Abstract
Lens epithelium-derived growth factor (LEDGF)/p75 is a cellular cofactor for HIV-1 DNA integration. It is well established that the simultaneous binding of LEDGF/p75 to chromatin and to HIV-1 integrase is required for its cofactor activity. However, the exact molecular mechanism of LEDGF/p75 in HIV-1 integration is not yet completely understood. Our hypothesis is that evolutionarily conserved regions in LEDGF/p75 exposed to solvent and harboring posttranslational modifications may be involved in its HIV-1 cofactor activity. Therefore, a panel of LEDGF/p75 deletion mutants targeting these protein regions were evaluated for their HIV-1 cofactor activity, chromatin binding, integrase interaction, and integrase-to-chromatin-tethering activity by using different cellular and biochemical approaches. The deletion of amino acids 267 to 281 reduced the cofactor activity of LEDGF/p75 to levels observed for chromatin-binding-defective mutants. This region contains a serine cluster (residues 271, 273, and 275) recurrently found to be phosphorylated in both human and mouse cells. Importantly, the conversion of these Ser residues to Ala was sufficient to impair the ability of LEDGF/p75 to mediate HIV-1 DNA integration, although these mutations did not alter chromatin binding, integrase binding, or the integrase-to-chromatin-tethering capability of LEDGF/p75. These results clearly indicated that serine residues 271, 273, and 275 influence the HIV-1 cofactor activity of integrase-to-chromatin-tethering-competent LEDGF/p75.
Lens epithelium-derived growth factor (LEDGF)/p75 is a cellular cofactor for HIV-1 DNA integration implicated in the efficiency and genomic location of the viral integration process (10, 20, 25, 35, 40). Cells lacking LEDGF/p75 showed a severe defect in HIV-1 infection characterized by decreased levels of integrated viral DNA in the absence of other preintegration defects in the viral life cycle (20, 35). In addition, in the absence of LEDGF/p75, HIV-1 DNA integration occurred less frequently in active transcription units and more often in close proximity to CpG islands (10, 25, 35). These data indicated that LEDGF/p75 is an important target for anti-HIV-1 drug development.
LEDGF/p75 is a ubiquitously expressed chromatin-bound protein evolutionarily conserved from bony fish to humans. The chromatin-binding domain of LEDGF/p75, located in the N-terminal region of the protein, was identified by an analysis of deletion mutants (22, 39). LEDGF/p75 mutants lacking the evolutionarily conserved PWWP and two AT hook motifs failed to bind to chromatin during both interphase and mitosis. However, the chromatin binding of mutants lacking only one of these protein regions was normal or minimally defective (22). These data indicated that LEDGF/p75 chromatin binding requires the functional interaction of the PWWP domain and two AT hook motifs (22). More recently, a mutagenesis analysis of evolutionarily conserved residues within the PWWP domain suggested that a hydrophobic cavity in this protein domain has a fundamental role in the chromatin binding of LEDGF/p75 (34).
The coimmunoprecipitation of HIV-1 integrase (IN) and LEDGF/p75 drove the interest of retrovirologists to this cellular protein (9). The interaction of LEDGF/p75 with HIV-1 integrase has been extensively characterized by using noninfected cells where integrase is expressed as a sole protein by stable or transient plasmid transfection and in in vitro systems with purified recombinant proteins (13, 29, 38, 42). The interaction of LEDGF/p75 with integrase occurs through a C-terminal region called the integrase-binding domain (IBD) (5, 8, 42). The IBD was identified by evaluating the interaction of HIV-1 integrase with LEDGF/p75 deletion mutants using different cellular and biochemical assays. LEDGF/p75 tethers HIV-1 integrase to host chromatin (21, 24), protects integrase from proteasome-mediated degradation (19), promotes integrase multimerization (26), and enhances its enzymatic activity in in vitro integration assays (6). The molecular basis of this protein-protein interaction was further defined after X-ray diffraction analysis of an IBD-integrase catalytic core domain complex (7, 16).
Although the molecular mechanism of the HIV-1 cofactor activity of LEDGF/p75 is not yet completely understood, it is well established that it requires its chromatin- and integrase-binding activities (20, 35). HIV-1 infection of LEDGF/p75-deficient cells was rescued upon the reexpression of the LEDGF/p75 wild type (WT) but not of deletion mutants lacking the chromatin- or the integrase-binding domain (20). These observations suggested that chromatin-bound LEDGF/p75 could tether lentiviral preintegration complexes to the host chromatin, allowing efficient viral integration (20). This tethering model was also supported by the role of LEDGF/p75 in HIV-1 DNA integration site distribution (10, 25, 35).
In order to evaluate if LEDGF/p75 regions not involved in the tethering mechanism are required for its HIV-1 cofactor activity, we studied a panel of deletion mutants. These mutants lacked different protein regions that are evolutionarily conserved and predicted to be solvent exposed as well as harboring different protein posttranslational modifications. HIV-1 cofactor activity and integrase-to-chromatin-tethering capacity were evaluated for each of these mutants. Using this systematic approach, we have identified that LEDGF/p75 serine residues 271, 273, and 275 are involved in its HIV-1 cofactor activity without influencing its integrase-to-chromatin-tethering activity. Our results indicate that in addition to its tethering activity, other molecular events seem to be implicated in the HIV-1 cofactor role of LEDGF/p75.
MATERIALS AND METHODS
Plasmids. (i) LEDGF/p75 expression plasmids.
pFLAG LEDGF/p75 was used for transient-expression experiments. This plasmid has a human cytomegalovirus (CMV) immediate-early gene promoter driving the transcription of a LEDGF/p75 cDNA containing seven synonymous mutations in the target site of 21-nucleotide small hairpin RNA (shRNA) 1340 (AACGGCAACACGAAGAGGCAA [changed nucleotides are underlined]) (21). This shRNA is present in all the LEDGF/p75-deficient cell lines used in this report. The LEDGF/p75 open reading frame was PCR amplified and BamHI and ApaI cloned into the expression plasmid pCMV-FLAG upstream of the FLAG sequence. pCMV-FLAG was derived from pCMV-Myc (21) by replacing Myc with the FLAG tag epitope. Myc was removed with ApaI/BglII, and a DNA linker containing the FLAG sequence flanked by ApaI/BglII sticky ends was inserted.
The stable expression of the LEDGF/p75 WT or mutants was achieved by retroviral transduction. LEDGF/p75 was expressed from the murine leukemia virus (MLV) expression plasmid pJZ308 (20). LEDGF/p75 mutants were generated by PCR with the Phusion site-directed mutagenesis kit (Finnzymes, Inc.) by using specific primers (primers sequences are available upon request) according to the manufacturer's instructions. All the constructs described above were verified by overlapping DNA sequencing of the complete LEDGF/p75 cDNA.
(ii) HIV-1 integrase expression plasmids.
pHIN-eGFP-IRES-P is a CMV-driven HIV-1 integrase-enhanced green fluorescent protein (EGFP) expression plasmid that was used to generate stable cell lines expressing this fusion protein. This plasmid was derived from pHIN (21) by replacing the Myc tag with EGFP and introducing an internal ribosome entry site (IRES)-puromycin resistance gene cassette downstream of the EGFP open reading frame. The IRES-puromycin resistance gene cassette was PCR amplified from pEFIRESP with primers 5′-TATAAGATCTAATTCACGCGTGGTACCTCTAG-3′ and 5′-TATAAGATCTGGTCGTGCGCTCCTTTCGGTCG-3′ and then introduced into a unique BglII site in pHINeGFP.
(iii) Retroviral vector plasmids.
Plasmids used for the production of retroviral vectors pHIVluc, JZluc, and pTSINcherry/p75 were previously described (20); pCMVΔR8.91 and pMD.G were a gift of D. Trono.
Cell lines. (i) LEDGF/p75-deficient cell lines.
Human CD4+ T-cell line LEDGF/p75-deficient TL3 cells (20) were used for the stable expression of LEDGF/p75 mutants. These cells were generated by the transduction of SupT1 cells with an HIV-derived vector expressing an shRNA against LEDGF/p75. As a control we used TC3 cells that were also derived by the transduction of SupT1 cells with an HIV-derived vector expressing a scrambled shRNA sequence (20). TL3 cells express 97% less LEDGF/p75 mRNA than TC3 cells, as determined by real-time PCR (20). To generate TL3 cells expressing different LEDGF/p75 mutants, cells were transduced with an MLV-derived vector expressing the LEDGF/p75 mutants (pJZ-LEDGF-FLAG) and selected in G418 (600 μg/ml) as described previously (20). Robust polyclonal G418-resistant cell lines were obtained and characterized by immunoblotting with an anti-LEDGF (catalog number 611714; BD Transduction Laboratories) or anti-FLAG (clone M2; Sigma) monoclonal antibodies (MAbs). The LEDGF/p75-deficient HEK293T-derived cell line si1340/1428 (21) was used for the transient expression of LEDGF/p75 proteins.
(ii) HIV-1 integrase cell lines.
LEDGF/p75-deficient HEK293T cells expressing Myc-tagged HIV-1 integrase (LH4 cells [19]) were used to evaluate the capacity of the LEDGF/p75 mutants to protect HIV-1 integrase from proteasome-mediated degradation. In addition, 2LKD-IN-eGFP cells were used to evaluate the integrase-to-chromatin-tethering capacity of LEDGF/p75 mutants. These cells are LEDGF/p75-deficient HEK293T cells stably expressing HIV-1 integrase C-terminally tagged with EGFP. To generate 2LKD IN-eGFP cells, HEK293T cells were plated at 3 × 106 cells in 75-cm2 flasks and calcium-phosphate transfected the next day with 20 μg of expression plasmid pCMV-IN-eGFP-IRES-P linearized at the prokaryotic backbone. Stably transfected cells were obtained after selection in the presence of puromycin (3 μg/ml), and integrase-EGFP expression was verified by immunoblotting with an anti-EGFP MAb and by fluorescence-activated cell sorting (FACS) analysis. Subsequently, a LEDGF/p75 deficiency in these cells was achieved by transduction with an HIV-based lentiviral vector expressing LEDGF/p75-specific shRNA 1340 (20). This lentiviral vector integrates into the host genome a cassette containing, in cis, a U6 small nuclear RNA promoter that drives the expression of a LEDGF/p75-specific shRNA as well as a CMV promoter directing the expression of an mCherry fluorescent protein. This expression system allows the selection of LEDGF/p75 knockdown cells based on their mCherry fluorescence levels (20). Knockdown levels of LEDGF/p75 were verified further by immunoblotting with an anti-LEDGF MAb.
SupT1-derived cell lines were grown in RPMI 1640 medium, while HEK293T-derived cells were grown in Dulbecco's modified Eagle's medium (DMEM), and both culture media were supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and 1% penicillin-streptomycin.
Generation of retroviral vectors.
Procedures previously described (20) were used for the production of the different retroviruses used here. Briefly, MLV-derived vectors were produced in Phoenix A packaging cells by calcium-phosphate cotransfection of 15 μg of the pJZLEDGF/p75 wild type or pJZLEDGF/p75 mutants or pJZluc and 5 μg of a vesicular stomatitis virus glycoprotein G (VSV-G) expression plasmid, pMD.G. Forty-eight hours after transfection, the viral supernatants were harvested and concentrated by ultracentrifugation at 124,750 × g for 2 h on a 20% sucrose cushion. HIV-derived vectors expressing anti-LEDGF shRNA were produced by calcium-phosphate cotransfection of HEK293T cells with 15 μg of pTSINcherry/p75 (20), 15 μg of pCMVΔR8.91, and 5 μg of pMD.G. The virus-containing supernatant was collected and concentrated by ultracentrifugation as described above. For single-round infection, luciferase-expressing HIV replication (HIVluc) was prepared by calcium-phosphate cotransfection of HEK293T cells with 15 μg of pHIVluc (20) and 5 μg of pMD.G. Viral supernatant was collected 48 h after transfection, and aliquots were stored at −80°C until use.
Single-round viral infectivity assay.
TL3, TC3, and LEDGF/p75-expressing TL3 cells were plated at 1 × 105 cells in 500 μl of RPMI 1640 culture medium in 24-well plates and infected with HIVluc or MLVluc viral supernatants. Five days postinfection, cells were collected by centrifugation at 1,000 × g for 6 min, and the pellet was lysed in 100 μl of phosphate-buffered saline (PBS)-1% Tween 20 for 15 min on ice. Cellular lysates were centrifuged at 22,000 × g for 2 min, and the supernatant was used for the quantification of luciferase activity. An aliquot of 20 μl of the cellular lysate supernatant was mixed with 45 μl of substrate (Bright-Glow luciferase assay system; Promega), and luciferase activity was quantified by using a microplate luminometer.
Immunoblotting.
Cellular lysates were resolved by SDS-PAGE and transferred overnight onto polyvinylidene difluoride (PDVF) membranes at 100 mA at 4°C. Membranes were blocked with Tris-buffered saline (TBS) containing 10% milk for 1 h and then incubated with the corresponding primary antibody diluted in TBS-5% milk-0.05% Tween 20 (antibody dilution buffer). FLAG-tagged LEDGF/p75 was detected with anti-FLAG MAb (1/500) (M2; Sigma), nontagged LEDGF/p75 was detected with anti-LEDGF MAb (1/250), and Myc-tagged HIV-1 integrase was detected with anti-Myc MAb (1/500) (clone 9E10; Covance). As a loading control, anti-alpha-tubulin MAb (clone B-5-1-2; Sigma) was used at a 1/4,000 dilution. Membranes were incubated overnight at 4°C with anti-FLAG, -LEDGF, and -Myc MAbs, whereas anti-alpha-tubulin MAbs were incubated for 2 h at 25°C. Primary antibody-bound membranes were washed in TBS-0.1% Tween 20, and bound antibodies were detected with goat anti-mouse Ig-horseradish peroxidase (HRP) (Sigma) diluted 1/2,000 in antibody dilution buffer followed by chemiluminescence detection.
Chromatin-binding assay.
Previously described procedures (22) were used, with minor modifications. Figure 3 in reference 22 shows a validation experiment where multiple proteins located in different cellular compartments were evaluated. Briefly, 18 × 106 TL3-derived cells expressing different LEDGF/p75 mutants were washed in PBS and distributed in three aliquots containing equal amounts of cells. Two of the samples were lysed for 15 min on ice in 100 μl of CSK I buffer {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.8), 100 mM NaCl, 1 mM EDTA, 300 mM sucrose, 1 mM MgCl2, 1 mM dithiothreitol (DTT), 0.5% Triton X-100} containing protease inhibitors (final concentrations of 2 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 μg/ml pepstatin A). The third sample was lysed for 15 min on ice in 100 μl CSK I buffer supplemented with 350 mM NaCl and protease inhibitors and centrifuged at 22,000 × g for 3 min at 4°C, and the supernatant was saved for further analysis (total fraction [T]). Cells lysed in CSK I buffer were centrifuged at 1,000 × g for 6 min at 4°C, and the supernatant was pooled (non-chromatin-bound fraction [S1]). S1 supernatants were clarified further by centrifugation at 22,000 × g for 3 min and supernatant transfer into a fresh tube while pellets were washed once in 200 μl of CSK I buffer. One of these pellets was resuspended in CSK I-350 mM NaCl buffer and incubated on ice for 15 min, followed by centrifugation at 22,000 × g for 3 min at 4°C, and the supernatant was collected for further analysis (chromatin-bound fraction [P1]). The other pellet, obtained after cell lysis in CSK I buffer, was resuspended in 100 μl of CSK II buffer supplemented with protease inhibitors, 4 units of Turbo DNase (Ambion), and 11 μl of 10× Turbo DNase reaction buffer. DNase treatment of this pellet was conducted at 37°C for 30 min and was then followed by extraction with 250 mM (NH4)2SO4 for 15 min at 37°C. The DNase-(NH4)2SO4-treated sample was centrifuged at 22,000 × g for 3 min, and the supernatant was saved for analysis (chromatin-bound fraction [S2]). The resulting pellet was further extracted with CSK I buffer-350 mM NaCl for 15 min on ice and centrifuged at 22,000 × g for 3 min, and the obtained supernatant (non-chromatin-bound fraction [P2]) was collected for analysis. A volume of 15.7 μl of S1, P1, P2, and T and 20 μl of S2 (amounts equivalent to 0.9 × 106 cells) was evaluated by immunoblotting using an anti-FLAG MAb.
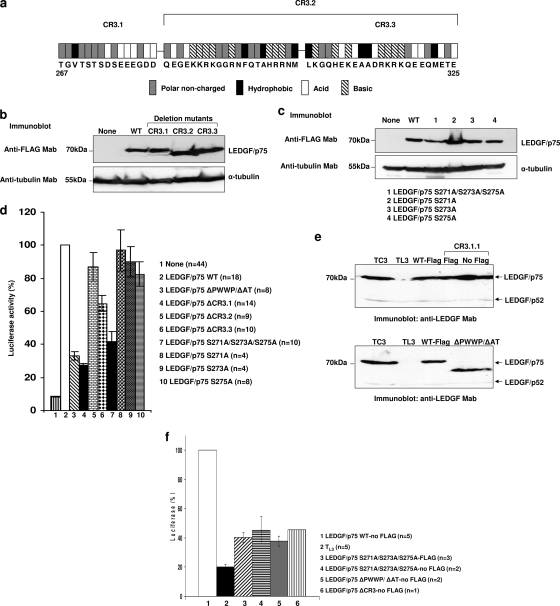
FIG. 3.
HIV-1 cofactor activity of different LEDGF/p75 CR3 mutants. (a) Analysis of charged residues in CR3 used to establish the boundaries of CR3.1, CR3.2, and CR3.3. (b and c) Expression of LEDGF/p75 mutants in TL3 cells evaluated by immunoblotting with an anti-FLAG MAb. Alpha-tubulin detection was used as a loading control. (d) Single-round infection of TL3 cells expressing LEDGF/p75 mutants. Cells evaluated in b and c were challenged with HIV-1 luciferase reporter viruses, and luciferase activity was analyzed 5 days later. Infectivity and error bars were calculated as described in the legend of Fig. 1d. (e) Immunoblotting detection of the reexpressed LEDGF/p75 WT and mutants in TL3 cells using an anti-LEDGF MAb. Detection of endogenous LEDGF/p52 was used as a loading control. (f) Single-round infection of TL3 cells expressing the nontagged LEDGF/p75 WT or mutants. Cells immunoblotted in e were challenged with HIVluc, and luciferase activity was analyzed 5 days later. Infectivity and error bars were calculated as described in the legend of Fig. 1d.
Salt extraction assay.
Approximately 36 × 106 TL3 cells or TL3 cells expressing LEDGF/p75 mutants were washed with PBS and distributed in six samples, each containing an equal amount of cells. One of the samples was resuspended in 100 μl of Laemmli sample buffer, boiled for 9 min, and centrifuged at 22,000 × g for 3 min, and the resulting supernatant was further analyzed (total fraction [T]). The other five cellular aliquots were lysed on ice for 15 min in 100 μl of CSK I buffer containing protease inhibitors and supplemented with increasing concentrations of NaCl (final concentrations of 100, 150, 200, 350, and 500 mM). Lysed cells were centrifuged at 22,000 × g for 3 min at 4°C, and the supernatant (salt-extracted fraction) was collected for analysis. The obtained pellets (salt-resistant fraction) were subsequently boiled in 100 μl of Laemmli sample buffer and centrifuged at 22,000 × g for 3 min at 4°C, and the resulting supernatant was collected. The presence of LEDGF/p75 in the total, salt-extracted, or salt-resistant fractions was analyzed by immunoblotting with an anti-FLAG MAb using 20 μl of cell lysate, equivalent to an amount of 6 × 104 cells.
Immunoprecipitation.
The interaction of the LEDGF/p75 ΔCR3 mutant with Myc-tagged HIV-1 integrase was evaluated by immunoprecipitation in the LEDGF/p75-deficient HEK293T-derived cell line si1340/1428 (19). Cells were plated at 0.45 × 106 cells/well in a six-well plate and cotransfected the next day by calcium-phosphate cotransfection with 2 μg of pFLAG-LEDGF/p75 ΔCR3, pFLAG-LEDGF/p75 WT, or pFLAG-LEDGF/p75 ΔIBD and 2 μg of HIV-1 integrase expression plasmid pHIN. Forty-eight hours after transfection, cells were lysed in 300 μl of radioimmunoprecipitation assay (RIPA) buffer (150 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% deoxycholate [DOC], 0.1% SDS, 1% NP-40) supplemented with protease inhibitors. Cell lysates were clarified by centrifugation at 22,000 × g for 3 min, and the supernatant was used for immunoprecipitation using goat anti-mouse Ig-coated magnetic beads (Pierce). Beads (100 μl) were previously incubated for 20 min on ice with 3 μg of anti-FLAG MAb diluted in RIPA buffer. The beads were then separated from the unbound antibodies, mixed with the cell lysate, and rotated for 3 h at 4°C. After this incubation, beads were washed three times in RIPA buffer, and bound proteins were eluted by boiling in 30 μl of Laemmli sample buffer. Immunoprecipitated proteins were analyzed for the presence of Myc-tagged HIV-1 integrase and LEDGF/p75-FLAG by immunoblotting with anti-Myc and anti-FLAG MAbs, respectively.
Integrase protection assay.
LEDGF/p75-deficient HEK293T cells expressing Myc-tagged HIV-1 integrase were used to evaluate the effect of different LEDGF/p75 deletion mutants on integrase protein stability. This assay is based on the integrase-binding-dependent capability of LEDGF/p75 to protect integrase from proteasome-mediated degradation (19). Integrase protein levels in these LEDGF/p75-deficient cells are very low, as detected by sensitive immunoblottings, and the reexpression of the LEDGF/p75 WT rescues integrase levels (19). Cells were plated at 0.45 × 106 cells per well in a six-well plate and transfected the next day by calcium-phosphate cotransfection with 2 μg of pFLAG-LEDGF/p75 WT or mutants. Forty-eight hours after transfection, cells were lysed in 300 μl of RIPA buffer supplemented with protease inhibitors. Cell lysates were clarified by centrifugation at 22,000 × g for 3 min, and the supernatant was used for immunoblotting with anti-Myc or anti-FLAG MAbs.
Integrase-to-chromatin-tethering assay.
The integrase-to-chromatin-tethering assay is based on the role of LEDGF/p75 in the chromatin tethering of HIV-1 integrase (21). Chromatin-bound LEDGF/p75 interacts with integrase through the IBD, tethering the viral enzyme to chromatin during all phases of the cell cycle. In LEDGF/p75-deficient cells, HIV-1 integrase lacks its exclusively nuclear localization during interphase and is not associated with chromatin during mitosis (21). 2LKD-IN-eGFP cells, lacking LEDGF/p75 and stably expressing HIV-1 integrase-eGFP, were used in this assay. Cells were plated at 2 × 105 cells in 2 ml of culture medium in LabTek II chambered coverglasses and were transfected the next day with 2 μg of pFLAG-LEDGF/p75 WT or mutants. Eighteen hours after transfection, fresh culture medium was added, and 48 h later, cells were washed three times in PBS and fixed with 4% formaldehyde-PBS for 10 min at 37°C. The cells were then washed twice with PBS and stained with DAPI (4′,6′-diamidino-2-phenylindole). The subcellular distribution of the HIV-1 IN-eGFP-LEDGF/p75 complex was analyzed by fluorescence microscopy.
HIV-1 DNA integration analysis.
Viral integration was quantified by the detection of Alu-long terminal repeat (LTR), and total HIV-1 cDNA was quantified by real-time PCR with TL3 and TL3 cells engineered to express the LEDGF/p75 WT or the S271A/S273A/S275A mutant. Two completely independent experiments using two different viral preparations were considered. In these experiments, 105 cells were infected with DNase-treated HΔEluc viral supernatant and cultured for 10 days. DNA was then extracted (High Pure PCR template preparation kit; Roche) from 106 infected cells, and 20 ng of DNA was used for the detection of total HIV-1 cDNA, mitochondrial DNA, and two-LTR (2LTR) circles, while 0.2 ng of DNA was used for the Alu-LTR junction PCR. Alu-LTR and total HIV-1 cDNA products were normalized for mitochondrial DNA and 2LTR circles, respectively. All these real-time PCRs were performed by use of a MiniOpticon system (Bio-Rad) with primers and conditions previously described (20). The fold change was calculated by using the ΔCT method as recommended in the thermocycler manual, and differences were expressed as percentages, considering the value for TL3 LEDGF/p75 WT cells to be 100%.
In silico analysis.
Mutations introduced into LEDGF/p75 were guided by a systematic bioinformatics analysis focused on evolutionary conservation across different species, the presence of posttranslational modifications, as well as the prediction of solvent accessibility. Evolutionary conservation, as referred to in this text, includes the presence of either identical or homologous residues in the compared protein sequences. LEDGF/p75 protein sequences were retrieved from the NCBI protein database and aligned using ClustalW2 (18), and sequence comparisons were performed with BLASTP 2.2.19+ (2). The LEDGF/p75 protein motif search was performed with the ELM server (31), and the prediction of relative solvent accessibility was performed with PaleAle (30).
RESULTS
Experimental strategy.
Two regions of LEDGF/p75, the chromatin-binding domain and the IBD, are required for its HIV-1 cofactor activity (20, 35). However, the involvement of other LEDGF/p75 regions in HIV-1 infection has not been previously studied. In order to evaluate if LEDGF/p75 protein regions not implicated in chromatin binding or integrase interactions are also necessary for HIV-1 infection, a panel of deletion mutants was analyzed. These mutants were stably expressed in a LEDGF/p75-deficient human CD4+ T-cell line (TL3 cells) (20), and their ability to rescue HIV-1 infection was evaluated. In addition, the chromatin- and integrase-binding activities of these mutants were assessed.
Although analyses of LEDGF/p75 deletion mutants were used previously to successfully map functional domains in LEDGF/p75 (8, 22, 23, 39, 41), this approach could lead to misinterpretations due to the global impact of mutations on the structure and, hence, the function of the analyzed protein. In order to prevent data overinterpretation, only deletion mutants causing a decrease in the HIV-1 cofactor activity of LEDGF/p75 similar to that observed following the deletion of the chromatin-binding or the integrase-binding domain were considered for further analysis. Finally, the phenotypes observed for deletion mutants were verified with LEDGF/p75 point mutants.
In order to facilitate the analysis of LEDGF/p75 mutants, C-terminally FLAG-tagged LEDGF/p75 was used. To exclude any artifactual observation due to the addition of this small tag to LEDGF/p75, tagged (WT-FLAG) and not-tagged (WT) versions of this protein were evaluated for their interactions with chromatin and HIV-1 integrase and for their HIV-1 cofactor activities (Fig. 1).
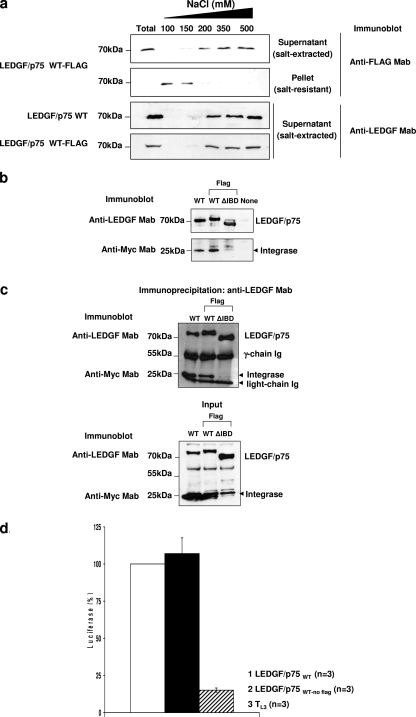
FIG. 1.
(a) Salt extraction assay. TL3 cells expressing the LEDGF/p75 WT or WT-FLAG were lysed in a buffer containing increasing NaCl concentrations. Cells were separated into a soluble fraction and an insoluble fraction by centrifugation, and the presence of LEDGF/p75 was evaluated by immunoblotting with an anti-LEDGF or anti-FLAG MAb, as indicated. An unfractionated total cellular fraction (T) was included as a control. (b) Integrase protection assay. LEDGF/p75-deficient HEK293T cells stably expressing Myc-tagged HIV-1 integrase were either transiently transfected or not transfected with plasmids expressing the LEDGF/p75 WT, WT-FLAG, or ΔIBD-FLAG. Expression levels of LEDGF/p75 proteins and HIV-1 integrase were determined by immunoblotting with anti-LEDGF and anti-Myc MAbs, respectively. (c) Coimmunoprecipitation of HIV-1 integrase with LEDGF/p75. LEDGF/p75-deficient HEK293T cells were cotransfected with plasmids expressing the LEDGF/p75 WT, WT-FLAG, or ΔIBD-FLAG and Myc-tagged HIV-1 integrase and subjected to immunoprecipitation with an anti-LEDGF MAb. Immunoprecipitated proteins were detected by immunoblotting with anti-Myc or anti-LEDGF MAbs. Mouse antibody heavy and light chains were used as a loading control for the immunoprecipitation. LEDGF/p75 and HIV integrase proteins were detected in the samples used for immunoprecipitation (input) by immunoblotting with anti-LEDGF and anti-Myc MAbs, respectively. (d) Single-round infection of TL3 cells expressing the LEDGF/p75 WT or WT-FLAG. Cells were challenged with HIVluc, and luciferase activity was analyzed 5 days later. Luciferase levels detected in TL3 cells expressing the LEDGF/p75 WT were considered to be 100%. Error bars indicate standard deviation values calculated for a number (n) of independent infection experiments performed on different days with different viral preparations.
The effect of this tag on the chromatin-binding strength of LEDGF/p75 was evaluated by using a chromatin salt extraction assay. This is based on the effect of increased salt concentrations on the binding of proteins to chromatin. TL3 cells expressing the LEDGF/p75 WT or WT-FLAG were lysed in 1% Triton X-100-CSK I buffer containing increasing concentrations of NaCl (100, 150, 200, 350, and 500 mM). Cellular lysates were centrifuged, and the supernatant (salt-extracted fraction) and pellet (salt-resistant fraction) were evaluated by immunoblotting for the presence of LEDGF/p75. Total LEDGF/p75 extraction was achieved by lysing unfractionated cells in Laemmli sample buffer (Fig. 1a). Using this method, we found that both LEDGF/p75 forms persisted bound to chromatin (salt-resistant fraction) at 100 and 150 mM NaCl and were fully extracted above 200 mM NaCl (Fig. 1a), indicating that the FLAG tag did not affect its binding to chromatin.
To evaluate the effect of the FLAG tag on the interaction of LEDGF/p75 with HIV-1 integrase, we compared the abilities of the LEDGF/p75 WT and WT-FLAG to protect integrase from proteasome-mediated degradation (integrase protection assay) (Fig. 1b). LEDGF/p75-deficient HEK293T cells stably expressing Myc-tagged HIV-1 integrase, LH4 cells, were transiently transfected with plasmids expressing the LEDGF/p75 WT, WT-FLAG, or the ΔIBD mutant, and integrase protein levels were evaluated by immunoblotting 48 h after transfection. Steady-state HIV-1 integrase levels were very low in the absence of LEDGF/p75 and dramatically increased upon the expression of the LEDGF/p75 WT or WT-FLAG but not of the ΔIBD mutant (Fig. 1b), indicating that the FLAG tag did not affect the interaction of LEDGF/p75 with HIV-1 integrase.
The influence of the FLAG tag on the direct interaction of LEDGF/p75 with HIV-1 integrase was also evaluated by coimmunoprecipitation. LEDGF/p75-deficient HEK293T cells coexpressing the LEDGF/p75 WT, WT-FLAG, or ΔIBD-FLAG and Myc-tagged HIV-1 integrase were lysed and immunoprecipitated with an anti-LEDGF/p75 MAb. The presence of integrase and LEDGF/p75 in the immunoprecipitated proteins was then evaluated by immunoblotting with anti-Myc or anti-LEDGF/p75 MAbs, respectively. In support of data shown in Fig. 1b, both the LEDGF/p75 WT and WT-FLAG very efficiently immunoprecipitated HIV-1 integrase, while the LEDGF/p75 ΔIBD mutant did not (Fig. 1c). These data also demonstrated that the FLAG tag on LEDGF/p75 did not affect its interaction with HIV-1 integrase.
Finally, we compared the HIV-1 cofactor activity of the LEDGF/p75 WT with that of WT-FLAG. TL3 cells stably expressing or not expressing each of these LEDGF/p75 proteins were infected with HIVluc, and 5 days after infection, luciferase levels were detected (Fig. 1d). In correlation with observations described in the legend of Fig. 1a to c, the C-terminal FLAG tag did not alter the LEDGF/p75 HIV cofactor activity.
In summary, the data in shown Fig. 1 clearly demonstrated that LEDGF/p75 WT-FLAG is functionally equivalent to its nonmodified variant, and unless otherwise specified, all the data described below were obtained with LEDGF/p75 FLAG tagged at the C terminus.
LEDGF/p75 deletion mutants.
Regions deleted from LEDGF/p75 are schematized in Fig. 2a. These regions were designated conserved/charged region 1 (CR1), CR2, CR3, CR4, and CR5. Region boundaries were established based on the evolutionary conservation of LEDGF/p75 across different species, the presence of predicted or demonstrated posttranslational modifications, and the existence of clusters of polar residues that correlate directly with protein solvent accessibility.
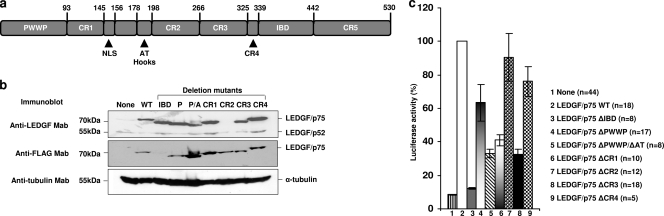
FIG. 2.
Evaluation of the HIV-1 cofactor activities of LEDGF/p75 deletion mutants. (a) Schematic representation of deleted regions. (b) Different LEDGF/p75 deletion mutants were stably expressed in LEDGF/p75 knockdown TL3 cells, and their protein levels were evaluated by immunoblotting using anti-LEDGF or anti-FLAG MAbs. Alpha-tubulin detection was used as a loading control. P and P/A represents PWWP and PWWP/AT hook, respectively. (c) Single-round HIV-1 infections of TL3-derived cells. Cells immunoblotted in b were challenged with HIVluc, and luciferase activity was determined 5 days later.
CR1 contains 52 amino acids and was selected based on its evolutionary conservation in mammals and based on its proposed role in the LEDGF/p75 PWWP domain chromatin-binding activity (22). This region is enriched for charged amino acids (54.8%), and 76.4% of them are identical or homologous residues (conserved) present from avian to human (data not shown). Interestingly, CR1 contains a cluster of eight Ser/Thr phosphosites between residues 105 and 135 (4, 12, 28, 33, 36, 43); this cluster represents 36.3% of the total identified phosphorylated Ser/Thr residues in LEDGF/p75. In addition, Thr141 has been found to be phosphorylated in both human and mouse cells (4, 12, 33). This phosphosite is contained in a predicted forkhead-associated (FHA) domain interaction motif, LEDGF/p75 residues 141 to 144. The FHA domain is a protein module present in prokaryotic and eukaryotic proteins involved in signaling, cell cycle control, and DNA repair that binds to specific threonine-phosphorylated sequences (31).
CR2 and CR3 boundaries were previously described (22). These regions were demonstrated to cooperate with the PWWP domain and AT hook motifs during LEDGF/p75 chromatin binding (22). CR2 is 68 amino acids long; 58.8% of the residues are charged, and 67% are conserved from amphibians to humans (data not shown). Evolutionary conservation and solvent exposure are particularly interesting for residues 212 to 249, where 68% are present in bony fish and 70.3% of them are charged and predicted to be exposed to the solvent (30). In addition, although CR2 represents only 12.8% of LEDGF/p75, it contains 29.4% of its lysine residues. Lysine is targeted by different posttranslational modifications including methylation, SUMOylation, ubiquitination, and glycosylation.
CR3 is 59 amino acids long, and 64% of these residues are charged (data not shown). The N-terminal region of CR3, residues 267 to 275, contains six demonstrated phosphorylated Ser/Thr sites, which represent 27.2% of the identified LEDGF/p75 Ser/Thr phosphosites (11, 12, 15, 17, 28, 36, 43, 44). These Ser/Thr residues are predicted to be targeted by protein kinase casein kinase 2 (PKCK2) (31). In addition, residues 271 to 275 contain two phosphorylated serines that are conserved from bony fish to humans. An evolutionarily conserved cluster of acidic residues follows these phosphosites.
The C-terminal part of LEDGF/p75 contains the IBD. N terminally to this domain, a region 14 amino acids long that is rich in conserved and charged residues was deleted for analysis (CR4 region). In addition, a region 88 amino acids long, designated CR5, that is located C terminally to the IBD was also independently deleted. Although 70% of the residues in CR5 are evolutionarily conserved, this region is not rich in charged amino acids. However, CR5 contains four demonstrated Ser/Thr phosphosites (12), and three of them are clustered in its C-terminal end.
Role of different protein regions of LEDGF/p75 in HIV-1 infection.
TL3 cells were used to evaluate the role of different LEDGF/p75 protein regions in HIV-1 infection. Deletion mutants were stably expressed by the retroviral transduction of TL3 cells. Similar levels of reexpressed LEDGF/p75 proteins of the expected relative molecular weights were verified by immunoblotting with anti-LEDGF/p75 and anti-FLAG MAbs (Fig. 2b). The non-FLAG-tagged LEDGF/p75 ΔIBD mutant was detected only with an anti-LEDGF/p75 MAb, while the FLAG-tagged LEDGF/p75 ΔCR2 mutant, lacking the epitope detected by the anti-LEDGF/p75 MAb, was detected only by the anti-FLAG antibody. The LEDGF/p75 ΔCR5 mutant failed to be expressed in TL3 cells, although robust polyclonal G418-resistant cell lines were obtained after two independent attempts. In addition, the treatment of these cell lines with the proteasome inhibitor MG132 did not rescue the expression of the LEDGF/p75 ΔCR5 mutant (data not shown); therefore, we excluded this region from further analysis.
TL3-derived cell lines expressing LEDGF/p75 deletion mutants were challenged with HIVluc (20), and 5 days later, luciferase activity was measured (Fig. 2c). Standard deviations and the numbers of independent experiments performed are indicated. Luciferase levels detected in TL3 cells expressing the LEDGF/p75 WT were considered to be 100% infectivity.
The reexpression of the FLAG-tagged LEDGF/p75 WT rescued infectivity in these cells, as previously reported for nontagged LEDGF/p75 (20). In addition, LEDGF/p75 proteins lacking the IBD, the PWWP domain (P), or the PWWP domain and the AT hook motifs (P/AT) exhibited 12.3%, 63.4%, and 32.9% of the HIV-1 cofactor activity of the LEDGF/p75 WT, respectively. These data were also in agreement with previously reported observations (20, 35). Interestingly, the deletion of CR2 or CR4 only slightly impaired the cofactor activity of LEDGF/p75, showing 90.5% and 76% of the wild-type activity, respectively. On the contrary, the deletion of CR1 reduced the cofactor activity to 41%, while the deletion of CR3 caused a greater defect, exhibiting only 32.4% of the wild-type activity. Importantly, the reduction in cofactor activity observed after deleting CR3 was comparable to that observed after the simultaneous deletion of PWWP and AT hooks (Fig. 2c).
In order to evaluate the role of CR3 of LEDGF/p75 in HIV-1 infection further, we generated three smaller deletion mutants within this region (Fig. 3a). The boundaries of the new deletion mutants were established considering the sequence aspects described above. A group of positively charged amino acids conserved from avian to human was considered to limit CR3.1, whereas CR3.2 was defined by the presence of two clusters of charged and conserved residues. CR3.3 is a deletion within CR3.2 containing the C-terminal cluster of conserved and charged residues present in CR3.2 (Fig. 3a). LEDGF/p75 proteins lacking CR3.1, CR3.2, and CR3.3 were expressed in TL3 cells (Fig. 3b), and their HIV-1 cofactor activities were evaluated (Fig. 3d). Although the deletion of CR3.2 and CR3.3 showed 86.9% and 64.7% of the LEDGF/p75 WT cofactor activity, respectively, the CR3.1 deletion decreased the activity of LEDGF/p75 to 27.3% of that observed for the wild-type protein. These data suggest that LEDGF/p75 residues 267 to 281 have a role in HIV-1 infection.
Role of Ser residues 271, 273, and 275 in HIV-1 infection.
Serine residues 271, 273, and 275 in LEDGF/p75 were previously reported to be phosphosites in global phosphoproteomics analyses of human and mouse cells (11, 12, 15, 17, 28, 36, 43, 44). In addition, these residues are predicted to be targeted by protein kinase casein kinase 2 (PKCK2) (31), an enzyme that phosphorylates serine/threonine residues that are followed by an acidic residue in position +3 from the C terminus of the phosphate acceptor (27). In order to evaluate the role of serine residues 271, 273, and 275 in HIV-1 infection, LEDGF/p75 S271A/S273A/S275A, S271A, S273A, and S275A mutants were stably expressed in TL3 cells (Fig. 3c), and HIV-1 infectivity was evaluated (Fig. 3d). Immunoblot analysis of transduced TL3 cells demonstrated similar levels of the reexpressed LEDGF/p75 mutants compared to cells expressing the wild-type protein (Fig. 3c). However, the HIV-1 cofactor activity of the LEDGF/p75 S271A/S273A/S275A mutant was only 41.5% of the activity of the wild-type protein, indicating the relevance of these phosphosites. These results were confirmed by evaluating two independently derived TL3 cell lines expressing the LEDGF/p75 S271A/S273A/S275A mutant in 10 independent infection experiments with HIVluc. Although HIV-1 infection was reduced in TL3 LEDGF/p75 S271A/S273A/S275A cells, its susceptibility to infection by a murine leukemia virus (MLV)-derived vector expressing luciferase was not altered compared to parental cells (data not shown). Since LEDGF/p75 is not required for MLV infection (20, 35), these data further confirmed the specificity of the observed effect, indicating that the reduced susceptibility to HIV-1 infection was not due to a global defect present in these two independently derived cell lines expressing the LEDGF/p75 S271A/S273A/S275A mutant.
Acidic residues in positions −1, +1, +2, +4, and +5 relative to the phosphate acceptor site are present in most of the physiological substrates of PKCK2 (27). Additional acidic residues are found at positions +5 of Ser271; +1, +4, and + 5 of Ser273; and −1, +1, +2, and + 5 of Ser275. This analysis predicts Ser275 to be a better substrate of PKCK2 than Ser271 or Ser273. To establish the implication of each of these residues in HIV-1 infection, TL3 cells expressing LEDGF/p75 S271A, S273A, or S275A single-point mutants were developed (Fig. 3c) and challenged with HIVluc. As shown in Fig. 3d, individual Ser-to-Ala mutations of these residues did not have any effect on the viral cofactor activity of LEDGF/p75, exhibiting 82.6% to 96.9% of the wild-type activity.
In all the HIV infection experiments described above (Fig. 2c and 3d), the LEDGF/p75 ΔCR3 and S271A/S273A/S275A mutants expressed in TL3 cells were FLAG tagged at their C termini. Although we demonstrated that FLAG did not affect the HIV cofactor activity of the LEDGF/p75 WT (Fig. 1d); we also evaluated the role of nontagged versions of the LEDGF/p75 S271A/S273A/S275A, ΔCR3, and ΔPWWP/ΔAT mutants in HIV infection. TL3 cells expressing the nontagged LEDGF/p75 WT or mutants (Fig. 3e) were challenged with HIVluc, and 5 days later, the luciferase activity was determined. As expected, a similar impairment in the HIV cofactor activity of LEDGF/p75 was observed for these mutants regardless of the presence or absence of the FLAG tag (Fig. 3f).
HIV-1 integration in TL3 cells expressing the LEDGF/p75 S271A/S273A/S275A mutant.
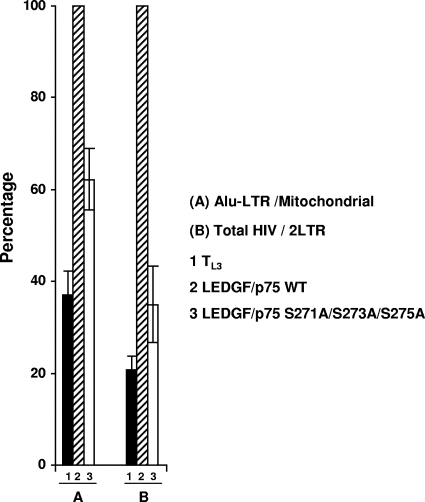
A role of LEDGF/p75 in HIV-1 DNA integration has been clearly established (20). In order to evaluate if the LEDGF/p75 motif S271/S273/S275 was involved in viral integration, TL3 cells expressing or not expressing the LEDGF/p75 WT or the S271A/S273A/S275A mutant were infected with HIVluc and analyzed 10 days later. Two completely independent experiments using two different viral preparations were analyzed. DNA was isolated from infected cells, and Alu-LTR junctions, mitochondrial DNA, HIV gag DNA (total HIV DNA), and HIV 2LTR circles were quantified by real-time PCR. Levels of Alu-LTR junctions were normalized to mitochondrial DNA to ensure that equal cell numbers were analyzed.
As reported above, Alu-LTR levels were decreased in TL3 cells compared to TL3 cells expressing the LEDGF/p75 WT, indicating that HIV-1 DNA integration is defective in the absence of LEDGF/p75 (Fig. 4). Importantly, TL3 cells expressing the LEDGF/p75 S271A/S273A/S275A mutant were also defective for viral integration, and the integration levels correlated to some extent with the degree of impairment in its cofactor activity, as shown in Fig. 3d and f.
FIG. 4.
HIV-1 DNA integration in TL3 cells expressing the LEDGF/p75 S271A/S273A/S275A mutant. Results from two independent experiments using different viral preparations are represented. DNA products were quantified by real-time PCR 10 days after infection. Bars represent Alu-LTR DNA levels after normalization to mitochondrial DNA (A) and total HIV DNA levels after normalization to 2LTR circle DNA (B). Normalized levels of Alu-LTR and total HIV DNA detected in TL3 LEDGF/p75 WT cells were considered to be 100%.
In order to estimate HIV-1 integration by an alternative approach, the level of total HIV DNA (gag DNA) was determined for these cells at 10 days postinfection. Since infected cells passed several generations before analysis, it is expected that only integrated HIV DNA forms will persist and that gag DNA will be a good indicator of viral integration. Nevertheless, to exclude the possibility of also detecting remnants of nonintegrated forms of the viral genome, the levels of total HIV DNA were normalized to levels of HIV 2LTR circles. Importantly, similar 2LTR levels were detected in the different infected cell lines at the time of analysis (data not shown). Results shown in Fig. 4 show that the total amount of HIV DNA was also reduced in infected TL3 LEDGF/p75 S271A/S273A/S275A cells compared to cells expressing the wild-type protein, indicating that HIV DNA integration is defective in TL3 cells expressing this LEDGF/p75 mutant.
Importantly, the total HIV DNA levels in the cells analyzed correlated better with their susceptibility to infection than did the levels of Alu-LTR junctions (compare data in Fig. 3d and 4). These results suggested that HIV-1 DNA integration in genomic regions distant from Alu sequences was also impaired in TL3 and TL3 LEDGF/p75 S271A/S273A/S275A cells.
Chromatin-binding activity of LEDGF/p75 mutants.
It was previously proposed that LEDGF/p75 acts as a molecular tether that links the preintegration complex-associated integrase to the host chromatin (10, 21). According to the tethering model, two functional LEDGF/p75 regions, the chromatin-binding domain and the IBD, are required for HIV-1 integration (20). In order to correlate the HIV-1 cofactor activity of LEDGF/p75 CR mutants with their tethering capacity, we evaluated their chromatin-binding and integrase interaction activities.
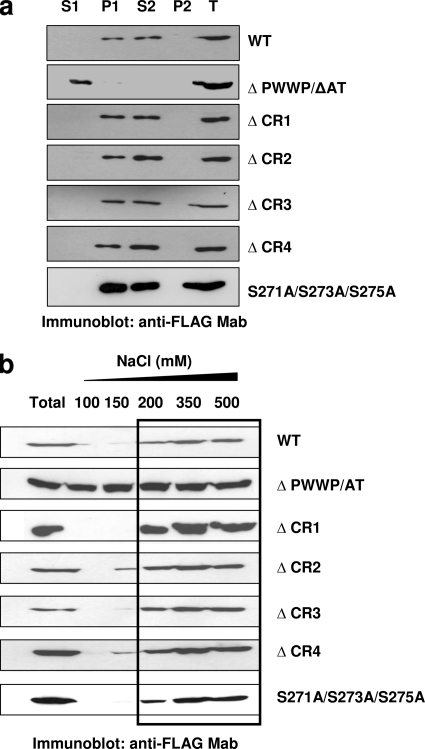
Chromatin binding was evaluated by using a previously reported chromatin-binding assay, with minor modifications (22) (Fig. 5a). This procedure is based on the resistance of chromatin-bound proteins to be extracted with isotonic buffers (i.e., CSK I buffer) containing 1% Triton X-100. In agreement with previously reported observations (22, 34), LEDGF/p75 persisted bound to the chromatin fraction (P1 fraction) after cellular lysis in 1% Triton X-100-CSK I buffer, and it was released from this compartment only by treatment with DNase and 250 mM (NH4)2SO4 (S2 fraction) (Fig. 5a). However, a LEDGF/p75 mutant lacking the chromatin-binding domains, the ΔPWWP/ΔAT mutant, was fully extracted by 1% Triton X-100-CSK I buffer (S1 fraction) (Fig. 5a). This assay revealed that all four LEDGF/p75 deletion mutants and the LEDGF/p75 S271A/S273A/S275A mutant were exclusively distributed to the chromatin-bound fractions (P1 and S2 fractions) as the wild-type LEDGF/p75 protein (Fig. 5a). Importantly, although the HIV-1 cofactor activity of the LEDGF/p75 ΔCR3 or LEDGF/p75 S271A/S273A/S275A mutant was impaired (Fig. 2c and 3d), the chromatin-binding activity of these mutants was unaffected. These results excluded defective chromatin binding as being the cause of deficient cofactor activity in these LEDGF/p75 mutants.
FIG. 5.
Chromatin-binding activity of LEDGF/p75 mutants. (a) Chromatin-binding assay. TL3 cells expressing different LEDGF/p75 mutants were fractionated into non-chromatin-bound fractions (S1 and P2) and chromatin-bound fractions (P1 and S2), and the presence of LEDGF/p75 in these fractions was evaluated by immunoblotting with an anti-FLAG MAb. An unfractionated total cellular fraction (T) was included as a control. (b) Salt extraction analysis of LEDGF/p75 mutants stably expressed in TL3 cells. The salt concentration that extracted the LEDGF/p75 WT from chromatin is marked with a rectangle.
Although the CR-deleted LEDGF/p75 mutants are chromatin bound, we evaluated the strength of their binding by the salt extraction method described in the legend of Fig. 1a. Using this method, we found that the LEDGF/p75 WT persisted bound to chromatin (salt-resistant fraction) at 100 and 150 mM NaCl and was fully extracted above 200 mM NaCl (Fig. 1a and 5b). Contrary to this, a mutant lacking the PWWP domain was partially extracted at 100 mM and fully extracted above 150 mM NaCl (data not shown), whereas the LEDGF/p75 ΔPWWP/ΔAT mutant, which failed to bind to chromatin, was fully extracted at 100 mM NaCl (Fig. 5b). Therefore, the strength of chromatin binding determined for the LEDGF/p75 WT and ΔPWWP/ΔAT mutant by the salt extraction method perfectly correlated with their chromatin-binding activities observed with the above described chromatin-binding assay (Fig. 5a).
The strength of chromatin binding of the different LEDGF/p75 CR deletion mutants was then evaluated by the salt extraction method. As shown in Fig. 5b, the deletion of CR1 or CR3 did not affect the LEDGF/p75 chromatin-binding strength since these mutant proteins were fully extracted only above 200 mM NaCl. In contrast to these results, the deletion of CR2 and CR4 slightly decreased LEDGF/p75 resistance to salt extraction, which was evidenced by a partial extraction of these mutants at 150 mM NaCl (Fig. 5b). These data suggested that CR2 and CR4 contribute to the number of charged interactions between LEDGF/p75 and chromatin. In addition, these results correlated with a data from previous report indicating a contribution of CR2 and the C-terminal region of LEDGF/p75 to chromatin binding (22).
Similarly to the LEDGF/p75 ΔCR3 mutant, the strength of the chromatin binding of the LEDGF/p75 S271A/S273A/S275A mutant was indistinguishable from that of the wild-type protein (Fig. 5b). These results, together with the wild-type phenotype observed for these mutants in the chromatin-binding assay (Fig. 5a), strongly indicated that their defective HIV-1 cofactor activity cannot be ascribed to an impaired chromatin-binding capacity.
Interaction of LEDGF/p75 mutants with HIV-1 integrase.
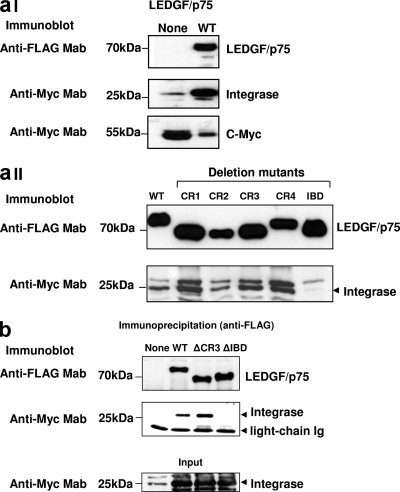
In order to analyze the functional interaction of LEDGF/p75 mutants with HIV-1 integrase, we evaluated their capacities to shield integrase from degradation (integrase protection assay) (Fig. 1b). LEDGF/p75-deficient HEK293T cells stably expressing Myc-tagged HIV-1 integrase were transiently transfected with plasmids expressing the LEDGF/p75 WT or mutants, and integrase protein levels were evaluated by immunoblotting 48 h later.
Steady-state HIV-1 integrase levels were very low in the absence of LEDGF/p75 and dramatically increased upon the expression of the LEDGF/p75 WT but not of the LEDGF/p75 ΔIBD mutant (Fig. 1b and 6aI), as previously reported (19). Using this reporter system, we evaluated the interactions of the different LEDGF/p75 deletion mutants with HIV-1 integrase. Significantly, the expression of the different LEDGF/p75 CR deletion mutants (Fig.6aII) and of the LEDGF/p75 S271A/S273A/S275A mutant (data not shown) prevented integrase degradation in LH4 cells. The levels of rescued integrase mirrored those of reexpressed LEDGF/p75 proteins, indicating that these mutations did not affect the capacity of LEDGF/p75 to interact with the viral protein. In addition, these data confirmed previously reported observations showing that the IBD was necessary and sufficient to protect integrase from degradation (19). Furthermore, these findings clearly indicated that the reduced cofactor activity observed for the LEDGF/p75 ΔCR3 mutant or the LEDGF/p75 S271A/S273A/S275A triple point mutant was not due to a defective interaction with HIV-1 integrase.
FIG. 6.
Interaction of LEDGF/p75 mutants with HIV-1 integrase. (a) Integrase protection assay. LEDGF/p75-deficient HEK293T cells stably expressing Myc-tagged HIV-1 integrase were either transiently transfected or not transfected with plasmids expressing the LEDGF/p75 WT (I) or mutants (II). Expression levels of LEDGF/p75 proteins and HIV-1 integrase were determined by immunoblotting with an anti-FLAG and anti-Myc MAbs, respectively. Detection of endogenous c-Myc was used as a loading control for these experiments. (b) Coimmunoprecipitation of HIV-1 integrase with the LEDGF/p75 ΔCR3 mutant. LEDGF/p75-deficient HEK293T cells were cotransfected with the FLAG-tagged LEDGF/p75 WT or different mutants and Myc-tagged HIV-1 and subjected to immunoprecipitation with an anti-FLAG MAb. Immunoprecipitated proteins were detected by immunoblotting with anti-Myc or anti-FLAG MAbs. Mouse antibody light chains were used as a loading control for the immunoprecipitation.
In order to evaluate the interaction of the LEDGF/p75 ΔCR3 mutant with HIV-1 integrase directly, we performed a coimmunoprecipitation experiment (Fig. 6b). The LEDGF/p75 WT or the ΔCR3 or ΔIBD mutant was transiently coexpressed with Myc-tagged HIV-1 integrase in a LEDGF/p75 knockdown HEK293T cell line, si1340/1428 cells (21). Forty-eight hours later, cells were lysed in RIPA buffer and incubated with anti-mouse Ig-coated magnetic beads loaded with anti-FLAG MAb. Immunoprecipitated proteins were later evaluated for the presence of HIV-1 integrase or LEDGF/p75 by immunoblotting with anti-Myc or anti-FLAG MAbs, respectively. The data shown in Fig. 6b clearly demonstrated that the deletion of CR3 did not alter the capacity of LEDGF/p75 to interact with HIV-1 integrase. These observations correlated with the data obtained with the integrase protection assay.
In summary, the results described above indicated that the defective HIV-1 cofactor activity detected for the LEDGF/p75 ΔCR3 or LEDGF/p75 S271A/S273A/S275A mutant is not due to an impaired interaction of these mutants with HIV-1 integrase.
Integrase-to-chromatin-tethering activity of LEDGF/p75 mutants.
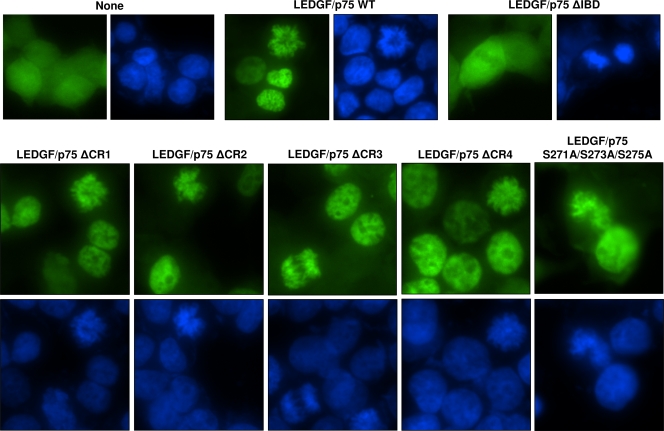
We next evaluated the capacity of the LEDGF/p75 mutants to tether HIV-1 integrase to chromatin (integrase-to-chromatin-tethering assay). In this assay, we used LEDGF/p75-deficient cells expressing HIV-1 integrase-EGFP (2LKD-IN-eGFP cells). HIV-1 integrase-EGFP was distributed pancellularly in 2LKD-IN-eGFP cells during interphase (Fig. 7), and it was not associated with condensed chromatin during mitosis (data not shown). However, upon the reexpression of the LEDGF/p75 WT, this fluorescent fusion protein was dramatically localized to the cell nuclei in interphase or to condensed chromatin during cell division (Fig. 7). However, and as expected, this striking subcellular redistribution was not observed after the expression of the LEDGF/p75 ΔIBD mutant in 2LKD-IN-eGFP cells (Fig. 7).
FIG. 7.
Integrase-to-chromatin-tethering assay. LEDGF/p75-deficient HEK293T cells stably expressing EGFP-tagged HIV-1 integrase were either transiently transfected or not transfected with plasmids expressing the LEDGF/p75 WT or mutants and analyzed by fluorescence microscopy. Chromatin was stained with DAPI.
Using the integrase-to-chromatin-tethering assay, we observed that the transient expression of the four LEDGF/p75 CR mutants or the LEDGF/p75 S271A/S273A/S275A mutant consistently tethered integrase-EGFP to the nuclear compartment and to mitotic chromosomes (Fig. 7). This effect on integrase-EGFP localization was identical to that observed with the LEDGF/p75 WT, indicating that mutations of these regions did not alter the tethering capacity of LEDGF/p75. These results were also in agreement with data obtained with the chromatin-binding and integrase protection assays.
Although a reduction in the strength of LEDGF/p75 chromatin binding was observed after the deletion of CR2 and CR4 (Fig. 5b), this defect did not impair the capacity of these mutants to tether integrase to chromatin in cells. Even more importantly, the defect of the LEDGF/p75 ΔCR3 or LEDGF/p75 S271A/S273A/S275A mutant in rescuing HIV-1 infection in TL3 cells cannot be attributed to an impairment of the tethering capacity of these mutants.
In sum, these results clearly demonstrated that the reduced HIV-1 cofactor activity observed for the LEDGF/p75 ΔCR3 and LEDGF/p75 S271A/S273A/S275A mutants is not due to an impairment of their chromatin-binding activity, HIV-1 integrase-binding activity, or integrase-to-chromatin-tethering capability. These data strongly emphasized the relevance of LEDGF/p75 serine residues 271, 273, and 275 in the HIV-1 cofactor activity of a tethering-competent LEDGF/p75 molecule.
DISCUSSION
LEDGF/p75 is a cellular cofactor of HIV-1 DNA integration (20). However, the molecular mechanism of this activity awaits further clarification. Analysis of LEDGF/p75 mutants indicated that the chromatin- and integrase-binding properties of this cellular protein are required for its cofactor activity (20, 35). These observations have led to the proposal of a tethering mechanism where chromatin-bound LEDGF/p75 interacts with HIV-1 integrase, tethering the PIC to the host chromatin (20). The role of LEDGF/p75 in the HIV-1 integration site distribution further supported this tethering model (10, 25, 35). In addition to tethering integrase to chromatin, LEDGF/p75 protects the viral enzyme from proteasome-mediated degradation and increases its enzymatic activity in vitro (6, 19). However, it is unknown if these LEDGF/p75 functions are also required for its HIV-1 cofactor activity.
Here we report molecular evidences of a new requirement for the HIV-1 cofactor activity of LEDGF/p75. We have found that serine residues 271, 273, and 275 of LEDGF/p75 importantly influence its HIV-1 cofactor activity. A mutation of these amino acids to alanine impairs the HIV-1 cofactor activity of LEDGF/p75 without altering its chromatin- or integrase-binding activities or its integrase-to-chromatin-tethering capacity, as measured by different cellular and biochemical approaches. The level of impairment in the cofactor activity of LEDGF/p75 caused by these point mutations is similar to levels observed for LEDGF/p75 mutants that are defective for chromatin binding. In addition, a preintegration defect in the HIV-1 viral life cycle was observed for TL3 cells expressing this mutant. These results suggest that serines 271, 273, and 275 mediate an unknown LEDGF/p75 function that is involved in HIV-1 DNA integration.
Beyond the identification of a role of serine residues 271, 273, and 275 in the HIV-1 cofactor activity of LEDGF/p75, our data formally demonstrate that the CRs of the protein are not required for chromatin or integrase interactions. Noticeably, chromatin-binding, integrase-binding, and integrase-to-chromatin-tethering functions of LEDGF/p75 tolerate significant structural changes, since none of the CR deletions importantly impaired them. In contrast, the HIV-1 cofactor activity of LEDGF/p75 was more sensitive to these structural changes. Although mutations affecting serines 271, 273, and 275 (ΔCR3, ΔCR3.1, and S271A/S273A/S275A) markedly reduced the cofactor activity of LEDGF/p75, the CR1 deletion also decreased its activity. These results indicated that the tethering of HIV-1 integrase to chromatin, while central in the molecular mechanism of LEDGF/p75 in HIV-1 infection, is not sufficient for the full HIV-1 cofactor activity of this cellular protein.
The molecular mechanism of serine residues 271, 273, and 275 in the HIV-1 cofactor activity of LEDGF/p75 is not yet known. This motif is evolutionarily conserved from avian to human and is located in a region of LEDGF/p75 predicted to be highly exposed to the solvent (30). Serine residues are targeted by different posttranslational modifications, including phosphorylation, O-acetylation, and O-glycosylation. Although these serines are not predicted to be O-acetylated or O-glycosylated, they have been found to be phosphorylated in different global phosphoproteomic analyses of human and mouse cells (11, 12, 15, 17, 28, 36, 37, 43, 44). Serines 271, 273, and 275 are predicted to be a substrate for phosphorylation by PKCK2. This kinase phosphorylates Ser or Thr residues in the motif S/T-X-X-E/D, where X is a nonbasic amino acid (27). PKCK2-dependent phosphorylation facilitates protein-protein interactions in different cellular processes (1, 3, 14, 27, 32). Based on all this previously reported evidence, we speculate that serine phosphorylation at this motif could influence the cofactor activity of LEDGF/p75 by facilitating its interaction with other cellular proteins implicated in HIV-1 integration.
Acknowledgments
This work was supported by Public Health Service grant 1SC2GM082301-01 from the NIH and also by University of Texas at El Paso (UTEP) start-up funds. J.A.G.-R. was supported by a fellowship from the UTEP RISE program, NIH grant 2R25GM069621-05. J.R.K. was supported by HHMI grant 52005908. UTEP core facilities are funded by BBRC grant 5G12RR008124.
We thank Eric Poeschla, Mayo Clinic, Rochester, MN, for providing us with important reagents; Janeth Cortez and Damaris Rosado for technical assistance; Igor Almeida and Luciane Ganiko, UTEP, for technical support with the microscopic analysis; and Jeremy Ross, UTEP, for important suggestions.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Abbott, K. L., M. B. Renfrow, M. J. Chalmers, B. D. Nguyen, A. G. Marshall, P. Legault, and J. G. Omichinski. 2005. Enhanced binding of RNAP II CTD phosphatase FCP1 to RAP74 following CK2 phosphorylation. Biochemistry 44:2732-2745. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arriagada, G., R. Paredes, J. Olate, A. van Wijnen, J. B. Lian, G. S. Stein, J. L. Stein, S. Onate, and M. Montecino. 2007. Phosphorylation at serine 208 of the 1alpha,25-dihydroxy vitamin D3 receptor modulates the interaction with transcriptional coactivators. J. Steroid Biochem. Mol. Biol. 103:425-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausoleil, S. A., M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley, and S. P. Gygi. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101:12130-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busschots, K., A. Voet, M. De Maeyer, J. C. Rain, S. Emiliani, R. Benarous, L. Desender, Z. Debyser, and F. Christ. 2007. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J. Mol. Biol. 365:1480-1492. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P., A. L. Ambrosio, S. Rahman, T. Ellenberger, and A. Engelman. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 102:17308-17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 11.Collins, M. O., L. Yu, I. Campuzano, S. G. Grant, and J. S. Choudhary. 2008. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell. Proteomics 7:1331-1348. [DOI] [PubMed] [Google Scholar]
- 12.Dephoure, N., C. Zhou, J. Villen, S. A. Beausoleil, C. E. Bakalarski, S. J. Elledge, and S. P. Gygi. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 105:10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman, A., and P. Cherepanov. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4:e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genovese, N. J., N. S. Banerjee, T. R. Broker, and L. T. Chow. 2008. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J. Virol. 82:4862-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, G., M. Ye, H. Zhou, X. Jiang, S. Feng, X. Jiang, R. Tian, D. Wan, H. Zou, and J. Gu. 2008. Large-scale phosphoproteome analysis of human liver tissue by enrichment and fractionation of phosphopeptides with strong anion exchange chromatography. Proteomics 8:1346-1361. [DOI] [PubMed] [Google Scholar]
- 16.Hare, S., M. C. Shun, S. S. Gupta, E. Valkov, A. Engelman, and P. Cherepanov. 2009. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5:e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J. E., S. R. Tannenbaum, and F. M. White. 2005. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J. Proteome Res. 4:1339-1346. [DOI] [PubMed] [Google Scholar]
- 18.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 19.Llano, M., S. Delgado, M. Vanegas, and E. M. Poeschla. 2004. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279:55570-55577. [DOI] [PubMed] [Google Scholar]
- 20.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461-464. [DOI] [PubMed] [Google Scholar]
- 21.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llano, M., M. Vanegas, N. Hutchins, D. Thompson, S. Delgado, and E. M. Poeschla. 2006. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 360:760-773. [DOI] [PubMed] [Google Scholar]
- 23.Maertens, G., P. Cherepanov, Z. Debyser, Y. Engelborghs, and A. Engelman. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421-33429. [DOI] [PubMed] [Google Scholar]
- 24.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, H. M., K. Ronen, C. Berry, M. Llano, H. Sutherland, D. Saenz, W. Bickmore, E. Poeschla, and F. D. Bushman. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKee, C. J., J. J. Kessl, N. Shkriabai, M. J. Dar, A. Engelman, and M. Kvaratskhelia. 2008. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 283:31802-31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 28.Molina, H., D. M. Horn, N. Tang, S. Mathivanan, and A. Pandey. 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 104:2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poeschla, E. M. 2008. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 65:1403-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollastri, G., A. J. Martin, C. Mooney, and A. Vullo. 2007. Accurate prediction of protein secondary structure and solvent accessibility by consensus combiners of sequence and structure information. BMC Bioinformatics 8:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puntervoll, P., R. Linding, C. Gemund, S. Chabanis-Davidson, M. Mattingsdal, S. Cameron, D. M. Martin, G. Ausiello, B. Brannetti, A. Costantini, F. Ferre, V. Maselli, A. Via, G. Cesareni, F. Diella, G. Superti-Furga, L. Wyrwicz, C. Ramu, C. McGuigan, R. Gudavalli, I. Letunic, P. Bork, L. Rychlewski, B. Kuster, M. Helmer-Citterich, W. N. Hunter, R. Aasland, and T. J. Gibson. 2003. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semplici, F., F. Meggio, L. A. Pinna, and S. Oliviero. 2002. CK2-dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TrCP and enhances beta-catenin degradation. Oncogene 21:3978-3987. [DOI] [PubMed] [Google Scholar]
- 33.Shu, H., S. Chen, Q. Bi, M. Mumby, and D. L. Brekken. 2004. Identification of phosphoproteins and their phosphorylation sites in the WEHI-231 B lymphoma cell line. Mol. Cell. Proteomics 3:279-286. [DOI] [PubMed] [Google Scholar]
- 34.Shun, M. C., Y. Botbol, X. Li, F. Di Nunzio, J. E. Daigle, N. Yan, J. Lieberman, M. Lavigne, and A. Engelman. 2008. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J. Virol. 82:11555-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shun, M. C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trinidad, J. C., C. G. Specht, A. Thalhammer, R. Schoepfer, and A. L. Burlingame. 2006. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5:914-922. [DOI] [PubMed] [Google Scholar]
- 37.Trinidad, J. C., A. Thalhammer, C. G. Specht, A. J. Lynn, P. R. Baker, R. Schoepfer, and A. L. Burlingame. 2008. Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7:684-696. [DOI] [PubMed] [Google Scholar]
- 38.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 39.Turlure, F., G. Maertens, S. Rahman, P. Cherepanov, and A. Engelman. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34:1653-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandekerckhove, L., F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw, and Z. Debyser. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118:1733-1743. [DOI] [PubMed] [Google Scholar]
- 42.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 31:98-105. [DOI] [PubMed] [Google Scholar]
- 43.Villen, J., S. A. Beausoleil, S. A. Gerber, and S. P. Gygi. 2007. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U. S. A. 104:1488-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, F., D. L. Stenoien, E. F. Strittmatter, J. Wang, L. Ding, M. S. Lipton, M. E. Monroe, C. D. Nicora, M. A. Gristenko, K. Tang, R. Fang, J. N. Adkins, D. G. Camp II, D. J. Chen, and R. D. Smith. 2006. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J. Proteome Res. 5:1252-1260. [DOI] [PubMed] [Google Scholar]