3T3 cells were infected with virus dilutions in the presence of Polybrene (Aldrich, Milwauke, WI) (4 μg/ml). After 3 days, cultures were irradiated with UV light from germicidal bulbs to kill the cells and were then overlaid with 106 rat XC cells per 60-mm-diameter dish. Plates were fixed and stained 3 days later and were then examined for plaques of XC cell syncytia to identify and enumerate foci of infected cells.
RNAi knockdown of Itch.
Cells were seeded at a density of 2 × 106 cells/ml (293T) and were transfected 5 h after plating using RNA interference (RNAi) oligonucleotides at a concentration of 20 μM for targeting cellular Itch (52) and Lipofectamine 2000 according to the manufacturer's instructions. The RNAi sequences used were 5′-CAAGAGCUAUGAGCAACUGAA-3′, 5′-AUGGGUAGCCUCACCAUGAAA-3′, and 5′-UGCCGCCGACAAAUACAAAUA-3′. After 24 h, cells were reseeded and transfected for a second time with RNAi oligonucleotides. At 24 h later, cells were harvested and lysed in lysis buffer, and their protein content was analyzed by SDS-PAGE and Western blotting using a rabbit anti-Itch antibody from Abcam.
MoMLV ubiquitination assays.
293T cells (2 × 106 cells per 25-cm2 dish) (80% confluent) were cotransfected using 1 μg of wild-type (wt) full-length MoMLV (pNCA) expression plasmid or the AAAY mutant version, 1.5 μg of HA-tagged ubiquitin expression plasmid (a generous gift from Arianna Calistri, University of Padova, Padova, Italy), a Flag-tagged Itch expression plasmid or the control empty vector, and Lipofectamine 2000 (Invitrogen) according to manufacturer instructions. At 40 h posttransfection, culture medium was filtered through 0.45-μm-pore-size syringe filters and virions were pelleted at 151,000 × g for 1 h on a 20% (wt/vol)-PBS sucrose cushion. Virion pellets were lysed in radioimmunoprecipitation assay (RIPA) buffer (0.5% NP-40, 50 mM HEPES [pH 7.3], 150 mM NaCl, 2 mM EDTA, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 0.5 μM dithiothreitol, and Complete protease inhibitor cocktail [Roche]) and incubated with protein A- and G-coated Sepharose beads (Pierce). Immunoprecipitations were carried out using a rat anti-MoMLV capsid antibody (ATCC) on virion lysates at 4°C. The beads were then washed five times with RIPA buffer, and protein complexes were eluted in 50 μl of sample buffer. Immunoprecipitates and cell lysates (input fractions) were analyzed by SDS-PAGE and immunoblotting using a mouse anti-HA antibody (Sigma) (1:10,000) to detect MoMLV Gag ubiquitination and with goat anti-MoMLV capsid to detect Gag and Gag-derived proteins.
RESULTS
Generation and analysis of MoMLV L domain mutants.
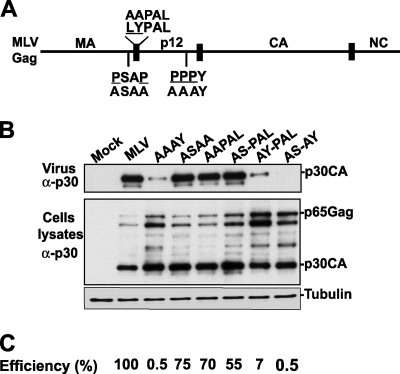
MoMLV contains a PPPY L domain that binds members of the Nedd4-like ubiquitin family and is considered to be the primary motif required for MoMLV budding and release (33, 62). MoMLV also harbors two additional L domain motifs, PSAP and LYPAL, which were found to play secondary roles in MoMLV release in the context of a green fluorescent protein (GFP)-tagged MoMLV Gag (46) (Fig. 1A). We examined the roles of PSAP and LYPAL motifs by the use of an infectious full-length proviral MoMLV construct (11) carrying mutations in either the PSAP motif (ASAA mutant) or the LYPAL motif (AAPAL mutant). These mutations are expected to prevent interactions with Tsg101 or Alix proteins, respectively. The ASAA and AAPAL mutations had only a modest effect on MoMLV release, as each mutant released p30CA proteins in amounts 20 to 30% lower than that seen with the wt virus (Fig. 1B and C). In contrast, and as expected, the MoMLV mutant carrying a disrupted PPPY motif (AAAY mutant) (Fig. 1A) exhibited a severe viral release defect (Fig. 1B and C). Mutations of both the PSAP and LYPAL L domains (AS-PAL double mutant) led to an approximately 45% decrease in virus production (Fig. 1C). These results indicate that the individual participation of the PSAP or LYPAL motif in MoMLV release from 293T cells makes only a modest contribution to virus release.
FIG. 1.
Generation and examination of MoMLV L domain mutants. (A) Schematic representation of MoMLV Gag. The positions of the three L domain motifs are indicated. The residues within each L domain that were replaced by alanines are underlined. (B) Release of MoMLV L domain mutants from 293T cells. Cells were mock transfected (Mock) or transfected with either wt MoMLV proviral DNA (lane 2) or a mutant virus carrying one mutated L domain (AAAY, ASAA, or AAPAL) or two mutated L domains (ASAA-AAPAL [designated AS-PAL for simplicity], AAAY-AAPAL [AY-PAL], or ASAA-AAAY [AS-AY]). Cells and viruses were collected 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using a goat anti-MoMLV CA antibody. (C) Percentages of release efficiency of wt MoMLV and of each MoMLV mutant examined in panel B [efficiency = virus-associated Gag/(virus-associated Gag + cell-associated Gag)].
Tsg101 and Alix overexpression failed to rescue MoMLV lacking the PPPY L domain.
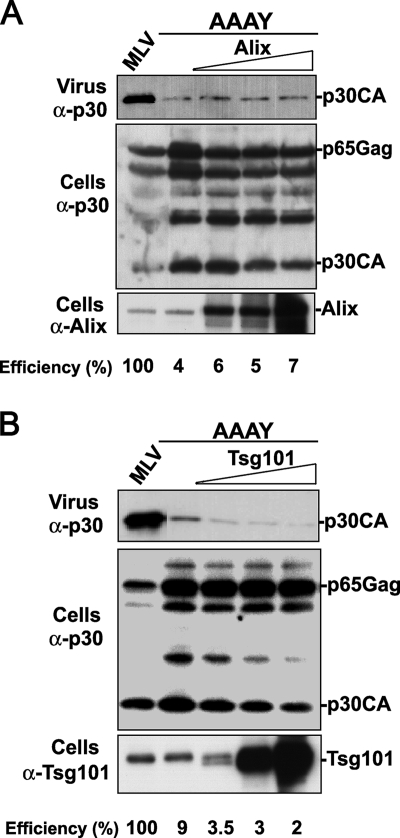
The results described above indicated that, together, the PSAP and LYPAL motifs are weak but active L domains that may constitute additional budding pathways for MoMLV. It has been previously shown that in HIV-1, budding defects caused by disruption of the PTAP/Tsg101 pathway were corrected by the overexpression of Alix (15, 54). These findings suggested that Alix, via interactions with the LYPXnL motif, compensated for the absent PTAP/Tsg101 budding pathway. We sought to examine whether the PSAP/Tsg101 or the LYPAL/Alix pathway could fulfill a similar function in the absence of PPPY, the primary MoMLV L domain. When overexpressed with the MoMLV AAAY mutant, neither Tsg101 nor Alix alleviated defects caused by the AAAY mutation, as both proteins failed to rescue the release of the MoMLV AAAY mutant (Fig. 2; see release efficiency values below panels). In fact, overexpression of Tsg101 appears to inhibit AAAY release even at the highest amount. Together, the data suggest that the recruitment of the cellular proteins Tsg101 and Alix by the PSAP and LYPAL secondary L domains, respectively, is not sufficient to stimulate the release of MoMLV in the absence of the PPPY primary L domain.
FIG. 2.
Overexpression of Tsg101 and Alix failed to rescue MoMLV lacking the PPPY L domain. 293T cells were transfected with either wt MoMLV proviral DNA (lane 1) or a mutant carrying the AAAY motif (lanes 2 to 5) alone or with HA-Alix (lanes 3 to 5) (A) or Myc Tsg101 (lanes 3 to 5) (B) expression plasmid. Cells and viruses were collected 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. Percentages of release efficiency of AAAY alone or in the presence of exogenously expressed Tsg101 or Alix [efficiency = virus-associated Gag/(virus-associated Gag + cell-associated Gag)] are shown under the panels.
Itch efficiently rescues MoMLV AAAY release defects.
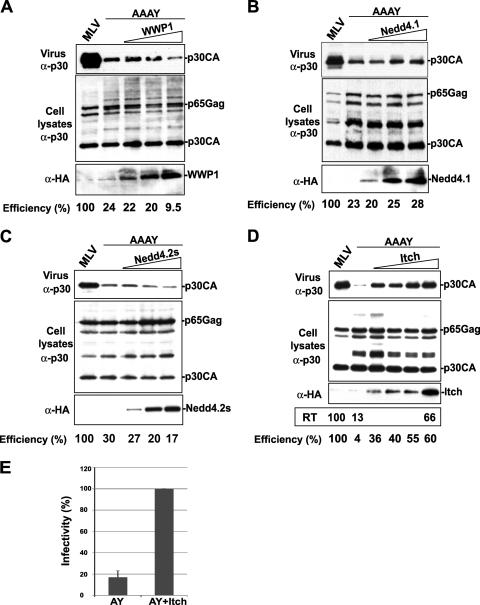
It has been previously reported that retroviral release defects caused by the disruption of primary L domains can be rescued by overexpression of members of the Nedd4-like ligase family (8, 10, 55). Interestingly, virus release stimulation by Nedd4-like ubiquitin ligases was previously shown to be independent of the presence of a PPPY L domain motif in the Gag protein (8, 10, 55). We thus hypothesized that MoMLV Gag lacking the PPPY Nedd4-like ligase binding motif might still be sensitive to the activity of these ubiquitin ligases. To test this hypothesis, we overexpressed the Nedd4-like ligase family members Nedd4.1, Nedd4.2s, WWP1, and Itch and examined their effect on the release of the MoMLV AAAY mutant. Overexpression of Itch rescued the release defects of MoMLV caused by disruption of the PPPY L domain (Fig. 3A, B, C, and D), while that of Nedd4.1, Nedd4.2 (data not shown), Nedd4.2s, and WWP1 did not. In fact, WWP1 exerted a slight inhibitory effect on AAAY mutant release, possibly because large amounts of WWP1, which is known to function in PPPY-mediated MoMLV release (33), might have depleted the cytoplasm from downstream acting ESCRT components necessary for the release-defective MoMLV. Itch-mediated rescue of the MoMLV AAAY mutant was remarkably efficient, as small amounts of Itch (∼100 ng of HA-Itch DNA expression plasmid was used to transfect a million cells) were sufficient for detection of rescue of release defects (Fig. 3D, lane 1). Up to ∼60% of the wt reverse transcriptase activity was restored when larger amounts of Itch (∼2 μg of HA-Itch expression plasmid DNA) were overexpressed (Fig. 3D; compare lanes 1 and 6). Additionally, Itch overexpression significantly (∼6-fold) increased the release of infectious MoMLV AAAY virus, as shown in Fig. 3E. Together, these findings demonstrate that Itch potently and specifically corrected release defects and enhanced infectivity of the MoMLV AAAY mutant.
FIG. 3.
Itch efficiently rescues MoMLV AAAY release defects. 293T cells were transfected with wt MoMLV proviral DNA (lane 1) or with a mutant carrying the AAAY motif (lanes 2 to 5) alone (lane 2) or with HA-WWP1 (A), HA-Nedd4.1 (B), HA-Nedd4.2s (C), or HA-Itch (D). Four different amounts of HA-Itch (increasing from 100 ng to 2 μg) were used (lanes 3 to 6). Cells and viruses were collected 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. Itch-mediated rescue of MoMLV AAAY release was assessed by measuring the reverse transcriptase (RT) activity of wt MoMLV, the AAAY mutant, and the rescued virus particles obtained in the presence of HA-Itch; values are shown in the box below panel D. (E) Itch overexpression enhances the release of infectious MoMLV AAAY mutant particles. Budding efficiency percentages [efficiency = virus-associated Gag/(virus-associated Gag + cell-associated Gag)] were calculated for each experiment and are shown under the panels. Infectivity of virus produced by 293T cells transfected with the AAAY mutant and empty vector or AAAY and Itch-expressing vector was measured using an XC overlay test by enumerating foci of infected cells. Values were obtained from three independent experiments and represent percentages of infectivity. The AY + Itch value was arbitrarily set to represent 100% infectivity; a value of 17% (infectivity percentage) was obtained with AY alone. AY, MoMLV AAAY mutant; AY+Itch, MoMLV AAAY coexpressed with HA-Itch.
Itch rescue of the MoMLV AAAY mutant is independent of the PSAP and LYPAL motifs.
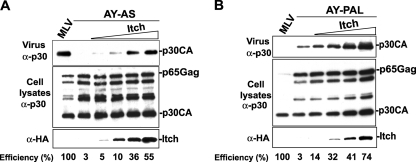
MoMLV Gag harbors three L domain motifs: PPPY, PSAP, and LYPAL. Although the PPPY motif was shown to play a predominant role in MoMLV release (33, 46, 62), the PSAP and LYPAL motifs appear to be secondary pathways that modestly contribute to MoMLV budding and release (see reference 46 and Fig. 1). Having identified the Nedd4-like ligase Itch as the host cell protein that restores MoMLV release when the PPPY motif is disabled, we next asked whether Itch stimulation of viral release involves the PSAP or LYPAL motif. To address this, we tested the effect of Itch overexpression on the release of a mutant MoMLV carrying proline-to-alanine substitutions in either the PPPY and PSAP motifs (AY-AS mutant) or the PPPY and LYPAL motifs (AY-PAL mutant) (Fig. 1A). Itch overexpression rescued the release of both MoMLV AY-AS and AY-PAL double mutants, demonstrating that mutation of PSAP or LYPAL has no effect on the ability of Itch to rescue MoMLV carrying an inactive PPPY motif (Fig. 4). The data indicate that the PSAP and LYPAL motifs are dispensable for Itch-mediated rescue of budding defects caused by an inactive PPPY motif in MoMLV.
FIG. 4.
Itch rescues the release defects of MoMLV AAAY independently of the PSAP and LYPAL motifs. 293T cells were transfected with either wt MoMLV proviral DNA (lane 1) or a double mutant carrying either the AS-AY motif (A) or AY-PAL motif (B) in the absence (lane 2) or presence of increasing amounts of HA-Itch (0.25 μg, 0.5 μg, 1 μg, and 2 μg; lanes 3 to 5). Cells and viruses were collected 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using either an anti-MoMLV p30CA antibody or an anti-HA antibody to detect Itch expression.
Itch sequence requirements for the rescue of MoMLV lacking PPPY.
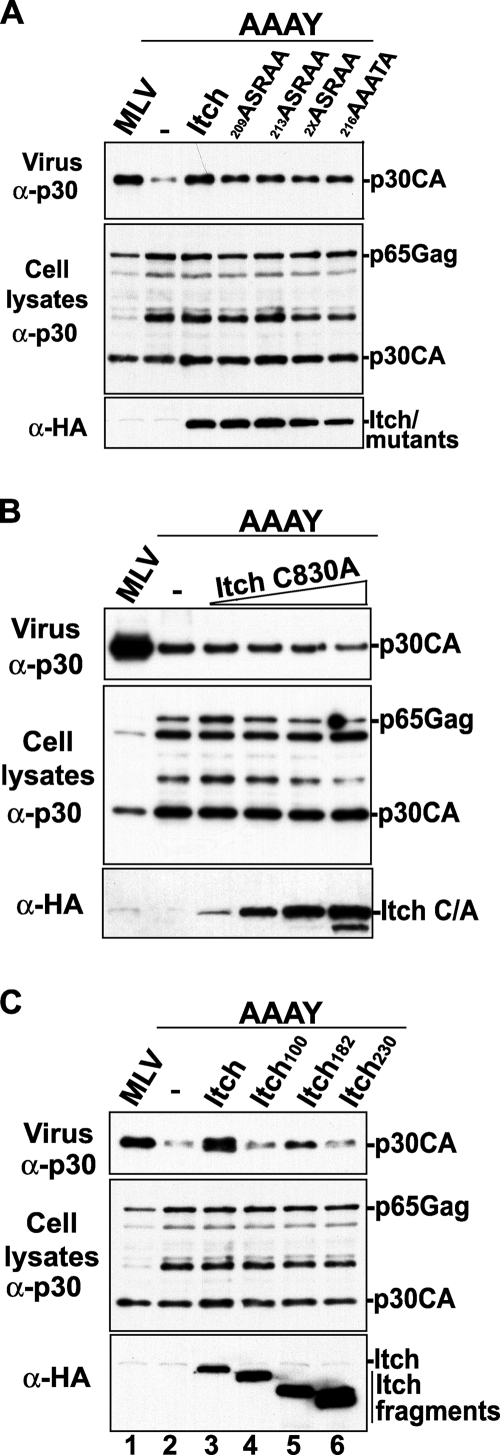
Itch is involved in multiple cellular pathways, including trafficking of cell signaling receptors through the endosomes (reviewed in reference 37). Unlike other members of the Nedd4-like ubiquitin ligase family, Itch contains a short proline-rich domain that mediates binding to endophilin (1), a cellular protein that functions in membrane trafficking at the endosome (12, 38). To examine a role for endophilin in Itch stimulation of MoMLV release, we introduced mutations (Fig. 5A) within the endophilin binding sequence in Itch (209PSRPPSRPPPTPRRP223) that were expected to interfere with its ability to bind endophilin. Three mutants were generated by replacing the proline residues with alanines in the 209PSRPP motif (209ASRAA mutant), in the 213PSRPP motif (213ASRAA mutant), within both adjacent motifs (2XASRAA), or within the proline-rich central 216PPPTP motif (216AAATA mutant). The results (Fig. 5A) showed that mutations in the endophilin binding site within Itch had little to no effect on its ability to rescue the release of MoMLV harboring an inactive PPPY motif. Consistent with this result, we found that overexpression of endophilin, which also binds MoMLV Gag directly via the matrix domain (59), failed to rescue the budding defects of the MoMLV AAAY mutant (data not shown). Collectively, these results suggest that Itch interaction with endophilin is not required for the rescue of MoMLV harboring a disabled PPPY motif.
FIG. 5.
(A) Interaction with endophilin is not required for Itch-mediated rescue of a PPPY defective-MoMLV. (A) 293T cells were transfected with the MoMLV expressing proviral DNA (lane 1) or with the MoMLV AAAY mutant DNA, either alone ([minus]) (lane 2) or with the wt HA-Itch plasmid or the indicated Itch mutation-expressing plasmids (lanes 3 to 7). (B) The catalytic activity of Itch is required for the rescue of MoMLV AAAY release defects. 293T cells were transfected with wt MoMLV proviral DNA (lane 1) or with a mutant carrying the AAAY motif (lanes 2 to 5), either alone ([minus]) (lane 2) or with the HA-Itch C830A mutant (lanes 3 to 6). Cells and viruses were collected 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. (C) The role of N-terminal regions of Itch in the rescue of MoMLV budding defects. The rescue experiment was performed as described above with HA-Itch or with the indicated Itch deletion mutant (Itch100, Itch182, or Itch230).
The catalytic activity of the ubiquitin ligase WWP1 was shown to be critical for promoting MoMLV release (33). Similarly, the enzymatic activity of the Nedd4-like ubiquitin ligase Nedd4.2s, which was recently shown to stimulate HIV-1 release, is required to correct the release defects caused by mutation of L domain motifs in HIV-1 (10, 55). To determine whether Itch-mediated rescue of the defective MoMLV AAAY mutant also requires enzymatic activity, we examined the effect of a catalytically inactive Itch (Itch C830A mutant) on the MoMLV AAAY mutant (1). We found that the Itch C830A mutant (carrying a replacement of the cysteine residue at position 830 with alanine) lost the ability to rescue the release defects of the AAAY mutant (Fig. 5B). This result demonstrates that the catalytic activity of Itch is required for the rescue of release defects caused by the disruption of the PPPY L domain in MoMLV Gag.
Itch is predominantly found associated with early and late endosomal compartments (1, 32), a localization that is mediated by the C2 domain of the protein (1). To examine the role of the Itch C2 domain in the rescue of the MoMLV AAAY mutant, we generated deletion mutants lacking the N-terminal residues of Itch that carry the C2 domain. Itch100 and Itch182 mutants lack the 100 and 182 N-terminal residues, respectively. Itch230 lacks not only the C2 domain but also the neighboring proline-rich region. Removal of the C2 domain (Itch100) ablated the ability of Itch to rescue the MoMLV AAAY mutant (Fig. 5C, lane 4). This indicated that in the context of the full-length Itch, the C2 domain is required for rescue of MoMLV AAAY release. In contrast, Itch182 exhibited about ∼30% of the wt Itch ability to rescue MoMLV release defects (Fig. 5C, lane 5). This deletion mutant lacks the entire C2 domain in addition to the following 82 residues, while the proline-rich region and the WW-Hect domain are intact. This result indicated that truncation of the 82 residues downstream of the C2 domain generated a partially active Itch fragment, possibly by allowing better protein folding and/or exposing an additional region(s) within Itch that is important for rescue activity. Finally, Itch230, which is N-terminally truncated to remove the C2 domain and the proline-rich region, resulted in a complete loss of activity in the rescue of MoMLV AAAY release (lane 6). This result suggested that in the context of Itch182, the proline-rich region that binds the endosomal protein endophilin is important for rescue activity. Collectively, these results suggest that regions that link Itch to endosomal compartments appear to be required for optimal rescue of MoMLV release defects.
Itch is incorporated into rescued MoMLV AAAY particles.
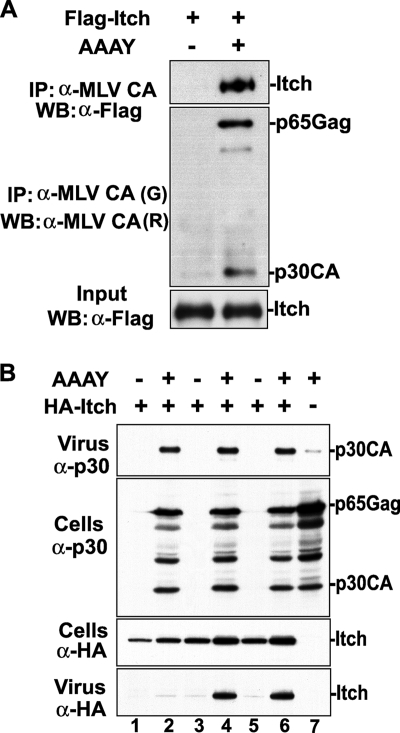
Retroviral Gag proteins recruit Nedd4-like ubiquitin ligases via interactions with the PPPY L domain motifs (6, 7, 21, 27, 33). At the same time, PPPY motif-independent interactions of retroviral Gag proteins with Nedd4-like ubiquitin ligases have been previously reported (8, 10, 55). We found that the MoMLV release defects caused by mutations in the PPPY motif were corrected by Itch overexpression in the cell, even though these mutations were found to prevent Itch binding of the L domain PPPY motif in yeast two-hybrid assays (data not shown). Moreover, Itch-mediated rescue of the MoMLV AAAY mutant was independent of the presence of PSAP and LYPAL motifs, since release defects were rescued when either motif was disabled simultaneously with the PPPY motif. These findings suggested that Itch might associate with MoMLV Gag in an L domain-independent manner. To test this hypothesis, we examined the ability of Flag-Itch to interact with MoMLV AAAY Gag by the use of immunoprecipitation assays. The data presented in Fig. 6A show that the anti-MoMLV CA antibody pulled down Flag-Itch when it was expressed with the AAAY Gag but not when Flag-Itch was expressed alone. These results indicated that Itch interacts with MoMLV Gag in the cell in a PPPY-independent manner.
FIG. 6.
Itch immunoprecipitates with MoMLV AAAY Gag and is incorporated in MoMLV AAAY-rescued particles. (A) 293T cells were transfected with the Flag-Itch expression plasmid or with Flag-Itch expression plasmid and the MoMLV AAAY proviral DNA. Cells were lysed in RIPA buffer, and cleared cell lysates were exposed to protein A and G Sepharose beads coated with a rat (R) anti-MLV CA antibody. Protein complexes captured on beads were washed with RIPA buffer, eluted, and analyzed by SDS-PAGE and Western blotting (WB) using a goat (G) anti-MLV CA antibody to detect Gag and an anti-Flag antibody to detect Itch. IP, immunoprecipitation. [minus], absent; +, present. (B) 293T cells were transfected with increasing amounts (0.5, 1, and 2 μg) of the HA-Itch expression plasmid alone (lanes 1, 3, and 5, respectively) or with mutant MoMLV AAAY proviral DNA (lanes 2, 4, and 6). HA-Itch proteins released in the culture media or incorporated into virus particles were collected by centrifugation at 32 h posttransfection, and the protein content of pelleted material was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. HA-Itch proteins were released only when coexpressed with MoMLV AAAY virus (lanes 2, 4, and 6).
Next, we expressed increasing amounts of Itch, either alone or with the MoMLV AAAY mutant, and reasoned that if Itch interacts with the MoMLV AAAY mutant Gag in order to achieve rescue, then the rescued MoMLV particles should incorporate Itch molecules. When Itch was expressed alone, very little to no Itch was found released in the cell culture media, including the results seen at the largest amount expressed (Fig. 6B, lanes 1, 3, and 5). Conversely, when Itch was expressed with the MoMLV AAAY mutant, the rescued virus particles contained Itch proteins (Fig. 6B, lanes 2, 4, and 6). Similarly, the rescued particles generated from the MoMLV AY-AS and AY-PAL double mutants incorporated Itch molecules as well (data not shown), demonstrating that the PSAP or the LYPAL is not involved in PPPY motif-independent Itch incorporation. Together, these data demonstrate that the MoMLV Gag protein can recruit the Nedd4-like ubiquitin ligase Itch via interaction(s) with regions outside of the L domain sequences.
Itch-mediated rescue of the MoMLV AAAY mutant requires the integrity of the host vps pathway.
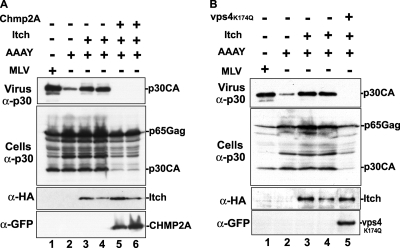
Like many other enveloped viruses, MoMLV requires a functional host vps machinery and the activity of the AAA ATPAse VPS4 for the release of virus (33). Mutations that prevent the recruitment of members of the Nedd4-like ubiquitin family via the PPPY L domain in MoMLV Gag lead to budding defects similar to those caused by dominant-negative versions of ESCRT-III or the VPS4 protein (34). These findings imply that MoMLV Gag recruits Nedd4-like ubiquitin ligases to gain access to members of the host vps machinery, including VPS4, which are critical for efficient viral release. MoMLV AAAY mutant release defects were corrected with overexpression of Itch (Fig. 3), suggesting that Itch may have linked the defective MoMLV mutant Gag to the host budding apparatus necessary for virus release. We reasoned that if this were the case, Itch-mediated rescue of MoMLV release should be sensitive to the dominant-negative versions of ESCRT members and the VPS4 AAA ATPase. To examine this hypothesis, we tested the effect of yellow fluorescent protein (YFP)-CHMP2A, an inhibitory version of CHMP2A that was previously shown to inhibit PPPY-driven MoMLV release (34). We found that expression of YFP-CHMP2A ablated Itch rescue of MoMLV AAAY (Fig. 7A). Similarly, expression of the GFP-VPS4aK173Q mutant, an inactive form of VPS4a that is known to interfere with the AAA ATPase activity (57), eliminated Itch-mediated release of the MoMLV AAAY mutant (Fig. 7B). These results demonstrate that Itch rescue of the MoMLV AAAY mutant requires functional vps machinery and suggest that MoMLV recruits Itch in an L domain-independent manner to serve as a link to members of the host budding pathway.
FIG. 7.
Itch-mediated rescue of the MoMLV AAAY mutant requires an intact ESCRT system and the activity of VPS4 ATPase. (A) 293T cells were transfected with the MoMLV proviral DNA alone (lane 1) or with the MoMLV AAAY mutant, alone (lane 2) or with the HA-Itch expression vector in the absence (lanes 3 and 4) or presence (lanes 5 and 6) of the dominant-negative YFP-CHMP2A mutant. [minus], absent; +, present. (B) 293T cells were transfected with the wt MoMLV proviral DNA (lane 1) or with the MoMLV AAAY mutant, alone (lane 2) or with HA-Itch-expressing vector in the absence (lanes 3 and 4) or presence (lane 5) of the dominant-negative mutant VPS4aK173Q plasmid. Two different amounts of HA-Itch were tested in the rescue assay, and the results are shown in lanes 3 and 4 of panels A and B. Cells and viruses were collected at 32 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using an anti-MoMLV p30CA antibody, an anti-HA antibody to detect Itch, or an anti-GFP antibody to detect VPS4aK173Q-GFP and YFP-CHMP2A expression.
RNAi knockdown of Itch inhibits MoMLV AAAY release.
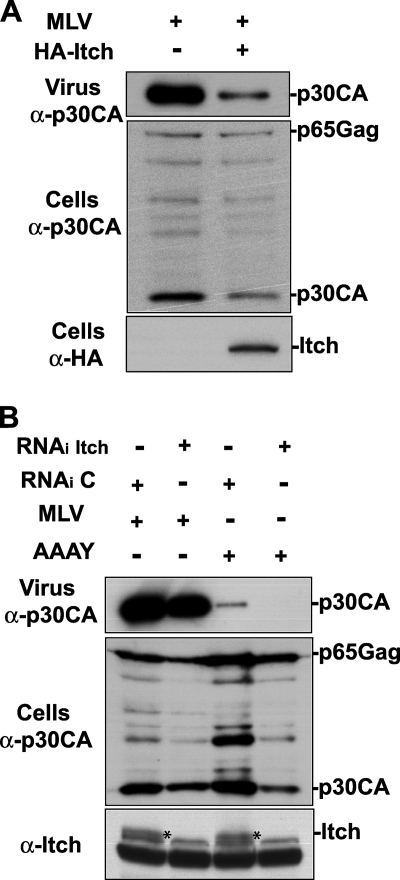
To examine the role of Itch in MoMLV budding, we first attempted an approach previously used with HIV-1, in which overexpression of Tsg101, a protein that plays a central role in HIV-1 budding, was shown to interfere with virus release from 293T cells (18). Overexpression of the full-length Itch, in amounts five times larger than those used to rescue the release of the MoMLV AAAY mutant (Fig. 3), led to a significant decrease of MoMLV release and Gag processing, as a smaller amount of mature p30CA was detected (Fig. 8A). This result suggested that Itch plays a role in MoMLV release. Second, we sought to examine whether Itch is required for MoMLV release. For this, we used RNAi to knock down Itch in the cell and tested the effect of this depletion on the release of both wt MoMLV and the AAAY mutant MoMLV. Whereas RNAi knockdown of cellular Itch led to a modest decrease in the level of MoMLV release, it resulted in a complete loss of MoMLV AAAY release (Fig. 8B). Together, these findings indicate that Itch is involved in but is not essential for the release of a MoMLV carrying an intact PPPPY L domain. Conversely, MoMLV requires Itch for release of virus from the cell in a PPPY-independent manner, revealing a secondary L domain-independent pathway by means of which MoMLV utilizes Itch to release particles.
FIG. 8.
(A) Overexpression of Itch reduces release of wt MoMLV. 293T cells were transfected with wt MoMLV proviral DNA in the absence (lane 1) or presence (lane 2) of HA-Itch-expressing vector. [minus], absent; +, present. (B) Depletion of endogenous Itch reduces the release of AAAY mutant MoMLV. 293T cells were transfected with wt MoMLV (lanes 1 and 2) or with AAAY mutant MoMLV (lanes 3 and 4) proviral DNA alone in the presence of control RNAi oligonucleotides (lanes 1 and 3) or RNAi oligonucleotides targeting cellular Itch (lanes 2 and 4). Cells and viruses were collected at 24 h post transfection, and their protein content was analyzed by SDS-PAGE and Western blotting using an anti-MoMLV p30CA antibody and either an anti-HA or anti-Itch antibody to detect Itch. Itch bands are marked with an asterisk.
Itch ubiquitinates MoMLV AAAY Gag.
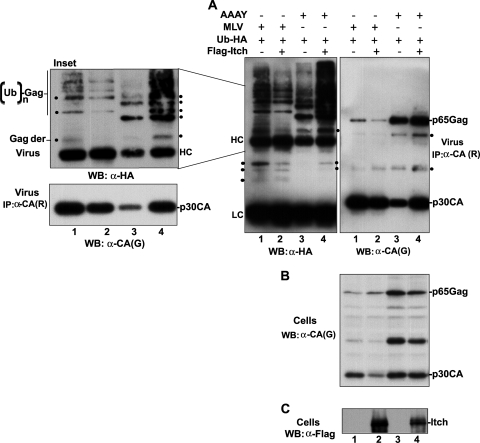
Previous studies suggested a role for Gag ubiquitination in virus release (8, 19, 22, 48). In addition, a recent report showed that ubiquitin attachment to EIAV Gag substituted for the lack of L domains and rescued viral budding (25), suggesting that ubiquitin molecules conjugated to Gag can signal the recruitment of the cell's budding machinery and drive virus release. We found that the catalytic activity of Itch is required for the rescue of release defects caused by the disruption of the PPPY L domain in MoMLV (Fig. 5), suggesting that ubiquitin transfer is critical for virus release. To examine whether Itch-mediated transfer of ubiquitin to Gag occurs during MoMLV release stimulation by Itch, we expressed wt MoMLV and AAAY mutant MoMLV with HA-tagged ubiquitin in the absence or presence of Flag-tagged Itch and checked the ubiquitination level of wt MoMLV Gag, comparing it to that of Gag AAAY mutant expressed alone or with Itch.
Gag protein expression was detected in all samples with the goat anti-MLV CA antibody (Fig. 9B). In the wt MoMLV virion fraction, the anti-HA antibody detected numerous proteins (Fig. 9A, lane 1 in the left panel), some of which were also detected with the anti-MLV CA antibody on a duplicate membrane (Fig. 9A, lane 1 in the right panel). This result indicated that wt MoMLV virions contain ubiquitinated Gag and Gag-derived proteins. Ubiquitinated Gag proteins were also detected in the fraction of virions released by the MoMLV AAAY mutant (Fig. 9A, lanes 3 in the left and right panels). Expression of Flag-Itch stimulated MoMLV AAAY release (Fig. 9C, lane 4 and lower panel in the inset), enhanced AAAY Gag ubiquitination, and led to the appearance of additional ubiquitinated Gag and Gag-processing products in AAAY-rescued virions (lane 4 in Fig. 9A and inset). Remarkably, these ubiquitinated Gag molecules were also detected in the wt MoMLV virions (lane 1 in Fig. 9A and inset), suggesting that ubiquitination of Gag proteins is involved in Itch-mediated rescue of the release-defective MoMLV AAAY mutant. In support of this notion, we found that inhibition of MoMLV release by overexpression of Flag-Itch (also see Fig. 8A) led to the decrease of virus release and Gag ubiquitination in virions (Fig. 9A and inset [compare lanes 1 and 2]). Together, these results demonstrate that ubiquitination of Gag accompanies Itch-mediated rescue of MoMLV AAAY release. The finding that Itch expression led to the release of ubiquitinated MoMLV AAAY Gag molecules that are naturally released in wt MoMLV virions suggests that AAAY MoMLV recruits Itch in the cell to ubiquitinate Gag, consequently facilitating virus release. In addition, these data strongly suggest a role for Gag ubiquitination in MoMLV release.
FIG. 9.
Itch increases Gag ubiquitination of the MoMLV AAAY Gag mutant. (A) 293T cells were co-transfected with constructs expressing wt MoMLV (lane 1 & 2) or AAAY mutant (lanes 2 & 3) proviral DNA and a HA-tagged ubiquitin expression vector (Ub-HA) in the presence (lane 2 & 4) or absence (lane 1& 3) of a Flag-tagged Itch expression plasmid. Viruses were collected 40 h posttransfection lysed in RIPA buffer and immunoprecipitated with a rat anti-MoMLV p30CA (R) antibody. Proteins complexes immunoprecipitated from virions were subsequently analyzed as duplicates by SDS-PAGE and Western blotting using a mouse anti-HA antibody to detect Gag ubiquitin modification (A, left panel) and a goat anti-MoMLV p30CA (G) antibody to detect Gag and Gag-derived (Gag-der) products (A, right panel), respectively. [minus], absent; +, present. Panel B shows analysis of Gag expression in the cell by Western blotting using a goat anti-MLV p30CA (G) antibody. Panel C shows analysis of Itch expression in cells using a mouse anti-Flag antibody. The amount of Flag-Itch expressed with wt MoMLV (lane 2) is 5 times higher than that used to rescue the release defects of MoMLV AAAY mutant (lane 4). A lighter exposure (and enlarged version) of the upper portion of panel A is shown in the inset. The enhancement of Gag ubiquitination levels and the newly ubiquitinated Gag proteins that appeared following co-expression with Flag-Itch (compare lane 3 and 4 to lane 1), are marked with dots (•) in the inset and panels in 9A. A lighter exposure of virus released is also shown under the inset. LC and HC: antibodies light and heavy chains, respectively. AAAY: MoMLV AAAY mutant.
DISCUSSION
The results presented here demonstrate that Itch corrects MoMLV budding defects caused by disruption of the PPPY motif (MoMLV AAAY mutant). In contrast, overexpression of Tsg101 and Alix fails to rescue the MoMLV AAAY mutation, despite the presence of intact L PSAP and LYPAL domains in Gag. Moreover, Itch stimulation of MoMLV AAAY release is independent of all known L domains, since disruption of the PSAP or LYPAL motif did not affect Itch-mediated virus rescue. However, since all three L domains were not simultaneously disrupted, our data do not unequivocally exclude the participation of the remaining secondary L domain (PSAP or LYPAL) in Itch rescue of the MoMLV double mutants. Nevertheless, our findings clearly demonstrate that although the PSAP-Tsg101 and LYPAL-Alix pathways do modestly participate in MoMLV release (see reference 46 and data in this study), their presence appears to be insufficient to promote MoMLV release in the absence of the PPPY L domain. Thus, even in the absence of the PPPY motif, MoMLV retains the requirement for interaction(s) with Itch (and possibly other Neddd4-like ligases) to drive viral release, suggesting that these interactions are a key global requirement for MoMLV release. Our data reveal a PPPY-independent recruitment of Itch by MoMLV Gag that might be an additional mechanism that synergistically functions with the PPPY-driven pathway to enhance virus release. Small amounts of Itch (∼100 ng of HA-Itch plasmid DNA was used to transfect a million cells) were sufficient to enhance MoMLV AAAY release and infectivity, suggesting that PPPY-independent Itch interactions with MoMLV Gag mediate a relatively efficient viral release mechanism. Similarly, a robust L domain-independent stimulation of HIV-1 release by expression of Nedd-4.2s was recently described (10, 55), suggesting that retroviral Gag proteins engage in L domain-independent interaction(s) with Nedd4-like ubiquitin ligases to efficiently release virus.
The budding-defective MoMLV AAAY mutant exhibited a clear preference for recruiting Itch to enhance virus release, as the other ubiquitin ligases tested, namely, WWP1, Nedd4.1, Nedd4.2, and Nedd4.2s (not shown), failed to function in the rescue assay. Interestingly, HA-tagged WWP1 failed to rescue the MoMLV AAAY mutant, although the YFP fusion version of WWP1 was previously shown to rescue viral release of a budding-attenuated MoMLV mutant (33). This finding suggested that, in contrast to Itch, WWP1 ligase might function only when it retains a residual binding mechanism via the PPPY motif. Therefore, overexpression of WWP1 did not correct budding defects as severe as those imposed by the AAAY mutation, possibly because of the complete loss of binding to the PPPY L domain. Although several attempts to demonstrate WWP1-mediated rescue were unsuccessful, we cannot rule out the possibility that under other experimental conditions, WWP1-WWP2 ligase may in fact rescue MoMLV budding defects in the absence of PPPY (i.e., in constructs that express higher amounts of protein or other fusion tags). Together, these observations support the notion that PPPY-independent Itch stimulation of MoMLV release is specific.
We found Itch to be incorporated in particles and released only when expressed with the MoMLV AAAY mutant or the AY-AS or AY-PAL double mutant. This finding indicates direct or indirect interactions between Gag and Itch via sequences located outside of the known L domains. Since mutants of Itch that lack the endophilin binding site retained activity in the virus rescue assay, we exclude endophilin, the mutual Itch and MoMLV Gag binding partner (1, 58), as a possible mediator for Itch enhancement of MoMLV AAAY release and infectivity. We have also excluded Tsg101 and Alix as possible mediators of Itch enhancement of MoMLV release, as RNAi depletion of either protein had no effect on Itch-mediated rescue of MoMLV AAAY (data not shown). However, these data do not exclude the involvement of another cellular protein(s) that might mediate Itch binding to MoMLV Gag. We have considered the possibility that Itch might carry a region(s) that interacts with MoMLV Gag in a PPPY domain-independent manner, because atypical interactions of Itch with cellular substrates, as well as its ability to recognize phospho-Ser and phospho-Thr residues, have been previously reported (31, 32, 44, 60). However, since interaction of MoMLV Gag lacking the PPPY domain with Itch is not detected in pull-down or yeast two-hybrid assays (reference 33 and data not shown), Itch incorporation in MoMLV AAAY-rescued particles is most likely mediated by a cellular factor that bridges Itch to MoMLV AAAY Gag. This hypothesis is supported by our finding that Itch was found in complex with MoMLV AAAY Gag in immunoprecipitation assays.
Itch-mediated rescue of MoMLV AAAY requires the protein's enzymatic activity, demonstrating that ubiquitin modification of a key protein(s) is critical to stimulate virus release. Similarly, the catalytic activity of Nedd4-like ligases has proven essential for PPPY domain-dependent and -independent release (10, 33, 55, 63), even though the ubiquitination of Gag itself is presumed to be dispensable in some contexts (63). However, a portion of PPPY motif-carrying Gag proteins is known to be ubiquitinated (41, 42). In agreement with these observations, we found that overexpression of Itch in amounts that efficiently rescue the release defects of the MoMLV AAAY mutant also led to the appearance of newly ubiquitinated Gag molecules in the rescued MoMLV virions. Remarkably, these ubiquitinated Gag proteins are present in the wt MoMLV virions, suggesting that MoMLV uses cellular Itch to release virus. In fact, RNAi knockdown of cellular Itch resulted in a complete loss of MoMLV AAAY release, supporting the notion that MoMLV recruits Itch in a PPPY-independent manner to enhance virus release.
Mapping of regions in Itch important for rescuing MoMLV budding defects showed the involvement of the C2 domain, as removal of the 100 residues that harbor this domain (Itch100) rendered Itch inactive in the virus rescue assay. Because the N-terminal C2 domain of Itch mediates its localization to endocytic compartments (1), we concluded that Itch interaction(s) with the endosomal pathway is important for the enhancement of MoMLV release. Interestingly, in the context of the WW-HECT fragment, residual activity in the virus rescue assay was detected only when WW-HECT was extended with the proline-rich motif that binds the endosomal protein endophilin (Itch182). Notably though, the proline-rich region is also present in the Itch100 mutant, which displayed no activity in the rescue assay. This suggests that the structural availability of this sequence, not only its presence, is key for function. Thus, Itch and its derived fragments function in the MoMLV rescue assay only when carrying regions that can mediate interactions with endosomal compartments, further supporting the notion that Itch links the PPPY-deficient MoMLV Gag to members of the vps machinery. In light of this observation, we propose a model in which MoMLV Gag interacts with Itch on endosomal membranes, possibly to recruit ESCRT proteins necessary to exit the cell. Interestingly, formation of MoMLV assembly complexes was previously reported to initiate on endosomal vesicles prior to their transport to the plasma membrane for budding (4), suggesting that MoMLV might use such a mechanism to recruit members of the ESCRT pathway.
Our studies suggest a model in which Itch-mediated ubiquitination of Gag enhances MoMLV release by signaling the recruitment of members of the host budding machinery. In this model, ubiquitin molecules conjugated to MoMLV AAAY Gag may substitute for the missing L domain and serve as a bridge that links the mutant Gag to the host budding apparatus. Indeed, ubiquitin fusion to EIAV Gag has been recently shown to compensate for the absence of the primary L domain motif (25), possibly by binding to motifs interacting with ubiquitin within members of the ESCRT pathway (17, 25, 47, 51, 53). We found that Itch-driven rescue of PPPY-independent MoMLV release is accompanied by the ubiquitination of MoMLV AAAY Gag and requires the integrity of the cellular ESCRT machinery as well as the activity of the VPS4 AAA ATPase. The finding that Itch expression leads to the release of ubiquitinated MoMLV AAAY Gag molecules that are naturally released in wt MoMLV virions suggests that AAAY MoMLV recruits Itch in the cell to ubiquitinate Gag, consequently facilitating virus release. Thus, it is possible that Itch links MoMLV Gag to ESCRT components of the vps pathway through its ubiquitin modification of Gag. This is in agreement with Itch's known colocalization with vps proteins in endosomal compartments (1, 32, 33) and its role in the ubiquitination of plasma membrane proteins to direct their trafficking through MVB compartments (32, 60).
In summary, we find that Itch potently enhances the release of MoMLV lacking the PPPY L domain via a mechanism that involves Gag ubiquitination. In addition, Itch interacts with Gag independently of the PPPY motif and incorporates into the rescued MoMLV virions. Thus, in addition to the budding pathway driven via the PPPY L domain, MoMLV can utilize a parallel, PPPY-independent route that specifically recruits Itch to exit the cell. Such a pathway might constitute an additional budding mechanism that MoMLV utilizes to enhance virus release.
Acknowledgments
We thank Annie Angers (University of Montreal) for the Itch expression plasmid, the Miyazano laboratory (Fred Hutchinson Cancer Research Institute) for the WWP1 expression plasmid, the Kazuza Institute (Chiba, Japan) for Nedd4-1 cDNA, Arianna Calistri (University of Padova) for the HA-ubiquitin plasmid, Steve Goff (Columbia University) for pNCA and endophilin expression vectors, Wes Sundquist (University of Utah) for CHMP2A, CHMP3, and VPS4 expression plasmids, Christine Kozak (Laboratory of Molecular Microbiology, NIAID) for help with the XC test, and Alicia Buckler-White and Ron Plishka (Laboratory of Molecular Microbiology, NIAID) for DNA sequencing. We are grateful to Melodi Javid, Alan Rein (Drug Resistance Program, NCI), Bernard Moss (Laboratory of Viral Diseases, NIAID), and Carol Carter (SUNY at Stony Brook, Stony Brook, NY) for critical readings of the manuscript and helpful discussions.
This work was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 28 October 2009.
REFERENCES
- 1.Angers, A., A. R. Ramjaun, and P. S. McPherson. 2004. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J. Biol. Chem. 279:11471-11479. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 6.Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 7.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calistri, A., C. Del Vecchio, C. Salata, M. Celestino, M. Celegato, H. Gottlinger, G. Palu, and C. Parolin. 2009. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J. Cell Physiol. 218:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908-1912. [DOI] [PubMed] [Google Scholar]
- 10.Chung, H. Y., E. Morita, U. von Schwedler, B. Muller, H. G. Krausslich, and W. I. Sundquist. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82:4884-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 12.de Heuvel, E., A. W. Bell, A. R. Ramjaun, K. Wong, W. S. Sossin, and P. S. McPherson. 1997. Identification of the major synaptojanin-binding proteins in brain. J. Biol. Chem. 272:8710-8716. [DOI] [PubMed] [Google Scholar]
- 13.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 14.Dussupt, V., M. P. Javid, G. Abou-Jaoude, J. A. Jadwin, J. de La Cruz, K. Nagashima, and F. Bouamr. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841-852. [DOI] [PubMed] [Google Scholar]
- 16.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 17.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77:9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottwein, E., S. Jager, A. Habermann, and H. G. Krausslich. 2006. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect. J. Virol. 80:6267-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidecker, G., P. A. Lloyd, F. Soheilian, K. Nagashima, and D. Derse. 2007. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J. Virol. 81:9769-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi, A., H. Garg, K. Nagashima, J. S. Bonifacino, and E. O. Freed. 2008. GGA and Arf proteins modulate retrovirus assembly and release. Mol. Cell 30:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi, A., U. Munshi, S. D. Ablan, K. Nagashima, and E. O. Freed. 2008. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic 9:1972-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzmann, D. J., S. Sarkar, T. Chu, A. Audhya, and S. D. Emr. 2004. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell 15:468-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung, J., A. Yueh, F. S. Appah, Jr., B. Yuan, K. de los Santos, and S. P. Goff. 2006. Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J. 25:2155-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325-1328. [DOI] [PubMed] [Google Scholar]
- 32.Marchese, A., C. Raiborg, F. Santini, J. H. Keen, H. Stenmark, and J. L. Benovic. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell 5:709-722. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melino, G., E. Gallagher, R. I. Aqeilan, R. Knight, A. Peschiaroli, M. Rossi, F. Scialpi, M. Malatesta, L. Zocchi, G. Browne, A. Ciechanover, and F. Bernassola. 2008. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 15:1103-1112. [DOI] [PubMed] [Google Scholar]
- 38.Micheva, K. D., B. K. Kay, and P. S. McPherson. 1997. Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J. Biol. Chem. 272:27239-27245. [DOI] [PubMed] [Google Scholar]
- 39.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 41.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 42.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu, L., C. Joazeiro, N. Fang, H. Y. Wang, C. Elly, Y. Altman, D. Fang, T. Hunter, and Y. C. Liu. 2000. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275:35734-35737. [DOI] [PubMed] [Google Scholar]
- 45.Sabo, Y., N. Laham-Karam, and E. Bacharach. 2008. Basal budding and replication of the murine leukemia virus are independent of the gag L domains. J. Virol. 82:9770-9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segura-Morales, C., C. Pescia, C. Chatellard-Causse, R. Sadoul, E. Bertrand, and E. Basyuk. 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280:27004-27012. [DOI] [PubMed] [Google Scholar]
- 47.Shields, S. B., A. J. Oestreich, S. Winistorfer, D. Nguyen, J. A. Payne, D. J. Katzmann, and R. Piper. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spidel, J. L., R. C. Craven, C. B. Wilson, A. Patnaik, H. Wang, L. M. Mansky, and J. W. Wills. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 50.Stuchell-Brereton, M. D., J. J. Skalicky, C. Kieffer, M. A. Karren, S. Ghaffarian, and W. I. Sundquist. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740-744. [DOI] [PubMed] [Google Scholar]
- 51.Sundquist, W. I., H. L. Schubert, B. N. Kelly, G. C. Hill, J. M. Holton, and C. P. Hill. 2004. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 13:783-789. [DOI] [PubMed] [Google Scholar]
- 52.Tao, M., P. C. Scacheri, J. M. Marinis, E. W. Harhaj, L. E. Matesic, and D. W. Abbott. 2009. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr. Biol. 19:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbé, S., M. Sachse, P. E. Row, C. Preisinger, F. A. Barr, G. Strous, J. Klumperman, and M. J. Clague. 2003. The UIM domain of Hrs couples receptor sorting to vesicle formation. J. Cell Sci. 116:4169-4179. [DOI] [PubMed] [Google Scholar]
- 54.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Usami, Y., S. Popov, E. Popova, and H. G. Gottlinger. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 58.Wang, G., J. M. McCaffery, B. Wendland, S. Dupre, R. Haguenauer-Tsapis, and J. M. Huibregtse. 2001. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell. Biol. 21:3564-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, M. Q., W. Kim, G. Gao, T. A. Torrey, H. C. Morse III, P. De Camilli, and S. P. Goff. 2003. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wegierski, T., K. Hill, M. Schaefer, and G. Walz. 2006. The HECT ubiquitin ligase AIP4 regulates the cell surface expression of select TRP channels. EMBO J. 25:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhadina, M., M. O. McClure, M. C. Johnson, and P. D. Bieniasz. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. USA 104:20031-20036. [DOI] [PMC free article] [PubMed] [Google Scholar]