Abstract
Retroviral genomes are dimeric, comprised of two sense-strand RNAs linked at their 5′ ends by noncovalent base pairing and tertiary interactions. Viral maturation involves large-scale morphological changes in viral proteins and in genomic RNA dimer structures to yield infectious virions. Structural studies have largely focused on simplified in vitro models of genomic RNA dimers even though the relationship between these models and authentic viral RNA is unknown. We evaluate the secondary structure of the minimal dimerization domain in genomes isolated from Moloney murine leukemia virions using a quantitative and single nucleotide resolution RNA structure analysis technology (selective 2′-hydroxyl acylation analyzed by primer extension, or SHAPE). Results are consistent with an architecture in which the RNA dimer is stabilized by four primary interactions involving two sets of intermolecular base pairs and two loop-loop interactions. The dimerization domain can independently direct its own folding since heating and refolding reproduce the same structure as visualized in genomic RNA isolated from virions. Authentic ex virio RNA has a SHAPE reactivity profile similar to that of a simplified transcript dimer generated in vitro, with the important exception of a region that appears to form a compact stem-loop only in the virion-isolated RNA. Finally, we analyze the conformational changes that accompany folding of monomers into dimers in vitro. These experiments support well-defined structural models for an authentic dimerization domain and also emphasize that many features of mature genomic RNA dimers can be reproduced in vitro using properly designed, simplified RNAs.
Retroviruses, including both simple model viruses and complex viruses like human immunodeficiency virus (HIV), contain genomes in the form of two coding RNA strands, noncovalently linked at their 5′ ends (11, 19, 20, 26, 38, 45). This 5′ linkage is termed the genomic RNA dimer. Packaging of RNA genomes into new virions is highly specific, even in the presence of a large background of cellular RNA (1, 7, 26). This packaging function is carried out by the Gag protein (14, 30, 43), which recognizes RNA sequences that overlap with the RNA dimerization domain (16, 26, 42, 44). The specific Gag-dimer interaction represents an elegant and direct mechanism by which exactly two RNA genomes are packaged into each nascent virion.
The genomic RNA dimer is initially assembled into an immature and noninfectious viral particle (9, 18-20). After the immature particle buds from the host cell, it undergoes extensive morphological changes to form the mature and infectious virion (21, 51). Maturation is initiated through cleavage of the Gag polyprotein by the viral protease to yield smaller Gag-derived proteins and also involves changes in the structure of the RNA dimer region. The RNA dimer structure appears to be more compact and, for many retroviruses, more thermostable in mature than in immature virions (18-20, 26).
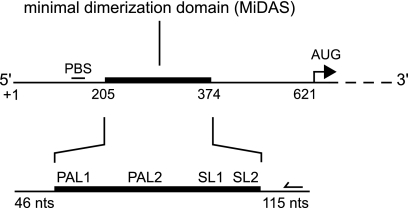
The closely related Moloney murine leukemia and sarcoma viruses (MuLV and MuSV, respectively) have served as important model systems for studying factors that impact retroviral infectivity. Recent work indicates that a compact region of ∼170 nucleotides (nt) in MuLV and MuSV is sufficient for dimerization in vitro (6). This dimerization region overlaps with sequences that were previously shown to be sufficient to allow heterologous RNAs to be packaged, albeit at reduced levels, as dimers into new virions (26). This region also forms high-affinity interactions with the viral Gag protein (16). Structural analysis of this minimal dimerization active sequence (the MiDAS RNA), performed with highly purified RNA under simplified solution conditions in vitro, supports a consensus model for the dimerization domain that involves three major structural components (Fig. 1) (4, 5, 22). Two conserved self-complementary sequences, PAL1 and PAL2 (5, 22, 24, 32, 39) (also called SL-B′ and SL-B, respectively), form extended intermolecular duplexes. A conserved double stem-loop domain, SL1-SL2 (29) (also called SLC-SLD), forms stable loop-loop and tertiary interactions with the same domain in the second RNA strand (4, 5, 22, 28).
FIG. 1.
Organization of structural elements in the MuLV dimerization domain. Major dimerization elements are indicated: PAL1 and PAL2, palindromic sequences; SL1-SL2, stem-loop domain. The minimal dimerization active sequence (MiDAS) is 170 nt; the in vitro transcript used in this work spans 331 nt. The binding site for the fluorescent DNA primers used for primer extension is represented by a backward-facing arrow.
To date, there are important gaps in our basic understanding of retroviral dimerization. First, with three exceptions (2, 40, 49), structural analysis of dimerization has focused on detailed characterization of simplified RNA transcripts in vitro. Dimerization in vitro is usually achieved by incubating an RNA at elevated temperatures, and it is unclear whether this appropriately mimics dimerization in vivo, which is facilitated by the chaperoning and strand-annealing activities of the viral nucleocapsid protein (31, 43). Thus, the correlation between RNA structures observed in vitro versus those that exist in virions is unknown.
Second, while there is an emerging consensus that a compact region in the MuLV and MuSV RNA genomes mediates dimerization both in vitro and in vivo, in vitro structural analyses have led to different, conflicting models for the dimerization domain. Key areas of disagreement include the boundaries of the PAL1 and PAL2 duplexes, the structures of the RNA elements between these helices, and the nature of the conformational changes that accompany formation of the authentic retroviral dimer (compare references 4-6, 13, 14, and 47). In addition, some studies have been performed with RNAs that either contain many mutations relative to the native sequence or span only a portion of the ∼170-nt dimerization element. Differences in these models and in the choice of simplified RNA are important because they influence whether the dimerization domain forms higher-order tertiary interactions (4, 5), make different predictions regarding how Gag and nucleocapsid bind (5, 13), and affect the choice of RNAs used in high-resolution structural studies (13, 14).
To address these issues, we use RNA SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) chemistry (35, 50) to map the structure of authentic genomic RNA that has been gently extracted from infectious virions (termed the ex virio RNA). SHAPE uses an electrophilic reagent, 1M7 (1-methyl-7-nitroisatoic anhydride) (37), that reacts selectively at the 2′-hydroxyl position of flexible nucleotides to form 2′-O-ester adducts. The reaction occurs preferentially at flexible RNA sites because these nucleotides are better able to adopt local conformations that enhance the reactivity of the 2′-hydroxyl group (23, 35). SHAPE reactivities thus correlate inversely with the probability that a nucleotide forms base pairing or tertiary interactions. SHAPE information can be used to create highly accurate models for an RNA secondary structure (12, 49).
Although not every base pairing interaction is predicted perfectly, benchmarking experiments indicate that SHAPE-constrained RNA secondary structure determination yields models for large RNAs that include 95% or greater of the accepted base pairs (12). SHAPE reactivity patterns typically yield quantitative information for nearly every nucleotide in an RNA. SHAPE profiles are thus helpful in ruling out incorrect or incomplete models. To provide a basis for interpreting the structure of authentic MuLV genomic RNA, we also compared the structure of the virion-derived dimer RNA with in vitro transcripts of both dimer and monomer states.
Secondary structure models derived from SHAPE measurements for the ex virio (genomic RNA isolated from mature MuLV particles) and in vitro (short, transcribed RNA, incubated at 60°C) dimers are highly similar but not identical. These data provide critical evidence that in vitro dimerization recapitulates most elements associated with the structure of the mature retroviral dimer. We also assess the structure of the dimerization domain in the context of the ∼8,300-nt ex virio RNA following denaturation and subsequent refolding by incubation at 60°C. The structure of this refolded genomic RNA is nearly identical to the structure of authentic genomic RNA as initially isolated from virions and is similar to the dimer generated from short transcripts in vitro. Collectively, these results provide a well-justified model for the structure of authentic MuLV genomic RNA and emphasize that the dimer created artificially in vitro closely mimics, but does not yet perfectly recapitulate, the structure of the mature dimer obtained from virions.
MATERIALS AND METHODS
Virus preparation.
Virions were obtained from 10 liters of clarified cell culture supernatant from a constitutively producing cell line infected with MuLV (cell line SL658 DN, the NB-tropic, IC isolate of Moloney MuLV propagated in 3T3FL cells) (17). Virus was recovered from culture fluids by single or double banding in sucrose gradients (10). Virus pellets were resuspended in TNE (0.01 M Tris-HCl [pH 7.2], 0.1 M NaCl, and 1 mM EDTA) buffer at final concentrations of 1,000× or 5,000×, relative to the starting culture volume, and aliquots were stored at −70°C.
Ex virio RNA dimer extraction and quantification.
For isolation of whole MuLV genomic RNA, concentrated virion preparations (equivalent to ∼2 × 1013 genomes) were collected by ultracentrifugation (Beckman SW-41 rotor; 235,000 × g at 4°C for 1 h). Pellets were resuspended in 1 ml of lysis buffer (50 mM Tris [pH 7.4], 10 mM EDTA, 1% [wt/vol] SDS, 100 mM NaCl, 100 μg/ml proteinase K, 120 μg/ml glycogen), incubated at 25°C for 45 min, and extracted three times with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with an equal volume of chloroform. Samples were precipitated in 70% ethanol with 0.3 M sodium acetate and stored at −20°C. RNA quantities were determined by real-time reverse transcriptase PCR (46), using an MuLV gag transcript from XhoI-linearized pJB2733 (GenBank accession no. J02255; nt 1 to 1560 contained in an XhoI-SpeI fragment, cloned into the homologous sites of pBluescript KS+ [Stratagene]) and quantifying the resulting gag cDNA target (nt 681 to 757; forward primer, 5′-CGG ATC GCT CAC AAC CAG T-3′; reverse primer, 5′-AAG GTT GGC CAT TCT GCA GA-3′; fluorescent probe, 5′-FAM-TAG ATG TCA AGA AGA GAC GTT GGG TTA CCT-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine).
Retroviral RNA transcripts.
The in vitro MuLV RNA construct contained 331 nt and spanned sequences corresponding to PAL1 through SL2, plus flanking 5′ and 3′ viral sequences of 46 and 115 nt, respectively (Fig. 1). The DNA template for in vitro transcription was generated by PCR from a full-length wild-type MuLV proviral plasmid (25), pRR88 (GenBank accession no. J02255). Transcription reactions (500 μl at 37°C for 6 h) contained 80 mM HEPES (pH 7.5), 40 mM dithiothreitol (DTT), 0.01% (vol/vol) Triton X-100, 2 mM spermidine, 10 mM MgCl2, 2 mM each nucleoside triphosphate (dNTP), ∼25 μg of DNA template, 250 U of SUPERase-In (Ambion), and 0.07 mg/ml T7 RNA polymerase. The RNA was purified by denaturing gel electrophoresis (6% [wt/vol] polyacrylamide, 7 M urea), excised from the gel, eluted overnight into 1/2× TBE (45 mM Tris-borate [pH 8], 1 mM EDTA) buffer at 4°C, concentrated by ethanol precipitation, and stored in 1× TE (10 mM Tris [pH 8], 1 mM EDTA) buffer at −20°C.
SHAPE analysis.
Authentic ex virio RNA, isolated from MuLV virions, was simply allowed to preequilibrate in dimerization buffer (50 mM HEPES [pH 7.5], 200 mM potassium acetate [pH 7.5], 5 mM MgCl2) for 15 min. For the renatured MuLV genomic RNA and for the in vitro generated RNAs, renaturation was accomplished by heating at 90°C for 3 min in water (2 pmol in 10 μl), snap-cooling on ice for 2 min, and treating with 3× dimerization buffer (6 μl; 150 mM HEPES [pH 7.5], 600 mM potassium acetate [pH 7.5], 15 mM MgCl2). Monomers were allowed to equilibrate for 2 min at 37°C prior to modification (see below); dimers were created by heating (30 min at 60°C). Use of our RNA construct and this refolding procedure yields very homogeneous dimer and monomer states as visualized by nondenaturing gel electrophoresis (data not shown). 2′-Hydroxyl modification for SHAPE was performed by treating MuLV RNA or transcript RNA (in 9 μl) with 1-methyl-7-nitroisatoic anhydride (1M7) (1 μl; at 20 mM) in anhydrous dimethyl sulfoxide (DMSO) (37) for 2 min (∼8 half-lives) at 37°C. Control reactions were obtained by the addition of DMSO, omitting 1M7. The reaction mixtures were recovered by ethanol precipitation and resuspended in 6 μl of 1/2× TE buffer.
Primer extension.
DNA primers (5′-GGU GCA CCA AAG AGU CCA AAA GC-3′, labeled with fluorophore Cy5, Cy5.5, or IR800) anneal with a complementary native viral sequence 47 nucleotides 3′ of the MuLV dimerization domain (nt 422 to 445). Primers (0.5 μl; 2 pmol) were annealed to the modified RNA (6 μl; 1 pmol in 1/2× TE buffer) by heating at 65°C and 45°C for 5 min. Reverse transcription buffer (3 μl; 167 mM Tris [pH 8.3], 250 mM KCl, 10 mM MgCl2, 1.67 mM each dNTP, 16.7 mM DTT) was added, and primer extension was performed with Superscript III reverse transcriptase (0.5 μl; 100 U) at 45°C (1 min), followed by incubation at 52°C and 65°C, for 5 min each. Reactions were quenched by cooling at 4°C and the addition of 3 M sodium acetate, pH 5.2 (1 μl). A dideoxy guanosine sequencing marker was generated by adding 0.25 μl ddCTP (10 mM) to the primer extension reaction mixture, using unmodified RNA. The three reaction mixtures (with and without 1M7 and dideoxy sequencing) were combined, precipitated with ethanol, and resuspended in deionized formamide (40 μl). cDNA fragments were resolved by capillary electrophoresis using a Beckman CEQ 2000XL capillary electrophoresis instrument.
Data processing and structure prediction.
Raw sequencer traces were corrected for dye variation and signal decay, and peak intensities were integrated using ShapeFinder (48). Reactivities were normalized by dividing the absolute peak intensities by the average of the 8% (14 nt) most reactive peaks after exclusion of the first 2% (3 nt). SHAPE reactivity information was used to impose a pseudo-free energy change constraint, in conjunction with nearest-neighbor thermodynamic parameters, using the algorithm in RNAstructure (12, 34).
RESULTS
SHAPE analysis of MuLV genomic RNA.
The overarching goal of this work was to develop a model for the global architecture and secondary structure of the dimerization domain for authentic MuLV retroviral RNA genomes isolated from virions. We therefore used single nucleotide resolution SHAPE chemistry to analyze the structure of the dimerization domain (Fig. 1) from genomic MuLV RNA purified from virions under nondenaturing conditions that preserve the dimeric and thermostable state of the RNA (20, 26). We term this RNA the ex virio dimer.
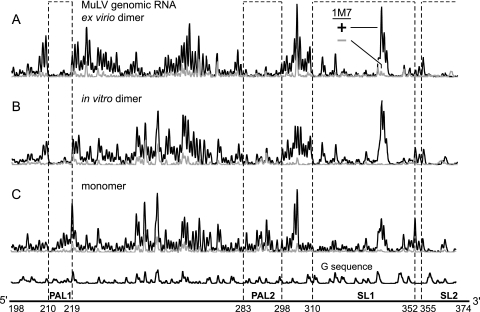
The output from a SHAPE experiment includes three components (Fig. 2): a trace of the reaction with reagent (plus-reagent trace)(Fig. 2, black trace), a background trace that omits the reagent (Fig. 2, gray trace), and a deoxynucleotide sequencing ladder which is used to assign the positions of sites reactive toward 1M7. Nucleotides tagged with 2′-O-adducts are detected as stops to reverse transcriptase-mediated primer extension. Extension reactions are performed using fluorescently labeled DNA primers and resolved by capillary electrophoresis (37, 48, 49) (Fig. 2). We computed the absolute reactivity at each nucleotide within the MuLV ex virio RNA by integrating individual peak areas and subtracting the background from the plus-reagent areas. Conformationally flexible nucleotides appear as tall peaks, whereas nucleotides that participate in strong base pairing or tertiary interactions have reactivities near the background (Fig. 3A, compare red and black columns).
FIG. 2.
SHAPE reactivities in the 190- to 375-nt region for the authentic ∼8,300-nt MuLV genomic RNA extracted from virions (A), the in vitro dimer (B), and the monomer state (C), resolved by capillary electrophoresis. 1M7 traces with and without reagent are shown in black and gray, respectively.
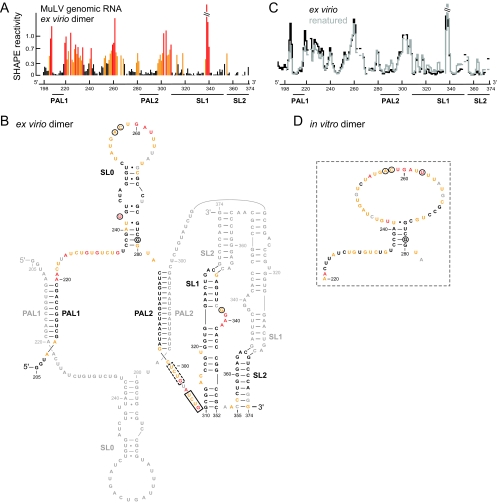
FIG. 3.
SHAPE reactivities and secondary structure model for the ex virio MuLV genomic RNA dimer. (A) Histogram of SHAPE reactivities for authentic ex virio MuLV genomic RNA isolated from virions. (B) Secondary structure model for the ex virio dimer. Nucleotides are colored according to their reactivity as shown in panel A. For clarity, only one of the two dimer strands is annotated in color; a small number of nucleotides in the colored strand (in gray) were not analyzed due to high background. Nucleotide differences between MuLV and MuSV are circled. The nucleocapsid binding site characterized by NMR (13) plus a similar site accessible in the SHAPE-constrained secondary structure are highlighted in solid and dashed boxes, respectively. (C) Comparison of the SHAPE profiles for the ex virio RNA with that of the same RNA that has been heated (to 90°C) and renatured in vitro. The y-axis scale is the same as that used in panel A. (D) Structure model for the region of the in vitro dimer that corresponds to the SL0 motif in the ex virio RNA.
Structure of the dimerization domain for MuLV.
We developed a secondary structure model for the dimerization region of ex virio MuLV genomic RNA using SHAPE reactivity information to impose a pseudo-free energy change constraint in a thermodynamics-based RNA structure prediction algorithm (12, 34). We show the resulting secondary structure model annotated by the nucleotide resolution SHAPE reactivity (Fig. 3B).
SHAPE analysis of the MuLV ex virio RNA supports a model in which the major structural domains are similar to a prior consensus model based on analysis of the closely related MuSV sequence (5). Sequences comprising the PAL1 and PAL2 intermolecular duplexes are unreactive. Similarly, base-paired positions in the SL1 and SL2 stem-loops are unreactive except at bulged nucleotides, as shown in our model. Modeling is also consistent with a large, previously unidentified, structured stem-loop region between PAL1 and PAL2, which we term SL0. Finally, the dimer structure contains three highly flexible elements. These lie immediately 3′ to PAL1 (positions 220 to 235) and PAL2 (positions 299 to 309) and in the large terminal loop in SL0 (positions 253 to 267) (Fig. 3B).
One common strategy for coaxing an RNA to dimerize has been using heating and quick-cooling to reduce the RNA to a monomeric state and then initiating dimerization by incubation at an elevated temperature. We subjected the authentic ex virio MuLV RNA to a heat denaturation and renaturation protocol (see Materials and Methods) and then analyzed the structure of the RNA by SHAPE. The resulting SHAPE profiles are shown using step histograms (Fig. 3C). The pattern of reactivity is almost identical to that for the gently purified native, heat-denatured and -renatured RNA. This experiment indicates that accurate RNA folding and dimerization can be induced by a simple heating and refolding procedure, at least in this region of the MuLV genome.
An in vitro RNA dimer folds similarly to the authentic dimer isolated from virions.
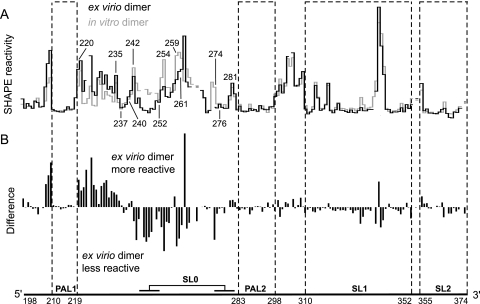
We next sought to assess how closely the authentic ex virio genomic RNA resembles a dimer formed in vitro using a simplified transcript RNA that spans the dimerization domain. We compared SHAPE reactivity profiles for the dimerization domain of genomic RNA extracted from virions with that of a 331-nt transcript (Fig. 4A, compare black and gray histograms). Differences between these two RNAs are readily visualized by subtracting individual nucleotide reactivities for the in vitro dimer from values of the ex virio dimer (Fig. 4B).
FIG. 4.
Comparison of SHAPE reactivities for MuLV ex virio dimers with dimers created in vitro using short (331 nt) RNA transcripts. (A) Reactivity histograms for the ex virio (black) and in vitro (gray) dimers. (B) Difference plot calculated by subtracting the in vitro dimer intensities from those of the ex virio dimer. Positive and negative amplitudes indicate nucleotides that are more flexible in the ex virio and in vitro dimers, respectively.
SHAPE profiles for the ex virio and in vitro dimers are nearly superimposable in all regions spanning the key elements—PAL1, PAL2, and the SL1-SL2 domain—that function to stabilize the dimer state (4, 22) (Fig. 4). These results imply that the structure of the in vitro dimer closely resembles that of the ex virio dimer.
Despite this overall pattern of nearly identical reactivities, important differences are observed between the ex virio and in vitro dimers (Fig. 4B, difference plot). Nucleotides 243 to 252 are unreactive in the ex virio dimer, whereas these nucleotides are reactive in the in vitro dimer. Other nucleotides (253, 254, and 259 to 261) are also less reactive in the dimer RNA extracted from virions. Immediately 5′ to this region (positions 220 to 235) the reverse occurs such that these nucleotides are more reactive in the ex virio RNA than in the in vitro dimer (Fig. 4B).
These data support the interpretation that the structure between PAL1 and PAL2 is different in the two RNAs and that this region forms the SL0 stem-loop only in the genomic RNA (structural differences between the ex virio and in vitro dimers are illustrated in Fig. 3B and D). The SHAPE reactivity pattern for the ex virio dimer is robust and persists even if the ex virio RNA is denatured by heat treatment and allowed to redimerize in vitro (Fig. 3C). These data suggest that this region may form long-range interactions in the ∼8,300-nt long genomic RNA or that folding and dimerization kinetics are altered in the intact RNA relative to short in vitro transcripts.
Structure of the in vitro MuLV monomer.
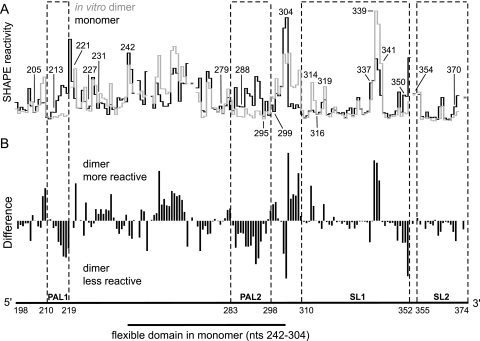
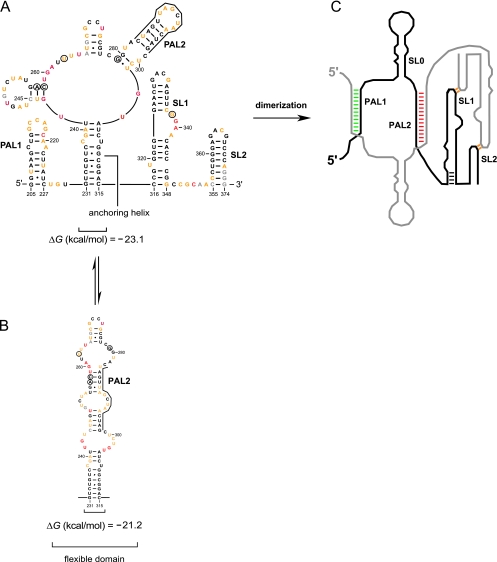
We next used SHAPE reactivity information (Fig. 2C) to establish a secondary structure model for the MuLV monomer state. In contrast to the general similarity between the ex virio genomic RNA and the in vitro dimer, there are extensive differences between the in vitro monomer and both the ex virio and in vitro dimers which occur throughout the RNA (Fig. 5). In PAL1, nucleotides 214 to 221 are highly reactive in the monomer while positions 205 to 213 and 222 to 227 are unreactive, consistent with formation of a stem-loop (Fig. 6A). Nucleotides 228 to 229 are flexible, supporting the existence of a linker that connects the PAL1 stem-loop to the rest of the monomer. Nucleotides in the SL1-SL2 domain (316 to 370) have a low reactivity overall, suggesting extensive base pairing and tertiary interactions in this region. SL1 is punctuated with a few flexible nucleotides (319 and 337 to 341), consistent with formation of two distinct bulges. Reactive nucleotides between positions 350 and 354 indicate that a flexible linker connects SL1 and SL2. Between the PAL1 stem-loop and the SL1-SL2 domain, nucleotides 231 to 241 and 305 to 315 are constrained while the majority of the positions in the intervening region (nt 242 to 304) are reactive. This pattern of reactivity suggests that nucleotides 242 to 304 form a flexible domain which is linked to the rest of the RNA via a stable anchoring helix (Fig. 6A). A similar architecture was identified and proposed previously for MuSV (6).
FIG. 5.
SHAPE reactivities for the RNA transcript in vitro dimer and monomer states. (A) Reactivity histograms for the in vitro dimer (gray) and monomer (black). Lines are interrupted for nucleotides that were not analyzed due to high background reactivity. (B) Difference plot calculated by subtracting the monomer intensities from those of the dimer. Positive and negative values indicate nucleotides that are more flexible in the dimer and the monomer, respectively.
FIG. 6.
Secondary structure of the MuLV MiDAS RNA monomer and its conversion to the dimer state. (A) Structure of the entire MiDAS monomer. Nucleotides are colored using the scale shown in Fig. 3A. (B) Alternate structure proposed to interconvert with the domain of nucleotides 231 to 315. Nucleotides that differ in MuLV compared to MuSV are circled. (C) Cartoon of the dimer state emphasizing the regions undergoing the largest conformational changes upon dimerization. The full nucleotide sequence is given in Fig. 3B.
Within the flexible domain, no single structure completely explains the overall pattern of reactivity. For example, in the most stable predicted structure for MuLV, PAL2 forms a stem-loop (supported by low reactivity at nt 279 to 287 and 295 to 299 and high reactivity for nt 288 to 294) (Fig. 5A and 6A). However, this structure contains local inconsistencies. For example, nucleotides 243 and 244 are reactive but are predicted to be paired in this contributing structure (Fig. 6A). We favor a model in which the flexible domain does not form a single structure but, instead, exists in equilibrium between multiple secondary structures (compare Fig. 6A and B). Two interconverting structures, which differ by only 2 kcal/mol, appear to be sufficient to account for the global pattern of SHAPE reactivity.
DISCUSSION
A structure for authentic MuLV genomic RNA in the dimer state.
The retroviral genomic RNA dimer structure was first visualized in electron micrographs of genomic RNA from several retroviruses. Subsequent work has made it clear that most retroviruses form a stable dimeric structure near their 5′ ends (8, 27, 33, 38). The dimer element appears to play a critical role in retroviral replication by creating the specific three-dimensional structure that is ultimately recognized by Gag during viral RNA packaging (9, 41, 45). The dimer state likely also functions to ensure that two copies of each genome are packaged into each virion and to facilitate recombination between genomes (3, 36).
Several convergent results, including this work, are now available to guide development of a model for the overall architecture and secondary structure of the dimerization domain in MuLV. First, the minimal RNA elements that are required to bind Gag (16) and are sufficient to direct a modest level of packaging in vivo (26) have become better defined. These elements overlap with the elements required to form a dimeric state in vitro (5, 6, 24, 32, 39, 47). As defined by all three experimental approaches, the minimal dimerization element appears to span ∼170 nt.
Second, using authentic genomic RNA purified from MuLV virions, we have obtained a comprehensive SHAPE reactivity profile for the entire dimerization domain (Fig. 3A). The SHAPE information places very strong constraints on possible secondary structure models and rules out many alternative models.
Finally, heating and then refolding the genomic RNA yields the same SHAPE reactivity profile as observed for the RNA as isolated from virions. Thus, although RNA folding may be facilitated by other factors and chaperone activities in vivo, the MuLV genomic RNA itself appears to contain all the information required for folding of the dimerization domain.
All elements previously proposed to be important for dimer formation are well defined in the structure of the MuLV dimerization domain (Fig. 3B). PAL1 and PAL2 form intermolecular duplexes of 10 and 16 bp, respectively. SL1 and SL2 form well-defined stem-loop structures. The GACG sequences at the termini of the stem-loops are unreactive, which we interpret in terms of formation of stable long-range loop-loop interactions (4, 22, 28) (Fig. 6C, indicated in orange).
The structure of authentic mature MuLV genomic RNA dimers, comprised of two strands of ∼8,300 nt, is very similar, but not identical, to the structure generated using short transcripts and a heating and refolding step in vitro (Fig. 4). Specific differences between the in vitro generated versus ex virio dimer are localized to sequences between PAL1 and PAL2 (Fig. 3B and D). We have interpreted these differences to indicate that the authentic ex virio dimer forms a stem-loop element that is not present in the in vitro dimer. Genome-specific differences likely involve additional long-range interactions with flanking RNA sequences not present in current in vitro models of the dimerization domain.
Conformational changes that characterize dimer formation.
Comparison of the overall reactivity profiles for the MuLV monomer and dimer states indicates that numerous conformational changes accompany the in vitro dimerization reaction (Fig. 5A). These changes are readily visualized in a difference analysis (Fig. 5B) and show that nucleotides in both PAL1 and PAL2 sequences change conformation and become unreactive during this structural transition (Fig. 3B and 6C, green and red areas). SHAPE reactivities also support the interpretation that the anchoring duplex melts and that the SL1-SL2 domain undergoes a local conformation change, consistent with elongation of SL1 by four base pairs during dimerization (Fig. 6C, indicated in black).
Our laboratory has analyzed two closely related model systems, MuLV (this work) and MuSV (4, 22). Consistent with their extensive sequence similarities, the major monomer and dimer structures fold identically in the two viruses. These elements also undergo similar conformational changes upon dimerization in vitro (Fig. 6A and C) (5). Although only five positions differ in sequence in the dimerization domains of MuLV and MuSV (circled in Fig. 3 and 6), these differences lead to clear and measurable SHAPE-detected structural differences in the both monomer and dimer states. These data both show the sensitivity of the SHAPE approach for detecting small changes in an RNA structure and also provide additional support for our overall model for the architecture of the dimerization domains. The sequence differences between MuLV and MuSV and the resulting structural changes occur precisely in the unique unstructured domains in these RNAs and therefore do not compromise the motifs that form the major stabilizing elements in the dimer.
Implications for future structural studies.
The secondary structure and overall architecture of the dimerization domain in the MuLV and MuSV gamma retroviruses now appear to be well defined. The structural models proposed here are broadly consistent with prior experimental chemical probing data reported for both authentic MuLV genomic RNA (2) and in vitro models of the dimerization domain (47). However, our interpretation of the SHAPE data differs from prior models. These differences reflect, in part, the greater coverage of the SHAPE data relative to conventional RNA mapping reagents and improvements in incorporating chemical modification data into secondary structure prediction methods.
The SHAPE-directed secondary structure models reported here are also the same as the pattern of base pairing seen in recent nuclear magnetic resonance (NMR) models of dimerization-related RNAs in the regions where the sequences of the two RNAs were identical (13, 14). However, the NMR studies were performed with RNAs that contained only a portion of the dimerization domain, spanning roughly PAL2 through the SL1-SL2 domain, and that also contained extra sequences 5′ of PAL2 and 3′ of SL2 and nucleotide changes in PAL2, SL1, and SL2 that clearly affect the global architecture of the dimer (14). In the regions in which the NMR studies used nonnative sequences, the secondary structures defined by SHAPE and by NMR are different.
The prior NMR work indicated that the MuLV nucleocapsid protein binds at the single stranded UCUG309 sequence (Fig. 3B, solid box) (13). The revised SHAPE-constrained model emphasizes that there are two UCUG motifs in the single-stranded linker between PAL2 and SL1, suggesting that nucleocapsid domains bound in tandem might bind the RNA in this region (Fig. 3B, dashed box). Both UCUG elements become single stranded in the dimer, consistent with a model in which selective exposure of these sequences plays a role in specific recognition of the dimer state by the nucleocapsid domain of Gag (13, 15). These differences in structure observed for the native MuLV sequence studied here and the mutated sequence studied by NMR emphasize that small changes in critical regions change the structure of the MuLV RNA in ways that are important for understanding viral protein-RNA interactions.
Dimerization in the MuLV/MuSV system is accompanied by large-scale and readily detected conformational changes in the RNA (Fig. 2, 5, and 6). This contrasts with the more difficult case of HIV type 1, where the monomer and dimer states have very similar overall SHAPE profiles (49). MuLV may therefore be the best model for understanding the process of retroviral RNA genome maturation, first, because the elements sufficient to recapitulate the biological function of this RNA are becoming well defined and, second, because dimerization yields clear structural changes as a function of RNA state. Important future objectives include indentifying the structural differences that distinguish mature from immature genomic RNA dimers and the role that these structures play in maturation of noninfectious, immature virions to infectious virions.
Acknowledgments
This work was supported by the U.S. National Institutes of Health (GM064803 to K.M.W.) and with federal funds from the National Cancer Institute (under contract N01-CO-12400 to R.J.G.).
We are indebted to Julian W. Bess, Jr., of the AIDS and Cancer Virus, Program Biological Products Core, for providing and purifying the MuLV genomic RNA.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford, R. L., S. Honda, C. B. Lawrence, and J. W. Belmont. 1991. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology 183:611-619. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. A., V. K. Pathak, and W. S. Hu. 2000. Effect of the murine leukemia virus extended packaging signal on the rates and locations of retroviral recombination. J. Virol. 74:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badorrek, C. S., C. M. Gherghe, and K. M. Weeks. 2006. Structure of an RNA switch enforces stringent retroviral genomic RNA dimerization. Proc. Natl. Acad. Sci. U. S. A. 103:13640-13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badorrek, C. S., and K. M. Weeks. 2006. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry 45:12664-12672. [DOI] [PubMed] [Google Scholar]
- 6.Badorrek, C. S., and K. M. Weeks. 2005. RNA flexibility in the dimerization domain of a gamma retrovirus. Nature Chem. Biol. 1:104-111. [DOI] [PubMed] [Google Scholar]
- 7.Banks, J. D., A. Yeo, K. Green, F. Cepeda, and M. L. Linial. 1998. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 72:6190-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, W., Y. H. Chien, S. Chattopadhyay, P. K. Vogt, M. B. Gardner, and N. Davidson. 1978. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 25:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 10.Bess, J. W. J., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J. Virol. 66:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 12.Deigan, K. E., T. W. Li, D. H. Mathews, and K. M. Weeks. 2009. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U. S. A. 106:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Souza, V., and M. F. Summers. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukemia virus. Nature 431:586-590. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza, V., A. Dey, D. Habib, and M. F. Summers. 2004. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 337:427-442. [DOI] [PubMed] [Google Scholar]
- 15.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643-655. [DOI] [PubMed] [Google Scholar]
- 16.Evans, M. J., E. Bacharach, and S. P. Goff. 2004. RNA sequences in the Moloney murine leukemia virus genome bound by the Gag precursor protein in the yeast three-hybrid system. J. Virol. 78:7677-7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischinger, P. J., C. O. Moore, and T. E. O'Connor. 1969. Isolation and identification of a helper virus found in the Moloney sarcoma-leukemia virus complex. J. Natl. Cancer Inst. 42:605-622. [PubMed] [Google Scholar]
- 18.Fu, W., Q. Dang, K. Nagashima, E. O. Freed, V. K. Pathak, and W.-S. Hu. 2006. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J. Virol. 80:1242-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganser-Pornillos, B. K., M. Yeager, and W. I. Sundquist. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gherghe, C., and K. M. Weeks. 2006. The SL1-SL2 (stem loop) domain is the primary determinant for stability of the gamma retroviral genomic RNA dimer. J. Biol. Chem. 281:37952-37961. [DOI] [PubMed] [Google Scholar]
- 23.Gherghe, C. M., Z. Shajani, K. A. Wilkinson, G. Varani, and K. M. Weeks. 2008. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 130:12244-12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girard, P. M., B. Bonnet-Mathoniere, D. Muriaux, and J. Paoletti. 1995. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry 34:9785-9794. [DOI] [PubMed] [Google Scholar]
- 25.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: Evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. U. S. A. 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibbert, C. S., J. Mirro, and A. Rein. 2004. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 78:10927-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoglund, S., A. Ohagen, J. Goncalves, A. T. Panganiban, and D. Gabuzda. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233:271-279. [DOI] [PubMed] [Google Scholar]
- 28.Kim, C., and I. Tinoco. 2000. A retroviral RNA kissing complex containing only two GC base pairs. Proc. Natl. Acad. Sci. U. S. A. 97:9396-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konings, D. A. M., M. A. Nash, J. V. Maizel, and R. B. Arlinghaus. 1992. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J. Virol. 66:632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E., A. Yeo, B. Kraemer, M. Wickens, and M. L. Linial. 1999. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J. Virol. 73:6282-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin, J. G., J. Guo, I. Rouzina, and K. Musier-Forsyth. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217-286. [DOI] [PubMed] [Google Scholar]
- 32.Ly, H., and T. G. Parslow. 2002. Bipartite signal for genomic RNA dimerization in Moloney murine leukemia virus. J. Virol. 76:3135-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangel, W. F., H. Delius, and P. H. Duesberg. 1974. Structure and molecular weight of the 60-70S RNA and the 30-40S RNA of the Rous sarcoma virus. Proc. Natl. Acad. Sci. U. S. A. 71:4541-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews, D. H., M. D. Disney, J. L. Childs, S. J. Schroeder, M. Zuker, and D. H. Turner. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101:7287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino, E. J., K. A. Wilkinson, J. L. Coughlan, and K. M. Weeks. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223-4231. [DOI] [PubMed] [Google Scholar]
- 36.Moore, M. D., W. Fu, O. Nikolaitchik, J. Chen, R. G. Ptak, and W. S. Hu. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 81:4002-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortimer, S. A., and K. M. Weeks. 2007. A fast acting reagent for accurate analysis of RNA secondary and tertiary structure for SHAPE chemistry. J. Am. Chem. Soc. 129:4144-4145. [DOI] [PubMed] [Google Scholar]
- 38.Murti, K. G., M. Bondurant, and A. Tereba. 1981. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J. Virol. 37:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oroudjev, E. M., P. C. E. Kang, and L. A. Kohlstaedt. 1999. An additional dimer linkage structure in Moloney murine leukemia virus RNA. J. Mol. Biol. 291:603-613. [DOI] [PubMed] [Google Scholar]
- 40.Paillart, J.-C., M. Dettenhofer, X.-F. Yu, C. Ehresmann, B. Ehresmann, and R. Marquet. 2004. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 279:48397-48403. [DOI] [PubMed] [Google Scholar]
- 41.Paillart, J. C., M. Shehu-Xhilaga, R. Marquet, and J. Mak. 2004. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2:461-472. [DOI] [PubMed] [Google Scholar]
- 42.Prats, A. C., C. Roy, P. A. Wang, M. Erard, V. Housset, C. Gabus, C. Paoletti, and J. L. Darlix. 1990. cis Elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 64:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 44.Sakuragi, J., A. Iwamoto, and T. Shioda. 2002. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J. Virol. 76:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakuragi, J., T. Shioda, and A. T. Panganiban. 2001. Duplication of the primary encapsidation and dimer linkage region of human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 75:2557-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, J. A., T. D. Gagliardi, W. G. Alvord, M. Lubomirski, W. J. Bosche, and R. J. Gorelick. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 353:41-51. [DOI] [PubMed] [Google Scholar]
- 47.Tounekti, N., M. Mougel, C. Roy, R. Marquet, J. Darlix, J. Paoletti, B. Ehresmann, and C. Ehresmann. 1992. Effect of dimerization on the conformation of the encapsidation psi domain of the Moloney murine leukemia virus. J. Mol. Biol. 223:205-220. [DOI] [PubMed] [Google Scholar]
- 48.Vasa, S. M., N. Guex, K. A. Wilkinson, K. M. Weeks, and M. C. Giddings. 2008. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14:1979-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson, K. A., R. J. Gorelick, S. Vasa, N. Guex, A. Rein, D. H. Matthews, M. C. Giddings, and K. M. Weeks. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson, K. A., E. J. Merino, and K. M. Weeks. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610-1616. [DOI] [PubMed] [Google Scholar]
- 51.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: Implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]